Abstract
Segment 5 of bluetongue virus (BTV) serotype 10, which encodes the outer capsid protein VP5, was tagged with glutathione S-transferase and expressed by a recombinant baculovirus. The recombinant protein was subsequently purified to homogeneity, and its possible biological role in virus infection was investigated. Purified VP5 was able to bind mammalian cells but was not internalized, which indicates it is not involved in receptor-mediated endocytosis. The purified VP5 protein was shown to be able to permeabilize mammalian and Culicoides insect cells, inducing cytotoxicity. Sequence analysis revealed that VP5 possesses characteristic structural features (including two amino-terminal amphipathic helices) compatible with virus penetration activity. To assess the role of each feature in the observed cytotoxicity, a series of deleted VP5 molecules were generated, and their expression and biological activity was compared with the parental molecule. VP5 derivatives that included the two amphipathic helices exhibited cytotoxicity, while those that omitted these sequences did not. To confirm their role in membrane destabilization two synthetic peptides (amino acids [aa] 1 to 20 and aa 22 to 41) encompassing the two helices and an additional peptide representing the adjacent downstream sequences were also assessed for their effect on the cell membrane. Both helices, but not the downstream VP5 sequence, exhibited cytotoxicity with the most-amino-terminal helix (aa 1 to 20) showing a higher activity than the adjacent peptide (aa 22 to 41). Purified VP5 was shown to readily form trimers in solution, a feature of many proteins involved in membrane penetration. Taken together, these data support a role for VP5 in virus-cell penetration consistent with its revelation in the entry vesicle subsequent to cell binding and endocytosis.
Infection of a cell by a virus involves a number of steps. The virus must attach to the cell surface, penetrate, and subsequently become sufficiently uncoated to make its genome accessible to viral or host machinery for transcription or translation. Virus attachment to cells in many instances leads to irreversible changes in the structure of virions, facilitating the penetration. While enveloped viruses (such as influenza viruses, paramyxoviruses, and retroviruses) rely predominantly on the fusion of their envelopes with the cell membrane during penetration, the mechanisms of penetration for nonenveloped viruses (such as picornaviruses, adenoviruses, and reoviruses) involve either protein-mediated rupture of endosomes, allowing the release of partially uncoated particles, or the formation of a protein-lined transmembrane pore through which the genome is transported to the cytoplasm (6, 26, 40, 62, 65). For nonenveloped viruses, separate coat proteins are often involved in the activities of virus attachment, entry, and penetration (10, 13, 14, 20, 22, 37, 39, 43, 52, 60). Understanding the mechanism of virus entry and uncoating for nonenveloped viruses has been greatly enabled by studies of small icosahedral viruses such as poliovirus (10, 20, 55) and of the larger and more complex icosahedral adenoviruses (50). Both viruses have true icosahedral symmetry, with capsid structures made up of equimolar amounts of proteins with relatively simple structures (see reviews in references 24 and 59). In contrast, the mechanisms by which large, spherical nonenveloped viruses with complex capsid structures, such as the members of the Reoviridae, penetrate the cell membrane are relatively poorly understood. To redress this imbalance, we have undertaken studies to define the entry and penetration mechanisms for Bluetongue virus (BTV), the type virus species of the genus Orbivirus within the family Reoviridae.
Although the overall morphological and physicochemical properties of orbiviruses are similar to those of the other members of the family, orbiviruses are distinct from reoviruses and rotaviruses in a number of ways. BTV and other orbiviruses are vectored to vertebrates by arthropod species, multiplying in both species. They are structurally unique in having proteins in their organization that bear no similarity in primary sequence to those of reovirus or rotaviruses (see reviews in references 56 and 57). The virions are also more fragile than reovirus, BTV infectivity being lost in mildly acidic conditions and on exposure to lipid solvents and detergents, although there is no evidence for the presence of a lipid component in purified virions (23).
