Abstract
B cell lymphoma 6 (BCL6) is a conserved multi-domain protein that functions principally as a transcriptional repressor. This protein regulates many pivotal aspects of immune cell development and function. BCL6 is critical for germinal center (GC) formation and the development of high-affinity antibodies, with key roles in the generation and function of GC B cells, follicular helper T (Tfh) cells, follicular regulatory T (Tfr) cells, and various immune memory cells. BCL6 also controls macrophage production and function as well as performing a myriad of additional roles outside of the immune system. Many of these regulatory functions are conserved throughout evolution. The BCL6 gene is also important in human oncology, particularly in diffuse large B cell lymphoma (DLBCL) and follicular lymphoma (FL), but also extending to many in other cancers, including a unique role in resistance to a variety of therapies, which collectively make BCL6 inhibitors highly sought-after.
Keywords: BCL6, B cell, cancer, germinal center, lymphoma, macrophage, repressor, T cell, transcription factor
1. Introduction
The B cell lymphoma 6 (BCL6, also called BCL6A) protein was first identified as the product of a gene involved in chromosomal translocations in the context of non-Hodgkin’s lymphoma (NHL) [1]. This protein has subsequently been identified as a multi-functional regulator with a conserved structure and function. BCL6 is particularly important in immune cell development, which is largely mediated by its strong transcriptional repressor functions. This includes its pre-eminent roles in germinal center (GC) reactions [2], being required for the formation and development of GC B cells [3] and the differentiation and maintenance of follicular helper T (Tfh) cells [4,5]. Its function also extends to a range of other immune cells, including other helper T (Th) and specific regulatory T (Treg) and memory T (Tm) cell populations [6,7,8], as well as macrophages [9,10]. Diverse additional roles have also been identified outside of the immune system. A key aspect of BCL6’s function is its impact on other transcriptional regulators, with the disruption of such regulatory networks linked to various malignancies [11]. This review provides an overview of BCL6 and its role in the normal development and function of various immune cells and other populations, as well as how this is conserved across diverse species. It also examines the contribution of BCL6 to the etiology of lymphomas and other hematological malignancies and solid tumors, with a consideration of its role in therapy resistance and its application as a therapeutic target.
2. Gene Conservation and Evolution
BCL6 homologues are highly conserved. Human BCL6 shares around 95% identity with mouse BCL6 [12] and approximately 60% identity with Bcl6 proteins from teleost fish, such as pufferfish and zebrafish [13,14], indicative of conserved function across vertebrates. BCL6 is highly related to the BCL6B protein, which is also conserved across a wide range of vertebrate species [15]. This extends to teleost fish, which possess Bcl6b and an additional related protein termed Bcl6ab [13,14]. Invertebrates also harbor proteins related to BCL6, typified by the fruit-fly Ken and Barbie (Ken) protein, which contains a subset of functional domains [16,17]. This suggests an evolutionary model in which a precursor of both the BCL6 and ken gene lineages existed in the common ancestor of invertebrates and vertebrates, which evolved into a precursor of the ken-related genes in extant invertebrates and a BCL6/BCL6B precursor gene in early vertebrates. The latter subsequently acquired additional sequences prior to duplication to yield distinct BCL6 and BCL6B gene lineages that ultimately gave rise to the present-day mammalian genes. A subsequent teleost-fish-specific duplication yielded an additional bcl6ab gene.
3. Gene Expression
The expression of BCL6 and related genes follows complex patterns throughout the lifespan of various organisms, with both immune and non-immune cell expression observed across different species, the latter notably including neurosensory tissues. During embryogenesis, zebrafish bcl6 is expressed during the primitive wave of hematopoiesis in both the anterior and posterior lateral plate mesoderm, consistent with the location of early hematopoietic precursors [14]. Its expression continues into the definitive wave, with a strong expression in the thymus of developing zebrafish [14], consistent with the robust Bcl6 expression observed in the fetal thymus and spleen of mice [18] and the BCL6 expression observed in human fetal thymocytes [19]. In adults, pufferfish bcl6 is expressed in the thymus as well as in the kidney, equivalent to teleost bone marrow [13]. The expression of mouse Bcl6 was also identified in the thymus [18], with human BCL6 expressed in peripheral blood leukocytes and lymph nodes [20]. Its strong expression was also noted across mouse and human pre-B and mature B cell lines, with a lower expression in T cell, myeloid, and erythrocyte lines derived from both humans and mice [21].
Outside of hematopoietic tissue, bcl6 expression was observed in the embryonic retina of zebrafish [22] and in their developing cerebellum and medulla [14], and Bcl6 expression was observed in the olfactory epithelium of prenatal mice [18], showing parallels with the expression of ken in fruit-fly cephalic furrows and larval eye antennae [17]. The expression of mouse Bcl6 was additionally observed in their pre-natal skeletal muscle, esophagus, upper airway lining, and skin [18], whilst fruit-fly ken was also expressed in their developing gut [17]. In adults, pufferfish bcl6 expression was identified in their skeletal muscle, intestine, ovary, brain, and nasal cavity [13]. Mouse Bcl6 was similarly expressed within skeletal muscle as well as in the cerebral cortex [18,23], with human BCL6 expression being described in adult skeletal muscle, thyroid, trachea, ovary, prostate, and spinal cord [20].
4. Structure and Function
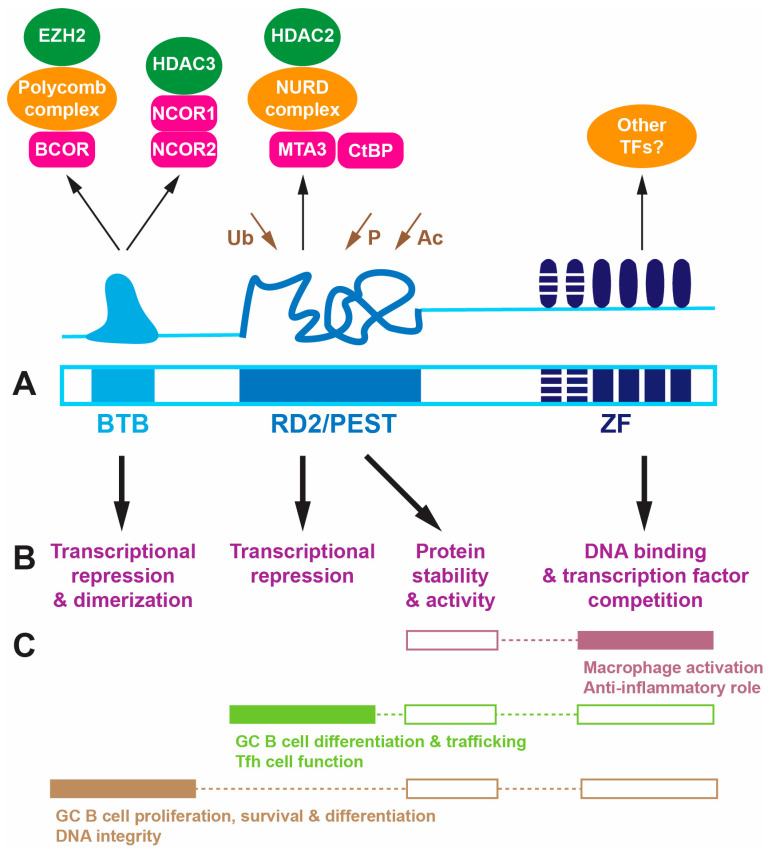
BCL6 is a 95 kDa multi-domain protein, which comprises a broad complex/tram track/bric-a-brac (BTB) domain at its N-terminus, a central unstructured region containing the so-called second repression domain (RD2) that overlaps with a proline (P), glutamic acid (E), serine (S) and threonine (T)-rich PEST domain, and a zinc finger (ZF) domain comprising an array of six C2H2 Krüppel-type zinc fingers at its C-terminus [24,25] (Figure 1A). An identical domain structure is seen across vertebrate BCL6 proteins [13,14], while Ken has fewer ZFs, with no RD2/PEST domain being identified [16].
Figure 1.
Structure and function of the BCL6 protein. (A). Domains of the BCL6 protein: BTB (light blue), RD2/PEST (blue), and ZF (dark blue, with striping for fingers not involved in DNA binding). Above is a schematic representation of the structure of each domain along with their interacting proteins, including co-repressors (pink) and associated transcriptional regulators (orange) and DNA modifying proteins (green), with sites of ubiquitination (Ub), phosphorylation (P), and acetylation (Ac) indicated (brown). (B). Major molecular function(s) of each domain. (C). Biological roles mapped to the molecular functions, with proven connections shown as filled boxes and assumed ones as unfilled boxes. Abbreviations: BTB: broad complex/tram track/bric-a-brac; RD2: repression domain 2; PEST: proline–glutamic acid–serine–threonine; ZF: zinc finger.
The BTB domain facilitates dimerization as well as interactions with distinct sets of co-repressors [26]. This includes BCL6 co-repressor (BCOR), which associates with Polycomb complex proteins that, in turn, recruit histone demethylases such as enhancer of zeste homolog 2 (EZH2) [27] as well as nuclear co-repressor 1 (NCOR1) and NCOR2/SMRT, which principally associate with histone deacetylase 3 (HDAC3) [28]. RD2 sequences interact with alternative co-repressor metastasis-associated protein 3 (MTA3), which associates with NURD complex proteins to also recruit HDAC2 [29,30] and CTBP [31]. The PEST sequences controls protein stability, mediated by multiple ubiquitination sites in this domain. The disordered RD2/PEST domain also regulates activity via the acetylation of a lysine residue by p300, which can ablate the ability of BCL6 to repress transcription [32], as well as via phosphorylation by mitogen-activated protein kinases (MAPKs) and other kinases [33]. Within the ZF domain, the four C-terminal fingers facilitate binding to specific DNA binding sites related to a core 5′-TTCCTAGAA sequence [34], but this domain has also been implicated in protein–protein interactions [34], particularly with other transcription factors and potentially additional HDACs [35].
Thus, BCL6 functions principally as a transcriptional repressor via the deacetylation and/or demethylation of histones in the vicinity of its target sites [24]. However, it can also impact transcription by directly blocking access of other transcription factors to adjacent or overlapping target sites within the genome [36]. Through these mechanisms, BCL6 is able to actively repress many different genes depending on the cellular context, such as those involved in DNA integrity, cell cycle regulation, and differentiation [24,25,37]. Notably, key targets include a number of transcriptional repressors, including itself, with the ‘repression of repressor’ mechanism leading to the upregulation of many genes indirectly [5].
