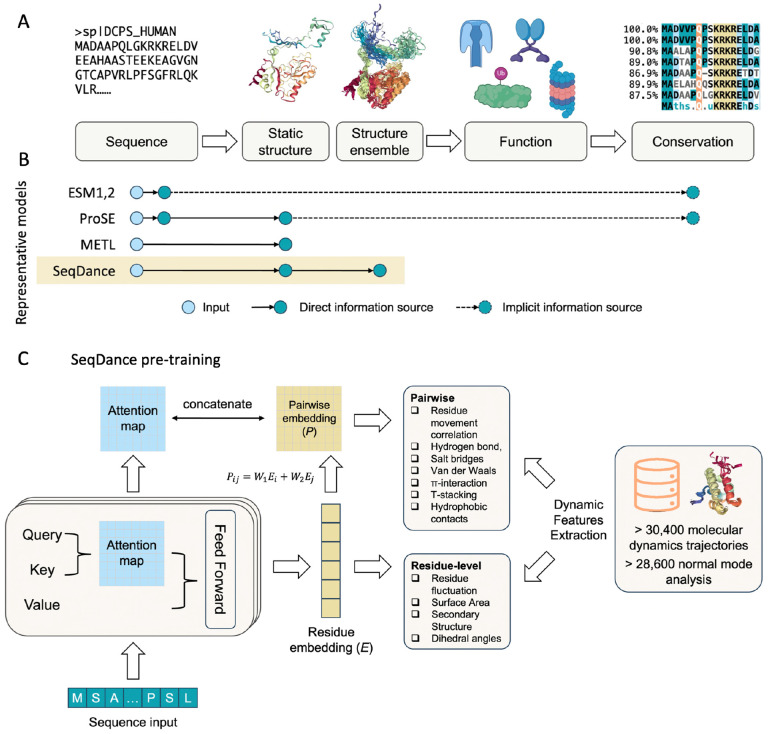
Abstract
Proteins perform their functions by folding amino acid sequences into dynamic structural ensembles. Despite the important role of protein dynamics, their complexity and the absence of efficient representation methods have limited their integration into studies on protein function and mutation fitness, especially in deep learning applications. To address this, we present SeqDance, a protein language model designed to learn representation of protein dynamic properties directly from sequence alone. SeqDance is pre-trained on dynamic biophysical properties derived from over 30,400 molecular dynamics trajectories and 28,600 normal mode analyses. Our results show that SeqDance effectively captures local dynamic interactions, co-movement patterns, and global conformational features, even for proteins lacking homologs in the pre-training set. Additionally, we showed that SeqDance enhances the prediction of protein fitness landscapes, disorder-to-order transition binding regions, and phase-separating proteins. By learning dynamic properties from sequence, SeqDance complements conventional evolution- and static structure-based methods, offering new insights into protein behavior and function.
Keywords: protein language model, molecular dynamics, normal mode analysis, deep learning, computational biology
Introduction
Deep learning has achieved considerable success in predicting various protein attributes such as structure (e.g., AlphaFold1, 2 and RoseTTAfold3), function, stability, localization, and interactions. A central challenge in developing effective deep learning models is choosing a representation that allows models to interpret and learn from easily. The simplest form of representation, one-hot encoding of amino acids, is unbiased and rooted in the idea that a protein’s sequence determines its properties. Yet, one-hot encoding often underperforms due to its simplicity. To overcome this, researchers have introduced more informative representations incorporating amino acid physicochemical properties, structural features, evolutionary profiles, and, more recently, embeddings from deep learning models. We generally classify these representations into two categories: evolution-based and biophysics-based4.
Evolution-based representation (EBR) introduces extra information of protein homologs in other species that experience similar selective pressures. A major source of EBR is multiple sequence alignment (MSA), which proves particularly valuable for identifying functional sites5, pathogenic mutations6, and predicting 3D structures1, 2. Recently, protein language models (pLMs) like ESM1,27, 8, ProtTrans9, and ProGen10 have emerged as powerful tools for generating implicit EBR. Trained in an unsupervised manner on large-scale protein sequence datasets, pLM has been shown to effectively memorize conserved patterns during the pre-training process11–15. pLM provides a computationally efficient alternative to MSA and has been successfully applied to various biological questions, including predicting 3D structures (e.g., ESMFold7), predicting signal peptides16, and even generating proteins10. However, the effectiveness of EBR relies on the quality and quantity of sequenced homologs. For instance, the prediction confidence scores from protein structure predictors are directly related to the number of homologous sequences1, 7. As a result, EBR is less effective for rapidly evolving viral proteins, immune proteins, and proteins from under-studied species such as extremophiles11, where homologous sequences are either sparse or highly divergent. Moreover, evolutionary profiles are consequences of functional protein behaviors rather than their causes (Figure 1A). Overreliance on EBR might bias the model toward the conservation pattern. For example, EBR-based pathogenic mutation predictors perform worse when evaluated solely on conserved or unconserved regions, as they can achieve good performance by simply predicting mutations in conserved regions to be pathogenic and those in unconserved regions to be benign.
Figure 1: Information flow in protein study, representative protein language models, and SeqDance pre-training.
A. Illustration of the “sequence - structure ensemble - function - evolution” paradigm. Sequences are the basic elements of proteins that fold into structural ensembles to perform specific functions. Functionally important regions exhibit conserved patterns across species. B. Representative protein language models (pLMs) and their information sources. ESM1, ESM2 and other pLMs trained in unsupervised manners memorize co-evolution information and conserved motifs during pre-training, thus implicitly relying on evolution data. ProSE was trained to predict masked residues, pairwise contact in static structures, and structure similarity. METL was trained to predict 55 biophysical terms calculated from static structures. SeqDance was trained on protein dynamic features from molecular dynamics (MD) simulations, experimental data, as well as normal mode analysis (NMA) of static structures. C. Diagram of the SeqDance pre-training process. SeqDance is a transformer encoder that takes a protein sequence as input and predicts residue-level and pairwise dynamic features extracted from over 30,400 MD trajectories and 28,600 NMA. For a protein of length L, the residue embedding dimension is L×480. A linear layer is added to this embedding to predict residue-level features. Pairwise embeddings derived from residue embeddings are concatenated with attention maps to predict pairwise features. After pre-training, the residue embeddings can be applied to study biological questions; users can also fine-tune all parameters for downstream tasks.
Biophysics-based representation (BBR), primarily derived from protein structures, avoids the limitations of EBR and typically maintains uniform performance across the entire protein space. Predicting protein behaviors from BBR aligns with the idea that sequence determines structure, which in turn dictates function (Figure 1A). Although the experimental determination of protein structures has historically been time-consuming and resource-intensive, recent advancements such as AlphaFold1, 2, RoseTTAfold3, and ESMFold7 have revolutionized static protein structure prediction, enabling proteome-wide analyses. BBR has been applied to a variety of biology questions. For example, predicted structures have been used to predict mutation effects and functional and binding sites17. The AAindex18 database, which compiles a number of physicochemical properties of amino acids derived from structures in the Protein Data Bank19 (PDB), is widely utilized in bioinformatics tools. Similarly, PScore20 and LLPhyScore21 leverage physical features from PDB structures to identify phase-separating proteins22. Additionally, there have been initiatives to integrate biophysical properties into protein language models (pLMs). For example, ProSE4 was trained to predict masked residues, contacts within static structures, and structural similarities, while METL23 was developed to predict 55 biophysical properties derived from Rosetta models of mutated structures.
However, current BBRs are derived exclusively from static protein structures. These structural snapshots lack crucial thermodynamic information and overlook the topological landscape of catalysis, allostery, and other long-range interactions. Moreover, static structures cannot describe the dynamic structure ensembles of intrinsically disordered regions (IDRs), which constitute more than 30% of the human proteome24. Despite lacking fixed structures, IDRs use their inherent flexibility to mediate essential biological processes such as signal transduction, transcriptional regulation, and phase separation25, 26.