BTV virions are architecturally complex and are composed of seven discrete proteins in specific but nonequimolar ratios that are organized into two capsids (for reviews, see references 27, 28, 29, 53, 56, 57, and 61). The virion proteins encapsidate the genome of 10 double-stranded RNA (dsRNA) segments. The outer capsid, which is composed of two major structural proteins, a larger 110-kDa protein, VP2, and a smaller 60-kDa protein, VP5, is involved with cell attachment and virus penetration during the initial stages of infection. Each protein is a product of a single gene and is not derived from a precursor protein, as in the case of outer capsid proteins of reovirus and rotaviruses (see reviews in references 17 and 51). After entry into cells, the virus is uncoated (VP2, VP5 removed) to yield a transcriptionally active core particle which is composed of two major proteins (VP7 and VP3) and three minor proteins (VP2, VP4, and VP6) in addition to the dsRNA genome (57). There is no evidence that any trace of the outer capsid remains associated with these cores, as has been described for reovirus (see reviews in references 16 and 51). Using cryo-electron microscopic analysis of unfixed and unstained virions and virus-like particles (VLPs), we have shown that the two outer shell proteins have distinctive shapes. One of these proteins is globular, almost spherical, with 360 molecules sitting neatly on each of the six-membered rings of the underlying VP7 trimers (27, 28, 29), and the other is sail-shaped with protruding triskelion spikes, located above 180 of the VP7 trimers. Together, the two proteins form around the core a continuous layer ca. 86 nm in diameter. VP2, which is believed to be the spike-like protein, has been shown to be responsible for eliciting virus neutralizing antibodies, hemagglutination activity, and serotype specificity (15, 25, 30, 31, 32, 34, 58). Antibody neutralization experiments supported by our recent studies on direct cell binding with VP2 and its subsequent internalization suggest that VP2 is the protein associated with the cell attachment and entry of virions (26). The second outer capsid protein VP5 may also play a role in virus neutralization activity as VP5 enhances the protective neutralization activity of VP2 in sheep (58).
Once BTV has attached to the surface of a susceptible cell, the virus is internalized by receptor-mediated endocytosis, forming clathrin-coated vesicles containing virus particles (see review in reference 16). Following internalization, the clathrin coats of endocytosed vesicles are rapidly lost, resulting in formation of large endocytic vesicles where VP2 is degraded. By a mechanism that has yet to be elucidated, the partly denuded BTV virion causes destabilization of vesicle membrane to allow the penetration of the newly uncoated core particles into the cytoplasm. The release of the BTV core has been shown to be dependent on acidic pH since the addition of compounds that raise the lysosomal or endosomal pH prevents endocytosed virus particles from entering the cytoplasm (16). Thus, although the outer capsid proteins are clearly responsible for virus entry and penetration, the precise mechanism of membrane traverse and the structural changes in the outer capsid proteins associated with it, remain unclear. The position of VP5 in the capsid would, however, be consistent with a role in the translocation of the core into the cytoplasm after cell entry.
To gain an understanding of the function of VP5, we have produced a tagged form of the molecule and examined its properties of cell binding, internalization, and membrane permeabilization. Our studies suggest that, although VP5 binds to the cell surface, it is not responsible for virion internalization but could have a role in the membrane destabilization required for core access to the cytoplasm. The data have further been substantiated by generating a series of VP5 deletion mutants and relevant peptides based on the predicted structural features of VP5 and by subsequent assessment of their effect on membrane permeabilization.
MATERIALS AND METHODS
Cells and viruses.
The cell lines used in this study were mouse fibroblasts (L929), BHK-21 (baby hamster kidney) cells, and Vero (African green monkey kidney) cells. The cells were grown in Leibovitz L-15 medium or in RPMI medium supplemented with HEPES, 1% l-glutamine and penicillin-streptomycin (Gibco), and 10% fetal bovine serum (FBS) and incubated at 37°C. BTV serotype 10 (BTV-10) was propagated in BHK-21 cells, and titration of the virus by plaque assay was conducted in Vero cells. Recombinant baculoviruses based on Autographa californica nuclear polyhedrosis virus (AcNPV) were propagated in Spodoptera frugiperda (Sf9) cells as described by King and Possee (36).
Cloning of segment M5 of BTV-10 into the GST pAcG2T baculovirus transfer vector.