Indeed, a hallmark of BCL6 and its modus operandi is its interplay with other transcription factors in a range of regulatory networks. These regulatory networks are typically specific for their role in particular cell populations and are often antagonistic. For example, BCL6 shows reciprocal antagonism with the transcription factor B-lymphocyte-induced maturation protein 1 (BLIMP-1, also known as PRDM1) [38], which also shows evolutionary conservation, with homologues in zebrafish and fruit-fly. BCL6 and BLIMP-1 each act to repress the transcription of the other, which serves as a cell fate switch in the differentiation of a range of immune cell lineages [39]. BCL6 also has a reciprocal antagonistic relationship with STAT5, which is activated by various cytokines to directly repress BCL6 transcription via binding to tetrameric sites in the BCL6 promoter [11,40]. Conversely, BCL6 negatively regulates a subset of STAT5-responsive genes via overlapping DNA binding motifs [41]. This reciprocal regulatory relationship is conserved, with almost identical tandem tetrameric STAT5 binding sites found within the zebrafish bcl6 promoter. Fruit-fly ken has also been demonstrated to be a target gene for its STAT homologue, Marelle (Stat92E) [42,43], while the Ken protein is able to block the expression of Stat92E target genes [44]. BCL6 also inhibits the actions of other transcription factors that drive differentiation down alternative pathways, such as TBX21, GATA3, and RORA [5].
5. Key Roles Played by BCL6
BCL6 has been shown to play important roles in the development and function of both lymphoid and myeloid immune cells, with additional roles outside the immune system.
5.1. B Cell Development and Function
BCL6 has been firmly established as a key regulator of GC B cells, which are fundamental for producing the high-affinity, class-switched antibodies essential for potent immunity [45]. BCL6 contributes to several key aspects of GC B cell development, including (i) promoting differentiation by suppressing genes such as BLIMP1; (ii) accommodating somatic hypermutation and class-switch recombination as part of the production of appropriate high-affinity antibodies, mediated by the repression of genes typically triggered by DNA damage, such as TP53 and Ataxia telangiectasia and Rad3-related (ATR); (iii) stimulating proliferation important for clonal expansion, involving genes like cyclin-dependent kinase inhibitor 1A (CDKN1A); and (iv) enabling cell death as part of clonal selection via the repression of anti-apoptotic genes such as BCL2 [3,24,37,46,47]. Bcl6-knockout mice failed to form germinal centers (GC) and produced high-affinity immunoglobulins associated with a distinct absence of GC B cells [2,48,49]. There was a significant reduction in bone marrow pre-B-cell self-renewal and differentiation [50], although plasma cell differentiation was maintained [51,52]. Interestingly, BCL6 downregulation was needed for the establishment and maintenance of GC B memory cells [53]. Mice harboring either a BTB domain mutant [37,54] or RD2 domain mutant [55,56] showed defective GC B cell formation, highlighting the key role for gene repression in mediating the function of BCL6 in cells along this developmental pathway. Such studies revealed that repression of particular genes was specific to different domains. For example, the BTB domain regulated ATR, TP53, and CDKN1A, while the RD2 domain regulated BLIMP1 [57].
5.2. T Cell Development and Function
BCL6 additionally plays a pivotal role in regulating the differentiation of specific T cell populations. In particular, it has been shown to drive the generation of Tfh cells, which facilitate antibody production by GC B cells, at the expense of other T helper (Th) subsets. Consequently, Bcl6-knockout mice displayed a significant decrease in the differentiation (and survival) of Tfh cells, with increased the differentiation of Th1 cells, which are involved in cell-mediated responses to intracellular infections, Th2 cells, which mediate humoral immune responses to extracellular parasites, and Th17 cells, which respond to extracellular infections at mucosal surfaces as well as tissue injury [2,4,58]. Key to this was the ability of BCL6 to inhibit key transcription factors driving the differentiation of Th1 (TBX21), Th2 (GATA3), and Th17 (RORA/RORC) cells [5]. Knockdown of Bcl6 in naive CD4+ T cells also resulted in increased differentiation of Th9 cells, which are involved in helminth infections and tumor immunity [59]. Conditional deletion of Bcl6 in T cells resulted in a 90% decrease in Tfh cells, with a 5-fold reduction in IgG, highlighting the impact on GC B cells [60]. BCL6 has also been shown to play a separate role in Tfh cell maintenance [61]. Interestingly, BTB mutant mice possessed normal Tfh cells [37,54], while RD2 mutants showed only partial impacts [55,56]. These studies revealed a more selective role for gene repression in the Tfh cell lineage, with direct competition with other transcription factors for DNA sites playing a significant role [8]. Other studies have identified additional functions for BCL6 in the generation of CD8+ central memory T (Tcm) cells, which underpin immune surveillance by lymph nodes [6], and the maintenance of long-term CD4+ memory T (Tm) cells, which provide a rapid response to previously encountered antigens [7], processes in which BLIMP1 repression is important [39]. BCL6 also contributes to Treg stability and functionality [62] and also promotes the development of follicular regulatory T (Tfr) cells, a Treg subset that promotes antigen-specific over self-reactive B cell clones in the GC [63,64], as well as various innate-like T cells [65]. A significant decrease in embryonic T lymphocytes was also observed in Bcl6-deficient zebrafish embryos [14], demonstrating conservation of the BCL6-mediated regulation of T cell development, although the impact on specific subsets remains to be delineated.
5.3. Macrophage Development and Function
BCL6 has separately been implicated in the regulation of macrophage development and function. Macrophages from Bcl6-knockout mice showed significantly enhanced M1 polarization but decreased macrophage motility and spreading [10,66], concomitant with disrupted chemokine production [67]. Significantly reduced macrophage numbers and reduced motility in response to wounding were also observed in Bcl6-deficient zebrafish embryos, which also displayed an enhanced susceptibility that was likely due to these macrophage defects [14]. In addition, Ken disruption impacted fruit-fly macrophage-like hemocytes cells [17], collectively suggesting an evolutionarily conserved role in macrophage cells. BCL6 has also been implicated in the development and function of dendritic cell (DC) subpopulations [68,69,70].
5.4. Inflammation
One of the most prominent phenotypes displayed by Bcl6-knockout mice was severe Th2-mediated inflammation with distinctive eosinophilic infiltration, affecting the heart, lungs, liver, and spleen, leading to profound myocarditis and vasculitis [2,52,71]. This is a likely major contributor to their poor survival, with most Bcl6-knockout mice not surviving past 9 weeks of age and some dying as early as 3 weeks [2,52,71]. No inflammatory phenotype was evident in mice harboring either a Bcl6 BTB mutant [37] or Bcl6 RD2 mutant [55], indicating that the repressive function of Bcl6 does not play a significant role in this phenotype, which is instead mediated by the ZF domain. The ability of BCL6 to inhibit inflammation has been largely attributed to its role in suppressing the activation of NLRP3 inflammasomes in macrophages, with Bcl6-deficient murine macrophages showing enhanced NLRP3 activation, leading to a heightened production of key chemokines driving Th2 inflammation [67,72]. Zebrafish embryos deficient in Bcl6 also showed an enhanced expression of the pro-inflammatory cytokine Il1b, also thought to lie downstream of NLRP3, with similar poor survival [14].
5.5. Growth and Other Aspects of Development
BCL6 has been demonstrated to impact growth and development more broadly, with evidence that this also represents a conserved function. Bcl6-knockout mice exhibited significantly reduced postnatal growth [2,71] in concert with a severe decrease in adipose mass [73]. Juvenile Bcl6-deficient zebrafish showed a similar growth retardation and reduced adiposity [14]. The background dysregulation of immunity and inflammation has complicated the interpretation of these phenotypes. However, defective skeletal muscle differentiation was reported in Bcl6-deficient mice [18,23,71], with skeletal-muscle-specific Bcl6 ablation resulting in a 30% reduction in muscle mass, concomitant with a disruption in the expression of genes involved in proteostasis, suggesting a direct effect on this tissue [74]. Altered hepatic lipid metabolism, including reduced expression of lipogenic enzyme genes, has been observed in Bcl6-deficient mice [73], with BCL6 separately identified as a regulator of early adipose commitment in mesenchymal stem cells, controlling expression of early and late adipogenic regulators [75]. However, adipose-specific Bcl6 ablation instead resulted in a specific increase in inguinal (but not perigonadal) adipocyte size and mass, attributed to altered insulin sensitivity and the prevention of steatosis [76].
BCL6 has also been identified as a pro-neurogenic factor during embryonic neocortex development [77], playing roles in progenitor cell differentiation [78] as well as in the survival of cortical neurons [79] and olfactory sensory neurons [80]. The ablation of Ken impacted behavioral responses to visual stimuli in fruit-flies, including escape behavior and synaptic function in the giant fiber system [81], suggesting some potential conservation. The knockdown of zebrafish Bcl6 was separately implicated in aberrant optic cup formation during embryogenesis [22], although this appeared to be unaffected in Bcl6-deficient zebrafish or Bcl6-deficient mice.
Finally, BCL6 can independently impact various aspects of reproduction. Bcl6-knockout mice showed 80% lower spermatozoa at 8 weeks of age, with only 15% being fertile, which correlated with enhanced spermatocyte apoptosis [82]. Ken has also been shown to maintain the self-renewal capacity of testis cells [44] and the proper development of external genitalia [16]. BCL6 has separately been shown to contribute to trophoblast migration/invasion during early placental development [83].
6. Role in Malignancy
The role of BCL6 in the etiology of cancer is complex. It can act as an oncoprotein in many types of cancer, but there are also examples where it performs a tumor-suppressor function. It is able to additionally influence tumor immunity that can be either promote or inhibit tumorigenesis. Finally, BCL6 facilitates therapy resistance across a range of cancer types.