To capture dynamic protein properties, molecular dynamics (MD) simulations are widely employed for both ordered structures and IDRs. MD simulations utilize Newton’s laws to update atomic coordinates based on interaction forces, generating ensembles of structures over a specified simulation time. However, all-atom MD simulations are computationally intensive, often requiring at least a week of GPU time to simulate a single protein at the microsecond scale. To mitigate computational demands, coarse-grained MD simulations simplify protein residues into pseudo-atoms and use specialized force fields to model interactions at the reduced scale. Another commonly used approach is normal mode analysis27, 28 (NMA), which describes protein vibrations (normal modes) around equilibrium conformations. Normal modes with varying frequencies represent distinct behaviors, with low-frequency modes capturing global movements. While these methods enable large-scale studies of protein dynamics, the data generated from MD and NMA are often high-dimensional and irregularly shaped. One current challenge is to represent these dynamic properties in a meaningful and efficient manner that can be integrated into deep learning models.
Here, we introduce SeqDance, a pLM designed to provide representation of protein dynamic properties. We first collected over 30,400 protein dynamics trajectories for ordered structures, membrane proteins, and IDRs, along with performing over 28,600 NMAs for proteins in the Protein Data Bank (PDB). From this dataset, we extracted rich residue-level and pairwise dynamic features and pre-trained SeqDance to predict these features from protein sequences (Figure 1B). SeqDance effectively learned both local and global dynamic properties in the pre-training process. These properties can be easily retrieved from SeqDance by inputting a protein sequence and can be applied to various biological questions. We also demonstrate that SeqDance provides informative dynamic embeddings for proteins that lack homologs in the pre-training set.
Results
Pre-training SeqDance with dynamics properties of over 59,000 proteins.
We collected high-resolution and low-resolution protein dynamics data to pre-train SeqDance (Table 1). High-resolution dynamics data includes experimental data and all-atom molecular dynamics (MD) simulation trajectories from ATLAS29, GPCRmd30, and PED31 (Table 1). ATLAS contains all-atom MD structure ensembles for over 1,500 representative non-membrane proteins, each simulated for 100 nanoseconds with three replicates. GPCRmd includes more than 500 MD simulations of G-protein-coupled receptors, with most proteins simulated for 500 nanoseconds in three replicates. PED provides ensembles of disordered proteins from both experiments and MD simulations, from which we filtered 382 ensembles. Since high-resolution dynamics data is limited, we augmented SeqDance pre-training with low-resolution dynamics data, including coarse-grained MD trajectories and normal mode analysis (NMA). We processed coarse-grained structure ensembles of 28,058 human disordered regions from IDRome24 and converted them to all-atom trajectories32 (see Methods for details). We also conducted NMA27, 28, 33 for over 28,600 representative structures in the PDB, covering single proteins, antibodies, and protein complexes34 (Table 1).
Table 1:
the protein dynamic datasets used to pre-train SeqDance
| Source | Description | Number | Method | |
|---|---|---|---|---|
| High resolution | ||||
| GPCRmd | Membrane proteins | 509 | All-atom MD, 3*500 ns | |
| Low resolution | ||||
| Proteinflow | Ordered structures in PDB | 28,631 | Normal mode analysis |
We extracted residue-level and pairwise dynamic features that describe the distribution of features in structure ensembles (Figure 1B, Supplementary Table 1, A detailed explanation can be found in the Discussion). Residue-level features include root mean square fluctuation (RMSF), surface area, eight-class secondary structures, and dihedral angles (phi, psi, chi1) which describe the rotation angles around bonds in the protein backbone and side chains. Pairwise features include the correlation of Cα movements and frequencies of hydrogen bonds, salt bridges, Pi-cation, Pi-stacking, T-stacking, hydrophobic interactions, and van der Waals interactions. For NMA data, we categorized normal modes of each structure into three frequency-based clusters. For each cluster, we calculated residue fluctuation and pairwise correlation maps (see Methods for details, Supplementary Table 1).
SeqDance is a transformer encoder model with 12 layers and 20 heads per layer, with 35 million parameters in total. SeqDance takes protein sequences as input and predicts residue-level and pairwise dynamic features (Figure 1B). For residue-level feature prediction, we added a linear layer to the last layer’s residue embeddings. For pairwise feature prediction, we transformed the residue embeddings into pairwise embeddings, concatenated them with SeqDance’s attention maps, and applied a linear layer to the concatenated matrix (A detailed explanation can be found in the Discussion). We randomly sampled approximately 95% of the protein dynamics data to pre-train SeqDance. We adjusted the weights for different data sources and features in the loss function (see Methods for details).
After pre-training, we observed a strong correlation between the weights for predicting co-movement in MD and NMA (Supplementary Figure 1). Specifically, the Pearson correlation between that for MD and low-frequency normal modes was 0.75, suggesting that NMA closely mimics dynamic movements captured by MD simulations. This finding supports the use of NMA to augment SeqDance pre-training.
Next, we evaluated whether SeqDance had effectively learned protein dynamic properties. Specifically, we examined if SeqDance’s self-attention mechanisms captured dynamic interactions and residue co-movement, and whether SeqDance embeddings encoded information about protein conformational properties.
SeqDance’s attention captures local dynamic residue interactions and co-movement.
Transformer model employs the self-attention mechanism35 to update the representation for each word by aggregating information from other words, with attention values representing the relationship between words (in this context, amino acid residues). Given that SeqDance’s attention maps were utilized to predict dynamic interactions and residue co-movement, we investigated whether SeqDance’s attention effectively capture these properties.
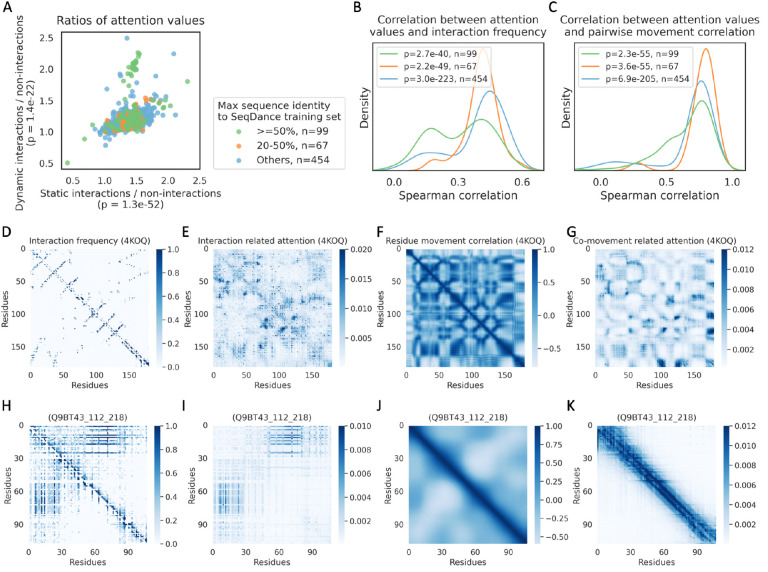
To analyze pairwise feature-related attention, we first selected the top 10 attention heads with the highest weights for predicting interactions out of 240 total attention heads (Supplementary Figure 1A). We then compared their averaged attention values with pairwise features across 620 held-out proteins from ATLAS, GPCRmd, PED, and IDRome. Interacting pairs were classified as either static interactions (observed in the first frame of the structural ensemble) or dynamic interactions (observed in subsequent frames). A subset of non-interacting control pairs was also sampled with the same distance distribution to account for the distance dependence of attention values. As shown in Figure 2A, SeqDance assigned significantly higher attention values to both static and dynamic interactions compared to non-interacting pairs (pairwise t-test P-values: 1.3e-52 for static interactions, 1.4e-22 for dynamic interactions). Moreover, we observed a significant positive correlation between attention values and interaction frequency (Figure 2B). For residue co-movement, we conducted the same analysis and found a significant positive relationship between attention values and pairwise movement correlations in held-out proteins, with a median Spearman correlation of 0.75 (Figure 2C). The same analysis on held-out NMA data also revealed significant positive correlations between attention values and co-movements from low- and medium-frequency normal modes (Supplementary Figure 2). These results underscore SeqDance’s ability to capture biologically meaningful dynamic interactions and co-movements across different datasets.
