Abstract
The envelope glycoproteins of human T-cell leukemia virus type 1 (HTLV-1) perform functions that are crucial for virus entry into cells. The surface glycoprotein (SU) is responsible for viral recognition of, and binding to, target cells through its interaction with an unknown cell surface receptor. To facilitate molecular analysis of the receptor-binding properties of SU and to characterize the cellular receptor employed by HTLV-1, we have expressed a recombinant SU fused to the Fc domain of human immunoglobulin G. Here, we demonstrate that this novel SU-immunoadhesin retains both the biochemical properties of Fc and the receptor-binding specificity of the HTLV-1 SU. We use this SU-immunoadhesin to demonstrate, by direct cell surface binding assays, that the receptor used by HTLV-1 has been conserved through vertebrate evolution. Moreover, using murine-human somatic cell hybrids we provide data that do not support the previously assigned location for the HTLV-1 receptor on human chromosome 17. Most importantly, we show that many cell lines that are resistant to HTLV-1 envelope-mediated infection and syncytium formation express functional receptors that are recognized by the HTLV-1 SU. Based on our results, we suggest that for some HTLV-1-resistant cell lines the block to viral entry occurs at a late post-receptor-binding step of the entry process. Our findings will be of value in developing new strategies to identify the cellular receptor used by HTLV-1.
Human T-cell leukemia virus (HTLV-1) is the etiologic agent of a rare but aggressive adult T-cell leukemia-lymphoma and a progressive demyelinating disease known as HTLV-1-associated myelopathy or tropical spastic paraparesis. HTLV-1 is endemic in Southern Japan, West Africa, Central and South America, and the Caribbean basin. Although uncommon in Europeans, HTLV-1 infections have been reported among indigenous and immigrant European populations and are prevalent among intravenous-drug users both in Europe and in the United States (reviewed in references 1, 4, 21, and 48). HTLV-1 primarily infects and immortalizes human CD4+ T cells in vivo, but in in vitro coculture systems HTLV-1 infection, viral replication, and virally induced syncytium formation can be supported by a variety of primate and nonprimate cell types (1, 4, 30, 45, 46, 49).
The promiscuous pattern of tropism observed for HTLV-1 in vitro has generated considerable interest in the molecular events that promote viral entry into cells, and a number of informative studies have highlighted the crucial role played by the viral envelope glycoproteins in the entry process (31–36, 44). The envelope is expressed as a 68-kDa precursor that is posttranslationally cleaved by a cellular protease to yield the 46-kDa surface glycoprotein (gp46, SU) and a 21-kDa transmembrane glycoprotein (gp21, TM) (4, 8, 31, 36). The gp46 surface glycoprotein remains associated with the transmembrane glycoprotein by noncovalent interactions following precursor cleavage, and this envelope complex is retained on the surfaces of virions or infected cells by the membrane-spanning region of TM. Accumulating evidence suggests that these HTLV-1 gene products function in a manner that is analogous to that of other retroviral SU and TM subunits (40, 42, 45). Consequently, it is thought that SU and TM carry out distinct but interdependent functions in the process of viral infection. The specificity for recognition of cells is attributed to SU, which binds to an as yet unidentified cell surface receptor (45, 46, 49), while TM is directly responsible for membrane fusion (25, 35, 36, 42). Models for envelope function suggest that receptor engagement by SU induces changes in envelope conformation that expose the N-terminal hydrophobic fusion domain of TM. Subsequently, the TM fusion domain inserts into the target cell membrane and catalyzes fusion of the viral and cellular membranes by a process that is only beginning to be understood (25, 42). Envelope-mediated membrane fusion may also occur between envelope-expressing HTLV-1-infected cells and adjacent uninfected cells bearing the cellular receptor recognized by HTLV-1. Such cell-to-cell fusion results in considerable cytopathic effect and the production of giant multinucleate cells or syncytia.
Syncytium formation and viral pseudotyping assays have been extensively used to study the cellular and molecular determinants that facilitate membrane fusion (7, 8, 19, 30–34, 49), cell-to-cell viral transfer (9, 10), and the host-encoded factors that inhibit viral entry into cells (11, 20, 38, 47). These studies reveal that cell-to-cell viral transfer is the principal route of transmission to target cells, as cell-free viral particles are poorly infectious (1, 4, 45). To date, a plethora of cell surface antigens have been implicated in the cell-to-cell transfer of HTLV-1; these include Hsc-70 (39), the tetraspan protein C33 (22), a 30- to 31-kDa surface antigen (16), and vascular-cell adhesion molecule 1 (19). Clearly, some of these molecules serve a supportive role in fusion, perhaps functioning as coreceptors or facilitating cell-to-cell contact. However, unequivocal evidence for a functional role as a primary receptor for HTLV-1 is lacking for these candidate antigens, and none of the factors so far described exhibits all of the characteristics predicted for a primary HTLV-1 receptor (reviewed in reference 45). Nevertheless, current evidence suggests that many cell types express receptors recognized by HTLV-1. In fact, remarkably few cell lines are resistant to envelope-induced syncytium formation or infection by viral particles pseudotyped with HTLV-1 envelope. Among these, murine cells do not readily form syncytia or permit envelope-mediated viral entry (17, 43). Consequently, murine-human somatic-cell hybrids have been used to map a gene encoding a putative cellular receptor for HTLV-1 to human chromosome 17 (17, 43). However, these mapping data have been questioned (46). To investigate these issues, we have generated a functional recombinant HTLV-1 SU for characterization of the HTLV-1 receptor.
Previously, Drosophila melanogaster cells have been used to express a variety of lentiviral surface glycoproteins (2, 3, 23), and it was found that in each case the recombinant envelope protein retained receptor-binding activity and that the affinity of these receptor-ligand interactions correlate with the biological properties of the viral isolates from which they were derived (2, 23). Moreover, the recombinant envelope proteins produced in this insect system faithfully recapitulate the immunological, biochemical, and receptor-binding properties of the native virally expressed envelope glycoprotein or envelope glycoproteins expressed in heterologous mammalian expression systems (2, 23). We have now used this system to express a recombinant epitope-tagged form of the HTLV-1 SU that retains the receptor binding properties of the native viral protein while providing a well-defined antigenic epitope for which immunological reagents are widely available.
Immunoadhesins represent fusions between a protein of interest and the Fc region of an immunoglobulin (Ig). Such chimeric proteins are ideal tools for studying receptor-ligand interactions, identifying unknown binding partners, and dissecting the molecular interactions that govern biological processes. In this study, we have expressed the HTLV-1 SU as a fusion to the Fc region of human IgG. We demonstrate that this hybrid molecule faithfully recapitulates both the known and the predicted biochemical properties of SU and the human IgG Fc domain. Using this SU-immunoadhesin we now directly demonstrate, by cell surface-binding assays, that the primary receptor for HTLV-1 is expressed on a variety of cell lines derived from both mammalian and nonmammalian vertebrates. Most importantly, we also demonstrate that many cell types, previously thought to be receptor negative, do in fact express functional receptors recognized by the SU of HTLV-1. Finally, using murine-human somatic-cell hybrids we reexamine the suggestion that the primary receptor for HTLV-1 maps to human chromosome 17.
MATERIALS AND METHODS
Cell culture and transformation.
D. melanogaster Schnieder 2 cells were maintained in Shields and Sang M3 medium supplemented with 10% fetal bovine serum (Gibco-BRL) (24). Cotransfection of cells with plasmid DNA and the selection vector pCOhygro and subsequent isolation of stable cell lines by the hygromycin B selection procedure have been described previously (3, 24). Each transfection cocktail contained 19 μg of envelope expression vector and 1 μg of pCOhygro. Addition of 500 μM CuSO4 to the medium and incubation of the cultures for 4 to 7 days were used to induce expression from the metallothionein promoter in cell cultures that had reached a cell density of 4 × 106 to 6 × 106 cells/ml. The results presented were confirmed with multiple independent stable cell lines.
Mammalian cells were cultured in Dulbecco's minimal essential medium (DMEM) or RPMI 1640–10% fetal bovine serum at 37°C and 5% CO2. Cell lines and their sources were as follows: SupT1, 293T, MolT4, and MT-2 cells were obtained from the National Institutes of Health, AIDS Research and Reference Reagent Program; Cos-1, NSO1, LM(TK−), XC, Madin-Darby bovine kidney (MDBK), and Madin-Darby canine kidney (MDCK) cells were from the European Culture Collection (ECACC); HeLa, Jurkat, and S2 cells were from SmithKline Beecham Pharmaceuticals; NIH 3T3, SV3T3, and MCF7 cells were from the Imperial Cancer Research Fund Culture Collection; GM00346, GM10321, GM10498, GM10502, and GM11941 cells were from the Corriel Cell Repository; and XTC and HuH7 cells were gifts from Brian McStay (University of Dundee) and Richard Elliot (University of Glasgow), respectively.
Construction of plasmids.