6.1. Oncogenic Functions
BCL6 was first identified as a frequent target gene for chromosomal translocations at chromosome 3q27 in NHL, particularly diffuse large B cell lymphoma (DLBCL) and follicular lymphoma (FL) [1,84], but it was subsequently found to be overexpressed by this and other mechanisms in a range of B cell malignancies extending across B cell lymphomas and leukemias [85]. It plays particularly key roles in more indolent forms of NHL, including FL, the DLBCL subtype primary mediastinal B cell lymphoma (PMBCL), as well as in intravascular large B cell lymphoma (IVLBCL), an aggressive high-grade B cell lymphoma that lies intermediately between DLBCL and Burkitt’s lymphoma [86,87]. It also forms part of the criteria for the revised WHO classification for high-grade B cell lymphomas, defined as those with mutations in MYC in concert with either BCL6 or BCL2, which show poor responses to standard therapy [88]. In the majority of cases, an increased BCL6 expression is mediated by translocations involving fusions of strong promoters active in B cells, such as the heavy and light immunoglobulin chain loci, to the BCL6 gene, leading to deregulated expression [89]. Additionally, deletions and other mutations are common in the 5′ non-coding exon disrupting auto-regulatory BCL6-binding sites [90] and/or repressive STAT5-binding sites [11], leading to enhanced BCL6 expression. In addition, mutations that disrupt factors involved in BCL6 regulation have been identified in B cell malignancies [91]. At the transcriptional level, these include activating mutations in the major positive regulator of BCL6, myocyte enhancer factor 2B (MEF2B) [92], and inactivating mutations in pathways involved in its repression, such as interferon regulatory factor 8 (IRF8) [93]. Post-translationally, this includes inactivating mutations in proteins involved in its acetylation, including CREBBP and EP300 [94], and ubiquitination, such as F box only protein 11 (FBXO11) [95]. Alternatively, enhanced upstream pathways can be involved. For example, BCL6 is highly expressed in B cell acute lymphoblastic leukemia (ALL) mediated by pre-BCR signaling [96] and Ikaros [97]. Regardless of the mechanism, high BCL6 expression ensues, resulting in an enhanced expression of oncogenic genes such as BCL2, MYC, CCND1, and BMI1 to facilitate lymphomagenesis [98].
BCL6 has additionally been identified as a driver of other hematological malignancies [25]. BCL6 is expressed at high levels in acute myeloid leukemia (AML) cell lines and primary AML cells [99], including aggressive AML [100]. It has further been shown to maintain the survival and self-renewal of primary AML by maintaining stem/progenitor cells [99]. It has additionally been demonstrated to facilitate leukemia initiation and self-renewal in chronic myeloid leukemia (CML) [101]. BCL6 also potentially contributes to HTLV-1-mediated adult T cell leukemia by enhancing cell cycle progression [102].
BCL6 has been implicated as an oncoprotein in solid tumors as well. In these cases, it is typically highly expressed in cancer cells, facilitating the enhanced expression of genes involved in proliferation, such as that encoding the cell cycle regulator cyclin D1 (CCND1), and decreasing genes involved in monitoring DNA integrity/repair, notably including TP53 [25]. Thus, BCL6 has been found to be highly expressed in breast cancer and associated with enhanced disease progression and decreased patient survival, with its overexpression increasing tumor growth and invasion in a xenograft model [103]. In ovarian cancer, high BCL6 expression correlated with high tumor burden [104] and poorer prognosis [104,105], with similar associations identified in the context of high-grade glioma [106]. However, the mechanisms involved in the enhanced expression are varied. In glioblastoma, BCL6 represents a key downstream target of MED12, with the increased BCL6 protein able to suppress TP53-mediated apoptosis [107], while it lies downstream of KRAS in lung cancer [108]. In colorectal carcinoma, hypoxia-induced long non-coding RNA (lncRNA) 00205 was shown to bind microRNA (miR)-10a and miR-34c, relieving their suppression of BCL6 mRNAs, with the elevated BCL6 protein then able to block TP53-mediated gene repression, thereby promoting metabolic changes that enhanced cancer progression [109]. In contrast, an increased BCL6 copy number has been observed in squamous cell carcinoma of the lower gastrointestinal tract [110]. Finally, a mechanism involving direct stabilization of the BCL6 protein by the tumorigenic (lncRNA) 00152 has been identified in ovarian cancer [111].
6.2. Tumor-Suppressor Functions
There is a growing recognition that in certain cancer types BCL6 instead functions as a tumor suppressor. Thus, in nasopharyngeal carcinoma, either a reduced BCL6 expression or the presence of a BCL6-SPECC1L fusion, which removes the repressive function of the protein, has been associated with enhanced growth [112]. It also suppressed the development of medulloblastoma in a mouse model through repression of the sonic hedgehog pathway [113]. In gastric cancer, a significantly decreased BCL6 expression has been observed, with a low expression associated with a more malignant clinical phenotype and poor patient prognosis [114]. BCL6 has also been shown to suppress non-alcoholic steatohepatitis (NASH)-induced liver injury associated with hepatic cancer [115].
6.3. Therapy Resistance
BCL6 has been separately demonstrated to inhibit the effectiveness of cancer therapies, with BCL6 often found to be upregulated by the therapeutic agent. This was originally described in the context of B cell lymphoma cell lines [116] but has subsequently been shown to be responsible for resistance to a broad range of inhibitors across multiple cancer types. Thus, BCL6 has been associated with resistance to HDAC inhibitors in DLBCL [117], BCR-ABL1 kinase inhibitors in ALL [118], cytarabine in AML [99], paclitaxel in breast cancer [119], and BET inhibitors in KRAS+ non-small-cell lung cancer [120]. BCL6 was induced by imatinib in gastrointestinal stromal tumor (GIST) [121]. It was also induced by epidermal growth factor receptor (EGFR) inhibitors in non-small-cell lung cancer (NSCLC), resulting in reduced apoptosis [122], while in glioblastoma, either chemotherapy or radiotherapy could upregulate BCL6 expression, which facilitated a blunting of the effectiveness of these therapies [123]. It has been suggested that this role for BCL6 is part of an evolutionarily conserved stress response that enables cancer cells to adapt to stressors, including therapeutic agents [124]. This involves the induction of BCL6 via the HSF-1 stress response, which is, in part, mediated by de-repression of the TOX gene [125]. However, there is evidence for other mechanisms inducing BCL6. For example, in CML, BCL6 upregulation by kinase inhibitors has been shown to be facilitated by interferon-gamma-induced STAT1 activation [126] and the relief of STAT5-mediated BCL6 gene repression [118]. Other studies have shown that BCL6 expression impacts the transcriptome in a similar manner to normal cells, such as the repression of TP53, leading to suppressed apoptosis [121].
6.4. Tumor Immunity
Distinct from its other roles in cancer cells, BCL6 separately influences tumor immunity in a variety of ways. BCL6 has been shown to promote the development and maintenance of tumor-associated stem/progenitor-like CD8+ T cells associated with the persistence of anti-tumor responses [127], providing sustained anti-tumor immunity in the context of lung cancer [128] and melanoma [128,129]. Conversely, BCL6 can also preserve the ability of Treg cells to restrict effector T cell function in the tumor microenvironment, with enhanced Treg expression of BCL6 correlating with poor prognosis in colorectal cancer and melanoma lymph node metastasis [130], while BCL6 has also been shown to suppress the infiltration of CD4+ T cells to promote hepatocellular carcinoma development [131]. Finally, BCL6 is able to induce a stem-like memory program in tumor-associated macrophages to promote long-lasting pro-tumor immunity [132].
7. BCL6-Based Therapeutics
The central role played by BCL6 in various aspects of biology and disease has made it an attractive target for therapeutic intervention. A number of therapeutic agents have been developed, which have been comprehensively reviewed [133]. The majority of these are small molecules that reversibly associate with the BTB domain to inhibit its interactions with co-repressors [134]. These include FX1 [135], WK369 [136], WK499 [137], 79-6 [138], CCT369260 [139], and the orally-available WK500B [140] and GSK137 [141], while FX1 has also been formulated as a pro-drug, AP-4-287, to improve water solubility [142]. TMX-2164 also targets the BTB domain but reacts covalently with a tyrosine residue to act as an irreversible inhibitor [143]. In addition, other molecule classes have also been developed, including an aptamer-based Apt48 [144] and a retro-inverso peptide inhibitor RI-BPI [145]. Other therapeutics targeting this domain act by promoting the proteasomal degradation of BCL6 via the ubiquitination pathway, either small molecules triggering polymerization [146] or so-called proteolysis targeting chimera (PROTAC) approaches that recruit E3 ligases [147]. Alternatively, siRNA [148] and miRNA [149] targeting BCL6 transcripts have also been developed.
BCL6 inhibitors have proven efficacious in a number of disease contexts. These include targeting the direct oncogenic role played by BCL6 in hematological malignancies. Efficacy in this context has been demonstrated for FX1 [135], RI-BPI [145], and WK500B [140] in BCL6-dependent DLBCL xenograft models. Such agents have also proven effective in the context of AML, with RI-BPI increasing apoptosis and reducing stem-ness [99] and WK500B inducing cell cycle arrest and apoptosis [137]. The utility of BCL6 inhibitors extends to solid tumors, with WK369 being shown to inhibit the growth and metastasis of ovarian cancer [136]. In a novel approach, the introduction of miR-144-3p, which targets BCL6, was able to suppress the proliferation and invasion of glioma cells [149]. Inhibitors have also been effective in situations where BCL6 mediates resistance to therapy. BI-3802 was shown to restore/enhance the effectiveness of imatinib, promoting increased apoptosis in the setting of GIST [121,150], and FX1 overcame resistance to the JAK1/2 inhibitor ruxolitinib in CRL2-rearranged ALL [151] and synergized with the EGFR inhibitor gefitinib in NSCLC [122], while WK499 synergized with chemotherapy in ALL [137]. Clinical trials are currently underway to evaluate PROTAC-based BCL6 inhibitors in the context of relapsed/refractory NHL, such as ARV-393 (Arvinas, phase 1, NCT06393738) and BMS-986458 (Bristol-Myers-Squibb, phase 1/2, NCT06090539).
Alternatively, BCL6 inhibitors have been utilized in suppressing the normal role of BCL6 in immunity, such as in the settings of immune rejection and autoimmunity. For example, the BCL6 inhibitor 79-6 protected against graft-versus-host disease in an allogeneic hematopoietic stem cell transplantation model, with a concomitant reduction in Tfh cells [138]. FX1 prolonged the long-term survival of cardiac grafts concurrently with a reduction in Tfh and other T cell populations [152]. This agent has also been shown to ameliorate symptoms in a mouse model of lupus-like autoimmunity through attenuating the Tfh, GC B, and Th1 cell responses [153]. Finally, an siRNA targeting BCL6 was able to ameliorate asthma symptoms in a mouse model [148].