Figure 2. SeqDance’s attention mechanism captures local dynamic residue interactions and co-movement.
A. Comparison of SeqDance’s attention values assigned to static interactions (present in the first frame of the structure ensemble), dynamic interactions (formed in subsequent frames), and a subset of non-interacting control pairs that matched to the distance distribution of interacting pairs. Each dot represents one of 620 held-out proteins. The legend on the right shows the numbers of proteins in three clusters with different sequence identities to training sequences. P-values on the x and y axes were calculated using pairwise t-tests. B-C. Distributions of Spearman correlation between SeqDance’s interaction-related attentions and interaction frequency (sum of nine types of interactions) (B), and between SeqDance’s co-movement related attentions and pairwise movement correlations (C), of three clusters of held-out proteins described above. P-values were calculated using a one-sample t-test with the null hypothesis that the mean value is zero. D-K. Visualization of pairwise features and related attention maps for a structured protein (PDB ID 4KOQ) and a disordered region (Q9BT43_112_218), including the interaction frequency (sum of nine types of interactions) map (D, H), the averaged attention map of the top 10 heads with the highest weights for interaction prediction (E, I), the residue movement correlation map (F, J), and the averaged attention map of the top 10 heads with the highest weights for co-movement prediction (G, K).
To assess SeqDance’s ability to capture pairwise relationships beyond homologous sequences, we categorized 620 held-out proteins based on their similarity to training sequences: 99 proteins had at least 50% sequence identity (with at least 80% coverage) to at least one training sequence, 67 proteins had at least 20% sequence identity (with at least 60% coverage), and 454 proteins were dissimilar to any training sequence. As shown in Figure 2A–C, the observed trends persisted for dissimilar held-out proteins. Additionally, we visualized two dissimilar held-out proteins: a structured protein (PDB ID: 4KOQ) from the ATLAS dataset (Figures 2D–G), and a disordered region (Q9BT43_112_218) from the IDRome dataset (Figures 2H–K). In both cases, we observed consistent patterns between interaction-related attention and interaction maps, as well as between co-movementrelated attention and residue movement correlations. Overall, these findings indicate that SeqDance can capture biologically meaningful relationships beyond homologous sequences.
SeqDance learns global protein conformation properties in the pre-training process.
Next, we investigated whether SeqDance embeddings encode additional protein conformational properties not included in the pre-training tasks. Since these dynamic features cannot be directly extracted from the embeddings, we applied supervised learning by using the mean-pooled embeddings for linear regression on protein conformational properties.
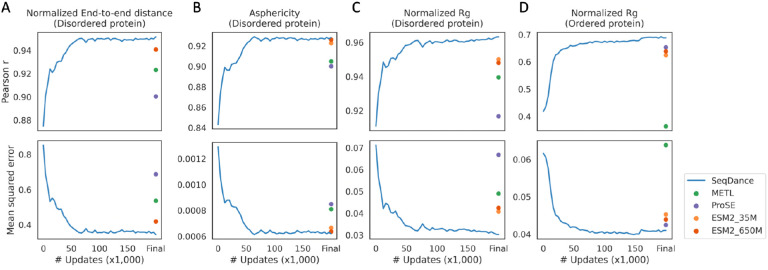
We first evaluated the models on structural ensembles of 18,415 Intrinsically disordered regions (IDRs) from coarse-grained MD simulations36 (see Methods for data filtering). These simulations employed a distinct IDR dataset and force field compared to IDRome. For evaluation, we used the average values of end-to-end distance, asphericity, and radius of gyration (Rg) within the ensembles. End-to-end distance reflects flexibility and motion range, asphericity quantifies deviation from a spherical shape, and Rg measures the distribution of atoms around the protein’s center of mass, indicating its compactness. To account for protein length, we used normalized values. The training and test sets were split using a 20% sequence identity cutoff to prevent information leakage. As shown in Figure 3A–C, SeqDance outperformed METL, ProSE, and ESM2 in predicting normalized end-to-end distance, asphericity, and Rg of IDRs, with performance improving as training progressed. To further assess SeqDance’s performance on proteins without homologs in the pre-training dataset, we removed IDRs with over 20% sequence identity (with at least 60% coverage) to any SeqDance training sequence. SeqDance maintained its performance on these dissimilar IDRs (Supplementary Figure 3), demonstrating its generalization capability.
Figure 3. SeqDance embeddings encode global protein conformational properties.
Performance comparison of embeddings from SeqDance, METL, ProSE, and ESM2 in predicting the normalized end-to-end distance of disordered proteins (A, two ESM2 models overlapped), asphericity of disordered proteins (B), normalized radius of gyration (Rg) of disordered proteins (C) and ordered proteins (D). The training and test split was 6:4 with a 20% sequence identity cutoff. The results presented are the averages of ten repeats. The x-axis represents the number of pre-training steps for SeqDance, “Final” on the x-axis represents the evaluation of released weights of the other methods, and 200k steps for SeqDance.
For ordered proteins, obtaining conformational properties from structure ensembles is more challenging. Therefore, we used normalized Rg values (see Methods for details) of over 11,000 static monomer structures in the PDB from the paper37. Since SeqDance was trained on NMA of nearly all representative PDB structures34, we did not exclude sequences with homologs in the SeqDance pre-training dataset. Using the same evaluation method as for disordered regions, we found that SeqDance outperformed METL, ProSE, and ESM2 in predicting normalized Rg of ordered proteins, with performance improving as training progressed (Figure 3D).
Overall, these results demonstrate that SeqDance learns both local and global dynamic properties for ordered proteins and IDRs in the pre-training process. We hypothesized that the dynamic features encoded in SeqDance embeddings are informative for understanding protein behavior and function. Thus, we further applied SeqDance to specific biological questions.
SeqDance enhances understanding of protein fitness landscapes.
We first applied SeqDance to predict protein fitness landscapes38, which are essential for interpreting disease mutations and guiding protein engineering. Mutation effects are determined by the residue context and the differences between wild-type and mutant amino acids39. Previous methods have leveraged sequence and structural contexts to study mutation effects6, 39–44, based on the hypothesis that mutations affecting linear motifs or 3D structures can alter protein behaviors. It is important to recognize that while evolution-based representations are effective at predicting mutation effects, they reflect the consequence, not the cause, of mutation effects. Previous studies have emphasized the role of protein dynamics as a causal context in understanding mutation effects45–49. For instance, mutations in residues that co-move with the catalytic core can disrupt its dynamics, thereby affecting enzyme activity. We hypothesized that SeqDance can help predicting protein fitness by providing additional protein dynamic-based representations.
