Abstract
Vaccines that can reduce the load of latent gammaherpesvirus infections are eagerly sought. One attractive strategy is vaccination against latency-associated proteins, which may increase the efficiency with which T cells recognize and eliminate latently infected cells. However, due to the lack of tractable animal model systems, the effect of latent-antigen vaccination on gammaherpesvirus latency is not known. Here we use the murine gammaherpesvirus model to investigate the impact of vaccination with the latency-associated M2 antigen. As expected, vaccination had no effect on the acute lung infection. However, there was a significant reduction in the load of latently infected cells in the initial stages of the latent infection, when M2 is expressed. These data show for the first time that latent-antigen vaccination can reduce the level of latency in vivo and suggest that vaccination strategies involving other latent antigens may ultimately be successfully used to reduce the long-term latent infection.
Many of the clinically relevant problems associated with gammaherpesvirus infection are a result of a long-lived latent infection. While these viruses are normally effectively contained by the immune system, they can cause a number of clinical problems when patients are immunocompromised. For example, patients with a latent Epstein-Barr virus (EBV) infection can develop lymphoproliferative disease when EBV-transformed B cells grow unchecked by the T-cell response. Similarly, immunocompromised human immunodeficiency virus patients can develop Kaposi's sarcoma, which is strongly associated with a latent infection with human herpesvirus 8. There is evidence to suggest that the risk of developing EBV-associated lymphoproliferative disease correlates with the latent virus burden (16, 17). A vaccine which could either reduce or eliminate the latent infection would therefore be of great clinical benefit.
T cells are known to be important in the control of EBV infection, particularly CD8+ cytotoxic T lymphocytes (12, 15). Many vaccines designed to stimulate T-cell immunity to EBV consist of antigens expressed during productive replication, in an effort to limit the initial infection with the virus (6, 14, 29). These proteins are downregulated during latency, so it is unclear what impact they would have on the latent infection. An alternative approach is to vaccinate with antigens which are expressed during latency (12). Some latency-associated EBV antigens can evade antigen processing and presentation (EBNA-1); however, others are immunogenic (e.g., EBNA-3 and LMP-2) and have been proposed as vaccine candidates (2, 12, 15). To date, no latent-antigen vaccines have been tested for their ability to limit the latent infection. This has mainly been due to the lack of tractable animal model systems to test such strategies.
In this study we used a small animal gammaherpesvirus model, murine gammaherpesvirus 68 (MHV-68), to test the effect of vaccination with a latency-associated antigen on the progression of the latent infection. This system is particularly powerful for immunological studies as several of the T-cell epitopes have been defined in both lytic and latent cycle proteins (5, 18). A CD8+ T-cell response to the latency-associated M2 protein of MHV-68 was previously identified (5), and it was shown that T cells specific for the M291–99/Kd epitope had the capacity to transiently reduce the titer of latently infected cells in vivo (24). This result was obtained using an adoptive-transfer system where large numbers of M291–99/Kd-specific CD8+ T cells were transferred into mice prior to virus infection. Although this shows the potential of these cells to control latency, it remains unclear whether a physiologically relevant number of M291–99/Kd-specific T cells are able to achieve the same effect. In addition it was not known how long the adoptively transferred T cells survived in the host, so it was possible that the transient effect was due to the transferred T cells dying several weeks after transfer. In this study we tested whether an active vaccination approach could induce a sufficiently strong endogenous M291–99/Kd-specific CD8+ T-cell response to reduce the level of latency in infected mice.
MATERIALS AND METHODS
Mice and virus.
MHV-68 virus (clone G2.4) was originally obtained from A. A. Nash (University of Edinburgh, Edinburgh, United Kingdom). Virus was propagated and titered as previously described (22). BALB/c mice were purchased from Jackson Laboratories (Bar Harbor, Maine) or bred at the Trudeau Institute Animal Breeding Facility. Mice were infected intranasally (i.n.) with 400 PFU of virus in 30 μl of Hanks balanced salt solution (HBSS). All animal experiments were approved by the Trudeau Institute Animal Care and Use Committee.
Chromium release assays.