Envelope expression plasmids were constructed by standard techniques (37) and consist of a pBR322-based vector carrying the Drosophila metallothionein promoter (PMtn) and the simian virus 40 (SV40) early polyadenylation signal to regulate expression of the HTLV-1 envelope-coding region. Plasmid pMtgp46-Fc was constructed by replacing the human immunodeficiency virus type 1 (HIV-1) gp120 envelope sequences in the vector pMT120Δ32 (2, 3) with a PCR-amplified DNA fragment encoding the gp46 SU open reading frame and a DNA fragment encoding the human IgG Fc region. The pMtgp46-Fc vector encodes the gp46 HTLV-1 SU starting at amino acid 5 (Ser) of mature gp46 (nucleotides 5274 to 6126, which include the sequence with accession number J02029 [41]), fused in frame at the carboxyl terminus to the hinge and Fc regions of human IgG. At the amino terminus, the envelope sequences are fused in frame to the 36-amino-acid signal sequence from human tissue plasminogen activator (tPA). In this type of fusion the tPA signal sequence is removed from the chimeric protein upon secretion and is predicted to yield a mature soluble SU-Fc fusion protein encoding four amino acids from tPA at the amino-terminal end. To ensure the fidelity of PCR-generated fragments and other recombinant sequences, all plasmids were sequenced by the dideoxynucleotide chain termination method using Sequenase version 2.0 (U.S. Biochemicals) or by automated sequencing on an ABI Prism 377 automated DNA sequencer (Perkin-Elmer).
Protein analysis and Western blotting.
Proteins were resolved on sodium dodecyl sulfate (SDS)–10 or 12% polyacrylamide gels and electrophoretically transferred overnight at 150 mA to nitrocellulose membranes (Schleicher & Schuell) as described previously (2, 14, 18). Nitrocellulose membranes were blocked (3, 14), and bound proteins were probed using the rabbit polyclonal anti-envelope antiserum 4077 or anti-human Fc sera at 1:6,000 dilutions and detected by an enhanced-chemiluminescence procedure employing horseradish peroxidase-conjugated secondary antibodies as directed by the manufacturer (Amersham).
Purification of SU-Fc.
Drosophila tissue culture cell-free medium supernatants (400 to 800 ml), harvested from cell lines expressing SU-Fc, were filtered through a 0.22-μm-pore-size filter and applied at 3 ml/min to a concanavalin A affinity column (5-ml bed volume) previously equilibrated in running buffer (50 mM Tris [pH 7.5], 400 mM NaCl, 1 mM CaCl2). Following application of the sample, the column was washed with 10 column volumes of running buffer and bound protein was specifically eluted with 3 column volumes of elution buffer (0.1 M Tris, 250 mM methyl α-d-mannopyranoside, 0.5 M NaCl, pH 7.5) at 1.2 ml/min. SU-Fc was also purified by affinity chromatography using protein A-agarose (Sigma). Drosophila cell-free culture medium supernatants or concanavalin A-purified fractions were applied to a 1-ml protein A-agarose column preequilibrated in wash buffer (50 mM Tris-HCl [pH 7.8], 140 mM NaCl). The column was washed with 10 column volumes of wash buffer, and bound protein was eluted with 3 column volumes of 0.2 M glycine HCl (pH 2.7). Samples were immediately neutralized with 1 M Tris-HCl (pH 7.8). Eluted protein was concentrated using a Vivaspin 6 (Sartorius) microconcentrator (10,000-molecular-weight cutoff), and the buffer was exchanged for storage buffer (0.1 M Tris [pH 7.5], 140 mM NaCl, 20% glycerol) as directed by the manufacturer. Total protein concentration was determined using the Bio-Rad protein assay, and SU-Fc concentrations were determined by comparative Western blotting and densitometry against a known standard. Purified SU-Fc samples were snap-frozen on dry ice and ethanol and stored in small aliquots at −80°C until required.
Flow cytometry.
Target cells (2.5 × 105) were incubated with fractions containing SU-Fc or control fractions lacking SU-Fc in RPMI 1640 medium supplemented with 10% fetal bovine serum in a total volume of 1 ml. Samples were incubated at room temperature on a rotary mixer for 1 h. The cells were pelleted (1,500 rpm for 5 min in an Eppendorf C5415C microcentrifuge), washed, and incubated with fluorescein isothiocyanate (FITC)-conjugated anti-human-Fc antiserum in RPMI medium at room temperature for 30 min in the dark. The cells were washed in phosphate-buffered saline (PBS)–0.1% sodium azide, fixed (0.5% paraformaldehyde in PBS, pH 7.4), and kept in the dark at 4°C until subjected to flow cytometry using FACScan (Becton Dickinson).
For competition binding experiments, 500 ml of conditioned cell-free medium supernatant from HTLV-1 infected MT-2 cells was concentrated 30-fold using Vivaspin 20 30,000-molecular-weight-cutoff concentrators. The concentrated supernatants contained 0.4 μg of Vgp46 per ml as determined by comparative analysis against a known standard. As a control, 3 ml of the concentrated MT-2 medium supernatant was immunodepleted of gp46 by affinity chromatography against rabbit anti-gp46 serum immobilized on a 1-ml protein A-Sepharose Fastflow column (Pharmacia). Control supernatant was also prepared from 150 ml of uninfected MolT4 cells. Target Jurkat cells were incubated for 30 min with control MolT4 supernatants lacking viral gp46, immunodepleted MT-2 supernatant, MT-2 supernatants containing viral gp46, or RPMI 1640 medium containing 1 μg of recombinant HIV-1JRFL gp120 per ml or 1 μg of purified human IgG (Sigma) per ml prior to the addition of 0.5 μg of SU-Fc. Cells were incubated with mixing at room temperature for 1 h, unbound SU-Fc was removed by a washing with fresh medium, and the bound SU-Fc was detected using FITC-conjugated anti-Fc antibody and flow cytometry as described above.
Syncytium interference assay.
Cos-1 cells (ECACC, CB2669) were transfected with pHTE-1 (12) using Fugene 6 as directed by the manufacturer (Boehringer Mannheim). The transfected cells were incubated for 18 h in a 37°C, 5% CO2 incubator. The monolayers of pHTE-1-transfected donor Cos-1 cells and target HeLa cells were resuspended using PBS–2 mM EDTA, washed in PBS, and resuspended in Dulbecco's minimal essential medium, supplemented with 10% fetal bovine serum, at 1.5 × 105 cells/ml. To examine syncytium interference by SU-Fc, HeLa target cells were incubated with partially purified SU-Fc fractions or control fractions lacking SU-Fc for 20 min at room temperature. One milliliter of donor Cos-1 pHTE-1 cells (1.5 × 105 cells/ml) and the pretreated HeLa target cells were mixed and used to plate the wells of six-well dishes. The cells were incubated in a 37°C, 5% CO2 incubator for 15 to 22 h. The cells were fixed (0.1 M phosphate buffer, pH 7.3, 2% formaldehyde, 0.2% glutaraldehyde, 5 mM EGTA) for 20 min and washed three times, each time for 10 min (PBS, 2 mM MgCl2), and then the syncytia were counted by low-power (×100 magnification) light microscopy using an Olympus LX50 microscope.
Pseudotyping assay.
To generate HIV particles pseudotyped with HTLV-1 envelope protein (27), we adapted the improved pseudotyping assay developed by Sutton and Littman (46). Briefly, 293T cells were cotransfected with 10 μg of pcREV (46) and 10 μg of the envelope-defective luciferase-transducing HIV proviral clone pNL4-3.LUC.R−E− (6) in the presence or absence of 10 μg of pHTLV-env-rre, using Fugene 6. Fifteen hours following transfection, 20 mM sodium butyrate was added. Forty-eight to 72 h later, the viral supernatatants were harvested by low-speed centrifugation (1,500 rpm in an MSE mistral 2000 centrifuge) and filtration through a 22-μm-pore-size filter to remove cells. Additional control stocks of pseudotyped HIV-1 particles were generated using pVPack-VSV-G (Stratagene), encoding the vesicular stomatitis virus G glycoprotein (VSV-G), in place of pHTLV-env-rre. Target human HeLa cells were resuspended (PBS–2 mM EDTA), washed, and finally resuspended at 3 × 105 cells/ml in medium containing 5 μg of Polybrene per ml. Triplicate samples of HeLa target cells were incubated in the presence or absence of partially purified sRgp46-Fc or control proteins as indicated for 20 min at room temperature. One milliliter of HeLa cells and 1 ml of HTLV-1 envelope-pseudotyped viral stock (0.2 ml for VSV-G-pseudotyped viral stocks) were mixed, plated into the wells of a six-well dish, and incubated for 8 h at 37°C and 5% CO2. The medium was replaced and the cells were incubated for 24 h, whereupon the cells were harvested and luciferase assays were performed using the Luciferase Assay System (Promega) and a Turner Designs TD-20/20 luminometer as directed by the manufacturers.
RESULTS
Expression of an HTLV-1 SU-immunoadhesin in Drosophila cells.