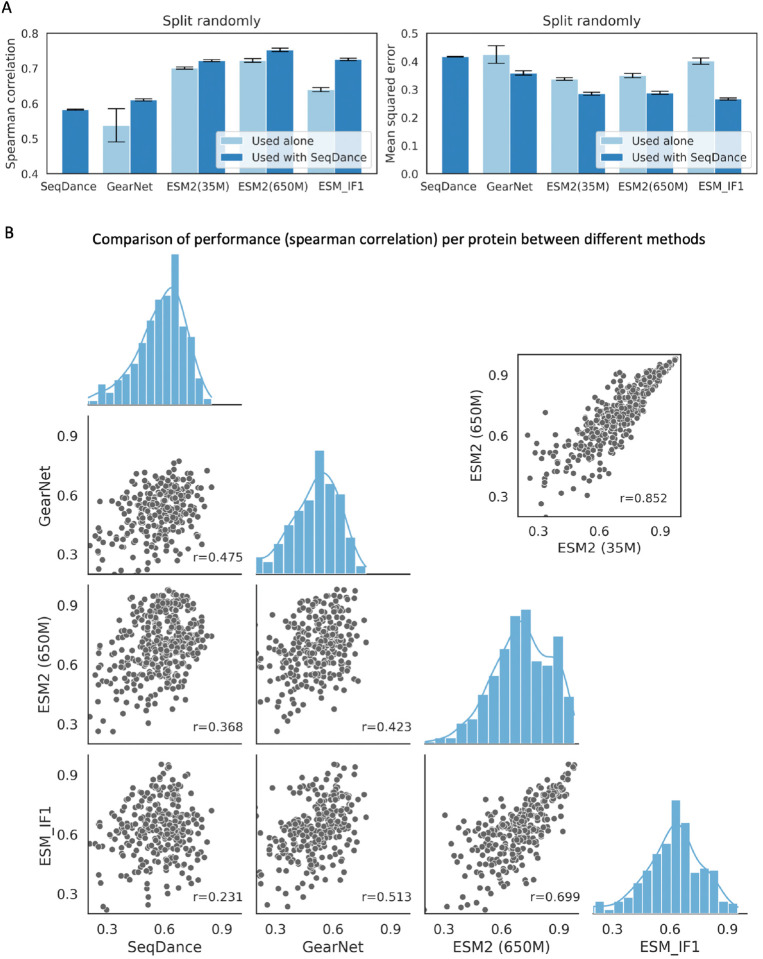
We used mutations on 20,955 residues from 412 proteins in a protein folding stability dataset38 (including both designed and PDB proteins, see Methods for details). SeqDance was compared with several published methods for predicting stability-related residue context (mean ddGs of all mutations on each residue), including GearNet50, a static structure-based pre-trained model that provides structural context; ESM27 that offers implicit evolution-based representations; and ESM_IF151, an inverse folding algorithm that provides evolution-based representations conditioned on structures. The training and test sets were divided either randomly or by protein. Our analysis revealed that, when evaluated individually, SeqDance’s dynamic-based representations consistently outperformed GearNet’s static structure-based representations, suggesting that static structures alone miss important information; ESM2 achieved the best overall performance, consistent with the fact that evolution-based representation is most effective in predicting mutation effects39 (Figure 4A, Supplementary Figure 4A). Although both ESM2 (35M) and ESM2 (650M) performed better than SeqDance individually, the combination of SeqDance and ESM2 (650M) outperformed the combination of two ESM2 models (Supplementary Figure 4B, SeqDance and ESM2 (35M) have the same embedding dimensions). Moreover, integrating SeqDance embeddings significantly improved the performance of all methods. Combining the results in Figure 4A and Supplementary Figure 4A, for GearNet, Spearman correlation increased by 13%, and mean squared error (MSE) decreased by 72%; for ESM_IF1, Spearman correlation rose by 12% and MSE dropped by 29%; for ESM2 (35M), Spearman correlation improved by 1.7% and MSE decreased by 13%; and for ESM2 (650M), Spearman correlation increased by 3.2% and MSE decreased by 14% (Figure 4A). Overall, these results indicate that SeqDance provides additional information that complements exiting methods.
Figure 4. SeqDance enhances the understanding of protein fitness landscapes.
A. Comparison of SeqDance, GearNet, ESM2, and ESM_IF1 embeddings in predicting mutation effects on protein folding stability. The training and test sets were divided randomly, and four-fold cross-validation was employed to determine Spearman correlation and mean squared error. The plots show the means and standard deviations of evaluation metrics across ten independent repeats. B. Performance comparison on individual proteins. Each dot represents the mean Spearman correlation for each protein in random split across ten repeats. The Pearson correlations were used to quantify the relationship between the performances of two methods across diverse proteins. The histograms illustrate the distribution of Spearman correlations for individual proteins for different methods.
We further analyzed SeqDance’s performance on individual proteins (see Methods for details). First, we observed that SeqDance’s performance was relatively orthogonal to both ESM2 and ESM_IF1 while it had a slightly higher correlation to GearNet, reflecting some overlap in structural context information (Figure 4B). In contrast, the performances of two ESM2 models, as well as between ESM2 and ESM_IF1, were highly correlated, due to their shared reliance on evolution-based information. Second, SeqDance performed particularly well on designed proteins (Supplementary Figure 5A) and showed comparable performance on PDB proteins, regardless of whether they had homologs in the pre-training set. (Supplementary Figure 5B), highlighting its generalizability across different protein types.
Finetuning SeqDance for protein disorder region-related tasks.
Intrinsically disordered regions (IDRs) are flexible protein segments essential for signal transduction, transcription regulation, and phase separation25. Since IDRs are less conserved than ordered regions, evolution-based predictors typically perform well in IDR prediction, while sequence-only predictors often underperform52. Given that SeqDance captures the local and global dynamics of IDRs, we evaluated its potential for IDR-related tasks.
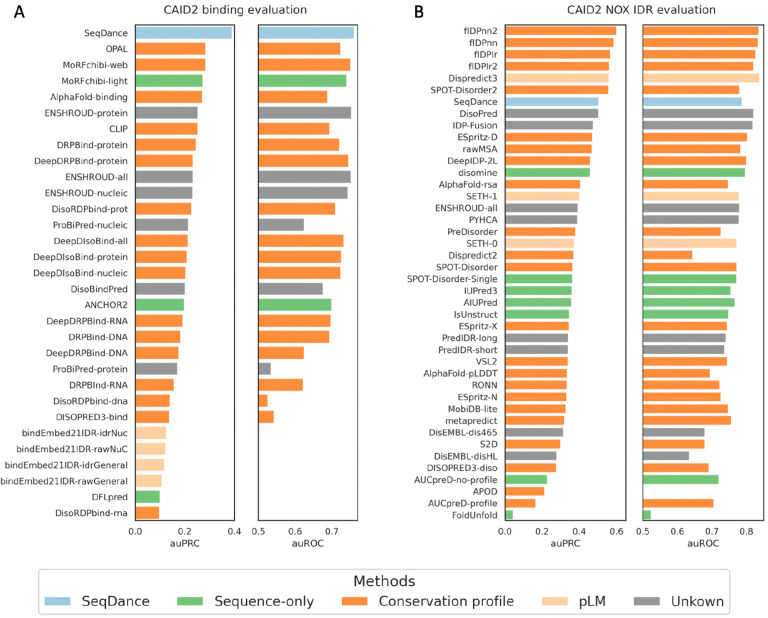
Using the Critical Assessment of Intrinsic Disorder (CAID2) benchmark52, which includes four tasks—the NOX IDR (missing residues in PDB), the PDB IDR (disordered residues in PDB), binding regions undergoing disorder-to-order transitions, and linker regions—we fine-tuned SeqDance on the training data of methods evaluated in CAID2 to prevent data leakage (see Methods for details). SeqDance achieved the best performance in predicting disorder-to-order transition binding regions and ranked among the top-performing sequence-only predictors for NOX IDR (Figure 5, Supplementary Figure 6). These results highlight that SeqDance learns valuable dynamic properties of IDRs, offering competitive performance as a sequence-only predictor for IDR-related tasks.
Figure 5. Fine-tuning SeqDance for predicting intrinsically disordered regions (IDRs) related tasks.
Performance comparison for predicting the binding regions (A) and NOX IDRs (missing residues in PDB structures) (B) in Critical Assessment of Intrinsic Disorder (CAID2). Performance is evaluated using the area under the Receiver Operating Characteristic curve (auROC) and the area under the Precision-Recall curve (auPRC). The auROC and auPRC for other methods were obtained from the CAID2 website. Methods evaluated in CAID2 are classified into four categories: sequence-only methods using features from single sequences; conservation profile-based methods; protein language model (pLM)-based methods; and methods with unknown inputs.
SeqDance augmented structure and sequence features in predicting phase-separating proteins.