Chromium release assays were performed as described previously (4). Briefly, spleen cells from DNA-vaccinated mice were restimulated in vitro with the M2-expressing S11 cell line (5, 24), which had been irradiated with 3,800 rads. Seven days later the cultures were harvested and added to target cells at effector-to-target ratios ranging from 100 to 1 to 6 to 1. BALB/c 3T3 target cells were loaded with 150 μCi of 51NaCrO4 with or without peptide for 18 h at 37°C. Target cells were washed and incubated with graded numbers of effector cells for 5 h at 37°C. Assays were performed in round-bottomed 96-well plates, and each well received 103 target cells. Spontaneous and maximum releases were measured by incubation with medium alone and 1% Triton X-100, respectively. The percentage of specific release was calculated using the following formula: % specific release = [(experimental − spontaneous)/(maximum − spontaneous)] × 100.
CFSE labeling and culture conditions.
Spleen cells from vaccinated mice were labeled by incubation with 0.5 μM carboxyfluorescein (diacetate) succinimidyl ester (CFSE) diluted in HBSS for 10 min in the dark. Cells were subsequently washed with HBSS or complete tumor medium (7) before use. These cells were restimulated in vitro at a 10:1 ratio with irradiated S11 cells (a tumor cell line expressing M2 [5, 25]) in the presence of human recombinant interleukin-2 (hrIL-2) at 10 U/ml (R&D Systems, Minneapolis, Minn.) at a cell density of 106 cells/ml in 24-well plates. Cultures were incubated for 4 days at 37°C and then harvested and stained with tetramer and antibody to CD8 followed by flow cytometric analysis as described below.
Major histocompatibility complex tetrameric reagents and analysis.
The construction of folded major histocompatibility complex class I-peptide complexes and their tetramerization have been described previously (1). Tetramers were generated by the Molecular Biology Core Facility at the Trudeau Institute. Two tetramers were used: Kd folded with peptide M291–99 (GFNKLRSTL) and Kd folded with peptide HA518–528 (IYSTVASSL) derived from influenza virus A/Puerto Rico/8/34. Tetramers were stored as aliquots at −20°C. Tetramers were titered using an M291–99/Kd-specific CD8+ T-cell line or spleen cells from mice transgenic for the Kd/HA518–528 epitope (11); no cross-reactivity between the two tetramers was detected. Cells were incubated with anti-CD16/CD32 Fc block (Pharmingen, San Diego, Calif.) for 10 min on ice; staining with tetrameric reagents took place for 1 h at room temperature, followed by staining with anti-CD8 tricolor (Caltag, Burlingame, Calif.) on ice for 20 min. Stained samples were analyzed using a FACScan flow cytometer and CELLQuest software (Becton Dickinson Immunocytometry Systems, San Jose, Calif.).
Plaque assay for detection of free virus in lungs.
Plaque assay was used to measure free virus in acutely infected lungs, as previously described (22, 26). Briefly, serial dilutions of homogenized lung tissue were added to 3T3 monolayers in a minimal volume and left to adsorb for 1 h before overlaying with carboxymethylcellulose. After 5 days of incubation at 37°C the assays were fixed with methanol and stained with Giemsa stain, and then the plaques were enumerated microscopically.
Limiting-dilution assays for free and latent virus.
Limiting-dilution assays were performed essentially as described previously (28). Latent-virus titers were evaluated by reactivation assay after culture of live spleen cells with a permissive-cell line. Cell-associated preformed virus in the samples was detected by rupturing the spleen cells by freeze-thaw prior to culture with permissive cells. Primary mouse embryo fibroblasts (MEF) were added to flat-bottomed 96-well plates (104 cells/well) and cultured overnight to allow adherence to the wells. A spleen cell suspension was prepared using organs from infected mice, and a graded number of cells (100,000 to 780 cells/well) were added to the wells containing MEF. Twenty-four replicate wells were used per dilution of spleen cells. The plates were cultured for 3 weeks at 37°C and then examined with a microscope to determine the proportion of wells exhibiting cytopathic effect (CPE). A duplicate sample was subjected to a freeze-thaw, and then dilutions of this sample (100,000 to 12,000 cell equivalents/well) were cultured with MEF as described above to measure the level of cell-associated preformed virus. To calculate the frequency of cells reactivating from latency, we plotted the log10 percentage of wells with no CPE against the number of cells per well. The Poisson distribution predicts that if an average of one cell per well is latently infected then 37% of wells will have no CPE. Thus, the frequency of latently infected cells was read from the number of cells per well, giving 37% of wells with no CPE. The frequency of preformed virus was <1 per 105 cell equivalents in most cases and never greater than 1 per 4.6 × 104 cell equivalents.
DNA vaccine vectors and gene gun vaccination.