8. Conclusions
The knowledge base regarding BCL6 is very large, with its functions in lymphoid cells and lymphoid malignancies being understood in intricate detail, often establishing key paradigms in their relevant fields. However, there is still much to learn, including the roles played by BCL6 across innate immune cell populations and in other aspects of biology, as well as the transcriptional networks it influences, extending to those involving BCL6B and other so-called ZBTB proteins. Deeper understanding is also required regarding the role of BCL6 in the non-malignant diseases where it has been implicated, such as pre-eclampsia [154], endometriosis [155], and ischemic stroke [156]. Moreover, the multiple roles played by BCL6 mean that therapeutic approaches need to be developed with an abundance of caution to ameliorate potential adverse effects. For example, in B cell lymphoma, the use of BCL6 inhibitors relieved the BCL6-mediated repression of BCL2, leading to addiction to this oncogene [157]. Moreover, in a mouse bronchopulmonary dysplasia model, the FX1 inhibitor was shown to worsen the pathology, which was attributed to the role of BCL6 in inhibiting inflammation [158]. Therefore, the continued advancement of therapeutic agents allowing for the precise manipulation of BCL6 depending on the disease context is essential.
Author Contributions
Conceptualization, A.C.W.; writing—original draft preparation, C.L., F.L.J.A. and A.C.W.; writing—review and editing, C.L., F.L.J.A. and A.C.W. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
F.L.J.A. was supported by the Higher Committee for Education Development in Iraq (HCED).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Ye B.H., Lista F., Lo Coco F., Knowles D.M., Offit K., Chaganti R.S., Dalla-Favera R. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 1993;262:747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- 2.Dent A.L., Shaffer A.L., Yu X., Allman D., Staudt L.M. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 3.Phan R.T., Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 4.Nurieva R.I., Chung Y., Martinez G.J., Yang X.O., Tanaka S., Matskevitch T.D., Wang Y.H., Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi J., Crotty S. Bcl6-mediated transcriptional regulation of follicular helper T cells (TFH) Trends Immunol. 2021;42:336–349. doi: 10.1016/j.it.2021.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ichii H., Sakamoto A., Kuroda Y., Tokuhisa T. Bcl6 acts as an amplifier for the generation and proliferative capacity of central memory CD8+ T cells. J. Immunol. 2004;173:883–891. doi: 10.4049/jimmunol.173.2.883. [DOI] [PubMed] [Google Scholar]
- 7.Ichii H., Sakamoto A., Arima M., Hatano M., Kuroda Y., Tokuhisa T. Bcl6 is essential for the generation of long-term memory CD4+ T cells. Int. Immunol. 2007;19:427–433. doi: 10.1093/intimm/dxm007. [DOI] [PubMed] [Google Scholar]
- 8.Hatzi K., Nance J.P., Kroenke M.A., Bothwell M., Haddad E.K., Melnick A., Crotty S. BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. J. Exp. Med. 2015;212:539–553. doi: 10.1084/jem.20141380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu R.Y., Wang X., Pixley F.J., Yu J.J., Dent A.L., Broxmeyer H.E., Stanley E.R., Ye B.H. BCL-6 negatively regulates macrophage proliferation by suppressing autocrine IL-6 production. Blood. 2005;105:1777–1784. doi: 10.1182/blood-2004-08-3171. [DOI] [PubMed] [Google Scholar]
- 10.Pixley F.J., Xiong Y., Yu R.Y., Sahai E.A., Stanley E.R., Ye B.H. BCL6 suppresses RhoA activity to alter macrophage morphology and motility. Pt 9J. Cell Sci. 2005;118:1873–1883. doi: 10.1242/jcs.02314. [DOI] [PubMed] [Google Scholar]
- 11.Walker S.R., Nelson E.A., Frank D.A. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene. 2007;26:224–233. doi: 10.1038/sj.onc.1209775. [DOI] [PubMed] [Google Scholar]
- 12.Fukuda T., Miki T., Yoshida T., Hatano M., Ohashi K., Hirosawa S., Tokuhisa T. The murine BCL6 gene is induced in activated lymphocytes as an immediate early gene. Oncogene. 1995;11:1657–1663. [PubMed] [Google Scholar]
- 13.Ohtani M., Miyadai T., Hiroishi S. Molecular cloning of the BCL-6 gene, a transcriptional repressor for B-cell differentiation, in torafugu (Takifugu rubripes) Mol. Immunol. 2006;43:1047–1053. doi: 10.1016/j.molimm.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 14.Almohaisen F.L., Heidary S., Sobah M.L., Ward A.C., Liongue C. B cell lymphoma 6A regulates immune development and function in zebrafish. Front. Cell. Infect. Microbiol. 2022;12:887278. doi: 10.3389/fcimb.2022.887278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okabe S., Fukuda T., Ishibashi K., Kojima S., Okada S., Hatano M., Ebara M., Saisho H., Tokuhisa T. BAZF, a novel Bcl6 homolog, functions as a transcriptional repressor. Mol. Cell. Biol. 1998;18:4235–4244. doi: 10.1128/MCB.18.7.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukacsovich T., Yuge K., Awano W., Asztalos Z., Kondo S., Juni N., Yamamoto D. The ken and barbie gene encoding a putative transcription factor with a BTB domain and three zinc finger motifs functions in terminalia development of Drosophila. Arch. Insect Biochem. Physiol. 2003;54:77–94. doi: 10.1002/arch.10105. [DOI] [PubMed] [Google Scholar]
- 17.Arbouzova N.I., Bach E.A., Zeidler M.P. Ken & barbie selectively regulates the expression of a subset of Jak/STAT pathway target genes. Curr. Biol. 2006;16:80–88. doi: 10.1016/j.cub.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 18.Bajalica-Lagercrantz S., Piehl F., Farnebo F., Larsson C., Lagercrantz J. Expression of the BCL6 gene in the pre- and postnatal mouse. Biochem. Biophys. Res. Commun. 1998;247:357–360. doi: 10.1006/bbrc.1998.8551. [DOI] [PubMed] [Google Scholar]
- 19.Hyjek E., Chadburn A., Liu Y.F., Cesarman E., Knowles D.M. BCL-6 protein is expressed in precursor T-cell lymphoblastic lymphoma and in prenatal and postnatal thymus. Blood. 2001;97:270–276. doi: 10.1182/blood.V97.1.270. [DOI] [PubMed] [Google Scholar]
- 20.Bajalica-Lagercrantz S., Piehl F., Lagercrantz J., Lindahl J., Weber G., Kerckeart J.P., Porwit-MacDonald A., Nordenskjold M. Expression of LAZ3/BCL6 in follicular center (FC) B cells of reactive lymph nodes and FC-derived non-Hodgkin lymphomas. Leukemia. 1997;11:594–598. doi: 10.1038/sj.leu.2400577. [DOI] [PubMed] [Google Scholar]
- 21.Allman D., Jain A., Dent A., Maile R.R., Selvaggi T., Kehry M.R., Staudt L.M. BCL-6 expression during B-cell activation. Blood. 1996;87:5257–5268. doi: 10.1182/blood.V87.12.5257.bloodjournal87125257. [DOI] [PubMed] [Google Scholar]
- 22.Lee J., Lee B.K., Gross J.M. Bcl6a function is required during optic cup formation to prevent p53-dependent apoptosis and colobomata. Hum. Mol. Genet. 2013;22:3568–3582. doi: 10.1093/hmg/ddt211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albagli-Curiel O., Dhordain P., Lantoine D., Aurade F., Quief S., Kerckaert J.P., Montarras D., Pinset C. Increased expression of the LAZ3 (BCL6) proto-oncogene accompanies murine skeletal myogenesis. Differentiation. 1998;64:33–44. doi: 10.1046/j.1432-0436.1998.6410033.x. [DOI] [PubMed] [Google Scholar]
- 24.Basso K., Dalla-Favera R. BCL6: Master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv. Immunol. 2010;105:193–210. doi: 10.1016/S0065-2776(10)05007-8. [DOI] [PubMed] [Google Scholar]
- 25.McLachlan T., Matthews W.C., Jackson E.R., Staudt D.E., Douglas A.M., Findlay I.J., Persson M.L., Duchatel R.J., Mannan A., Germon Z.P., et al. B-cell lymphoma 6 (BCL6): From master regulator of humoral immunity to oncogenic driver in pediatric cancers. Mol. Cancer Res. 2022;20:1711–1723. doi: 10.1158/1541-7786.MCR-22-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melnick A., Carlile G., Ahmad K.F., Kiang C.L., Corcoran C., Bardwell V., Prive G.G., Licht J.D. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol. Cell. Biol. 2002;22:1804–1818. doi: 10.1128/MCB.22.6.1804-1818.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huynh K.D., Fischle W., Verdin E., Bardwell V.J. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000;14:1810–1823. doi: 10.1101/gad.14.14.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmad K.F., Melnick A., Lax S., Bouchard D., Liu J., Kiang C.L., Mayer S., Takahashi S., Licht J.D., Prive G.G. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol. Cell. 2003;12:1551–1564. doi: 10.1016/S1097-2765(03)00454-4. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H., Okada S., Hatano M., Okabe S., Tokuhisa T. A new functional domain of Bcl6 family that recruits histone deacetylases. Biochim. Biophys. Acta. 2001;1540:188–200. doi: 10.1016/S0167-4889(01)00128-8. [DOI] [PubMed] [Google Scholar]
- 30.Fujita N., Jaye D.L., Geigerman C., Akyildiz A., Mooney M.R., Boss J.M., Wade P.A. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004;119:75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 31.Mendez L.M., Polo J.M., Yu J.J., Krupski M., Ding B.B., Melnick A., Ye B.H. CtBP is an essential corepressor for BCL6 autoregulation. Mol. Cell. Biol. 2008;28:2175–2186. doi: 10.1128/MCB.01400-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bereshchenko O.R., Gu W., Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nat. Genet. 2002;32:606–613. doi: 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- 33.Moriyama M., Yamochi T., Semba K., Akiyama T., Mori S. BCL-6 is phosphorylated at multiple sites in its serine- and proline-clustered region by mitogen-activated protein kinase (MAPK) in vivo. Oncogene. 1997;14:2465–2474. doi: 10.1038/sj.onc.1201084. [DOI] [PubMed] [Google Scholar]