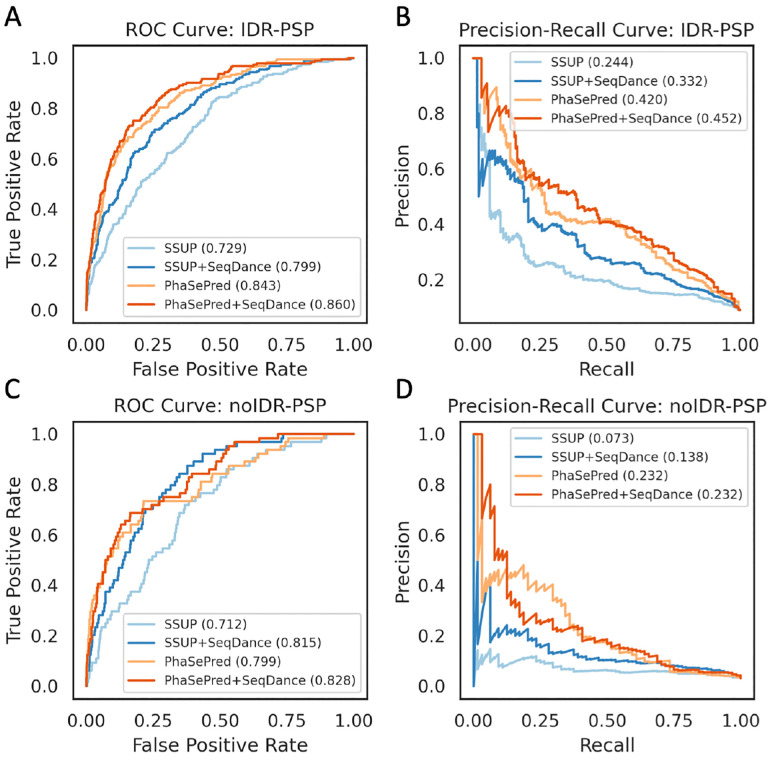
Phase separation is a crucial cellular process in which biomolecules assemble into membrane-less organelles to regulate cellular organization, metabolism, and stress responses53. Phase-separating proteins (PSPs) are driven by dynamic interactions between IDRs and/or interacting surface patches on ordered regions. Previous methods, such as PhaSePred-8feat54 (sequence-based) and SSUP55 (static structure-based), have been used to predict PSPs. We hypothesized that the dynamic properties learned by SeqDance could improve PSP prediction.
Using the datasets of PSPs with IDR (IDR-PSPs) and without IDR (noIDR-PSPs) established by Hou et al.55, we found that SeqDance embeddings significantly enhanced the performance of both PhaSePred and SSUP in predicting IDR-PSP and noIDR-PSP. The improvement was particularly notable for static structure-based SSUP: for IDR-PSP, SeqDance embeddings increased the area under the receiver operating characteristic curve (auROC) by 9.6% (from 0.729 to 0.799) and the area under the precision-recall curve (auPRC) by 36.1% (from 0.244 to 0.332). For noIDR-PSP, SeqDance embeddings improved SSUP’s auROC by 14.5% (from 0.712 to 0.815) and the auPRC by 89.0% (from 0.073 to 0.138).
Discussion
In this work, we started from the information flow in protein study (Figure 1A) and developed SeqDance, a novel protein language model (pLM) pre-trained on dynamic properties derived from molecular dynamics (MD) simulations and normal mode analysis (NMA). SeqDance captures both local dynamic interactions, co-movement, and global conformational features, complementing traditional evolution- and static structure-based methods. Our results demonstrate that SeqDance improves predictions in several biological questions, including protein fitness landscapes, intrinsically disordered regions (IDRs), and phase-separating proteins (PSPs).
In the pre-training process, we did not directly train SeqDance on full structure ensembles due to the immense size of the dataset (over 50 million frames) and the complexity of modeling entire ensembles. In fact, we are still unable to predict a static structure from a single sequence without relying on conservation profiles, let alone predict entire structure ensembles. To address this, we used simplified dynamic feature descriptors such as the mean, standard deviation, and interval distribution of ensemble-derived properties. Prior study has demonstrated that mean values from structure ensembles provide significantly more information than static structural values56. This strategy enables SeqDance to learn from simplified but informative representations of protein dynamics, without the overwhelming computational demand of full ensemble modeling.
During SeqDance pre-training, we employed attention maps and pairwise embeddings to predict dynamic interactions and residue co-movements (Supplementary Figure 1). Although using attention maps increases memory usage, it does not slow down training speed. The use of attention maps in predicting these features helped constrain SeqDance to focus on interacting and co-moving residue pairs, reducing random or irrelevant attention. This approach is crucial for learning biophysically meaningful representations and improves the model’s ability to extrapolate to unseen proteins. Besides, using attention maps and pairwise embeddings as input can capture the distinct characteristics of pairwise features: the mean value of interaction feature depends on sequence length, as the maximum number of interactions a residue can form is fixed, while the mean value of correlation feature is length-independent. By combining length-dependent attention values (which sum to one after SoftMax operation in attention calculation) with length-independent pairwise embeddings, the model can effectively capture both pairwise features. In Supplementary Figure 2, we observed a negative correlation between attention values and co-movements derived from high-frequency normal modes. We attribute this to the smaller absolute values of high-frequency features, which contribute less to the training loss, thus underrepresented in the pre-training process.
SeqDance embedding effectively captures global conformational properties of both ordered and disordered proteins, which are essential for understanding protein shape and flexibility. In comparison, METL23 underperformed in predicting the radius of gyration (Rg) for ordered proteins (Figure 3A), despite having Rg prediction as a pre-training task. This may be due to an overabundance of pre-training tasks and limited training set diversity. ProSE4 performed well in predicting the conformational properties of ordered regions but struggled with disordered regions, likely because its pre-training focused on contact prediction in ordered PDB structures. SeqDance, on the other hand, was pre-trained on dynamic properties of both ordered and disordered proteins, providing a comprehensive representation of protein dynamics. Evolution-based representations from ESM27 also performed well, as conformational properties are conserved among homologs36.
SeqDance enhances understanding of protein fitness landscapes by providing dynamic context information that complements traditional evolution-based and static structure-based representations. SeqDance embedding significantly improved GearNet’s ability to predict protein fitness, this suggests that static structures alone miss important information, consistent with the fact that static structure-based predictors often underperform in mutation-related tasks40. Combining representations from static and dynamic structure ensembles could be especially valuable for studying novel or rapidly evolving proteins where evolutionary profiles are limited or misleading. Further studies are needed to validate these findings across diverse experimental and clinical mutation datasets.
Furthermore, SeqDance yielded promising results in tasks related to IDRs and PSPs. SeqDance excelled in predicting disorder-to-order binding regions, likely because these regions exhibit specific dynamic patterns in the MD simulation that SeqDance captures in pre-training. In PSP prediction, SeqDance improved performance by integrating dynamic features with existing structure- and sequence-based features. The greater enhancement seen with SSUP55 compared to PhaSePred54 may be due to PhaSePred already utilizing biophysical features, such as predicted pi-interactions and physicochemical properties, which have overlap with information encoded in SeqDance embeddings.
We envision several directions to further improve SeqDance. First, expanding protein dynamic data: while high-resolution dynamic data is scarce compared to the vast number of sequenced proteins, lower-resolution data, like coarse-grained MD simulations and NMA, have proven valuable. More dynamic data from these faster methods could be generated to further pre-train SeqDance. Second, incorporating more detailed features that can describe higher-order relationships and time dependence. Third, scaling up model size: as seen in deep learning field, larger models with more parameters could capture more complex relationships.
In conclusion, SeqDance represents a significant advancement in the field of protein representation. By learning representations of protein dynamics, we gain valuable insights into protein behaviors that were previously reliant on extensive MD simulations. This capability has the potential to reduce our dependence on computationally expensive MD simulations, offering a more efficient approach to study protein behaviors and functions.