The pcDNA3.1 expression vector was obtained from Invitrogen (Carlsbad, Calif.). The full-length M2 gene was inserted between the BamHI and EcoRI restriction enzyme sites, and the resulting plasmid was designated pcDNA3.1/M2. Purified preparations of pcDNA3.1 and pcDNA3.1/M2 plasmids were used to transform Escherichia coli DH5α cells using standard techniques. Cultures of transformed cells were grown in Luria broth, and then plasmid DNA was extracted using a Qiagen Plasmid Maxi Kit (Valencia, Calif.). Plasmid DNA was precipitated onto 1.6-μm gold beads and used to coat the inside of Tefzel tubing (McMaster-Carr, Chicago, Ill.) which was cut into 1.3-cm lengths to form cartridges, as described previously (3). Each cartridge was then confirmed to contain approximately 0.5 to 1 μg of plasmid DNA. Mice to be vaccinated were anesthetized with 2,2,2-tribromoethanol, and then their abdomens were shaved. Two cartridges were administered to each shaved abdomen at nonoverlapping sites using a Helios Gene Gun delivery system (Bio-Rad, Hercules, Calif.). Mice were boosted a further 2 to 3 times at approximately 2- to 3-week intervals by following the same immunization protocol.
QF-PCR.
DNA was extracted from spleen or lung tissue using a Qiagen DNeasy kit, and then it was quantitated using a UV spectrophotometer. DNA (250 to 1,000 ng) was subjected to quantitative fluorescent (QF)-PCR using Taqman Universal PCR Mastermix (Applied Biosystems, Foster City, Calif.), 500 nM primers complementary to the sequence of the open reading frame (ORF) 50 gene (27) (3′ primer, CCCTGAGGCTCAACAATTGG; 5′ primer, GGATACGCCTGTCCAGCATATT), and 200 nM [(3′, 6′-dipivaloylfluoresceinyl)-6-carboxamido-hexyl]-6-O-2(2-cynoethyl)-(N,N-diisopropyl)-phosphoramidite (FAM)/Black Hole Quencher-1-labeled probe complementary to the ORF 50 sequence (TGCAATCTGGCTCAACGCCCG; BioSearch Technologies, Inc., Novato, Calif.). Samples were subjected to 2 min at 50°C (reaction of AmpErase uracil-N-glycosylase), 10 min at 95°C (activation of AmpliTaq Gold), and 40 cycles of 15 s at 95°C and 1 min at 60°C. QF-PCR was performed using an ABI 7700 Sequence Detection system (Applied Biosystems). To construct a standard curve, a graded number of copies of the pTW-27 (FLAG-Rta-genomic) plasmid (containing the genomic ORF 50 gene, kind gift from T.-T. Wu, University of California at Los Angeles) were mixed with 250 ng of DNA from the spleens of normal BALB/c mice and subjected to QF-PCR. A graph was constructed comparing threshold cycle (Ct) with copy number, and this was used to convert the Ct of the test sample into genome copy number. The assay was able to detect fewer than 10 viral genomes per sample. All reactions were performed on two separate occasions with similar results. Similar results were obtained using a second set of primers and probe complementary to the Mta gene of MHV-68 (ORF 57) (3′ primer, AGTGCAGCTTTCAATAGGGTTATACA; 5′ primer, GGGACCCTCTGCTGACACA; FAM/Black Hole Quencher-1-labeled probe, TGTGGTCCTAAAGTATCAGCCAGGCGA). No template controls containing 250 ng of normal BALB/c spleen DNA were negative for both primer-probe sets.
RESULTS
DNA vaccination induced a potent CD8+ T-cell response to the M291–99/Kd epitope.