To express an HTLV-1 SU-immunoadhesin in Drosophila cells, we constructed the vector pMtgp46-Fc (Fig. 1). Plasmid Mtgp46-Fc carries the gp46 envelope coding sequence, starting at the coding sequence for amino acid 5 (Ser) of the mature SU, fused at the carboxyl terminus to the hinge and Fc regions (CH2 and CH3 heavy-chain constant domains) of human IgG via a short spacer arm. In this fusion, the Fc domain replaces a four-amino-acid arginine-rich protease cleavage signal that is found at the carboxyl terminus of the SU (Fig. 1), thereby preventing posttranslational proteolytic removal of SU from the Fc epitope tag. To facilitate secretion from Drosophila cells, the coding sequences of the chimeric protein were fused at the amino terminus to the secretion signal from the human tPA gene. Expression of this transcription unit is regulated by the inducible Drosophila metallothionein promoter and terminated by the SV40 polyadenylation signal (Fig. 1). Drosophila melanogaster S2 cells were cotransfected with pMtgp46-Fc and the selection vector pCOhygro, and stable cell lines were obtained by hygromycin B selection.
FIG. 1.
Constructs used to express SU-Fc. Shown is a simplified diagram of the HTLV-1 genome illustrating the major genes and highlighting the region used to generate the SU expression constructs. Plasmid pMtgp46-Fc, used to express the HTLV-1 SU-immunoadhesin, and the nucleotide coordinates of the env region used (nucleotide numbering follows that of Seiki et al. [41]) are shown below. At the carboxyl terminus four amino acid codons (RSRR) of SU were replaced by the human IgG Fc domain-coding region. Transcription was driven by the inducible Drosophila metallothionein promoter (PMtn), and the transcripts were terminated by use of the SV40 early polyadenylation sequences. For expression and secretion of the recombinant SU-immunoadhesin, the signal sequence and first four amino acids of the mature SU-coding region were replaced with the 36-amino-acid signal sequence from the human tPA gene. Speckled rectangle, tPA sequences; open rectangle, env sequences; hatched rectangle, human IgG Fc sequences; thin line, vector sequences. The predicted structure of the mature protein products expressed are shown in one-letter amino acid code; three amino acids (GAR), derived from the tPA signal sequence, replace the first four amino acids of SU, and the SU sequences are fused at the carboxyl terminus to sequences derived from Fc.
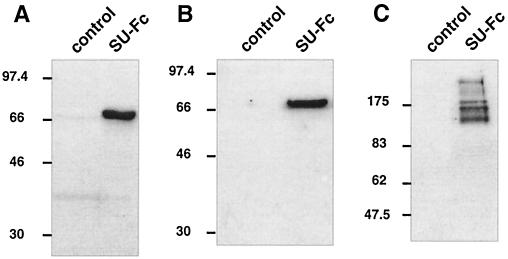
Following induction, Drosophila cells stably transfected with pMtgp46-Fc secreted a 68-kDa antigen that accumulated in the tissue culture medium. This 68-kDa antigen was reactive with the anti-HTLV-1 SU antiserum R4077, as demonstrated by Western blotting of cell-free tissue culture supernatants (Fig. 2A). Importantly, the 68-kDa antigen was also cross-reactive with antibodies raised against the Fc region of human IgG (Fig. 2B). Using these antibodies, antigens of similar electrophoretic mobility were not detected in the tissue culture supernatants from untransfected cells. The apparent molecular mass of the antigen detected by the anti-gp46 and anti-Fc sera was consistent with the predicted molecular mass of the SU-Fc fusion protein, allowing for glycosylation of the recombinant protein. Since the heavy chains of IgG normally exist as disulfide-linked dimers, we suspected that the SU-Fc fusion protein would also form dimers. We therefore compared the mobilities of the recombinant antigen under reducing (Fig. 2B) and nonreducing SDS-polyacrylamide gel electrophoresis (PAGE) conditions by Western blot analysis using anti-Fc sera. Under nonreducing conditions (Fig. 2C), the antigen detected by the anti-Fc sera exhibited a greatly reduced electrophoretic mobility (120 to 170 kDa) that was consistent with the formation of disulfide-linked dimers. The broad smearing of the antigen detected under nonreducing conditions (Fig. 2C) is likely to be due to heterogeneity in the glycosylation pattern of the recombinant protein and to anomalous migration of partially denatured conformations of the fusion protein. Our Western blot analysis indicates that Drosophila cells stably transfected with pMtgp46-Fc efficiently express an HTLV-1 SU-Fc chimeric protein and suggests that this polypeptide is secreted into the culture medium as a disulfide-linked homodimeric fusion protein.
FIG. 2.
Expression of an HTLV-1 SU-immunoadhesin in transfected Drosophila cells. Cell-free culture supernatants from control untransfected Drosophila S2 cells or cells transfected with pMtgp46-Fc (SU-Fc) were assayed for HTLV-1 SU expression by Western blotting. The SU-Fc fusion protein was detected using the anti-gp46 polyclonal rabbit sera 4077 (A) or goat anti-human IgG sera (B) and goat anti-human IgG sera (nonreducing SDS-PAGE) (C).
Characterization of the SU-Fc fusion protein.
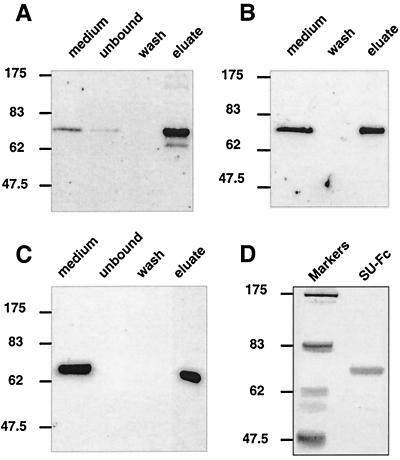
We wished to determine if the recombinant SU-Fc fusion protein retained the biochemical properties of both IgG-Fc and HTLV-1 SU. First, we determined if SU-Fc was glycosylated, as assessed by antigen capture on concanavalin A-Sepharose beads. We found that SU-Fc could be efficiently recovered from tissue culture supernatants by affinity capture on concanavalin A-Sepharose and that the bound fusion protein could be specifically eluted by 250 mM methyl α-d-mannopyranoside (Fig. 3A), indicating that the SU-Fc was extensively glycosylated in Drosophila cells. Moreover, SU-Fc also bound to and was precipitated by protein A-Sepharose (Fig. 3B), indicating that the Fc region of the fusion protein was correctly folded and appropriately presented for recognition by the IgG-binding Fc receptor, protein A (similar data were obtained for protein G; data not shown). In keeping with these observations, we also found that the SU-Fc chimera could be bound to an anti-IgG agarose affinity column and eluted under acid conditions (Fig. 3C). Based on these results, the SU-Fc fusion protein was purified from cell-free and serum-free tissue culture supernatants (Fig. 3D) in two standard chromatographic steps comprising concanavalin A-Sepharose followed by protein A-Sepharose affinity chromatography. The SU-Fc recovered by this procedure (Fig. 3D) was approximately 90% pure and represented a recovery of approximately 60 to 65% as judged by SDS-PAGE and comparative Western blot analysis (data not shown).
FIG. 3.
The SU-immunoadhesin is glycosylated and the Fc domain is functional. Western blots of cell-free medium supernatants and fractions from concanavalin A-Sepharose chromatography (A), protein A-Sepharose precipitation (B), and anti-Fc-Sepharose affinity chromatography (C) are shown. SU-Fc was detected by Western blotting using anti-human IgG sera. (D) Coomassie blue-stained SDS-polyacrylamide gel of purified SU-Fc.
SU-Fc binds to cells and inhibits syncytium formation.
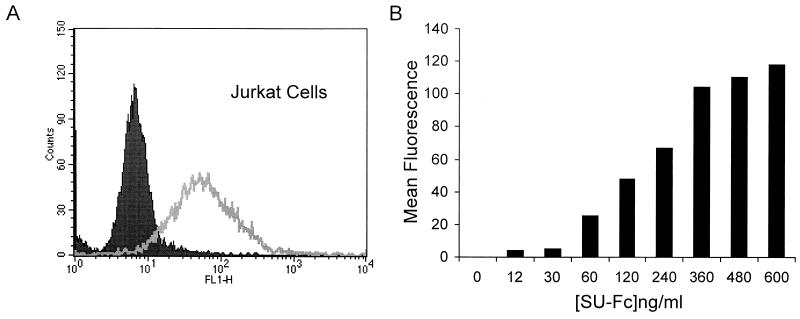
Having expressed and purified the epitope-tagged SU and demonstrated that the Fc domain was immunologically and biochemically functional, we wished to determine if the fusion protein retained the receptor-binding properties of the native virally expressed SU. However, because the primary receptor for HTLV-1 has not been identified, we were unable to examine direct binding of SU-Fc to the isolated receptor in vitro. Instead, we developed a flow cytometry-based assay to monitor direct binding of recombinant SU-Fc to receptor-positive HTLV-1-sensitive T cells. Since Jurkat T cells are permissive for HTLV-1 infection and express an endogenous receptor that is recognized by HTLV-1, we examined the ability of SU-Fc to bind to these cells. Jurkat cells were incubated with affinity-purified SU-Fc, after which the cells were washed, probed for bound SU-Fc using FITC-conjugated anti-human IgG serum, and detected by flow cytometry. The SU-Fc fusion protein bound efficiently to Jurkat cells, as shown by a higher mean fluorescence intensity of cells incubated with SU-Fc than that of cells incubated in the absence of SU-Fc (Fig. 4A). Importantly, binding of SU-Fc to cells was both dose dependent and saturable (Fig. 4B).