- 34.Mascle X., Albagli O., Lemercier C. Point mutations in BCL6 DNA-binding domain reveal distinct roles for the six zinc fingers. Biochem. Biophys. Res. Commun. 2003;300:391–396. doi: 10.1016/S0006-291X(02)02873-5. [DOI] [PubMed] [Google Scholar]
- 35.Lemercier C., Brocard M.P., Puvion-Dutilleul F., Kao H.Y., Albagli O., Khochbin S. Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor. J. Biol. Chem. 2002;277:22045–22052. doi: 10.1074/jbc.M201736200. [DOI] [PubMed] [Google Scholar]
- 36.Meyer R.D., Laz E.V., Su T., Waxman D.J. Male-specific hepatic Bcl6: Growth hormone-induced block of transcription elongation in females and binding to target genes inversely coordinated with STAT5. Mol. Endocrinol. 2009;23:1914–1926. doi: 10.1210/me.2009-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang C., Hatzi K., Melnick A. Lineage-specific functions of Bcl-6 in immunity and inflammation are mediated by distinct biochemical mechanisms. Nat. Immunol. 2013;14:380–388. doi: 10.1038/ni.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baxendale S., Davison C., Muxworthy C., Wolff C., Ingham P.W., Roy S. The B-cell maturation factor Blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to Hedgehog signaling. Nat. Genet. 2004;36:88–93. doi: 10.1038/ng1280. [DOI] [PubMed] [Google Scholar]
- 39.Crotty S., Johnston R.J., Schoenberger S.P. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat. Immunol. 2010;11:114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kikuchi M., Miki T., Kumagai T., Fukuda T., Kamiyama R., Miyasaka N., Hirosawa S. Identification of negative regulatory regions within the first exon and intron of the BCL6 gene. Oncogene. 2000;19:4941–4945. doi: 10.1038/sj.onc.1203864. [DOI] [PubMed] [Google Scholar]
- 41.Lin G., LaPensee C.R., Qin Z.S., Schwartz J. Reciprocal occupancy of BCL6 and STAT5 on growth hormone target genes: Contrasting transcriptional outcomes and promoter-specific roles of p300 and HDAC3. Mol. Cell. Endocrinol. 2014;395:19–31. doi: 10.1016/j.mce.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flaherty M.S., Zavadil J., Ekas L.A., Bach E.A. Genome-wide expression profiling in the Drosophila eye reveals unexpected repression of notch signaling by the JAK/STAT pathway. Dev. Dyn. 2009;238:2235–2253. doi: 10.1002/dvdy.21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon W.H., Meinhardt H., Montell D.J. miRNA-mediated feedback inhibition of JAK/STAT morphogen signalling establishes a cell fate threshold. Nat. Cell Biol. 2011;13:1062–1069. doi: 10.1038/ncb2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Issigonis M., Matunis E. The Drosophila BCL6 homolog Ken and Barbie promotes somatic stem cell self-renewal in the testis niche. Dev. Biol. 2012;368:181–192. doi: 10.1016/j.ydbio.2012.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matz H.C., McIntire K.M., Ellebedy A.H. Persistent germinal center responses: Slow-growing trees bear the best fruits. Curr. Opin. Immunol. 2023;83:102332. doi: 10.1016/j.coi.2023.102332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baron B.W., Anastasi J., Thirman M.J., Furukawa Y., Fears S., Kim D.C., Simone F., Birkenbach M., Montag A., Sadhu A., et al. The human programmed cell death-2 (PDCD2) gene is a target of BCL6 repression: Implications for a role of BCL6 in the down-regulation of apoptosis. Proc. Natl. Acad. Sci. USA. 2002;99:2860–2865. doi: 10.1073/pnas.042702599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parekh S., Prive G., Melnick A. Therapeutic targeting of the BCL6 oncogene for diffuse large B-cell lymphomas. Leuk. Lymphoma. 2008;49:874–882. doi: 10.1080/10428190801895345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye B.H., Cattoretti G., Shen Q., Zhang J., Hawe N., de Waard R., Leung C., Nouri-Shirazi M., Orazi A., Chaganti R.S., et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat. Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 49.Toyama H., Okada S., Hatano M., Takahashi Y., Takeda N., Ichii H., Takemori T., Kuroda Y., Tokuhisa T. Memory B cells without somatic hypermutation are generated from Bcl6-deficient B cells. Immunity. 2002;17:329–339. doi: 10.1016/S1074-7613(02)00387-4. [DOI] [PubMed] [Google Scholar]
- 50.Duy C., Yu J.J., Nahar R., Swaminathan S., Kweon S.M., Polo J.M., Valls E., Klemm L., Shojaee S., Cerchietti L., et al. BCL6 is critical for the development of a diverse primary B cell repertoire. J. Exp. Med. 2010;207:1209–1221. doi: 10.1084/jem.20091299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niu H. The proto-oncogene BCL-6 in normal and malignant B cell development. Hematol. Oncol. 2002;20:155–166. doi: 10.1002/hon.689. [DOI] [PubMed] [Google Scholar]
- 52.Cattoretti G., Pasqualucci L., Ballon G., Tam W., Nandula S.V., Shen Q., Mo T., Murty V.V., Dalla-Favera R. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 53.Shehata L., Thouvenel C.D., Hondowicz B.D., Pew L.A., Pritchard G.H., Rawlings D.J., Choi J., Pepper M. Interleukin-4 downregulates transcription factor BCL6 to promote memory B cell selection in germinal centers. Immunity. 2024;57:843–858.e845. doi: 10.1016/j.immuni.2024.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nance J.P., Belanger S., Johnston R.J., Takemori T., Crotty S. T follicular helper cell differentiation is defective in the absence of Bcl6 BTB repressor domain function. J. Immunol. 2015;194:5599–5603. doi: 10.4049/jimmunol.1500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang C., Gonzalez D.G., Cote C.M., Jiang Y., Hatzi K., Teater M., Dai K., Hla T., Haberman A.M., Melnick A. The BCL6 RD2 domain governs commitment of activated B cells to form germinal centers. Cell Rep. 2014;8:1497–1508. doi: 10.1016/j.celrep.2014.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nance J.P., Belanger S., Johnston R.J., Hu J.K., Takemori T., Crotty S. Bcl6 middle domain repressor function is required for T follicular helper cell differentiation and utilizes the corepressor MTA3. Proc. Natl. Acad. Sci. USA. 2015;112:13324–13329. doi: 10.1073/pnas.1507312112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parekh S., Polo J.M., Shaknovich R., Juszczynski P., Lev P., Ranuncolo S.M., Yin Y., Klein U., Cattoretti G., Dalla Favera R., et al. BCL6 programs lymphoma cells for survival and differentiation through distinct biochemical mechanisms. Blood. 2007;110:2067–2074. doi: 10.1182/blood-2007-01-069575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mondal A., Sawant D., Dent A.L. Transcriptional repressor BCL6 controls Th17 responses by controlling gene expression in both T cells and macrophages. J. Immunol. 2010;184:4123–4132. doi: 10.4049/jimmunol.0901242. [DOI] [PubMed] [Google Scholar]
- 59.Bassil R., Orent W., Olah M., Kurdi A.T., Frangieh M., Buttrick T., Khoury S.J., Elyaman W. BCL6 controls Th9 cell development by repressing Il9 transcription. J. Immunol. 2014;193:198–207. doi: 10.4049/jimmunol.1303184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hollister K., Kusam S., Wu H., Clegg N., Mondal A., Sawant D.V., Dent A.L. Insights into the role of Bcl6 in follicular Th cells using a new conditional mutant mouse model. J. Immunol. 2013;191:3705–3711. doi: 10.4049/jimmunol.1300378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alterauge D., Bagnoli J.W., Dahlstrom F., Bradford B.M., Mabbott N.A., Buch T., Enard W., Baumjohann D. Continued Bcl6 expression prevents the transdifferentiation of established Tfh cells into Th1 cells during acute viral infection. Cell Rep. 2020;33:108232. doi: 10.1016/j.celrep.2020.108232. [DOI] [PubMed] [Google Scholar]
- 62.Sawant D.V., Wu H., Yao W., Sehra S., Kaplan M.H., Dent A.L. The transcriptional repressor Bcl6 controls the stability of regulatory T cells by intrinsic and extrinsic pathways. Immunology. 2015;145:11–23. doi: 10.1111/imm.12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fu W., Liu X., Lin X., Feng H., Sun L., Li S., Chen H., Tang H., Lu L., Jin W., et al. Deficiency in T follicular regulatory cells promotes autoimmunity. J. Exp. Med. 2018;215:815–825. doi: 10.1084/jem.20170901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trujillo-Ochoa J.L., Kazemian M., Afzali B. The role of transcription factors in shaping regulatory T cell identity. Nat. Rev. Immunol. 2023;23:842–856. doi: 10.1038/s41577-023-00893-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gioulbasani M., Galaras A., Grammenoudi S., Moulos P., Dent A.L., Sigvardsson M., Hatzis P., Kee B.L., Verykokakis M. The transcription factor BCL-6 controls early development of innate-like T cells. Nat. Immunol. 2020;21:1058–1069. doi: 10.1038/s41590-020-0737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Q., Zhou L., Wang L., Li S., Xu G., Gu H., Li D., Liu M., Fang L., Wang Z., et al. Bcl6 modulates innate immunity by controlling macrophage activity and plays critical role in experimental autoimmune encephalomyelitis. Eur. J. Immunol. 2020;50:525–536. doi: 10.1002/eji.201948299. [DOI] [PubMed] [Google Scholar]