Methods
Molecular dynamic data collection and processing
The ATLAS29 database (v1) contains 1,516 molecular dynamics (MD) simulations, each conducted using the CHARMM36m force field for 100 ns with three replicates. Trajectory files comprising 10,000 frames were downloaded. The Protein Ensemble Database31 (PED) provides conformational ensembles for intrinsically disordered proteins, primarily derived from experiments, with some from simulations or predictions. All available ensembles were retrieved, and sequences shorter than 16 residues were excluded. GPCRmd30 is a community-driven database of MD simulations of G-protein-coupled receptors (GPCRs), with most proteins simulated for 500 ns in three replicates. IDRome24 contains conformational ensembles of human disordered regions generated via the coarse-grained residue-level CALVADOS model. All coarse-grained trajectories were downloaded and converted to all-atom trajectories using cg2all32. All data was obtained in January 2024. For each trajectory, the first 20% of frames were discarded (except for IDRome, where the first 10 frames were excluded as described in the original paper24). All frames were aligned to the first frame based on Cα atoms using MDTraj57, and trajectories of the same protein were merged.
Feature extraction from MD trajectories
GetContacts (https://getcontacts.github.io) was used to extract nine types of interactions from MD trajectories: backbone-to-backbone hydrogen bonds, side-chain-to-backbone hydrogen bonds, side-chain-to-side-chain hydrogen bonds, salt bridges, Pi-cation, Pi-stacking, T-stacking, hydrophobic interactions, and van der Waals interactions. Default definitions of these interactions were used as described in https://getcontacts.github.io/interactions.html. For each residue pair, nine interaction frequencies were calculated, resulting in a matrix with the size of L×L×9 for a protein of length L.
MDTraj57 was used to extract residue-level features. Root mean square fluctuations (RMSF) were calculated using mdtraj.rmsf; Eight-class secondary structure was determined using mdtraj.compute_dssp; Surface area per residue was computed using the Shrake-Rupley algorithm (mdtraj.shrake_rupley(mode=‘residue’)), and the mean and standard deviation of surface areas were recorded; For dihedral angles, mdtraj.compute_phi, mdtraj.compute_psi, and mdtraj.compute_chi1 were employed to extract the phi, psi, and chi1 angles, respectively. Dihedral angles across all frames were partitioned into 12 bins (30° intervals), and the percentage of frames falling into each bin was calculated for each residue. Collectively, this yielded a residue-level feature matrix of size L×(1+8+2+3×12) for a protein of length L, the dimension corresponds to RMSF (1), secondary structure (8), surface area (2), and dihedral angle distributions (3×12), respectively.
For the calculation of pairwise residue movement correlations, we first computed the covariance matrix for the x, y, and z coordinates of all Cα atoms:
| (1) |
Where p is the number of trajectory frames, Xi represents the positions (x, y, z) of all Cα atoms in frame i, and is the mean position of the Cα atoms over all frames. The matrix C3L has a dimension of 3L×3L where L is the number of Cα atoms (protein length).
To reduce this 3D covariance matrix to residue level, the trace over the spatial dimensions is taken:
| (2) |
Here, CL is the reduced covariance matrix, and the diagonal of CL corresponds to the squared fluctuation of L residues. The correlation matrix RL is then computed by normalizing the covariance matrix:
| (3) |
| (4) |
where CL,ij is the covariance between residues i and j, σi and σj are their respective standard deviations. This correlation matrix describes the linear relationship between the displacements of residue pairs, independent of their absolute motion magnitude.
Normal mode analysis
For normal mode analysis (NMA), PDB structures in ProteinFlow34 were used. Structures containing sequence gaps or exceeding 5,000 residues were excluded. Terminal missing residues were removed. For the 20230102_stable dataset, MMseqs258 clustering at 90% sequence identity yielded 26,670 representative structures. For the 20231221_sabdab dataset, MMseqs2 clustering at 100% identity resulted in 2,097 structures. Structures of complexes were also used, ‘X’ was added between chains for MMseqs2 clustering.
NMA was conducted using the Gaussian Network Model (GNM)27 and the Anisotropic Network Model (ANM)28, both implemented in ProDy33. These models represent macromolecules as elastic node-and-spring networks, where Cα atoms serve as nodes, and springs connect residues within a defined cutoff distance. A distance-dependent spring force constant was applied as in ProDy website (http://www.bahargroup.org/prody/tutorials/enm_analysis/gamma.html): for Cα atoms 10–15 Å apart, a unit force constant was used; for atoms 4–10 Å apart, a force constant twice as strong was used; and for atoms within 4 Å (i.e., connected residue pairs), a force constant 10 times stronger was employed. GNM, which models isotropic motion, was used to compute residue-level features, while ANM, which captures anisotropic motion, was employed to calculate pairwise features.
After building the elastic network (Kirchhoff matrix for GNM or Hessian matrix for ANM), normal modes were computed by eigenvalue decomposition. Eigenvalues (λm) and eigenvectors (vm) were used to describe the collective motions of residues in mode m. The individual contribution of each mode is the proportion of the inverse eigenvalue to all modes:
| (5) |
Where M is the number of total modes (L-1 for GNM and 3L-6 for ANM, L is protein length). Modes of GNM and ANM were first ranked by contribution, then partitioned into three ranges separately. The ranges were selected such that the first set of modes accounts for ~33% of the dynamics, the second set for ~33–66%, and the final set for ~66–100%. This ensures that the slow, intermediate, and fast modes are separated.
For each set of modes, the mean-square fluctuation (MSF) of each residue was calculated from GNM modes as:
| (6) |
Where vmi is the eigenvector component corresponding to mode m and residue i, and λm is the corresponding eigenvalue. This calculation was repeated for three mode ranges.
The residue covariance matrix was calculated using ANM modes. Firstly, the covariance matrix with the size of 3L×3L was first computed as:
| (7) |
Where V is the matrix of eigenvectors and Λ−1 is the inverse diagonal matrix of eigenvalues. To reduce the 3D covariance matrix to the residue-level, we used the same method as described in the calculation of pairwise residue movement correlations (equation 2–4). Readers can also read the paper59 for the calculation of pairwise correlation in NMA.
SeqDance model architecture
SeqDance is a transformer encoder based on the ESM27 architecture of 35 million parameters, using the same sequence tokenizer. All parameters were randomly initialized as described in the paper7. ESM2 weights were not used to avoid incorporating conservation information implicitly. The model consists of 12 layers with 20 attention heads each and employs rotational positional embeddings. The final embedding dimension is 480. A linear layer is added to predict residue-level features from the final residue embedding. For pairwise feature prediction, we compute pairwise embeddings from residue embeddings as:
| (8) |
where Ei and Ej represent the final residue embeddings of residues i and j, and W1 and W2 are learnable transformation matrices. The pairwise embeddings Pij, along with attention values from 240 attention heads, are passed through a linear layer to predict pairwise features.
Pre-training procedure
The model was implemented and trained using PyTorch60 (version 2.2.0). SeqDance was pre-trained on over 95% of all data, and randomly selected 600 proteinflow34 PDB structures, 500 IDRome trajectories, 75 ATLAS trajectories, 25 GPCRmd trajectories, and 20 PED ensembles were held out for evaluation. The batch size was set to 72, and parameters were updated every 10 batches: one batch from high-resolution data, four batches from IDRome, four batches from the 20230102_stable dataset, and one batch from the 20231221_sabdab dataset. Different weights were assigned to training data from different sources in the loss function, after adjustments to the batch numbers, considering the different resolutions and relative confidences: 0.5 for high-resolution data, 0.2 for IDRome, and 0.2 for NMA data. Due to limited computational resources, we did not experiment with adjusting the batch size or the weights assigned to different data sources.
The model was optimized using the AdamW optimizer, with a peak learning rate of 1×10−4, an epsilon value of 1×10−8, and betas of (0.9, 0.98). A weight decay of 0.01 was applied, and the learning rate followed a schedule with 5,000 warm-up steps, followed by a linear decay to 1×10−5. A dropout rate of 0.1 was applied, and the maximum input sequence length was set to 512. For sequences longer than this, random peptides of 512 residues were selected in each epoch. Training lasted for a total of 200,000 updates, for ten days, using six Nvidia A6000 GPUs.