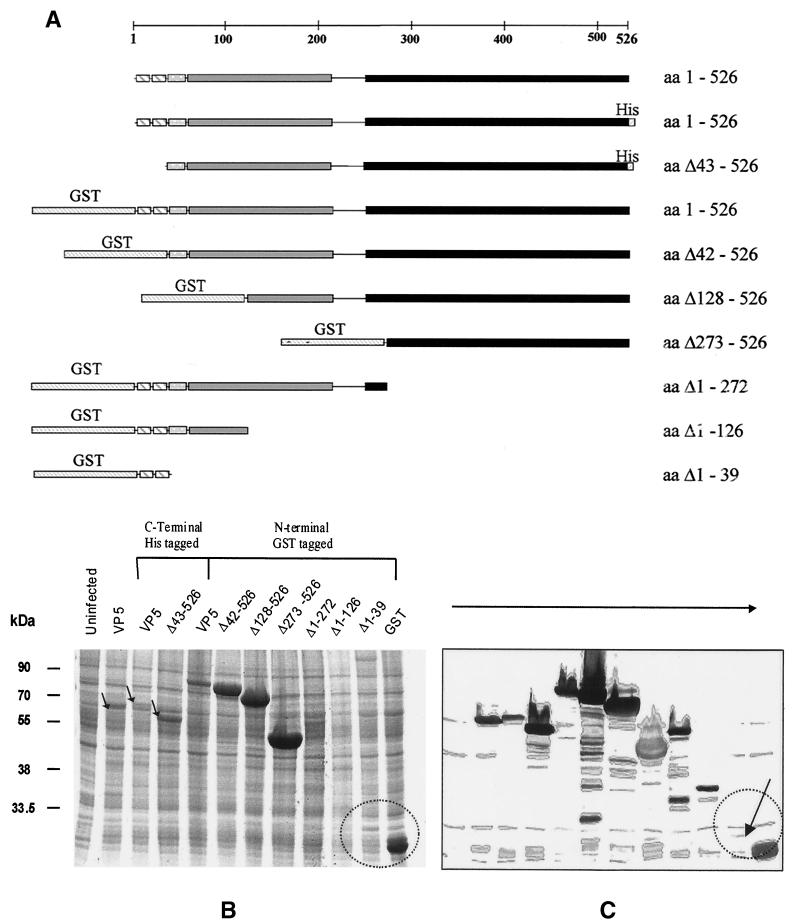
The baculovirus transfer plasmid pAcYM1-10.5 encoding full-length VP5 of BTV serotype 10 has been described previously (45). To generate a baculovirus glutathione S-transferase (GST)–VP5 expression plasmid, the full-length coding sequence of VP5 was amplified and cloned into the baculovirus transfer vector pAcG2T (Pharmacia). The orientation of inserts were characterized by restriction mapping and DNA sequencing. Recombinant baculoviruses were obtained by transfecting Sf9 cells with a mixture of the newly generated transfer vector (pAcG2T-VP5) DNA and a linearized AcNPV DNA (BacPAK6; Clontech) as described by King and Possee (36). The expression of full-length GST-VP5 was analyzed by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS–10% PAGE), followed by detection by Coomassie blue staining and the identity of VP5 was confirmed by Western blotting using BTV-10 antisera as described previously (26). Other plasmid constructions encoding variants of the VP5 coding sequence were prepared using a combination of the PCR and subcloning. A GST-VP5 fusion protein lacking amino acids (aa) 1 to 41 was amplified with primers to delete the 5′ end of the VP5 coding region, followed by cloning into pAcG2T as before. To remove the region encoding aa 127 to 526 of VP5, a construct expressing full-length GST-VP5 was digested with SacI and SmaI, blunt ended with Klenow polymerase, and religated. Similarly, to remove the region encoding aa 273 to 526, the NdeI-SmaI fragment was removed, and the resulting plasmid was religated. To express only aa 128 to 526 of VP5, the SacI-BamHI fragment was excised from pAcYM1-10.5, blunt ended, and cloned into the BamHI site of pAcG2T. Similarly, to express aa 273 to 526 as a fusion protein with GST, an NdeI-BamHI fragment was excised from pAcYM1-10.5 and cloned into the BamHI site of pAcG2T. GST fusion proteins encoding the first 39 amino acids of VP5 were produced by amplification of the relevant coding region, followed by cloning in pAcGST2T as before. A full-length VP5 fused to a C-terminal His tag and a VP5 deletion mutant lacking the first 41 aa was produced by amplification of the coding region of VP5, followed by cloning into baculovirus transfer vector pBac2cp (Novagen). All transfer vectors were subject to confirmation by DNA sequencing before their use to produce recombinant baculoviruses. A representation of each of the constructions used in this work is shown in Fig. 6A.
FIG. 6.