- 67.Toney L.M., Cattoretti G., Graf J.A., Merghoub T., Pandolfi P.P., Dalla-Favera R., Ye B.H., Dent A.L. BCL-6 regulates chemokine gene transcription in macrophages. Nat. Immunol. 2000;1:214–220. doi: 10.1038/79749. [DOI] [PubMed] [Google Scholar]
- 68.Pantano S., Jarrossay D., Saccani S., Bosisio D., Natoli G. Plastic downregulation of the transcriptional repressor BCL6 during maturation of human dendritic cells. Exp. Cell Res. 2006;312:1312–1322. doi: 10.1016/j.yexcr.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 69.Zhang T.T., Liu D., Calabro S., Eisenbarth S.C., Cattoretti G., Haberman A.M. Dynamic expression of BCL6 in murine conventional dendritic cells during in vivo development and activation. PLoS ONE. 2014;9:e101208. doi: 10.1371/journal.pone.0101208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiao H., Ulmert I., Bach L., Huber J., Narasimhan H., Kurochkin I., Chang Y., Holst S., Morbe U., Zhang L., et al. Genomic deletion of Bcl6 differentially affects conventional dendritic cell subsets and compromises Tfh/Tfr/Th17 cell responses. Nat. Commun. 2024;15:3554. doi: 10.1038/s41467-024-46966-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoshida T., Fukuda T., Hatano M., Koseki H., Okabe S., Ishibashi K., Kojima S., Arima M., Komuro I., Ishii G., et al. The role of Bcl6 in mature cardiac myocytes. Cardiovasc. Res. 1999;42:670–679. doi: 10.1016/S0008-6363(99)00007-3. [DOI] [PubMed] [Google Scholar]
- 72.Zhang S., Lin S., Tang Q., Yan Z. Knockdown of miR-205-5p alleviates the inflammatory response in allergic rhinitis by targeting B-cell lymphoma 6. Mol. Med. Rep. 2021;24:818. doi: 10.3892/mmr.2021.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.LaPensee C.R., Lin G., Dent A.L., Schwartz J. Deficiency of the transcriptional repressor B cell lymphoma 6 (Bcl6) is accompanied by dysregulated lipid metabolism. PLoS ONE. 2014;9:e97090. doi: 10.1371/journal.pone.0097090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ramachandran K., Futtner C.R., Sommars M.A., Quattrocelli M., Omura Y., Fruzyna E., Wang J.C., Waldeck N.J., Senagolage M.D., Telles C.G., et al. Transcriptional programming of translation by BCL6 controls skeletal muscle proteostasis. Nat. Metab. 2024;6:304–322. doi: 10.1038/s42255-024-00983-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.He M., Chen K., Li S., Zhang S., Zheng J., Hu X., Gao L., Chen J., Song X., Zhang W., et al. Clinical significance of “double-hit” and “double-protein” expression in primary gastric B-cell lymphomas. J. Cancer. 2016;7:1215–1225. doi: 10.7150/jca.15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Senagolage M.D., Sommars M.A., Ramachandran K., Futtner C.R., Omura Y., Allred A.L., Wang J., Yang C., Procissi D., Evans R.M., et al. Loss of transcriptional repression by BCL6 confers insulin sensitivity in the setting of obesity. Cell Rep. 2018;25:3283–3298.e3286. doi: 10.1016/j.celrep.2018.11.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tiberi L., van den Ameele J., Dimidschstein J., Piccirilli J., Gall D., Herpoel A., Bilheu A., Bonnefont J., Iacovino M., Kyba M., et al. BCL6 controls neurogenesis through Sirt1-dependent epigenetic repression of selective Notch targets. Nat. Neurosci. 2012;15:1627–1635. doi: 10.1038/nn.3264. [DOI] [PubMed] [Google Scholar]
- 78.Cossard A., Stam K., Smets A., Jossin Y. MKL/SRF and Bcl6 mutual transcriptional repression safeguards the fate and positioning of neocortical progenitor cells mediated by RhoA. Sci. Adv. 2023;9:eadd0676. doi: 10.1126/sciadv.add0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wiegreffe C., Wahl T., Joos N.S., Bonnefont J., Liu P., Britsch S. Developmental cell death of cortical projection neurons is controlled by a Bcl11a/Bcl6-dependent pathway. EMBO Rep. 2022;23:e54104. doi: 10.15252/embr.202154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Otaki J.M., Hatano M., Matayoshi R., Tokuhisa T., Yamamoto H. The proto-oncogene BCL6 promotes survival of olfactory sensory neurons. Dev. Neurobiol. 2010;70:424–435. doi: 10.1002/dneu.20786. [DOI] [PubMed] [Google Scholar]
- 81.Allen M.J., Drummond J.A., Sweetman D.J., Moffat K.G. Analysis of two P-element enhancer-trap insertion lines that show expression in the giant fibre neuron of Drosophila melanogaster. Genes Brain Behav. 2007;6:347–358. doi: 10.1111/j.1601-183X.2006.00263.x. [DOI] [PubMed] [Google Scholar]
- 82.Kojima S., Hatano M., Okada S., Fukuda T., Toyama Y., Yuasa S., Ito H., Tokuhisa T. Testicular germ cell apoptosis in Bcl6-deficient mice. Development. 2001;128:57–65. doi: 10.1242/dev.128.1.57. [DOI] [PubMed] [Google Scholar]
- 83.Ritter A., Safdar B.K., Jasmer B., Kreis N.N., Friemel A., Roth S., Solbach C., Louwen F., Yuan J. The function of oncogene B-cell lymphoma 6 in the regulation of the migration and invasion of trophoblastic cells. Int. J. Mol. Sci. 2020;21:8393. doi: 10.3390/ijms21218393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kerckaert J.P., Deweindt C., Tilly H., Quief S., Lecocq G., Bastard C. LAZ3, a novel zinc-finger encoding gene, is disrupted by recurring chromosome 3q27 translocations in human lymphomas. Nat. Genet. 1993;5:66–70. doi: 10.1038/ng0993-66. [DOI] [PubMed] [Google Scholar]
- 85.Capello D., Fais F., Vivenza D., Migliaretti G., Chiorazzi N., Gaidano G., Ferrarini M. Identification of three subgroups of B cell chronic lymphocytic leukemia based upon mutations of BCL-6 and IgV genes. Leukemia. 2000;14:811–815. doi: 10.1038/sj.leu.2401766. [DOI] [PubMed] [Google Scholar]
- 86.Wagner S.D., Ahearne M., Ko Ferrigno P. The role of BCL6 in lymphomas and routes to therapy. Br. J. Haematol. 2011;152:3–12. doi: 10.1111/j.1365-2141.2010.08420.x. [DOI] [PubMed] [Google Scholar]
- 87.Green M.R., Vicente-Duenas C., Romero-Camarero I., Long Liu C., Dai B., Gonzalez-Herrero I., Garcia-Ramirez I., Alonso-Escudero E., Iqbal J., Chan W.C., et al. Transient expression of Bcl6 is sufficient for oncogenic function and induction of mature B-cell lymphoma. Nat. Commun. 2014;5:3904. doi: 10.1038/ncomms4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Davies A. Double-hit lymphoma: So what? Hematol. Oncol. 2019;37((Suppl. S1)):19–23. doi: 10.1002/hon.2581. [DOI] [PubMed] [Google Scholar]
- 89.Akasaka H., Akasaka T., Kurata M., Ueda C., Shimizu A., Uchiyama T., Ohno H. Molecular anatomy of BCL6 translocations revealed by long-distance polymerase chain reaction-based assays. Cancer Res. 2000;60:2335–2341. [PubMed] [Google Scholar]
- 90.Pasqualucci L., Migliazza A., Basso K., Houldsworth J., Chaganti R.S., Dalla-Favera R. Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma. Blood. 2003;101:2914–2923. doi: 10.1182/blood-2002-11-3387. [DOI] [PubMed] [Google Scholar]
- 91.Yang H., Green M.R. Epigenetic programing of B-cell lymphoma by BCL6 and its genetic deregulation. Front. Cell Dev. Biol. 2019;7:272. doi: 10.3389/fcell.2019.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ying C.Y., Dominguez-Sola D., Fabi M., Lorenz I.C., Hussein S., Bansal M., Califano A., Pasqualucci L., Basso K., Dalla-Favera R. MEF2B mutations lead to deregulated expression of the oncogene BCL6 in diffuse large B cell lymphoma. Nat. Immunol. 2013;14:1084–1092. doi: 10.1038/ni.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li H., Kaminski M.S., Li Y., Yildiz M., Ouillette P., Jones S., Fox H., Jacobi K., Saiya-Cork K., Bixby D., et al. Mutations in linker histone genes HIST1H1 B, C, D, and E.; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood. 2014;123:1487–1498. doi: 10.1182/blood-2013-05-500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pasqualucci L., Dominguez-Sola D., Chiarenza A., Fabbri G., Grunn A., Trifonov V., Kasper L.H., Lerach S., Tang H., Ma J., et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Duan S., Cermak L., Pagan J.K., Rossi M., Martinengo C., di Celle P.F., Chapuy B., Shipp M., Chiarle R., Pagano M. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90–93. doi: 10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Geng H., Hurtz C., Lenz K.B., Chen Z., Baumjohann D., Thompson S., Goloviznina N.A., Chen W.Y., Huan J., LaTocha D., et al. Self-enforcing feedback activation between BCL6 and pre-B cell receptor signaling defines a distinct subtype of acute lymphoblastic leukemia. Cancer Cell. 2015;27:409–425. doi: 10.1016/j.ccell.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ge Z., Zhou X., Gu Y., Han Q., Li J., Chen B., Ge Q., Dovat E., Payne J.L., Sun T., et al. Ikaros regulation of the BCL6/BACH2 axis and its clinical relevance in acute lymphoblastic leukemia. Oncotarget. 2017;8:8022–8034. doi: 10.18632/oncotarget.14038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hatzi K., Melnick A. Breaking bad in the germinal center: How deregulation of BCL6 contributes to lymphomagenesis. Trends Mol. Med. 2014;20:343–352. doi: 10.1016/j.molmed.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kawabata K.C., Zong H., Meydan C., Wyman S., Wouters B.J., Sugita M., Goswami S., Albert M., Yip W., Roboz G.J., et al. BCL6 maintains survival and self-renewal of primary human acute myeloid leukemia cells. Blood. 2021;137:812–825. doi: 10.1182/blood.2019001745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Alhumaid M.S., Dasouki M.J., Ahmed S.O., AbalKhail H., Hagos S., Wakil S., Hashmi S.K. Comprehensive genomic analysis of Noonan syndrome and acute myeloid leukemia in adults: A review and future directions. Acta Haematol. 2020;143:583–593. doi: 10.1159/000505715. [DOI] [PubMed] [Google Scholar]