Loss function
Mean squared error (MSE) was used as the loss function for SeqDance pre-training. For secondary structure and dihedral angles, where the values for each feature sum to one, the predicted values were first passed through a softmax function to ensure they sum to one before calculating the MSE. The loss for pairwise interaction frequency was calculated as:
| (9) |
Where p is the prediction from SeqDance and f is the interaction frequency. As most interaction frequencies are zero, we balanced the MSEs for interacting and non-interacting pairs. Additionally, some interacting pairs exhibit interaction frequencies close to zero, which are biologically meaningful but are treated similarly to zero in the MSE. To address this, we applied the transformation f1/3 to scale up low-frequency interactions.
SeqDance was pre-trained on multiple tasks of varying scales. To balance the losses across these tasks, we first used a baseline model that predicts the mean value for each feature of each protein to calculate the training set losses. These baseline losses were then employed to adjust the weights for each task. Since pairwise features are more abundant than residue-level features, we set the pairwise-to-residue loss ratio at 4:1. The loss weights for solvent accessible surface area (SASA), root mean square fluctuation (RMSF), secondary structure, and dihedral angles were set to be equal. Additionally, the cumulative loss for nine interaction features was balanced against the pairwise correlation loss, maintaining a 4:1 ratio. Due to limited computational resources, we did not experiment further with adjusting these ratios.
Evaluation of attention map
For the evaluation of attention maps, we selected the top 10 attention heads based on their weights from the pairwise feature prediction layer (Supplementary Figure 1A). To compute the sequence identity of the held-out sequences compared to the training set, we used MMseqs2 for iterative best-hit identification. The following command was applied: mmseqs search qDB tDB rDB tmp --start-sens 1 --sens-steps 3 -s 7 -a 1. The coverage of the training sequences was calculated based on the held-out proteins.
Evaluation of Conformational Properties for IDRs and Ordered Structures
We analyzed normalized conformational properties from MD trajectories of over 40,000 IDR sequences36. IDRs with a normalized Rg value below 1 were excluded, as they were considered too compact to be truly disordered. Additionally, only IDRs ranging from 32 to 320 residues in length were included, resulting in 18,415 IDRs for downstream analysis. MMseqs2 search was used to identify homologous sequences in SeqDance’s training set as in the evaluation of attention map. IDRs with over 20% sequence identity (at least 60% coverage) to any SeqDance training sequence were removed to further assess SeqDance’s performance on dissimilar sequences.
Rg values for PDB structures were downloaded from the paper37, and normalized as described in the paper37:
| (10) |
Where L is the sequence length.
For both IDRs and ordered structures, MMseqs2 easy-cluster was used to cluster sequences for supervised learning with parameters: --min-seq-id 0.2 -c 0.6 --cov-mode 0. Sequences from 60% of randomly selected clusters were used as the training set, and the remaining sequences formed the test set. To ensure a fair comparison, we used the first 200 principal components of the embedding from each method. Linear regression (sklearn.linear_model.LinearRegression with default parameters) was applied to predict conformational properties. This process was repeated five times, and the mean values of the evaluation metrics were reported to assess the model’s performance.
Evaluation of protein folding stability dataset
For the evaluation of the protein folding stability dataset38, single-point replacement mutations from the dataset “Tsuboyama2023_Dataset2_Dataset3_20230416.csv” were utilized. Residues with at least 10 mutations that have “ddG_ML” values were used, resulting in a final dataset of 20,955 residues from 412 proteins. The mean value across all mutations was calculated for each residue for evaluation. AlphaFold-predicted structures were downloaded and used as input for both ESM_IF151 and GearNet (pre-trained weight in mc_gearnet_edge.pth was used)50 models.
A multi-layer perceptron (MLP) with two layers was employed to predict the average mutation effect, utilizing the MLPRegressor from scikit-learn with parameters set to hidden_layer_sizes=(10,) and max_iter=1000. Four-fold cross-validation was conducted to evaluate model performance, with the validation repeated ten times. The predicted values from all ten repeats in random split were saved to calculate the performance for each protein. It should be noted that the performance of individual proteins was not solely based on training and testing within that protein, the training process also included other proteins.
Evaluation of intrinsically disordered regions and phase separation proteins
For the tasks involving intrinsically disordered regions (IDRs), the datasets provided by CAID252 were used for evaluation. For the PDB, NOX, and linker prediction tasks, SeqDance was fine-tuned and validated on DisProt (version: DisProt_2023_12)61. We removed DisProt entries where the protein appeared in the CAID2 test set or could not be matched to the UniProt sequence. For the binder task, SeqDance was fine-tuned and validated on the training sequences from two methods (MoRFchibi62 and DeepDISOBind63) that were also evaluated in CAID2. For fine-tuning, we applied a balanced cross-entropy loss function. The peak learning rate was set to 1×10−4, with a warm-up phase of 1,000 steps and a dropout rate of 0.3. Model weights were saved every 100 steps, and the best model based on validation set performance was used for evaluation.
For the tasks of predicting phase-separating proteins (PSPs), the dataset from the paper55 was used, and separate models were trained and tested for noIDR-PSP and IDR-PSP categories. Two-fold random negative protein samples were used. The XGBoost Python package (https://xgboost.readthedocs.io/) was used for training the models. The following parameters were applied: the objective function was set to binary logistic classification (objective: binary:logistic), the maximum tree depth was set to 3 (max_depth: 3), the learning rate was 0.3 (eta: 0.3), and the evaluation metric was the area under the curve (AUC) (eval_metric: auc). The models were trained for 100 boosting rounds (num_round: 100).
Supplementary Material
Figure 6. SeqDance augmented structure and sequence-based features in predicting phase-separating proteins.
A-D. Performances for predicting intrinsically disordered phase-separating proteins (IDR-PSPs) and non-intrinsically disordered phase-separating proteins (noIDR-PSPs) using PhaSePred or SSUP as input, either alone or combined with SeqDance embeddings. The results presented are the averages of ten independent repeats.
Acknowledgment
This work was supported by NIH grants (R35GM149527) and Simons Foundation SFARI #1019623. We thank Dr. Guojie Zhong, Dr. Haiqing Zhao (UTMD), and Aziz Zafar for helpful discussions and suggestions.
Footnotes
Competing interests
The authors declare no competing interests.
Data and code availability
All the training data and evaluation data are publicly available. Codes used in model training and analysis can be found at GitHub: https://github.com/ShenLab/SeqDance, we also provide the pre-trained weight at https://zenodo.org/records/13909695.