FIG. 4.
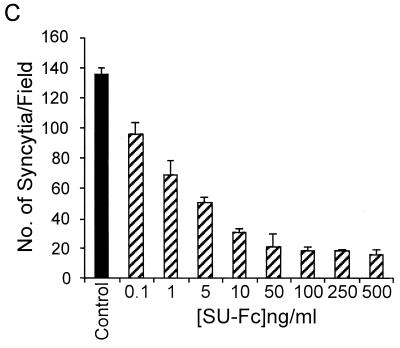
The chimeric SU-Fc protein retains HTLV-1 receptor binding activity. Jurkat cells were incubated with control fractions lacking SU-Fc (solid histogram) or fractions containing SU-Fc (500 ng/ml) (open histogram); cells were probed for bound SU-Fc protein using anti-Fc FITC-conjugated antibody and detected by fluorescence-activated cell sorter analysis. (B) Dose-dependent binding of SU-Fc to Jurkat T cells. Cells were incubated with increasing concentrations of SU-Fc, and bound protein was detected as described above. Control samples were incubated with a protein fraction that lacked SU-Fc, and the basal mean fluorescence intensity from these controls was subtracted from each of the data points shown. (C) Inhibition of syncytium formation by SU-Fc. HeLa target cells expressing endogenous receptor were cocultured with donor Cos-1 cells transfected with the HTLV-1 envelope-expressing vector pHTE-1 in the presence of a control fraction lacking SU-Fc (control) or with increasing concentrations of SU-Fc as indicated. Cultures were scored for syncytium formation per low-power field; the data represent the means and standard deviations from five random fields per culture from three independent cultures.
We predicted that a functional SU-immunoadhesin should be able to inhibit HTLV-1 envelope-mediated syncytium formation when added to the tissue culture medium of cocultured envelope-expressing cells and receptor-expressing target cells. This biological assay is an important test of the specificity of binding for recombinant HTLV-1 SUs. We therefore examined the ability of the SU-Fc fusion protein to prevent HTLV-1 envelope-mediated cell-to-cell fusion in syncytium interference assays. Receptor-positive HeLa cells treated with or without SU-Fc were cocultured with Cos-1 cells transfected with the HTLV-1 envelope expression vector HTE-1. After 24 h of coculture, the numbers of syncytia in cultures containing SU-Fc were scored and compared to those in control cultures lacking SU-Fc. As predicted, exogenous addition of SU-Fc potently blocked syncytium formation in a dose-dependent manner (Fig. 4C), suggesting that the recombinant SU-Fc recognizes and competitively binds to a cell surface protein that is important for HTLV-1-induced cell-to-cell fusion.
Importantly, a variety of cell surface proteins support HTLV-1-induced cell-to-cell fusion but are not themselves primary receptors for HTLV-1 (7, 19, 20). To exclude the possibility that SU-Fc merely blocked syncytium formation by interfering with these nonreceptor interactions, we examined the ability of SU-Fc to block infection of cells by free viral particles. To this end, we adapted the improved pseudotyping assay of Sutton and Littman (46). The envelope-defective recombinant HIV-1 proviral clone pNL4-3.LUC.R−E− (6), which encodes luciferase in place of the viral nef gene, was transfected into 293T cells along with the HTLV-1 envelope expression vector pHTLV-env and the HIV Rev expression vector pRev. Cell-free supernatants containing HIV-1 virions pseudotyped with HTLV-1 envelope proteins were subsequently collected from the transfected 293T cell cultures and used to infect human HeLa cells. Infection of the HeLa cells was monitored by assaying cells for the transduced luciferase marker encoded by the recombinant HIV-1 provirus.
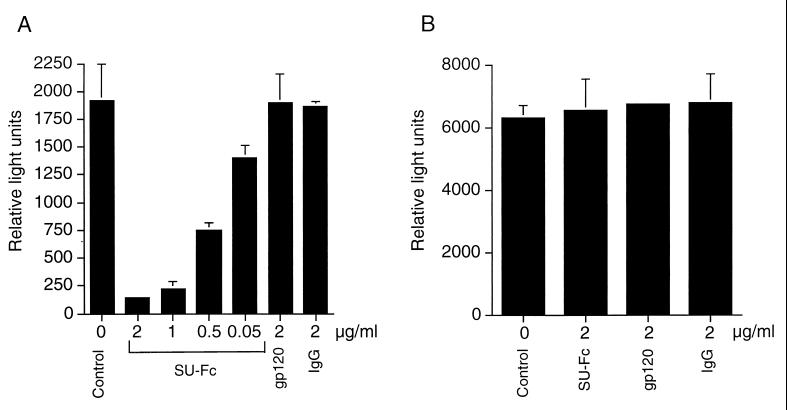
Following infection with HTLV-1 envelope-pseudotyped virions, HeLa cells expressed substantial levels of luciferase activity (Fig. 5A). Significantly, pretreatment of the target cells with SU-Fc resulted in a dramatic drop in the level of luciferase detected (Fig. 5A), indicating that the SU-Fc fusion protein is an effective competitive inhibitor of cell-free viral infection. In contrast, control proteins including recombinant gp120 or irrelevant pooled and purified human IgG did not inhibit entry of HTLV-1 envelope-pseudotyped virions. Moreover, SU-Fc did not block entry of HIV-1 particles pseudotyped with the VSV G-glycoprotein (Fig. 5B), indicating that the SU-Fc-mediated block to viral entry was specific for HTLV-1 and that SU-Fc did not affect the entry of viruses that use alternative receptors. Taken together, the results presented above strongly imply that SU-Fc competes with native viral SU protein for binding to a cell surface receptor that fulfills an essential function in envelope-mediated membrane fusion and viral infection.
FIG. 5.
Incubation of target cells with SU-Fc inhibits infection by virus pseudotyped with HTLV-1 envelope. HeLa cells were incubated in the absence of envelope proteins (Control) or in the presence of SU-Fc (SU-Fc), HIV-1 gp120 (gp120), or pooled and purified human IgG (IgG) at the concentrations indicated. Subsequently, the target cells were infected with luciferase-transducing cell-free HIV-1 stocks pseudotyped with HTLV-1 envelope (A) or VSV-G protein (B). The cells were incubated for 24 h, and luciferase activity was determined. Results represent means and standard deviations from three independent assays.
Viral gp46 competes with SU-Fc for cell binding.
The ability of SU-Fc to inhibit syncytium formation and viral infection of cells suggests that the SU domain of the fusion protein mediates cell surface binding and that the chimeric protein likely binds to the receptor used by HTLV-1 for entry into human T cells. It was therefore important to determine if binding of SU-Fc to cells could be competitively blocked by native virally expressed envelope protein. Conditioned medium supernatants from HTLV-1-infected MT-2 cells contain significant amounts of virus-associated or free soluble gp46 SU (5, 29). Since the viral gp46 is relatively dilute, we concentrated MT-2 medium supernatants 30-fold for these experiments. Also, concentrated control supernatants, lacking Vgp46, were prepared from uninfected MolT4 cells. Jurkat cells were incubated in the presence of control medium supernatants that lacked gp46 or with Vgp46 obtained from the supernatants of MT-2 cells. Subsequently, SU-Fc was added and allowed to bind, and the bound fusion protein was detected using FITC-conjugated anti-Fc antibody and flow cytometry.
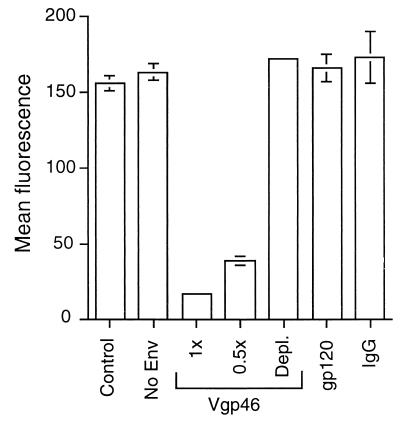
We found that concentrated MT-2 medium supernatants containing Vgp46 potently inhibited binding of the chimeric SU-Fc fusion protein to cells (Fig. 6). In contrast, control Molt 4 supernatants lacking Vgp46 or control proteins, including recombinant gp120 (2) and purified human IgG, had no effect on the binding of SU-Fc to cells (Fig. 6). Moreover, immunodepletion of Vgp46 from the MT-2 conditioned medium supernatants abolished the ability of the supernatants to block binding of SU-Fc to target cells. Our findings that SU-Fc binds to HTLV-1-permissive cells, inhibits virally induced syncytium formation, and competes with virally expressed gp46 for cell surface binding strongly suggest that SU-Fc recognizes and binds to the primary cell surface receptor that is employed by HTLV-1 for entry into cells. Thus, SU-Fc displays the immunological and biochemical properties predicted for an HTLV-1-derived SU-immunoadhesin, having the Fc receptor binding characteristics of the Fc domain of human IgG and the receptor-binding characteristics of the HTLV-1 SU.