- 101.Hurtz C., Hatzi K., Cerchietti L., Braig M., Park E., Kim Y.M., Herzog S., Ramezani-Rad P., Jumaa H., Muller M.C., et al. BCL6-mediated repression of p53 is critical for leukemia stem cell survival in chronic myeloid leukemia. J. Exp. Med. 2011;208:2163–2174. doi: 10.1084/jem.20110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ishikawa C., Mori N. FX1, a BCL6 inhibitor, reactivates BCL6 target genes and suppresses HTLV-1-infected T cells. Investig. New Drugs. 2022;40:245–254. doi: 10.1007/s10637-021-01196-1. [DOI] [PubMed] [Google Scholar]
- 103.Wu Q., Liu X., Yan H., He Y.H., Ye S., Cheng X.W., Zhu G.L., Wu W.Y., Wang X.N., Kong X.J., et al. B-cell lymphoma 6 protein stimulates oncogenicity of human breast cancer cells. BMC Cancer. 2014;14:418. doi: 10.1186/1471-2407-14-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhu L., Feng H., Jin S., Tan M., Gao S., Zhuang H., Hu Z., Wang H., Song Z., Lin B. High expressions of BCL6 and Lewis y antigen are correlated with high tumor burden and poor prognosis in epithelial ovarian cancer. Tumour. Biol. 2017;39:1010428317711655. doi: 10.1177/1010428317711655. [DOI] [PubMed] [Google Scholar]
- 105.Wang Y.Q., Xu M.D., Weng W.W., Wei P., Yang Y.S., Du X. BCL6 is a negative prognostic factor and exhibits pro-oncogenic activity in ovarian cancer. Am. J. Cancer Res. 2015;5:255–266. [PMC free article] [PubMed] [Google Scholar]
- 106.Xu L., Chen Y., Dutra-Clarke M., Mayakonda A., Hazawa M., Savinoff S.E., Doan N., Said J.W., Yong W.H., Watkins A., et al. BCL6 promotes glioma and serves as a therapeutic target. Proc. Natl. Acad. Sci. USA. 2017;114:3981–3986. doi: 10.1073/pnas.1609758114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Srivastava S., Makala H., Sharma V., Suri V., Sarkar C., Kulshreshtha R. MED12 is overexpressed in glioblastoma patients and serves as an oncogene by targeting the VDR/BCL6/p53 axis. Cell. Mol. Life Sci. 2022;79:104. doi: 10.1007/s00018-021-04056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li K., Liu Y., Ding Y., Zhang Z., Feng J., Hu J., Chen J., Lian Z., Chen Y., Hu K., et al. BCL6 is regulated by the MAPK/ELK1 axis and promotes KRAS-driven lung cancer. J. Clin. Investig. 2022;132:e161308. doi: 10.1172/JCI161308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Meng X., Peng J., Xie X., Yu F., Wang W., Pan Q., Jin H., Huang X., Yu H., Li S., et al. Roles of lncRNA LVBU in regulating urea cycle/polyamine synthesis axis to promote colorectal carcinoma progression. Oncogene. 2022;41:4231–4243. doi: 10.1038/s41388-022-02413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Astaras C., De Vito C., Chaskar P., Bornand A., Khanfir K., Sciarra A., Letovanec I., Corro C., Dietrich P.Y., Tsantoulis P., et al. The first comprehensive genomic characterization of rectal squamous cell carcinoma. J. Gastroenterol. 2023;58:125–134. doi: 10.1007/s00535-022-01937-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang S., Weng W., Chen T., Xu M., Wei P., Li J., Lu L., Wang Y. LINC00152 promotes tumor progression and predicts poor prognosis by stabilizing BCL6 from degradation in the epithelial ovarian cancer. Front. Oncol. 2020;10:555132. doi: 10.3389/fonc.2020.555132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fang S.G., Xia T.L., Fu J.C., Li T., Zhong Q., Han F. BCL6-SPECC1L: A novel fusion gene in nasopharyngeal carcinoma. Technol. Cancer Res. Treat. 2022;21:15330338221139981. doi: 10.1177/15330338221139981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tiberi L., Bonnefont J., van den Ameele J., Le Bon S.D., Herpoel A., Bilheu A., Baron B.W., Vanderhaeghen P. A BCL6/BCOR/SIRT1 complex triggers neurogenesis and suppresses medulloblastoma by repressing Sonic Hedgehog signaling. Cancer Cell. 2014;26:797–812. doi: 10.1016/j.ccell.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 114.Guo S., Deng J., Wang P., Kou F., Wu Z., Zhang N., Zhao Z., Nie Y., Yang L. The malignancy suppression and ferroptosis facilitation of BCL6 in gastric cancer mediated by FZD7 repression are strengthened by RNF180/RhoC pathway. Cell Biosci. 2023;13:73. doi: 10.1186/s13578-023-01020-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kamiya A., Ida K. Liver injury and cell survival in non-alcoholic steatohepatitis regulated by sex-based difference through B cell lymphoma 6. Cells. 2022;11:3751. doi: 10.3390/cells11233751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kurosu T., Fukuda T., Miki T., Miura O. BCL6 overexpression prevents increase in reactive oxygen species and inhibits apoptosis induced by chemotherapeutic reagents in B-cell lymphoma cells. Oncogene. 2003;22:4459–4468. doi: 10.1038/sj.onc.1206755. [DOI] [PubMed] [Google Scholar]
- 117.Fan G., Zhang Y., Li Q., Rong R., Chen S., He L., Li B., Zhuang W. BCL6 confers resistance to HDAC inhibitors in DLBCL. Biochem. Pharmacol. 2024;227:116466. doi: 10.1016/j.bcp.2024.116466. [DOI] [PubMed] [Google Scholar]
- 118.Duy C., Hurtz C., Shojaee S., Cerchietti L., Geng H., Swaminathan S., Klemm L., Kweon S.M., Nahar R., Braig M., et al. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature. 2011;473:384–388. doi: 10.1038/nature09883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sultan M., Nearing J.T., Brown J.M., Huynh T.T., Cruickshank B.M., Lamoureaux E., Vidovic D., Dahn M.L., Fernando W., Coyle K.M., et al. An in vivo genome-wide shRNA screen identifies BCL6 as a targetable biomarker of paclitaxel resistance in breast cancer. Mol. Oncol. 2021;15:2046–2064. doi: 10.1002/1878-0261.12964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Guo J., Liu Y., Lv J., Zou B., Chen Z., Li K., Feng J., Cai Z., Wei L., Liu M., et al. BCL6 confers KRAS-mutant non-small-cell lung cancer resistance to BET inhibitors. J. Clin. Investig. 2021;131:e133090. doi: 10.1172/JCI133090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zeng X., Zhao F., Jia J., Ma X., Jiang Q., Zhang R., Li C., Wang T., Liu W., Hao Y., et al. Targeting BCL6 in gastrointestinal stromal tumor promotes p53-mediated apoptosis to enhance the antitumor activity of imatinib. Cancer Res. 2023;83:3624–3635. doi: 10.1158/0008-5472.CAN-23-0082. [DOI] [PubMed] [Google Scholar]
- 122.Zhou Tran Y., Minozada R., Cao X., Johansson H.J., Branca R.M., Seashore-Ludlow B., Orre L.M. Immediate adaptation analysis implicates BCL6 as an EGFR-TKI combination therapy target in NSCLC. Mol. Cell. Proteomics. 2020;19:928–943. doi: 10.1074/mcp.RA120.002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fabre M.S., Stanton N.M., Slatter T.L., Lee S., Senanayake D., Gordon R.M.A., Castro M.L., Rowe M.R., Taha A., Royds J.A., et al. The oncogene BCL6 is up-regulated in glioblastoma in response to DNA damage, and drives survival after therapy. PLoS ONE. 2020;15:e0231470. doi: 10.1371/journal.pone.0231470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fernando T.M., Marullo R., Pera Gresely B., Phillip J.M., Yang S.N., Lundell-Smith G., Torregroza I., Ahn H., Evans T., Gyorffy B., et al. BCL6 evolved to enable stress tolerance in vertebrates and is broadly required by cancer cells to adapt to stress. Cancer Discov. 2019;9:662–679. doi: 10.1158/2159-8290.CD-17-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tribe A.K.W., Peng L., Teesdale-Spittle P.H., McConnell M.J. BCL6 is a context-dependent mediator of the glioblastoma response to irradiation therapy. Pt 1Int. J. Biol. Macromol. 2024;270:131782. doi: 10.1016/j.ijbiomac.2024.131782. [DOI] [PubMed] [Google Scholar]
- 126.Madapura H.S., Nagy N., Ujvari D., Kallas T., Krohnke M.C.L., Amu S., Bjorkholm M., Stenke L., Mandal P.K., McMurray J.S., et al. Interferon gamma is a STAT1-dependent direct inducer of BCL6 expression in imatinib-treated chronic myeloid leukemia cells. Oncogene. 2017;36:4619–4628. doi: 10.1038/onc.2017.85. [DOI] [PubMed] [Google Scholar]
- 127.Sun Q., Cai D., Liu D., Zhao X., Li R., Xu W., Xie B., Gou M., Wei K., Li Y., et al. BCL6 promotes a stem-like CD8(+) T cell program in cancer via antagonizing BLIMP1. Sci. Immunol. 2023;8:eadh1306. doi: 10.1126/sciimmunol.adh1306. [DOI] [PubMed] [Google Scholar]