Expression of VP5 deletion mutants by recombinant baculoviruses in insect cells. (A). Schematic representation of VP5 and various deletion mutants. The different structural features and domains of VP5 are indicated by different shadings. The constructs are drawn to scale to the full-length VP5 represented by a bar at the top. Numbers to the right indicate the amino acids of VP5 that are contained in the recombinant proteins. N-terminal GST tags and C-terminal His6 tags are indicated. (B) SDS–12% PAGE analysis of Sf9 cell lysates recovered after 48 h of infection by each of the recombinant baculoviruses. Proteins were stained with Coomassie blue. (C) Western analysis of SDS-PAGE gels by probing with an anti-VP5 polyclonal antiserum. Labels on the top indicate the recombinant baculoviruses used for infection. Lane 1 contained an uninfected cell lysate as control. The low-level expression of the deletion mutant Δ1–39, in contrast to GST expression, is indicated. The sizes of molecular mass markers are indicated on the left in kilodaltons.
Expression and purification of recombinant VP5.
Sf9 cells at 2 × 106 cells/ml cultivated as suspension cultures in TC-100 medium containing 5% FBS and antibiotics were infected with the recombinant VP5 baculovirus at a multiplicity of infection (MOI) of 10 to 25 PFU/cell. The cells were harvested by centrifugation after 42 h of incubation at 28°C and processed further to obtain the soluble extract as described previously (26). The purification of the VP5 was performed using a glutathione-Sepharose column as suggested by the manufacturer (Pharmacia). Briefly, the GST-VP5 fusion protein was bound to high-affinity GST-beads (Pharmacia) and subsequently cleaved from the beads using a biotinylated thrombin preparation. The biotinylated thrombin was then removed from the purified VP5 by using streptavidin-agarose beads.
Metabolic-labeling of BTV and recombinant baculovirus-infected cells.
Radiolabeling of BTV and VP5 proteins with [35S]methionine was conducted as described previously (26).
Preparation of antibodies against the expressed purified VP5.
A monospecific polyclonal antibody (PAb) against the purified VP5 was prepared in mice using standard procedures for antibody production. The immunoglobulin fraction of the antisera was purified using the monoclonal antibody (MAb) Trap GII system (Pharmacia) according to the manufacturer's instructions. The protein concentration was estimated using the Bradford protein assay method (4).
Immunoprecipitation assay.
This assay was conducted according to the methods of Hussy et al. (33). The anti-VP5 PAb (described above) was bound to protein A-Sepharose for 2 h at 4°C. Controls included normal rabbit serum, anti-BTV-10 antibody, or the polyclonal anti-VP2 antibody against the VP2. The antibody-coated beads were incubated with 10 μl (2 μg) of purified [35S]methionine-labeled VP5, 50 μl of BTV-infected cell lysates, or mock-infected cells for 16 h at 4°C. Samples were washed, and the complexes were heated for 10 min in SDS-PAGE sample buffer prior to separation on SDS–10% PAGE, followed by autoradiography.
Binding and internalization assays using indirect immunofluorescence.
A Vero cell line permissive for BTV was used. For immunofluorescence studies, cells were grown on coverslips and incubated with 15 μg of purified VP5 and VP2. Binding was carried out at 4°C for 1 h and internalization was at 37°C for an additional 1 h. Surface membrane immunofluorescence and internal staining was conducted as described previously (26).
Generation of purified VP2, VP7 proteins, and particles.
The recombinant VP2 and VP7 were purified as described previously (2, 26). BTV core-like particles (CLPs) synthesized by a recombinant baculovirus were purified by sucrose density gradient centrifugation as described previously (19). The purity of the proteins and particles was confirmed by SDS-PAGE and electron microscopy. The protein concentration was estimated using the Bradford protein assay method (4).
Cytotoxicity assay.