FIG. 6.
Native virally expressed gp46 competes with SU-Fc for binding to T cells. Jurkat cells were incubated with 1 ml of unconditioned fresh medium (Control) or 1 ml of concentrated MolT 4 conditioned medium supernatants lacking Vgp46 (No Env) or with 1 or 0.5 ml of concentrated MT-2 medium supernatants containing 0.4 μg of viral gp46 (Vgp46) per ml. Cells were also incubated with 1 ml of concentrated MT-2 medium supernatants from which the Vgp46 had been depleted by passage over an anti-gp46 affinity column (Depl.) or with 1 ml of fresh medium containing 1 μg of recombinant gp120 (gp120) per ml or 1 μg of purified irrelevant human IgG (IgG) per ml. Subsequently, 0.5 μg of SU-Fc was added to each sample, and bound SU-Fc was detected by flow cytometry. The basal mean fluorescence of control cells incubated in the absence of SU-Fc was subtracted from the data presented. Assays were performed in triplicate, and the mean fluorescence and standard deviation are shown.
Murine cells express the HTLV-1 receptor.
Having generated a functional SU-immunoadhesin that retains the receptor specificity of HTLV-1 SU, and which is easily detected and purified by virtue of the Fc epitope tag, we wished to use this recombinant protein to examine the distribution of the HTLV-1 receptor among a variety of cells. Previous studies have suggested that some mammalian cell lines, and in particular murine LM(TK−) cells, lack the primary receptor recognized by HTLV-1 (17, 43); this conclusion is based upon the observation that such cells do not readily support syncytium formation or entry of virus pseudotyped with the HTLV-1 envelope glycoprotein. Using pseudotyping techniques and murine-human somatic-cell hybrids, a putative primary receptor facilitating envelope-mediated infection of murine cells has been mapped to human chromosome 17 (17, 43). We therefore examined LM(TK−) cells and LM(TK−) murine-human somatic-cell hybrids retaining human chromosome 17 for the ability to bind the HTLV-1 SU-immunoadhesin.
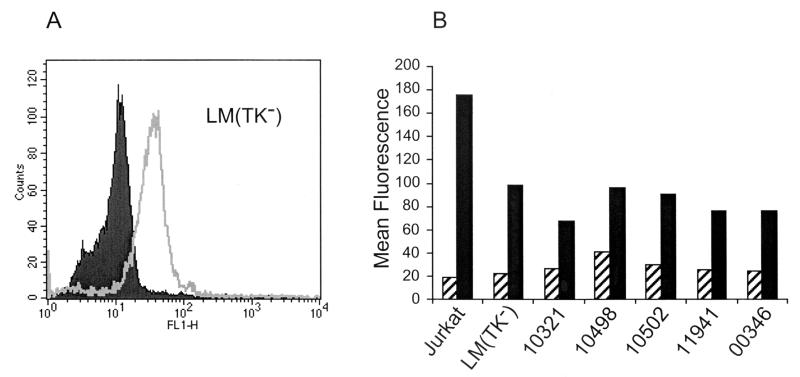
Murine LM(TK−) cells were incubated with SU-Fc, unbound immunoadhesin was washed away, and bound SU-Fc was detected using FITC-conjugated anti-human IgG Fc sera and flow cytometry. Surprisingly, LM(TK−) cells bound significant amounts of SU-Fc (Fig. 7A), an observation consistent with the notion that LM(TK−) cells, previously considered to be receptor negative, do in fact express functional HTLV-1 receptors. Moreover, LM(TK−) somatic-cell hybrids carrying human chromosome 17 (cell lines GM10321 and GM10498) or a fragment of human chromosome 17 (cell line GM10502) also bound SU-Fc to levels that were similar to that of the parental LM(TK−) cell line (Fig. 7B). Importantly, no enhanced binding of SU-Fc to these murine-human chromosome 17 somatic-cell hybrids was observed. Similar SU-Fc binding data were obtained for the control murine A-9 (GM00346) and A-9-human chromosome 11 hybrid (GM11941) cell lines (Fig. 7B). Thus, murine LM(TK−) and murine A9 cells express receptors recognized by HTLV-1 envelope, and binding of SU to these cells is not enhanced by the expression of genes on human chromosome 17.
FIG. 7.
Murine cells express receptors recognized by the HTLV-1 SU. (A) Murine LM(TK−) cells were incubated with SU-Fc (open histogram) or control fractions lacking SU-Fc (solid histogram). Bound protein was detected using FITC-conjugated anti-human Fc antisera and flow cytometry. (B) Jurkat control cells, murine cells, and murine-human somatic-cell hybrids were incubated with SU-Fc (black bars) or without SU-Fc (hatched bars), and the bound protein was detected by flow cytometry. LM(TK−) are control parental murine cells; GM10321 and GM10498 are murine-human somatic-cell hybrids retaining human chromosome 17; GM10502 cells retain a q-terminal fragment of human chromosome 17. Control cell lines include GM00346, a murine A-9 parental cell line, and GM11941, an A-9-derived murine-human somatic-cell hybrid retaining human chromosome 11. The data presented are typical results from at least three independent assays.
The HTLV-1 receptor is widely expressed.
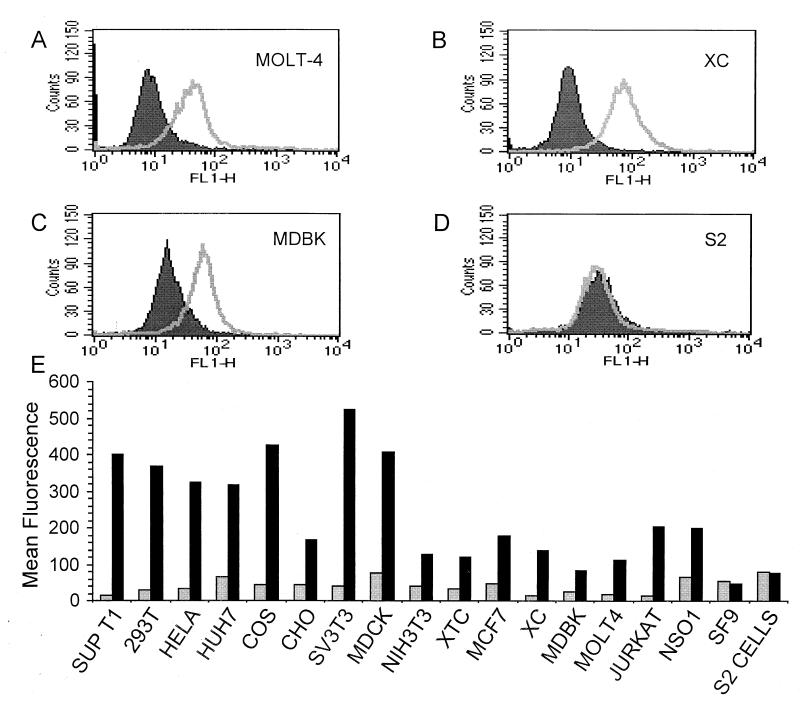
Syncytium formation assays and infection of cells by viral particles pseudotyped with HTLV-1 envelope have been used extensively to monitor the presence of receptors used by HTLV-1 on cells. However, these biological assays, though highly sensitive, represent a relatively indirect method of assessing primary receptor activity. Therefore, we used flow cytometry to directly examine binding of SU-Fc to a variety of human and nonhuman cell types. Cell lines derived from several different animal species were each incubated with SU-Fc or control fractions that lacked the fusion protein. Subsequently, the SU-immunoadhesin that bound to cells was probed using FITC-conjugated anti-Fc sera and detected by flow cytometry (Fig. 8). As anticipated, HTLV-1-permissive cells, such as the human T-cell line MolT4 (Fig. 8A and E) and the rat cell line XC (Fig. 8B), bound SU-Fc efficiently, as did the vast majority of the cell lines examined (Fig. 8E). Thus, most of the cell lines examined express a receptor that is recognized by the HTLV-1 SU. In particular, all of the primate and mammalian cell lines tested bound sRgp46. Moreover, the vast majority of cell lines that are positive for SU-Fc binding also form syncytia when cocultured with cells expressing HTLV-1 envelope proteins (30; our unpublished results). Our results demonstrate, by direct cell surface-binding assays, that the HTLV-1 receptor is widely expressed on a diverse array of vertebrate-derived cell lines. These results confirm and extend previously published data secured from biological assays of receptor activity.