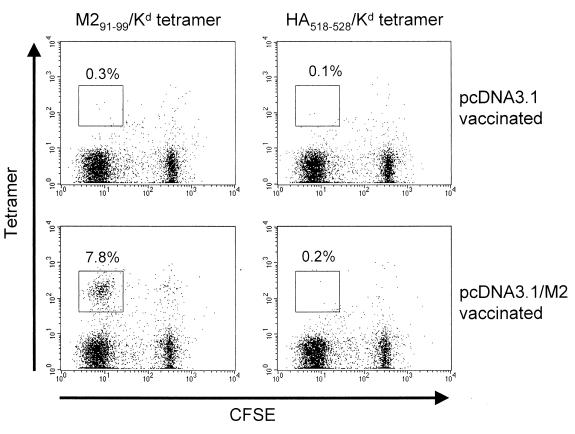
BALB/c mice were vaccinated using a gene gun to introduce plasmid DNA into the shaved skin on the abdomen. One group of mice received the pcDNA3.1/M2 plasmid, containing the full-length M2 gene, and the other received the pcDNA3.1 vector alone. Mice were boosted two to three times after the initial vaccination, at approximately 2- to 3-week intervals, as previous work had shown this to be the most effective boosting regimen (3). To confirm that vaccination was successful, we isolated spleen cells 2 to 3 weeks after the last immunization and assayed for a response to the M291–99/Kd epitope. Thus, spleen cells were labeled with CFSE, a fluorescent dye that is progressively diluted out as cells divide, and then cultured with irradiated S11 cells (a latently infected cell line which expresses M2 [5, 25]) and hrIL-2 in vitro. Cultures were then stained with a tetrameric reagent specific for the M291–99/Kd epitope (24) and anti-CD8 antibody to specifically identify CD8+ M291–99/Kd-specific cells. As shown in Fig. 1, spleen cells from mice vaccinated with pcDNA3.1/M2 contained a substantial population of M291–99/Kd-specific cells (7.8% of CD8+ cells) which expressed low levels of CFSE, indicating they had divided in response to in vitro restimulation. In contrast, in cultures derived from mice vaccinated with the pcDNA3.1 vector alone, no M291–99/Kd-specific cells were detected (0.2% of CD8+ cells). As a specificity control we also stained the cultures with a tetrameric reagent folded with an irrelevant peptide, derived from influenza virus hemagglutinin (HA518–528/Kd). No staining was detected using this tetramer (Fig. 1).
FIG. 1.
DNA vaccination elicits a response specific for the M291–99/Kd epitope from BALB/c mice. Spleen cells from mice vaccinated with either pcDNA3.1 vector or pcDNA3.1/M2 were labeled with CFSE and then cultured in vitro for 4 days with irradiated S11 cells and hrIL-2. Cultures were stained with anti-CD8 and tetramers specific for M291–99/Kd or HA518–528/Kd as a specificity control. The graphs were gated on live, CD8+ T lymphocytes. Numbers refer to the proportion of cells in the gate indicated as a percentage of total CD8+ cells. Data are representative of two experiments.
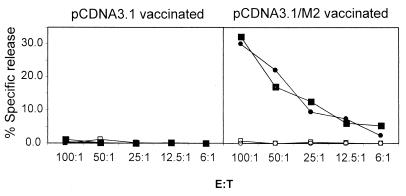
To test whether the M291–99/Kd-specific CD8+ cells in these restimulated cultures were cytolytic, we performed a 51Cr release assay using target cells pulsed with M291–99 peptide. As shown in Fig. 2, in vitro-restimulated cells from mice vaccinated with pcDNA3.1/M2 lysed M291–99 peptide-pulsed targets but not unpulsed target cells. Cells from pcDNA3.1 vector-vaccinated mice lysed neither target cell. Therefore, vaccination with the pcDNA3.1/M2 construct induced a potent CD8+ T-cell response specific for the M291–99/Kd epitope and these cells were cytolytic in vitro.
FIG. 2.
M291–99/Kd-specific CD8+ T cells elicited by DNA vaccination kill M291–99 peptide-pulsed cells in vitro. Spleen cells from vaccinated mice were restimulated in vitro with irradiated S11 cells for 7 days and then added to 51Cr-labeled target cells at various effector/target ratios. Data from two individual mice are shown, one represented by circles and the other by squares. Closed symbols, specific lysis of M291–99 peptide-pulsed targets; open symbols, specific lysis of nonpulsed targets. Data are representative of two experiments.
Vaccination with M2 had no effect on acute lung infection.
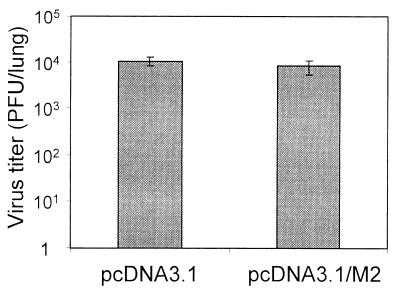
Vaccination with lytic-phase proteins of MHV-68 dramatically reduces the virus titer during acute infection in the lungs after i.n. infection (10, 19). As M2 is latency specific and not expressed in the lytic cycle (5), we would not expect M2 vaccination to have an effect on acute infection. To test this hypothesis, we infected BALB/c mice vaccinated with either pcDNA3.1 or pcDNA3.1/M2 3 weeks after the last boost. A 3-week interval was chosen to ensure that the mice had memory T cells specific for M291–99/Kd at the time of infection, rather than a population of preexisting effector cells. Seven days after infection the lungs were removed from both groups of mice, and the virus titers were measured using a standard plaque assay. As shown in Fig. 3, there was no difference in the virus titers of the two groups, indicating that vaccination with M2 had no effect on the acute lung infection.
