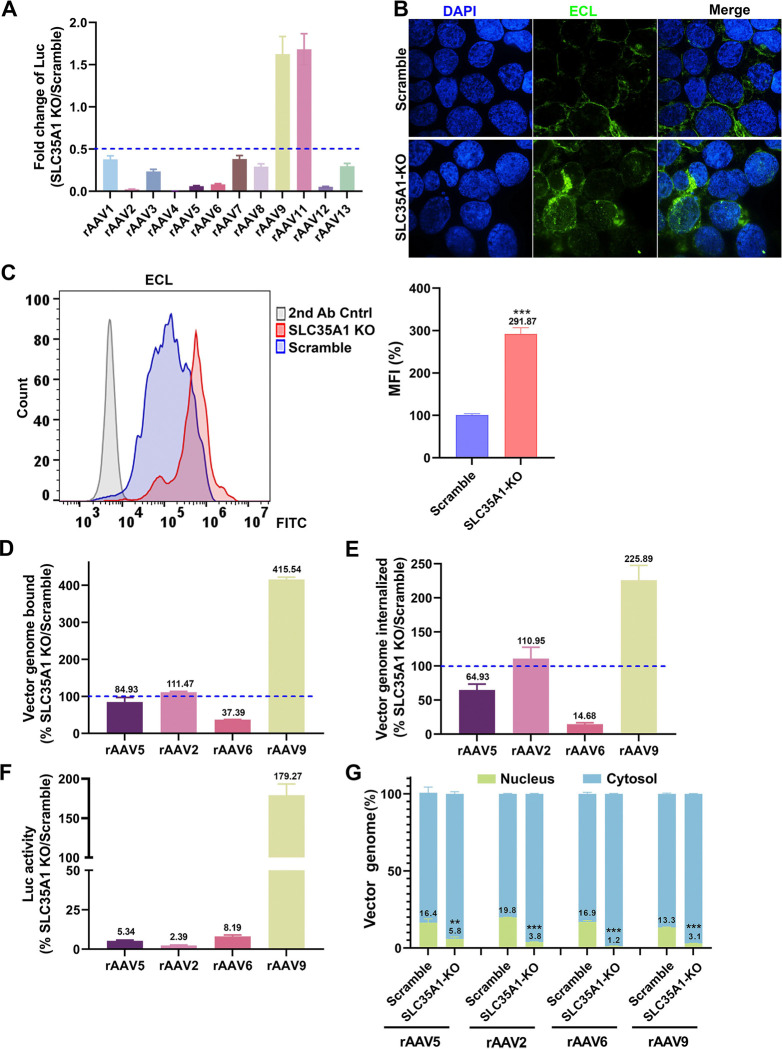
Figure 6. SLC35A1 KO significantly decreases the transduction efficiency of rAAV1–8, 12 and 13, but increases the transduction efficiency of rAAV9 and rAAV11, and causes a significant decrease in the nuclear import of rAAV.
(A) Luciferase activities. HEK293Scramble and HEK293SLC35A1-KO cells were respectively transduced with various serotypes of rAAV vectors as indicated at an MOI of 20,000 DRP/cell. At 3 dpt, the luciferase activities were measured and normalized to Scramble cells (set as 1). The fold changes of luciferase activities in SLC35A1-KO vs Scramble are shown with means and an SD from at least three replicates. (B&C) Lectin staining. HEK293Scramble and HEK293SLC35A1-KO cells were respectively stained with Erythrina cristagalli lectin (ECL) for analyses by confocal microscopy (B) and by flow cytometry (C). The mean fluorescence intensity (MFI) values were calculated and normalized to the Scramble cells as percentages (%), which are shown with a mean and SD from three replicates, and were analyzed by the Student’s t-test. (D-F) rAAV binding, internalization, transduction. HEK293Scramble and HEK293SLC35A1-KO cells were respectively transduced with four selected representative vectors, rAAV5, rAAV2, rAAV6, and rAAV9, in parallel. Vector binding (D), Internalization (E), and transduction (F) are assessed and relative fold changes in SLC35A1-KO vs Scramble as percentages (%) are shown with a mean and SD from at least three replicates. (G) Nuclear import assays. HEK293Scramble and HEK293SLC35A1-KO cells were respectively transduced with four selected representative vectors, rAAV5, rAAV2, rAAV6, and rAAV9, in parallel. At 12 hpt, the cytoplasm and nucleus were fractionated, and the percentage of vector genome copies in the cytoplasm and nucleus fractions were quantified. The data shown are means with an SD from at least three replicates. P value was determined by using the Student t-test for the comparison of the vector genome copies in the nucleus between the KO cell group and the Scramble cell group.