FIG. 8.
Many vertebrate cell lines express receptors for HTLV-1. Human MolT4 T cells (A), Rat XC (B), bovine MDBK (C), and Drosophila S2 (D) cells were incubated with SU-Fc (open histograms) or without SU-Fc (solid histograms). (E) Cells from a variety of species and tissues were examined for receptor expression; cells were incubated with control fractions lacking SU-Fc (grey bars) or with SU-Fc (black bars). Bound protein was detected as described in Materials and Methods and in the legend to Fig. 7. Included are human SupT1, 293T, HeLa, HuH7, Jurkat, and MolT 4 cells; murine SV3T3, NIH 3T3, MCF7, and NSO1 cells; rat XC cells; Chinese hamster ovary (CHO) cells; African green monkey Cos cells; canine MDCK cells; bovine MDBK cells; frog XTC cells; Spodoptera SF9 cells; and Drosophila S2 cells.
Of particular interest, we also examined the binding of SU-Fc to cell lines that do not, or only poorly, support HTLV-1-mediated cell fusion and syncytium formation. As discussed above, murine cells and the cell lines HuH7, MDBK, and MDCK do not readily form syncytia or permit infection by pseudotyped viral particles (30, 49); we therefore examined each of these cell types for SU-Fc binding. In each case, we found that these cell lines, previously reported to be incompetent for HTLV-1-mediated cell fusion, were able to bind SU-Fc to various degrees. Bovine MDBK cells do not undergo syncytium formation, yet these cells bound significant amounts of SU-Fc (Fig. 8C); similar results were obtained for the dog and human cell lines MDCK and HuH7, respectively (Fig. 8E). In addition, we also observed binding of the SU-immunoadhesin to the murine cell lines NIH 3T3, SV3T3, NSO1 and MCF7 (Fig. 8E). Our data indicate that all of these cell types express a functional cell surface receptor that is recognized by the HTLV-1 SU.
Finally, we examined the ability of SU-Fc to bind to nonmammalian cells. Surprisingly, the Xenopus somatic-cell line XTC bound significant amounts of the SU-immunoadhesin (Fig. 8E), suggesting that the cell surface antigen recognized by HTLV-1 has counterparts in nonmammalian vertebrates. In contrast, insect cells SF9 (Spodoptera frugiperda) (Fig. 8E) and S2 (D. melanogaster) (Fig. 8D and E) did not bind SU-Fc to any detectable level. Our results strongly suggest that insect cells do not possess cell surface antigens that are recognized as receptors by HTLV-1.
DISCUSSION
HTLV-1 SU plays a key role in the events that promote viral entry into human cells. SU is directly responsible for recognition of target cells and anchoring of the virion or virus-producing cell to the target cell membrane. By binding to an unknown cell surface receptor, SU brings the viral and target cell membranes into close apposition, orientates the envelope glycoprotein complex into a fusion-competent configuration, and induces conformational changes in the envelope that stimulate the membrane fusion activity of TM. To investigate the envelope-orchestrated events that culminate in viral entry, we have expressed a functional form of SU fused to the Fc domain of human IgG. We have used this recombinant protein to develop direct-binding assays for primary receptor activity and have used these assays to examine a diverse array of cell types for expression of the HTLV-1 receptor.
To generate the SU-Fc fusion, we replaced four carboxyl-terminal amino acids of gp46, constituting the arginine-rich protease cleavage signal, with the hinge and Fc regions (CH2 and CH3 heavy-chain constant domains) of human IgG. The recombinant fusion protein is secreted from Drosophila cells and accumulates as a soluble product in the conditioned medium of transfected cells. As predicted, the Fc epitope is not removed by cellular proteases during the secretion process but is retained on the mature protein product. The recombinant fusion protein is efficiently glycosylated, retains the protein A-binding properties of IgG, and is recognized by anti-Fc antibodies. Moreover, SU-Fc is expressed as a disulfide-linked homodimer. These observations indicate that the Fc region is appropriately folded and retains the known biochemical properties of the Fc domain of human IgG.
Importantly, the SU-Fc fusion protein is recognized by anti-gp46 sera and faithfully recapitulates both the known and the predicted properties of the native HTLV-1 SU. In particular, SU-Fc binds to HTLV-1-permissive target cells, such as human T cells, in a dose-dependent manner; and native viral gp46 competes with the SU-Fc fusion protein for cell surface binding. Moreover, exogenous addition of SU-Fc to permissive target cells potently inhibits cell-to-cell fusion and syncytium formation with HTLV-1 envelope-expressing cells. Our results indicate that SU-Fc binds to cells in a manner that disrupts an interaction that is critical to envelope-mediated membrane fusion, and they strongly imply that SU-Fc and viral gp46 compete for binding to a common cell surface receptor. Thus, we have demonstrated that the HTLV-1 SU can accommodate the addition of a relatively large protein domain to the carboxyl terminus of mature SU without loss of receptor-binding specificity. The ability to modify SU in this way may facilitate the development of novel envelope protein chimeras for retargeting or expanding the cellular tropism of viral vectors used in gene therapy protocols. Importantly, our binding and competition data also indicate that amino acid sequences within gp46 are the principal determinants of primary receptor binding activity. Previous studies have suggested that regions of TM interact with cell surface factors that support syncytium formation (34, 38). However, our data demonstrate that sequences within TM are not required for recognition of target cells by SU and that amino acid sequences contained within SU are both necessary and sufficient for the primary receptor-binding activity of the envelope.
Based on syncytium formation (30) and viral pseudotyping assays (46, 49), it has been suggested that the receptor for HTLV-1 is expressed on a variety of cell types. We find that this is indeed the case. Using a highly sensitive flow cytometry-based assay, we have directly investigated binding of the HTLV-1 SU-Fc fusion protein to cells and found that the majority of the cell lines examined bound significant amounts of SU-Fc. Significantly, most of the cell lines that bound SU-Fc also support efficient syncytium formation with envelope-expressing cells (30; our unpublished results). Our SU-Fc-binding analysis directly confirms the widespread expression of the HTLV-1 receptor in mammals and further indicates that some nonmammalian vertebrates express a functional homologue of this cell surface antigen.
Although the receptor for HTLV-1 is expressed on cells of diverse origin, some cell types do not support syncytium formation or infection by virus particles pseudotyped with HTLV-1 envelope (17, 30, 43, 49). The simplest explanation for these results has been that syncytium-resistant cells lack the receptor recognized by HTLV-1. In particular, murine cells do not readily permit infection by HTLV-1 envelope-pseudotyped virus and poorly support envelope-mediated syncytium formation. Using murine-human somatic-cell hybrids, a factor facilitating infection of murine cells by pseudotyped virus was mapped to human chromosome 17 (17, 43). Using our direct-binding assays, we have reexamined murine cells and murine-human somatic-cell hybrids retaining human chromosome 17 for the expression of a receptor for HTLV-1. Surprisingly, all of the murine cells examined, i.e., LM(TK−), A9, NIH 3T3, and MCF7 cells, expressed a cell surface receptor that was capable of binding to the SU-Fc protein. Moreover, we did not observe enhanced binding of our SU fusion protein to murine-human somatic cell hybrids that retain human chromosome 17. Therefore, it is unlikely that human chromosome 17 encodes a factor that binds directly to the HTLV-1 SU. These data are supported by recent results from improved highly sensitive pseudotyping assays, which reveal that murine cells do, in fact, support low levels of infection by virus particles pseudotyped with HTLV-1 envelope (46). In addition, murine-human chromosome 17 somatic-cell hybrids did not exhibit improved infection kinetics for HTLV-1 envelope-pseudotyped virus (46). Thus, while not conclusive, the accumulating evidence does not support the genetic data that map the primary HTLV-1 receptor to a gene on human chromosome 17.
Perhaps the most important finding of our study is that a number of cell lines previously thought to be receptor negative do, in fact, express cell surface factors capable of binding the SU of HTLV-1. Using syncytium formation and virus pseudotyping assays, previous studies have suggested that HUH7, LM(TK−), NIH 3T3, MDCK, and MDBK cells lack receptors for HTLV-1 (17, 30, 43, 49). In particular, the cell lines LM(TK−) and MDBK have repeatedly been scored as receptor negative. In our study, association of the SU-immunoadhesin with cells was monitored by a highly sensitive flow cytometry-based direct binding assay. Using this assay, we found that each of the syncytium-resistant mammalian cell lines bound significant amounts of the recombinant HTLV-1 SU. These results indicate that some cell lines express functional receptors for HTLV-1 but are resistant to HTLV-1 envelope-mediated membrane fusion and viral entry. Therefore, we suggest that for these receptor-positive nonpermissive cells the blocks to membrane fusion and viral entry occur at a post-receptor-binding step of the entry process.