A colorimetric cell cytotoxicity assay (Cytotox 96; Promega) was adopted for the determination of cell leakage. This quantitative assay measures levels of lactate dehydrogenase (LDH), a stable cytoplasmic enzyme released on cell lysis. This assay reveals early, low-level damage to cell membranes that may be missed with other methodologies. Furthermore, this technology has been used for measuring natural cytotoxicity and has been shown to give similar values as determined in parallel 51Cr release assays (12, 38). In this assay, a concentration of cells with absorbance values at least two times above the background absorbance of the control were used. The concentration of serum used to maintain the cells' viability was kept to a minimum (i.e., 2%) since serum contains LDH at low levels. Control samples were therefore set up to correct for LDH in the medium and from spontaneous release of LDH from uninfected cells. The maximum release of LDH from uninfected cells was measured after lysis of the cells with Triton X-100 at a final concentration of 0.8% in the medium. Either L929 or Vero cells were used as target cells and incubated with the purified BTV proteins and particles or peptides at 37°C for 4 h in V-bottom plates. Quadruple samples were used for each, and the average of LDH release was calculated. Three to five sets of experiments were performed, and the readings varied no more than 5% for individual sample. Synthetic peptides used, were dissolved in dimethyl sulfoxide (DMSO) and diluted with cell culture medium. The plates were then centrifuged, and the supernatant was harvested and incubated with the assay substrate at room temperature for 30 min before the reaction was stopped using 1 M acetic acid as described by the manufacturer (Promega). The controls were included in quadruplicate. The percent toxicity values were calculated by substitution of the mean absorbance values at 492 nm into the following equation: percent cytotoxicity = [(experimental culture medium background) − (spontaneous culture medium background)/(maximum release − volume correction control)] × 100.
SDS-PAGE analysis of the oligomeric nature of VP5.
Preliminary studies on the oligomeric nature of the purified [35S]methionine radiolabeled VP5 were performed by analyzing the protein by PAGE. The protein was resuspended in sample buffer (1% SDS; 15% glycerol; 10 mM Tris-HCl, pH 6.8) with or without 1% β-mercaptoethanol and with or without heat treatment at 100°C for 2 min. The samples were analyzed by electrophoresis on 7% PAGE gels followed by autoradiography. Apparent molecular weights were estimated using a mixture of molecular weight markers (Sigma).
Glycerol gradient sedimentation.
The purified VP5 (ca 0.5 mg) was sedimented through 25 to 40% glycerol gradients, in 20 mM Tris (pH 8.4)–100 mM NaCl–1 mM dithiothreitol–0.5 mM EDTA for 30 h at 35,000 rpm in an SW41 rotor at 4°C. A mixture of marker proteins containing bovine serum albumin (BSA; 66 kDa), β-amylase (200 kDa), and apoferritin (440 kDa) was also centrifuged using a parallel gradient. Gradients were fractionated in 0.5-ml volumes beginning at the top of the gradient (fraction 1) and continuing until 24 fractions had been collected. Aliquots of 20 μl of each fraction were resolved using SDS-PAGE, followed by Western blot analysis.
Size exclusion column chromatography of VP5 oligomers.
Sf9 cells were infected at an MOI of 2 with a recombinant baculovirus expressing untagged VP5 (45) and harvested at 24 h postinfection. The cell pellet was lysed in 10 ml of TENT (150 mM NaCl; 50 mM Tris, pH 7.5; 1 mM EDTA; 1% Triton X-100) buffer. The lysate was subsequently centrifuged for 30 min at 11,000 rpm in a JA12 rotor, filtered through a 0.22-μm (pore-size) filter, and 1/3 loaded on a 1-ml HitrapQ column equilibrated in 100 mM NaCl–50 mM Tris (pH 7.5). The column was developed with a linear gradient from 100 mM to 1 M NaCl for 20 min at 0.5 ml/min. Fractions containing VP5 were pooled and 45% (wt/vol) NH4SO4 in 25 mM Tris (pH 7.5) was added to adjust the NH4SO4 concentration to 15%. The sample was then loaded onto a 1-ml Hitrap phenyl-Sepharose high-performance column. To elute bound proteins, the column was developed with 20 column volumes of a linear gradient from 15% NH4SO4 to 0% NH4SO4 in 25 mM Tris-HCl (pH 7.5) at 0.5 ml/min. Fractions containing VP5 were pooled and applied on a Superdex 200 column equilibrated in 25 mM Tris (pH 7.5)–0.5 M CH3COONH4. Proteins were eluted at 0.5 ml/min, and 2-ml fractions were collected.
RESULTS
High-level expression and rapid purification of VP5.