FIG. 3.
Comparable lung virus titers in mice vaccinated with M2 or vector only. Mice vaccinated with pcDNA3.1 vector or pcDNA3.1/M2 were infected i.n. with MHV-68 3 weeks after the last immunization, and then the lungs were removed to determine virus titers at 7 days postinfection. Data show the mean lung titers of four mice per group; error bars show one standard deviation. Data are representative of two experiments.
Vaccination with M2 caused a transient reduction in latency.
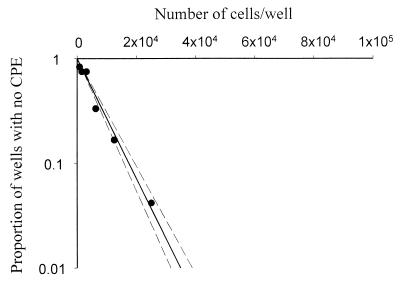
To measure the effect of M2 vaccination on the latent infection with MHV-68, we infected mice vaccinated with either pcDNA3.1/M2 or pcDNA3.1 vector and measured the frequency of latently infected cells in the spleen. We utilized a limiting dilution reactivation assay (28) to detect the frequency of cells capable of reactivating from a latent state and releasing infectious virus. Data from this assay conformed to the Poisson distribution, giving a straight line on a graph plotting the number of cells/well against the log10 of the proportion of wells with no CPE (Fig. 4), showing that the assay measured single-hit kinetics and validating its use for estimating the frequency of cells reactivating from latency. In these assays we distinguished between latent virus and cell-associated preformed virus by performing repeat assays using freeze-thawed cells in which all cells were disrupted but free virus was intact. Free virus was below the limit of detection in the large majority of samples and never present at a frequency of >1 per 4.6 × 104 cell equivalents, indicating that our assays accurately reflected the titers of latent virus in the samples.
FIG. 4.
Representative graph of data obtained from limiting dilution infective center assays. Limiting dilution infective center assays were performed as described in Materials and Methods, and the proportion of wells without CPE was plotted against the number of cells per well. Filled circles, data obtained using live spleen cells from a vaccinated MHV-68-infected mouse. Dashed lines, 95% confidence limits.
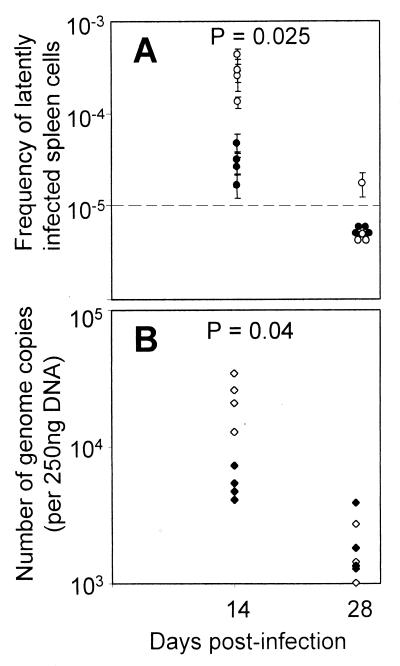
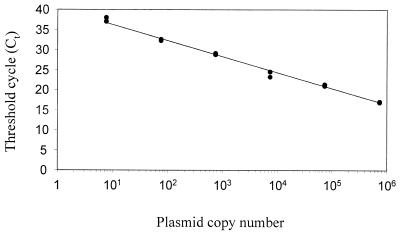
At 14 days postinfection, there was approximately a 10-fold reduction in the frequency of latently infected cells in the pcDNA3.1/M2-vaccinated mice compared with the pcDNA3.1 vector-vaccinated mice (Fig. 5A). At 28 days postinfection the frequency of latently infected cells was much reduced in both groups, and most samples were below the limit of detection in the reactivation assay (Fig. 5A). We therefore used an alternative assay, QF-PCR, to quantitate the viral DNA load. DNA was extracted from spleen cells, and then a known amount of DNA was subjected to QF-PCR. To obtain absolute quantitation of viral DNA load, a standard curve was constructed using serial dilutions of plasmid DNA encoding the ORF 50 gene in a background of normal cellular DNA (see Materials and Methods). When the threshold cycle was plotted against plasmid copy number, we reproducibly obtained a straight line (Fig. 6), validating its use as a calibrator in our experiments. By comparing the threshold cycle of test samples with this standard curve, we could accurately measure the number of viral genomes per unit of input DNA.