References
- 1.Tunyasuvunakool K. et al. Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abramson J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baek M. et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bepler T. & Berger B. Learning the protein language: Evolution, structure, and function. Cell Syst 12, 654–669 e653 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panchenko A.R., Kondrashov F. & Bryant S. Prediction of functional sites by analysis of sequence and structure conservation. Protein science : a publication of the Protein Society 13, 884–892 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H., Xu M.S., Fan X., Chung W.K. & Shen Y. Predicting functional effect of missense variants using graph attention neural networks. Nat Mach Intell 4, 1017–1028 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023). [DOI] [PubMed] [Google Scholar]
- 8.Rives A. et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc Natl Acad Sci U S A 118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elnaggar A. et al. ProtTrans: Toward Understanding the Language of Life Through Self-Supervised Learning. IEEE Trans Pattern Anal Mach Intell 44, 7112–7127 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Madani A. et al. Large language models generate functional protein sequences across diverse families. Nat Biotechnol 41, 1099–1106 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding F. & Steinhardt J. Protein language models are biased by unequal sequence sampling across the tree of life. bioRxiv, 2024.2003.2007.584001 (2024). [Google Scholar]
- 12.Zhang Z. et al. Protein language models learn evolutionary statistics of interacting sequence motifs. bioRxiv, 2024.2001.2030.577970 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayes T. et al. Simulating 500 million years of evolution with a language model. bioRxiv, 2024.2007.2001.600583 (2024). [DOI] [PubMed] [Google Scholar]
- 14.Gordon C., Lu A.X. & Abbeel P. Protein Language Model Fitness Is a Matter of Preference. bioRxiv, 2024.2010.2003.616542 (2024). [Google Scholar]
- 15.Hermann L., Fiedler T., Nguyen H.A., Nowicka M. & Bartoszewicz J.M. Beware of Data Leakage from Protein LLM Pretraining. bioRxiv, 2024.2007.2023.604678 (2024). [Google Scholar]
- 16.Hou C., Li Y., Wang M., Wu H. & Li T. Systematic prediction of degrons and E3 ubiquitin ligase binding via deep learning. BMC Biol 20, 162 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akdel M. et al. A structural biology community assessment of AlphaFold2 applications. Nat Struct Mol Biol 29, 1056–1067 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawashima S. et al. AAindex: amino acid index database, progress report 2008. Nucleic Acids Res 36, D202–205 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berman H.M. et al. The Protein Data Bank. Nucleic Acids Research 28, 235–242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vernon R.M. et al. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife 7, e31486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai H., Vernon R.M. & Forman-Kay J.D. An Interpretable Machine-Learning Algorithm to Predict Disordered Protein Phase Separation Based on Biophysical Interactions. Biomolecules 12, 1131 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou C. et al. PhaSepDB in 2022: annotating phase separation-related proteins with droplet states, co-phase separation partners and other experimental information. Nucleic Acids Res 51, D460–D465 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gelman S. et al. Biophysics-based protein language models for protein engineering. bioRxiv (2024). [Google Scholar]
- 24.Tesei G. et al. Conformational ensembles of the human intrinsically disordered proteome. Nature (2024). [DOI] [PubMed] [Google Scholar]
- 25.Holehouse A.S. & Kragelund B.B. The molecular basis for cellular function of intrinsically disordered protein regions. Nat Rev Mol Cell Biol 25, 187–211 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi M. et al. Quantifying the phase separation property of chromatin-associated proteins under physiological conditions using an anti-1,6-hexanediol index. Genome Biol 22, 229 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haliloglu T., Bahar I. & Erman B. Gaussian Dynamics of Folded Proteins. Physical Review Letters 79, 3090–3093 (1997). [Google Scholar]
- 28.Atilgan A.R. et al. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys J 80, 505–515 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vander Meersche Y., Cretin G., Gheeraert A., Gelly J.C. & Galochkina T. ATLAS: protein flexibility description from atomistic molecular dynamics simulations. Nucleic Acids Res 52, D384–D392 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez-Espigares I. et al. GPCRmd uncovers the dynamics of the 3D-GPCRome. Nat Methods 17, 777–787 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Ghafouri H. et al. PED in 2024: improving the community deposition of structural ensembles for intrinsically disordered proteins. Nucleic Acids Res 52, D536–D544 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pang Y.T., Yang L. & Gumbart J.C. From simple to complex: Reconstructing all-atom structures from coarse-grained models using cg2all. Structure 32, 5–7 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakan A., Meireles L.M. & Bahar I. ProDy: Protein Dynamics Inferred from Theory and Experiments. Bioinformatics 27, 1575–1577 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozlova E., Valentin A., Khadhraoui A. & Nakhaee-Zadeh Gutierrez D. ProteinFlow: a Python Library to Pre-Process Protein Structure Data for Deep Learning Applications. bioRxiv, 2023.2009.2025.559346 (2023). [Google Scholar]
- 35.Vaswani A. et al. in Advances in neural information processing systems 5998–6008 (2017).
- 36.Lotthammer J.M., Ginell G.M., Griffith D., Emenecker R.J. & Holehouse A.S. Direct prediction of intrinsically disordered protein conformational properties from sequences. Nat Methods (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanner J.J. Empirical power laws for the radii of gyration of protein oligomers. Acta Crystallogr D Struct Biol 72, 1119–1129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuboyama K. et al. Mega-scale experimental analysis of protein folding stability in biology and design. Nature 620, 434–444 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong G., Zhao Y., Zhuang D., Chung W.K. & Shen Y. PreMode predicts mode of action of missense variants by deep graph representation learning of protein sequence and structural context. bioRxiv, 2024.2002.2020.581321 (2024). [Google Scholar]
- 40.Cheng J. et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science (2023). [DOI] [PubMed] [Google Scholar]
- 41.Gao H. et al. The landscape of tolerated genetic variation in humans and primates. Science 380, eabn8153 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brandes N., Goldman G., Wang C.H., Ye C.J. & Ntranos V. Genome-wide prediction of disease variant effects with a deep protein language model. Nat Genet 55, 1512–1522 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uyar B., Weatheritt R.J., Dinkel H., Davey N.E. & Gibson T.J. Proteome-wide analysis of human disease mutations in short linear motifs: neglected players in cancer? Mol Biosyst 10, 2626–2642 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y. et al. A multiscale functional map of somatic mutations in cancer integrating protein structure and network topology. bioRxiv (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ose N.J. et al. Dynamic coupling of residues within proteins as a mechanistic foundation of many enigmatic pathogenic missense variants. PLoS Comput Biol 18, e1010006 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ose N.J., Campitelli P., Patel R., Kumar S. & Ozkan S.B. Protein dynamics provide mechanistic insights about epistasis among common missense polymorphisms. Biophysical Journal 122, 2938–2947 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodrigues C.H.M., Pires D.E.V. & Ascher D.B. DynaMut2: Assessing changes in stability and flexibility upon single and multiple point missense mutations. Protein science : a publication of the Protein Society 30, 60–69 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Witham S., Takano K., Schwartz C. & Alexov E. A missense mutation in CLIC2 associated with intellectual disability is predicted by in silico modeling to affect protein stability and dynamics. Proteins 79, 2444–2454 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ose N.J. et al. (eLife Sciences Publications, Ltd, 2024).
- 50.Zhang Z. et al. Protein Representation Learning by Geometric Structure Pretraining. bioRxiv (2022). [Google Scholar]
- 51.Hsu C. et al. Learning inverse folding from millions of predicted structures. bioRxiv, 2022.2004.2010.487779 (2022). [Google Scholar]
- 52.Conte A.D. et al. Critical assessment of protein intrinsic disorder prediction (CAID) - Results of round 2. Proteins 91, 1925–1934 (2023). [DOI] [PubMed] [Google Scholar]
- 53.Alberti S., Gladfelter A. & Mittag T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 176, 419–434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Z. et al. Screening membraneless organelle participants with machine-learning models that integrate multimodal features. Proc Natl Acad Sci U S A 119, e2115369119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hou S. et al. Machine learning predictor PSPire screens for phase-separating proteins lacking intrinsically disordered regions. Nat Commun 15, 2147 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gu S. et al. Can molecular dynamics simulations improve predictions of protein-ligand binding affinity with machine learning? Brief Bioinform 24 (2023). [DOI] [PubMed] [Google Scholar]
- 57.McGibbon R.T. et al. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys J 109, 1528–1532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steinegger M. & Soding J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol 35, 1026–1028 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Chiang Y., Hui W.-H. & Chang S.-W. Encoding protein dynamic information in graph representation for functional residue identification. Cell Reports Physical Science 3 (2022). [Google Scholar]
- 60.Paszke A. et al. in Proceedings of the 33rd International Conference on Neural Information Processing Systems Article 721 (Curran Associates Inc., 2019). [Google Scholar]
- 61.Aspromonte M.C. et al. DisProt in 2024: improving function annotation of intrinsically disordered proteins. Nucleic Acids Res 52, D434–d441 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Malhis N., Jacobson M. & Gsponer J. MoRFchibi SYSTEM: software tools for the identification of MoRFs in protein sequences. Nucleic Acids Res 44, W488–493 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang F., Zhao B., Shi W., Li M. & Kurgan L. DeepDISOBind: accurate prediction of RNA-, DNA- and protein-binding intrinsically disordered residues with deep multi-task learning. Brief Bioinform 23 (2022). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the training data and evaluation data are publicly available. Codes used in model training and analysis can be found at GitHub: https://github.com/ShenLab/SeqDance, we also provide the pre-trained weight at https://zenodo.org/records/13909695.