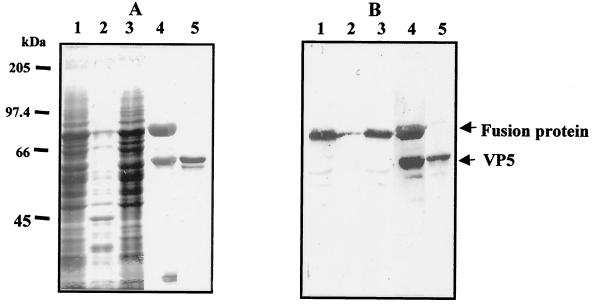
To obtain sufficient purified VP5 protein for biochemical analysis, we prepared a recombinant baculovirus that expressed BTV VP5 in Sf9 cells as a GST-VP5 fusion protein. When recombinant virus-infected cells were examined by SDS-PAGE, the recombinant fusion protein was expressed at a maximum level between 36 to 48 h postinfection, following which the expression level declined gradually. The level of expression of fusion protein was higher than that of untagged VP5 protein, as reported previously (40). Band intensity, as measured by densitometry of infected cell extracts at 36 h postinfection, suggested that about 20% of the total infected cellular protein was GST-VP5 (Fig. 1A, lane 1). The protein was found in the detergent-soluble fraction of infected cells (Fig. 1A, compare lane 2 to lane 3) and was identified as VP5 by Western blotting (Fig. 1B) with the anti-VP5 MAb 10AE12 (46). GST-VP5 was purified from extracts by glutathione affinity chromatography (Fig. 1A, lane 4) and released by cleavage of the immobilized fusion protein with thrombin (Fig. 1A, lane 5). Some free GST was also present in the soluble lysate but not in the final VP5 preparation (Fig. 1A, compare lane 4 to lane 5). The final yield of purified soluble VP5 was 0.3 to 0.5 mg per 108 infected cells, an amount sufficient for functional characterization. The recombinant VP5 protein generated has two additional non-VP5 residues at its N terminus (Gly and Ser), due to the construction of GST VP5 fusion vector.
FIG. 1.
Expression and purification of VP5. (A) Coomassie blue-stained SDS–10% PAGE gels (A) and Western blot of expressed VP5 using anti-VP5 MAb (B). Lane 1, whole-cell lysates of Sf9 cells infected by a recombinant GST-VP5 baculovirus at 42 h postinfection; lane 2, cell lysate pellet; lane 3, supernatant of cell lysate; lane 4, fusion product, cleaved VP5, and the released GST band; lane 5, purified VP5 recovered from the GST-beads. A contaminant cellular protein is visible on the stained gel that was not recognized by anti-VP5 antisera.
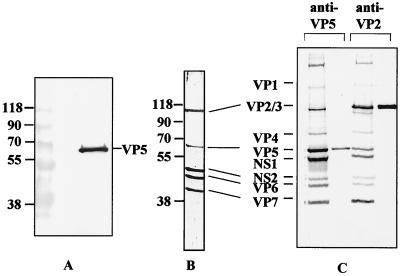
The authenticity of the soluble recombinant purified VP5 protein produced as a GST fusion and purified as described above was further confirmed by the generation of sera in guinea pigs, followed by assessment of its ability to recognize the authentic BTV virion protein. The serum reacted with only VP5 by Western blot of BTV particles (Fig. 2A). When used for immunoprecipitation assays, the serum was able to precipitate BTV particles from the supernatant of BTV-infected BHK-21 cells as efficiently as a VP2-specific serum used as a control (Fig. 2C). The position of all BTV structural proteins was identified by reaction of an anti-BTV-10 polyvalent serum with purified virions by Western blot (Fig. 2B).
FIG. 2.
Western blot analysis and immunoprecipitation of BTV proteins by antiserum raised against the purified VP5. (A) Western blot of purified BTV virions probed with an anti-VP5 VP5 antibody. (B) Western blot of purified BTV virion probed with anti-BTV-10 antiserum. (C) Immunoprecipitation of purified and recombinant BTV proteins by anti-VP5 (lanes 1 and 2) and anti-VP2 (lanes 3 and 4) PAbs. BHK-21 cells were infected with BTV-10 virus at an MOI of 5 and radiolabeled with [35S]methionine for 2 h at 60 h postinfection, followed by immunoprecipitation (lanes 1 and 3). Sf9 cells infected with recombinant baculoviruses expressing either VP5 (lane 2) or VP2 (lane 4) were similarly labeled and immunoprecipitated as controls. Cells were lysed with RIPA buffer, and the supernatant was immunoprecipitated with the two antibodies. Lanes 1 and 3, BTV proteins in BHK-21 cell lysates precipitated by anti-VP5 and anti-VP2 antibodies, respectively; lanes 2 and 4, recombinant VP5 and VP2 precipitated with anti-VP5 and anti-VP2 antibodies, respectively.