FIG. 5.
M2-vaccinated mice have significantly lower latent-virus loads at day 14 postinfection. Spleens were taken from mice vaccinated with either pcDNA3.1 or pcDNA3.1/M2 and then used for either a limiting dilution reactivation assay (A) or the viral genome copy number quantitated by QF-PCR (B) at the times indicated. Open circles, mice vaccinated with pCDNA3.1 vector; closed circles, mice vaccinated with pCDNA3.1/M2; dashed line, limit of detection of the assay. Each point represents the titer obtained for a single mouse; error bars, 95% confidence limits. Data are representative of two experiments. The P value shown represents the probability that the pcDNA3.1 and pcDNA3.1/M2 data come from the same population.
FIG. 6.
A representative standard curve for QF-PCR analysis. QF-PCR was performed using a serial dilution of ORF 50 plasmid DNA in a background of 250 ng of normal cellular DNA as described in Materials and Methods. Each reaction was performed in duplicate; points correspond to the threshold cycle (Ct) for the number of input plasmid molecules indicated.
This assay confirmed our findings at day 14 postinfection, as there was a significant (P = 0.04) difference in the amount of viral DNA between the pcDNA3.1/M2- and pcDNA3.1 vector-vaccinated groups (Fig. 5B). At day 28 postinfection the amount of viral DNA in the spleen was reduced relative to that of day 14; however, there was no significant difference between M2-vaccinated and vector only-vaccinated groups. By 5 months postinfection viral DNA in the spleen was below the limit of detection in some animals and at comparable levels in control and M2-vaccinated mice (data not shown). Therefore, vaccination with M2 caused a transient reduction in viral load; however, this did not result in a reduction of long-term viral latency. MHV-68 also establishes a latent lung infection (21), so we used QF-PCR to quantitate the load in the lungs at 5 months postinfection. Comparable quantities of viral genome were detected in the lungs from both control and M2-vaccinated mice at 5 months postinfection (data not shown). Therefore, DNA vaccination with the M2 gene caused a significant decrease in latency at 14 days postinfection but did not reduce the level of latency in the long term.
DISCUSSION
This report represents the first time that active vaccination with a latent gammaherpesvirus antigen has been shown to have an effect on viral latency. It was previously shown that the adoptive transfer of an M2-specific CD8+ T-cell line resulted in a lower level of latency at 14 days postinfection (24); however, it was unclear whether this effect was merely due to the large numbers of T cells transferred. In this report we now show that mice immunized with M2 could also significantly reduce the initial latent-virus burden compared with nonimmunized animals. Vaccination with lytic-cycle antigens can also cause a transient reduction in the latent-virus titer (10, 19, 20); however, unlike latent-antigen vaccination, this vaccination strategy also reduces the virus titer during acute lung infection. It could therefore be envisaged that an inhibition of virus replication during acute infection causes less virus to seed a latent infection, resulting in lower initial latent-virus titers in the spleen. In contrast, the effect we observe after M2 vaccination is likely the result of M2-specific T cells directly killing latently infected cells in the spleen.
The reduction in latent-virus titer in vaccinated mice did not extend into long-term latency. This was not unexpected, as the M2 gene is expressed transiently around 14 days postinfection and mRNA cannot be detected consistently thereafter (24), suggesting that latently infected cells may be susceptible to T-cell attack only during this time period. Nevertheless, it could be argued that if M2 is expressed in all latently infected cells during this time an aggressive T-cell response may be able to eliminate enough cells to result in a lower long-term latent infection. One explanation for the observed results is that all M2-expressing cells are eliminated but there remains another subset of M2-negative cells which can then persist in the animal for a long time. This would imply that M2 expression does not switch off in some latently infected cells after 14 days postinfection, but rather that all cells that express this gene are eliminated. Interestingly, there is a rapid rise in latently infected cell numbers between days 7 and 14 postinfection (22, 23) and a subsequent sharp decrease in the next 2 weeks. This may imply that M2 expression is part of a viral gene expression program designed to expand the numbers of latently infected cells, analogous to EBV latency III or the growth program.