- 128.Gong Y., Suzuki T., Kozono H., Kubo M., Nakano N. Tumor-infiltrating CD62L+PD-1-CD8 T cells retain proliferative potential via Bcl6 expression and replenish effector T cells within the tumor. PLoS ONE. 2020;15:e0237646. doi: 10.1371/journal.pone.0237646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Czerwinska P., Rucinski M., Wlodarczyk N., Jaworska A., Grzadzielewska I., Gryska K., Galus L., Mackiewicz J., Mackiewicz A. Therapeutic melanoma vaccine with cancer stem cell phenotype represses exhaustion and maintains antigen-specific T cell stemness by up-regulating BCL6. Oncoimmunology. 2020;9:1710063. doi: 10.1080/2162402X.2019.1710063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li Y., Wang Z., Lin H., Wang L., Chen X., Liu Q., Zuo Q., Hu J., Wang H., Guo J., et al. Bcl6 preserves the suppressive function of regulatory T cells during tumorigenesis. Front. Immunol. 2020;11:806. doi: 10.3389/fimmu.2020.00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li J., Feng J., Li Z., Ni Y., Liu L., Lei X., Chai Z., Zhuang N., Xu J., He Y., et al. B cell lymphoma 6 promotes hepatocellular carcinoma progression by inhibiting tumor infiltrating CD4(+)T cell cytotoxicity through ESM1. NPJ Precis. Oncol. 2024;8:139. doi: 10.1038/s41698-024-00625-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang W., Han Q., Ding Y., Zhou H., Chen Z., Wang J., Xiang J., Song Z., Abbas M., Shi L. Bcl6 drives stem-like memory macrophages differentiation to foster tumor progression. Cell. Mol. Life Sci. 2022;80:14. doi: 10.1007/s00018-022-04660-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gu H., He J., Li Y., Mi D., Guan T., Guo W., Liu B., Chen Y. B-cell lymphoma 6 inhibitors: Current advances and prospects of drug development for diffuse large B-cell lymphomas. J. Med. Chem. 2022;65:15559–15583. doi: 10.1021/acs.jmedchem.2c01433. [DOI] [PubMed] [Google Scholar]
- 134.Ai Y., Hwang L., MacKerell A.D., Jr., Melnick A., Xue F. Progress toward B-cell lymphoma 6 BTB domain inhibitors for the treatment of diffuse large B-cell lymphoma and beyond. J. Med. Chem. 2021;64:4333–4358. doi: 10.1021/acs.jmedchem.0c01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cardenas M.G., Yu W., Beguelin W., Teater M.R., Geng H., Goldstein R.L., Oswald E., Hatzi K., Yang S.N., Cohen J., et al. Rationally designed BCL6 inhibitors target activated B cell diffuse large B cell lymphoma. J. Clin. Investig. 2016;126:3351–3362. doi: 10.1172/JCI85795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wu M., Xie J., Xing Y., Zhang L., Chen H., Tang B., Zhou M., Lv S., Huang D., Jian S., et al. Selectively targeting BCL6 using a small molecule inhibitor is a potential therapeutic strategy for ovarian cancer. Int. J. Biol. Sci. 2024;20:486–501. doi: 10.7150/ijbs.86303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang L., Wu M., Guo W., Zhu S., Li S., Lv S., Li Y., Liu L., Xing Y., Chen H., et al. A small molecule BCL6 inhibitor as chemosensitizers in acute myeloid leukemia. Biomed. Pharmacother. 2023;166:115358. doi: 10.1016/j.biopha.2023.115358. [DOI] [PubMed] [Google Scholar]
- 138.Chen X., Wang Y., Huang X., Geng S., Li C., Zeng L., Huang L., Du X., Weng J., Lai P. Targeting Bcl-6 prevents sclerodermatous chronic graft-versus-host disease by abrogating T follicular helper differentiation in mice. Int. Immunopharmacol. 2023;117:109746. doi: 10.1016/j.intimp.2023.109746. [DOI] [PubMed] [Google Scholar]
- 139.Bellenie B.R., Cheung K.J., Varela A., Pierrat O.A., Collie G.W., Box G.M., Bright M.D., Gowan S., Hayes A., Rodrigues M.J., et al. Achieving in vivo target depletion through the discovery and optimization of benzimidazolone BCL6 degraders. J. Med. Chem. 2020;63:4047–4068. doi: 10.1021/acs.jmedchem.9b02076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Xing Y., Guo W., Wu M., Xie J., Huang D., Hu P., Zhou M., Zhang L., Zhang Q., Wang P., et al. An orally available small molecule BCL6 inhibitor effectively suppresses diffuse large B cell lymphoma cells growth in vitro and in vivo. Cancer Lett. 2022;529:100–111. doi: 10.1016/j.canlet.2021.12.035. [DOI] [PubMed] [Google Scholar]
- 141.Pearce A.C., Bamford M.J., Barber R., Bridges A., Convery M.A., Demetriou C., Evans S., Gobbetti T., Hirst D.J., Holmes D.S., et al. GSK137, a potent small-molecule BCL6 inhibitor with in vivo activity, suppresses antibody responses in mice. J. Biol. Chem. 2021;297:100928. doi: 10.1016/j.jbc.2021.100928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cai Y., Poli A.N.R., Vadrevu S., Gyampoh K., Hart C., Ross B., Fair M., Xue F., Salvino J.M., Montaner L.J. BCL6 BTB-specific inhibitor reversely represses T-cell activation, Tfh cells differentiation, and germinal center reaction in vivo. Eur. J. Immunol. 2021;51:2441–2451. doi: 10.1002/eji.202049150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Teng M., Ficarro S.B., Yoon H., Che J., Zhou J., Fischer E.S., Marto J.A., Zhang T., Gray N.S. Rationally designed covalent BCL6 inhibitor that targets a tyrosine residue in the homodimer interface. ACS Med. Chem. Lett. 2020;11:1269–1273. doi: 10.1021/acsmedchemlett.0c00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zacharchenko T., Kalverda A.P., Wright S.C. Structural basis of Apt48 inhibition of the BCL6 BTB domain. Structure. 2022;30:396–407.e393. doi: 10.1016/j.str.2021.10.010. [DOI] [PubMed] [Google Scholar]
- 145.Cerchietti L.C., Yang S.N., Shaknovich R., Hatzi K., Polo J.M., Chadburn A., Dowdy S.F., Melnick A. A peptomimetic inhibitor of BCL6 with potent antilymphoma effects in vitro and in vivo. Blood. 2009;113:3397–3405. doi: 10.1182/blood-2008-07-168773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Slabicki M., Yoon H., Koeppel J., Nitsch L., Roy Burman S.S., Di Genua C., Donovan K.A., Sperling A.S., Hunkeler M., Tsai J.M., et al. Small-molecule-induced polymerization triggers degradation of BCL6. Nature. 2020;588:164–168. doi: 10.1038/s41586-020-2925-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.McCoull W., Cheung T., Anderson E., Barton P., Burgess J., Byth K., Cao Q., Castaldi M.P., Chen H., Chiarparin E., et al. Development of a novel B-cell lymphoma 6 (BCL6) PROTAC to provide insight into small molecule targeting of BCL6. ACS Chemical. Biology. 2018;13:3131–3141. doi: 10.1021/acschembio.8b00698. [DOI] [PubMed] [Google Scholar]
- 148.Zhou C.Z., Xiong X., Tan W.J., Wang Y.F., Yang Z., Li X.Y., Yang X.W., Liu X.F., Yu S.F., Wang L.C., et al. Inhibition of Bcl-6 expression ameliorates asthmatic characteristics in mice. Curr. Med. Sci. 2024;44:110–120. doi: 10.1007/s11596-023-2800-z. [DOI] [PubMed] [Google Scholar]
- 149.Zhou J., Liu R. Upregulation of miR-144-3p expression attenuates glioma cell viability and invasion by targeting BCL6. Exp. Ther. Med. 2021;22:1157. doi: 10.3892/etm.2021.10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang X., Zhu L., Ying S., Liao X., Zheng J., Liu Z., Gao J., Niu M., Xu X., Zhou Z., et al. Increased RNA editing sites revealed as potential novel biomarkers for diagnosis in primary Sjogren’s syndrome. J. Autoimmun. 2023;138:103035. doi: 10.1016/j.jaut.2023.103035. [DOI] [PubMed] [Google Scholar]
- 151.Tsuzuki S., Yasuda T., Goto H., Maeda N., Akahane K., Inukai T., Yamamoto H., Karnan S., Ota A., Hyodo T., et al. BCL6 inhibition ameliorates resistance to ruxolitinib in CRLF2-rearranged acute lymphoblastic leukemia. Haematologica. 2023;108:394–408. doi: 10.3324/haematol.2022.280879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xia Y., Jin S., Wu Y. Small-molecule BCL6 inhibitor protects chronic cardiac transplant rejection and inhibits T follicular helper cell expansion and humoral response. Front. Pharmacol. 2023;14:1140703. doi: 10.3389/fphar.2023.1140703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Venkatadri R., Sabapathy V., Dogan M., Mohammad S., Harvey S.E., Simpson S.R., Grayson J.M., Yan N., Perrino F.W., Sharma R. Targeting Bcl6 in the TREX1 D18N murine model ameliorates autoimmunity by modulating T-follicular helper cells and germinal center B cells. Eur. J. Immunol. 2022;52:825–834. doi: 10.1002/eji.202149324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ren Z., Gao Y., Gao Y., Liang G., Chen Q., Jiang S., Yang X., Fan C., Wang H., Wang J., et al. Distinct placental molecular processes associated with early-onset and late-onset preeclampsia. Theranostics. 2021;11:5028–5044. doi: 10.7150/thno.56141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sansone A.M., Hisrich B.V., Young R.B., Abel W.F., Bowens Z., Blair B.B., Funkhouser A.T., Schammel D.P., Green L.J., Lessey B.A., et al. Evaluation of BCL6 and SIRT1 as non-invasive diagnostic markers of endometriosis. Curr. Issues Mol. Biol. 2021;43:1350–1360. doi: 10.3390/cimb43030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wei P., Chen H., Lin B., Du T., Liu G., He J., You C. Inhibition of the BCL6/miR-31/PKD1 axis attenuates oxidative stress-induced neuronal damage. Exp. Neurol. 2021;335:113528. doi: 10.1016/j.expneurol.2020.113528. [DOI] [PubMed] [Google Scholar]
- 157.Dupont T., Yang S.N., Patel J., Hatzi K., Malik A., Tam W., Martin P., Leonard J., Melnick A., Cerchietti L. Selective targeting of BCL6 induces oncogene addiction switching to BCL2 in B-cell lymphoma. Oncotarget. 2016;7:3520–3532. doi: 10.18632/oncotarget.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chen D., Zhao H.M., Deng X.H., Li S.P., Zhou M.H., Wu Y.X., Tong Y., Yu R.Q., Pang Q.F. BCL6 attenuates hyperoxia-induced lung injury by inhibiting NLRP3-mediated inflammation in fetal mouse. Exp. Lung Res. 2024;50:25–41. doi: 10.1080/01902148.2024.2320665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.