Purified VP5 binds metabolically active mammalian cells but is not internalized.
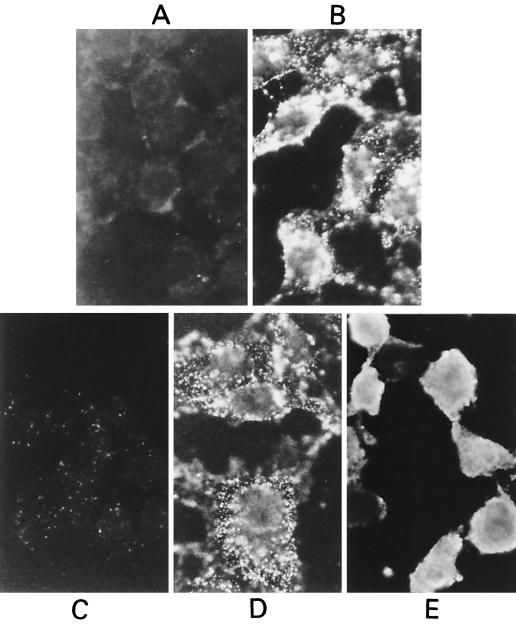
To determine whether VP5 is able to attach to cells and subsequently be internalized by receptor-mediated endocytosis, purified VP5 protein was used in cell binding and internalization assays as described previously (26). In this assay, VP5 was bound to Vero cells at 4°C and detected by indirect immunofluorescent antibody staining as described in Materials and Methods. VP5 was found to efficiently attach to the cell surface, exhibiting a ring of fluorescence on the surface of the cells (Fig. 3B) that was absent from mock preparations (Fig. 3A). Cells with VP5 bound were allowed a further incubation at 37°C for 1 h to allow internalization of the bound antigen, but no change in the fluorescent pattern was observed (Fig. 3D). In contrast, purified VP2 gave a clear homogeneous fluorescent staining of the cytoplasm (see Fig. 3E) when incubated at 37°C for 1 h to allow internalization, as previously reported (26). No such cytoplasmic staining was observed when BTV proteins were not included (Fig. 3C). These data suggest that both VP5 and VP2 proteins, present in the outer capsid of the virion, are able to bind to the cell surface, but only VP2 can be internalized. Thus, unlike VP2, VP5 membrane binding is unlikely to be part of a receptor-mediated endocytotic uptake mechanism but may reflect intrinsic hydrophobicity.
FIG. 3.
Indirect immunofluorescence staining showing binding to, and internalization of, VP5 and VP2 by L929 cells. (A and C) Controls of binding and internalization in the absence of purified VP5 protein. (B) VP5 bound to cell surface at 4°C. (D) No change of staining with further incubation at 37°C for internalization. (E) The control VP2 protein, however, shows strong internalization by L929 cells. Magnification, ×3,200.
Membrane permeabilization by VP5.
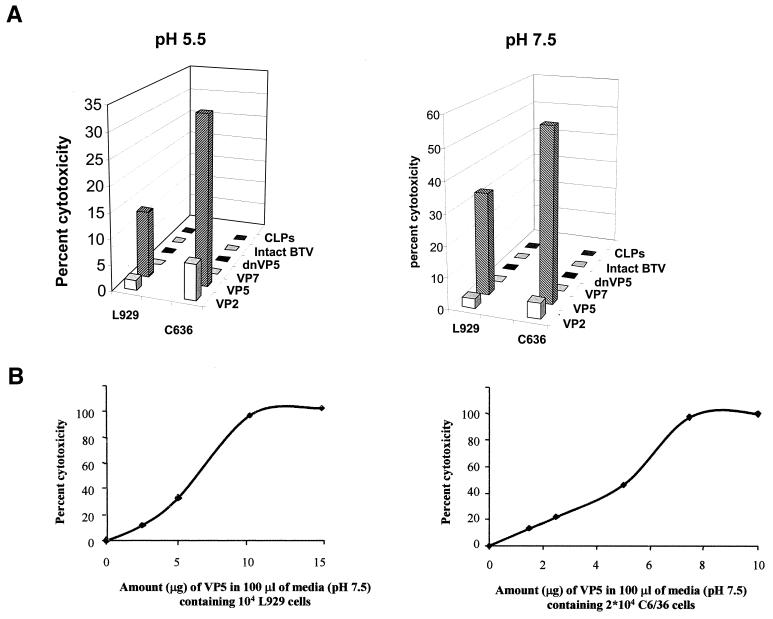
The expression of BTV VP5 alone by recombinant baculoviruses has been noted previously to result in early cell death (40, 42). Moreover, the membrane-binding capability of purified VP5 suggests that toxicity may be related to membrane disruption. To examine this possibility, we adapted a commercially available cytotoxicity assay that measures the release of an intracellular enzyme, LDH, to assess the effect of exogenously applied VP5. Similar assays have been successfully used when other types of virus-induced cytotoxicity were studied (12–14, 38). VP5 