Given the success of M2 vaccination in reducing viral latency early in the infection, vaccination with latency-associated antigens that are expressed for longer periods in infected cells may cause a more prolonged reduction in latent viral load. This may prove problematic for some EBV antigens such as EBNA-1, as it has evolved a mechanism to inhibit processing by the proteosome. However, another EBV antigen, LMP-2, is clearly immunogenic (8, 9) and there is evidence that it is expressed during long-term latency (13), making it a more attractive vaccine candidate. An alternative approach is to use a multivalent vaccine, incorporating antigens from both the lytic and latent stages of the infection. This approach should both limit the amount of virus available to seed a latent infection and target latently infected cells directly, and it may therefore intervene at two different stages of the virus life cycle.
ACKNOWLEDGMENTS
We thank Scottie Adams and Tim Miller of the Trudeau Institute Molecular Biology Core Facility for the generation of tetrameric reagents.
This work was supported by NIH grants AI37597 (D.L.W.) and AI42927 (M.A.B.), the Trudeau Institute, the Cancer Research Campaign (United Kingdom), and The Royal Society. J.P.S. is a Royal Society University Research Fellow.
REFERENCES
- 1.Altman J D, Moss P H, Goulder P R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 2.Burrows S R, Sculley T B, Misko I S, Schmidt C, Moss D J. An Epstein-Barr virus-specific cytotoxic T cell epitope in EBV nuclear antigen 3 (EBNA 3) J Exp Med. 1990;171:345–349. doi: 10.1084/jem.171.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y, Usherwood E J, Surman S L, Hogg T L, Woodland D L. Long-term CD8+ T cell memory to Sendai virus elicited by DNA vaccination. J Gen Virol. 1999;80:1393–1399. doi: 10.1099/0022-1317-80-6-1393. [DOI] [PubMed] [Google Scholar]
- 4.Cole G A, Clements V K, Garcia E P, Ostrand-Rosenberg S. Allogeneic H-2 antigen expression is insufficient for tumor rejection. Proc Natl Acad Sci USA. 1987;84:8613–8617. doi: 10.1073/pnas.84.23.8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Husain S M, Usherwood E J, Dyson H, Coleclough C, Coppola M A, Woodland D L, Blackman M A, Stewart J P, Sample J T. Murine gammaherpesvirus M2 gene is latency-associated and its protein a target for CD8(+) T lymphocytes. Proc Natl Acad Sci USA. 1999;96:7508–7513. doi: 10.1073/pnas.96.13.7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackman W T, Mann K A, Hoffmann H J, Spaete R R. Expression of Epstein-Barr virus gp350 as a single chain glycoprotein for an EBV subunit vaccine. Vaccine. 1999;17:660–668. doi: 10.1016/s0264-410x(98)00248-5. [DOI] [PubMed] [Google Scholar]
- 7.Kappler J W, Skidmore B, White J, Marrack P. Antigen-inducible, H-2-restricted, interleukin-2-producing T cell hybridomas. Lack of independent antigen and H-2 recognition. J Exp Med. 1981;153:1198–1214. doi: 10.1084/jem.153.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S P, Thomas W A, Murray R J, Khanim F, Kaur S, Young L S, Rowe M, Kurilla M, Rickinson A B. HLA A2.1-restricted cytotoxic T cells recognizing a range of Epstein- Barr virus isolates through a defined epitope in latent membrane protein LMP2. J Virol. 1993;67:7428–7435. doi: 10.1128/jvi.67.12.7428-7435.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S P, Tierney R J, Thomas W A, Brooks J M, Rickinson A B. Conserved CTL epitopes within EBV latent membrane protein 2: a potential target for CTL-based tumor therapy. J Immunol. 1997;158:3325–3334. [PubMed] [Google Scholar]
- 10.Liu L, Usherwood E J, Blackman M A, Woodland D L. T-cell vaccination alters the course of murine herpesvirus 68 infection and the establishment of viral latency in mice. J Virol. 1999;73:9849–9857. doi: 10.1128/jvi.73.12.9849-9857.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morgan D J, Liblau R, Scott B, Fleck S, McDevitt H O, Sarvetnick N, Lo D, Sherman L A. CD8(+) T cell-mediated spontaneous diabetes in neonatal mice. J Immunol. 1996;157:978–983. [PubMed] [Google Scholar]
- 12.Moss D J, Schmidt C, Elliott S, Suhrbier A, Burrows S, Khanna R. Strategies involved in developing an effective vaccine for EBV-associated diseases. Adv Cancer Res. 1996;69:213–245. doi: 10.1016/s0065-230x(08)60864-7. [DOI] [PubMed] [Google Scholar]