When monitoring primary-receptor activity, why should there be such a marked difference in the results obtained from direct-binding assays and the biological assays? One possible explanation is that the direct-binding assays are more sensitive. This explanation seems unlikely to us, since many of the recently described biological assays for receptor activity are very sensitive techniques (30, 46, 49). Instead, we believe that the answer may reside in the assays themselves. Using the biological assays for receptor function, it is not possible to distinguish primary-receptor binding from the other envelope-dependent and -independent events that accompany membrane fusion. For example, cell-to-cell fusion may require the cooperative and supportive activity of nonreceptor host-encoded factors such as cell adhesion molecules (7, 19). Therefore, cell-to-cell membrane fusion is likely to be highly sensitive to the confounding effects of cell-specific expression of these factors or to cell-specific expression of fusion inhibitors. An interesting precedent for cell-specific blocks to receptor recognition is provided by the cat endogenous virus RD114. Murine cells are resistant to RD114 infection (26). However, treatment of murine cells with the glycosylation inhibitor tunicamycin renders these cells permissive for RD114 infection (26). Thus, murine cells express the receptor for RD114 but the use of this receptor for viral entry is blocked by glycosylation. In addition, traditional assays of syncytium formation may not be a good indicator of receptor expression. Previous studies have demonstrated that for HTLV-1, and other retroviruses, envelope-dependent cell-to-cell viral transfer may occur efficiently with strains that form only greatly reduced syncytia or even in the absence of overt syncytium formation (9, 51). Thus, syncytium formation may be an extreme example of virally induced membrane fusion, and cell-to-cell viral transfer may occur without these dramatic effects on cellular morphology. Finally, in the case of free-virus infection of cells, infection may not correlate directly with the level of primary receptor expression. Cell lines may be resistant to infection despite the presence of excess primary receptors on the cell surface, as observed for ecotropic murine retrovirus (50) and, most notably, for HIV (15, 28, 40, 45). Therefore, resistance to infection may reflect the need for a coreceptor or another host-encoded factor that supports viral entry into cells.
Based on the premise that receptor-negative cell lines may facilitate expression cloning and characterization of the HTLV-1 receptor, considerable effort has been invested in attempts to identify such cells. The data presented here, and supported by other studies, indicate that HTLV-1-receptor-negative mammalian cell lines are extremely rare. In fact, we have observed SU-binding activity on a somatic-cell line derived from the amphibian Xenopus laevis. In keeping with these results, other groups have reported receptor activity on cell lines from nonmammalian vertebrates that include reptilian and avian species (44, 46). The widespread expression of receptors that support binding of recombinant HTLV-1 SU, or infection by envelope-pseudotyped virus, clearly raises questions regarding the nature of the cellular receptor employed by HTLV-1. To date, the accumulating evidence is consistent with the view that the receptor is a cell surface protein, as binding of recombinant SU to cells is a process that is readily saturated, indicating that there are a limited number of binding sites for SU on permissive cells, and protease treatment of target cells blocks envelope-mediated viral infection of cells (49). If the receptor for HTLV-1 is indeed a surface protein and not, for example, a cell surface polysaccharide component, then the primary sequence of this receptor protein must be highly conserved throughout vertebrate evolution. However, rigid conservation of primary sequence over such an evolutionary distance would be surprising. An alternative view is that gp46 may exhibit considerable functional flexibility, perhaps recognizing a structural motif common to a family of conserved proteins rather than a defined linear amino acid sequence. A precedent for this type of functional plasticity is observed in the case of HIV-1; here some viral strains are able to recognize and use multiple members of the chemokine receptor family for viral entry (13, 45). If a similar situation existed for the HTLV-1 primary receptor, then the topology of the primary receptor relative to the target cell membrane and to membrane-associated cofactors would determine the functional competence of the receptor for viral entry. Whatever the explanation, it is not yet clear why the receptor for HTLV-1 should be so widespread and why some receptor-positive cells should be resistant to infection. Resolution of these issues must await the definitive identification and characterization of the primary receptor used by HTLV-1.
Finally, we have demonstrated that insect cells do not bind SU-Fc to any detectable level, suggesting that these cells lack the requisite surface-antigens recognized as receptors by the HTLV-1 SU. Therefore, the observed conservation of receptor function does not extend to nonvertebrate metazoans. While significant technical limitations currently hinder their use, receptor-negative insect cells may ultimately be of utility in the characterization of the HTLV-1 receptor. In conclusion, in our study we have expressed a functional SU-immunoadhesin and have used this recombinant protein to show that the HTLV-1 receptor is expressed on a wide range of vertebrate cell types. Most importantly, we have demonstrated that many syncytium-resistant cell lines express functional HTLV-1 receptors. Based on our results, we suggest that there exist postbinding blocks to viral entry in nonpermissive cell lines. Our findings will be of considerable importance in designing new strategies to identify the cellular receptor employed by HTLV-1.
ACKNOWLEDGMENTS
We thank Brian MacStay and members of the HTLV-1 European Research Network (HERN) for reagents and helpful discussions, Richard Elliot for cell lines, and Nathaniel Landau for proviral clones.
This work was generously supported by grants to D.W.B. from The Misses Barrie Charitable Trust and The Leukaemia Research Fund.
REFERENCES
- 1.Anonymous. Human T-cell lymphotropic viruses. IARC Monogr Eval Carcinog Risks Hum. 1996;67:261–334. [PMC free article] [PubMed] [Google Scholar]
- 2.Brighty D W, Rosenberg M, Chen I S Y, Ivey-Hoyle M. Envelope proteins from clinical isolates of human immunodeficiency virus type 1 that are refractory to neutralization by soluble CD4 possess high affinity for the CD4 receptor. Proc Natl Acad Sci USA. 1991;88:7802–7805. doi: 10.1073/pnas.88.17.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brighty D W, Rosenberg M. A cis-acting repressive sequence that overlaps the Rev-responsive element of human immunodeficiency virus type 1 regulates nuclear retention of env mRNAs independently of known splice signals. Proc Natl Acad Sci USA. 1994;91:8314–8318. doi: 10.1073/pnas.91.18.8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cann A J, Chen I S Y. Human T-cell leukemia virus type I and II. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1501–1527. [Google Scholar]
- 5.Carrington C V, Weiss R A, Schulz T F. A truncated HTLV-I envelope protein, lacking the hydrophobic membrane anchor domain, is associated with cellular membranes and virions. Virology. 1994;202:61–69. doi: 10.1006/viro.1994.1322. [DOI] [PubMed] [Google Scholar]
- 6.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 7.Daenke S, McCracken S A, Booth S. Human T-cell leukaemia/lymphoma virus type 1 syncytium formation is regulated in a cell-specific manner by ICAM-1, ICAM-3 and VCAM-1 and can be inhibited by antibodies to integrin beta2 or beta7. J Gen Virol. 1999;80:1429–1436. doi: 10.1099/0022-1317-80-6-1429. [DOI] [PubMed] [Google Scholar]
- 8.Delamarre L, Pique C, Pham D, Tursz T, Dokhelar M C. Identification of functional regions in the human T-cell leukemia virus type I SU glycoprotein. J Virol. 1994;68:3544–3549. doi: 10.1128/jvi.68.6.3544-3549.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delamarre L, Rosenberg A R, Pique C, Pham D, Dokhelar M C. A novel human T-leukemia virus type 1 cell-to-cell transmission assay permits definition of SU glycoprotein amino acids important for infectivity. J Virol. 1997;71:259–266. doi: 10.1128/jvi.71.1.259-266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delamarre L, Pique C, Rosenberg A R, Blot V, Grange M P, Le Blanc I, Dokhelar M C. The Y-S-L-I tyrosine-based motif in the cytoplasmic domain of the human T-cell leukemia virus type 1 envelope is essential for cell-to-cell transmission. J Virol. 1999;73:9659–9663. doi: 10.1128/jvi.73.11.9659-9663.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desgranges C, Souche S, Vernant J C, Smadja D, Vahlne A, Horal P. Identification of novel neutralization-inducing regions of the human T-cell lymphotropic virus type I envelope glycoproteins with human HTLV-1-seropositive sera. AIDS Res Hum Retrovir. 1994;10:163–173. doi: 10.1089/aid.1994.10.163. [DOI] [PubMed] [Google Scholar]