- 13.Qu L, Rowe D T. Epstein-Barr virus latent gene expression in uncultured peripheral blood lymphocytes. J Virol. 1992;66:3715–3724. doi: 10.1128/jvi.66.6.3715-3724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 15.Rickinson A B, Moss D J. Human cytotoxic T lymphocyte responses to Epstein-Barr virus infection. Annu Rev Immunol. 1997;15:405–431. doi: 10.1146/annurev.immunol.15.1.405. [DOI] [PubMed] [Google Scholar]
- 16.Riddler S A, Breinig M C, McKnight J L. Increased levels of circulating Epstein-Barr virus (EBV)-infected lymphocytes and decreased EBV nuclear antigen antibody responses are associated with the development of posttransplant lymphoproliferative disease in solid-organ transplant recipients. Blood. 1994;84:972–984. [PubMed] [Google Scholar]
- 17.Savoie A, Perpete C, Carpentier L, Joncas J, Alfieri C. Direct correlation between the load of Epstein-Barr virus-infected lymphocytes in the peripheral blood of pediatric transplant patients and risk of lymphoproliferative disease. Blood. 1994;83:2715–2722. [PubMed] [Google Scholar]
- 18.Stevenson P G, Belz G T, Altman J D, Doherty P C. Changing patterns of dominance in the CD8+ T cell response during acute and persistent murine gamma-herpesvirus infection. Eur J Immunol. 1999;29:1059–1067. doi: 10.1002/(SICI)1521-4141(199904)29:04<1059::AID-IMMU1059>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 19.Stevenson P G, Belz G T, Castrucci M R, Altman J D, Doherty P C. A gamma-herpesvirus sneaks through a CD8(+) T cell response primed to a lytic-phase epitope. Proc Natl Acad Sci USA. 1999;96:9281–9286. doi: 10.1073/pnas.96.16.9281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart J P, Micali N, Usherwood E J, Bonina L, Nash A A. Murine gamma-herpesvirus 68 glycoprotein 150 protects against virus-induced mononucleosis: a model system for gamma-herpesvirus vaccination. Vaccine. 1999;17:152–157. doi: 10.1016/s0264-410x(98)00190-x. [DOI] [PubMed] [Google Scholar]
- 21.Stewart J P, Usherwood E J, Ross A, Dyson H, Nash T. Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J Exp Med. 1998;187:1941–1951. doi: 10.1084/jem.187.12.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sunil-Chandra N P, Efstathiou S, Arno J, Nash A A. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J Gen Virol. 1992;73:2347–2356. doi: 10.1099/0022-1317-73-9-2347. [DOI] [PubMed] [Google Scholar]
- 23.Usherwood E J, Brooks J W, Sarawar S R, Cardin R D, Young W D, Allen D J, Doherty P C, Nash A A. Immunological control of murine gammaherpesvirus infection is independent of perforin. J Gen Virol. 1997;78:2025–2030. doi: 10.1099/0022-1317-78-8-2025. [DOI] [PubMed] [Google Scholar]
- 24.Usherwood E J, Roy D J, Ward K, Surman S L, Dutia B M, Blackman M A, Stewart J P, Woodland D L. Control of gammaherpesvirus latency by latent antigen-specific CD8(+) T cells. J Exp Med. 2000;192:943–952. doi: 10.1084/jem.192.7.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Usherwood E J, Stewart J P, Nash A A. Characterization of tumor cell lines derived from murine gammaherpesvirus-68-infected mice. J Virol. 1996;70:6516–6518. doi: 10.1128/jvi.70.9.6516-6518.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Usherwood E J, Stewart J P, Robertson K, Allen D J, Nash A A. Absence of splenic latency in murine gammaherpesvirus 68-infected B cell-deficient mice. J Gen Virol. 1996;77:2819–2825. doi: 10.1099/0022-1317-77-11-2819. [DOI] [PubMed] [Google Scholar]
- 27.Virgin H W, IV, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weck K E, Barkon M L, Yoo L I, Speck S H, Virgin H I. Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J Virol. 1996;70:6775–6780. doi: 10.1128/jvi.70.10.6775-6780.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson A D, Lovgren-Bengtsson K, Villacres-Ericsson M, Morein B, Morgan A J. The major Epstein-Barr virus (EBV) envelope glycoprotein gp340 when incorporated into Iscoms primes cytotoxic T-cell responses directed against EBV lymphoblastoid cell lines. Vaccine. 1999;17:1282–1290. doi: 10.1016/s0264-410x(98)00351-x. [DOI] [PubMed] [Google Scholar]