- 12.Dokhelar M C, Pickford H, Sodroski J, Haseltine W A. HTLV-1 p27Rex regulates gag and env protein expression. J Aquir Immune Defic Syndr. 1989;2:431–440. [PubMed] [Google Scholar]
- 13.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 14.Fasken M B, Saunders R, Rosenberg M, Brighty D W. A leptomycin B-sensitive homologue of human CRM1 promotes nuclear export of nuclear export sequence-containing proteins in Drosophila cells. J Biol Chem. 2000;275:1878–1886. doi: 10.1074/jbc.275.3.1878. [DOI] [PubMed] [Google Scholar]
- 15.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 16.Gavalchin J, Fan N, Lane M J, Papsidero L, Poiesz B J. Identification of a putative cellular receptor for HTLV-I by a monoclonal antibody, Mab 34-23. Virology. 1993;194:1–9. doi: 10.1006/viro.1993.1228. [DOI] [PubMed] [Google Scholar]
- 17.Gavalchin J, Fan N, Waterbury P G, Corbett E, Faldasz B D, Peshick S M, Poiesz B J, Papsidero L, Lane M J. Regional localization of the putative cell surface receptor for HTLV-I to human chromosome 17q23.2–17q25.3. Virology. 1995;212:196–203. doi: 10.1006/viro.1995.1468. [DOI] [PubMed] [Google Scholar]
- 18.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 19.Hildreth J E, Subramanium A, Hampton R A. Human T-cell lymphotropic virus type 1 (HTLV-1)-induced syncytium formation mediated by vascular cell adhesion molecule-1: evidence for involvement of cell adhesion molecules in HTLV-1 biology. J Virol. 1997;71:1173–1180. doi: 10.1128/jvi.71.2.1173-1180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hildreth J E. Syncytium-inhibiting monoclonal antibodies produced against human T-cell lymphotropic virus type 1-infected cells recognize class II major histocompatibility complex molecules and block by protein crowding. J Virol. 1998;72:9544–9552. doi: 10.1128/jvi.72.12.9544-9552.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The HTLV-1 European Research Network. Seroepidemiology of the human T-cell leukaemia/lymphoma viruses in Europe. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13:68–77. doi: 10.1097/00042560-199609000-00010. [DOI] [PubMed] [Google Scholar]
- 22.Imai T, Fukudome K, Takagi S, Nagira M, Furuse M, Fukuhara N, Nishimura M, Hinuma Y, Yoshie O. C33 antigen recognized by monoclonal antibodies inhibitory to human T-cell leukemia virus type 1-induced syncytium formation is a member of a new family of transmembrane proteins including CD9, CD37, CD53, and CD63. J Immunol. 1992;149:2879–2886. [PubMed] [Google Scholar]
- 23.Ivey-Hoyle M, Culp J S, Chaikin M A, Hellmig B D, Matthews T J, Sweet R W, Rosenberg M. Envelope glycoproteins from biologically diverse isolates of immunodeficiency viruses have widely different affinities for CD4. Proc Natl Acad Sci USA. 1991;88:512–516. doi: 10.1073/pnas.88.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansen H, van der Straten A, Sweet R, Otto E, Maroni G, Rosenberg M. Regulated expression at high copy number allows production of a growth-inhibitory oncogene product in Drosophila Schneider cells. Genes Dev. 1989;3:882–889. doi: 10.1101/gad.3.6.882. [DOI] [PubMed] [Google Scholar]
- 25.Kobe B, Center R J, Kemp B E, Poumbourios P. Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc Natl Acad Sci USA. 1999;96:4319–4324. doi: 10.1073/pnas.96.8.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koo H M, Parthasarathi S, Ron Y, Dougherty J P. Pseudotyped REV/SRV retroviruses reveal restrictions to infection and host range within members of the same receptor interference group. J Virol. 1994;205:345–351. doi: 10.1006/viro.1994.1651. [DOI] [PubMed] [Google Scholar]
- 27.Landau N R, Page K A, Littman D R. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J Virol. 1991;65:162–169. doi: 10.1128/jvi.65.1.162-169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 29.Morozov V A, Weiss R A. Two types of HTLV-1 particles are released from MT-2 cells. Virology. 1999;255:279–284. doi: 10.1006/viro.1998.9578. [DOI] [PubMed] [Google Scholar]
- 30.Okuma K, Nakamura M, Nakano S, Niho Y, Matsuura Y. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology. 1999;254:235–244. doi: 10.1006/viro.1998.9530. [DOI] [PubMed] [Google Scholar]
- 31.Pique C, Tursz T, Dokhelar M C. Mutations introduced along the HTLV-I envelope gene result in a non-functional protein: a basis for envelope conservation? EMBO J. 1990;9:4243–4248. doi: 10.1002/j.1460-2075.1990.tb07872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pique C, Pham D, Tursz T, Dokhelar M C. Human T-cell leukemia virus type I envelope protein maturation process: requirements for syncytium formation. J Virol. 1992;66:906–913. doi: 10.1128/jvi.66.2.906-913.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pique C, Pham D, Tursz T, Dokhelar M C. The cytoplasmic domain of the human T-cell leukemia virus type I envelope can modulate envelope functions in a cell type-dependent manner. J Virol. 1993;67:557–561. doi: 10.1128/jvi.67.1.557-561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poon B, Chen I S Y. Identification of a domain within the human T-cell leukemia virus type 2 envelope required for syncytium induction and replication. J Virol. 1998;72:1959–1966. doi: 10.1128/jvi.72.3.1959-1966.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenberg A R, Delamarre L, Pique C, Pham D, Dokhelar M C. The ectodomain of the human T-cell leukemia virus type 1 TM glycoprotein is involved in postfusion events. J Virol. 1997;71:7180–7186. doi: 10.1128/jvi.71.10.7180-7186.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenberg A R, Delamarre L, Preira A, Dokhelar M C. Analysis of functional conservation in the surface and transmembrane glycoprotein subunits of human T-cell leukemia virus type 1 (HTLV-1) and HTLV-2. J Virol. 1998;72:7609–7614. doi: 10.1128/jvi.72.9.7609-7614.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Sagara Y, Inoue Y, Shiraki H, Jinno A, Hoshino H, Maeda Y. Identification and mapping of functional domains on human T-cell lymphotropic virus type 1 envelope proteins by using synthetic peptides. J Virol. 1996;70:1564–1569. doi: 10.1128/jvi.70.3.1564-1569.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sagara Y, Ishida C, Inoue Y, Shiraki H, Maeda Y. 71-kilodalton heat shock cognate protein acts as a cellular receptor for syncytium formation induced by human T-cell lymphotropic virus type 1. J Virol. 1998;72:535–541. doi: 10.1128/jvi.72.1.535-541.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider-Scaulies J. Cellular receptors for viruses: links to tropism and pathogenesis. J Gen Virol. 2000;81:1413–1429. doi: 10.1099/0022-1317-81-6-1413. [DOI] [PubMed] [Google Scholar]
- 41.Seiki M, Hattori S, Hirayama Y, Yoshida Y. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci USA. 1983;80:3618–3622. doi: 10.1073/pnas.80.12.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sodroski J G. HIV-1 inhibitors in the side pocket. Cell. 1999;99:243–246. doi: 10.1016/s0092-8674(00)81655-4. [DOI] [PubMed] [Google Scholar]
- 43.Sommerfelt M A, Williams B P, Clapham P R, Solomon E, Goodfellow P N, Weiss R A. Human T cell leukemia viruses use a receptor determined by human chromosome 17. Science. 1988;242:1557–1559. doi: 10.1126/science.3201246. [DOI] [PubMed] [Google Scholar]
- 44.Sommerfelt M A, Weiss R A. Receptor interference groups of 20 retroviruses plating on human cells. Virology. 1990;176:58–69. doi: 10.1016/0042-6822(90)90230-o. [DOI] [PubMed] [Google Scholar]
- 45.Sommerfelt M A. Retrovirus receptors. J Gen Virol. 1999;80:3049–3064. doi: 10.1099/0022-1317-80-12-3049. [DOI] [PubMed] [Google Scholar]
- 46.Sutton R E, Littman D R. Broad host range of human T-cell leukemia virus type 1 demonstrated with an improved pseudotyping system. J Virol. 1996;70:7322–7326. doi: 10.1128/jvi.70.10.7322-7326.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka Y, Zeng L, Shiraki H, Shida H, Tozawa H. Identification of a neutralization epitope on the envelope gp46 antigen of human T cell leukemia virus type I and induction of neutralizing antibody by peptide immunization. J Immunol. 1991;147:354–360. [PubMed] [Google Scholar]
- 48.Taylor G P The HTLV-1 European Research Network. The epidemiology and clinical impact of HTLV infections in Europe. AIDS Rev. 1999;1:195–204. [Google Scholar]
- 49.Trejo S R, Ratner L. The HTLV receptor is a widely expressed protein. Virology. 2000;268:41–48. doi: 10.1006/viro.2000.0143. [DOI] [PubMed] [Google Scholar]
- 50.Wang H, Paul R, Burgeson R E, Keene D R, Kabat D. Plasma membrane receptors for ecotropic murine retroviruses require a limiting accessory factor. J Virol. 1991;65:6468–6477. doi: 10.1128/jvi.65.12.6468-6477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiss C D, Barnett S W, Cacalano N, Littman D R, White J M. Studies of HIV-1 envelope glycoprotein-mediated fusion using a simple fluorescence assay. AIDS. 1996;10:241–246. doi: 10.1097/00002030-199603000-00001. [DOI] [PubMed] [Google Scholar]