activity (as a fixed concentration) was compared to that exhibited by several other BTV reagents, including purified VP2, purified VP7, wild-type BTV particles, and baculovirus-derived CLPs. A high concentration of soluble VP5 was used in order to mimic the locally high and oriented concentrations of VP5 that occur during infection. The LDH release assay was performed at both pH 5.5 and pH 7.5. Cell substrates included L929 cells and C6/36 cells as BTV replicates in both mammalian and insect cells such as Culicoides cells and mosquito-derived cells (C6/36). Purified VP5 induced a substantial release of LDH in both cell types at either pH, indicating a lack of specificity in the protein's ability to disrupt the cellular plasma membrane (Fig. 4A). Heat-denatured VP5 (65°C for 30 min) abrogated the observed cell membrane disruption, indicating a role for the tertiary structure of the protein in activity. Purified VP2 protein showed a very low permeabilization of both cell types (2 to 4%), while the other reagents tested showed no ability to cause LDH release.
FIG. 4.
Cytotoxicity of purified VP5. (A) Cell cytotoxicity was determined at pH 5.5 and 7.5 of L15 media using 5 μg of purified VP5. As controls, two other purified BTV proteins, VP2 and VP7, and particles, virions, and CLPs were used in parallel. In addition, VP5 denatured by heating at 65°C for 30 min (dnVP5) was used as a negative control. The percent cytotoxicity was measured by the amount of LDH release per 104 L929 cells or per 2 × 104 C6/36 cells induced by each preparation as described in Materials and Methods. (B) Effect of increasing amounts of VP5 added onto L929 cells and C6/36 cells at pH 7.5. Readings from quadruple wells of each preparation were used to calculate the percent cytotoxicity. The percent cytotoxicity was calculated by the substitution of the mean absorbance values at 492 nm divided by the maximum release of LDH as described in Materials and Methods.
LDH release was found to correlate with the amount of VP5 added for both C6/36 and L929 cell lines, reaching a plateau at approximately 10 μg of VP5 per 104 L929 cells and 8 μg per 104 C636 cells (Fig. 4B). These results show that VP5-induced cytotoxicity is dose dependent and varies little between cell types.
Domain structure of VP5 revealed by secondary structure analysis.
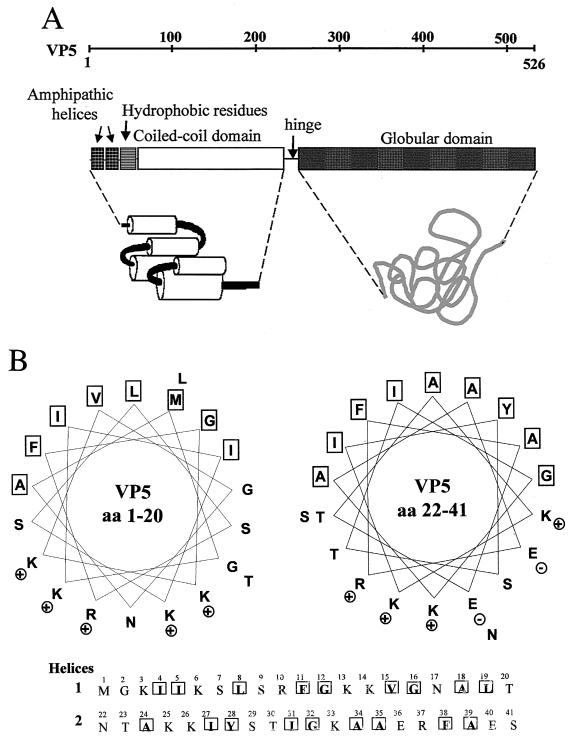
Three-dimensional reconstruction of BTV virion particles following cryoelectron microscopy suggests that VP5 may be a globular protein with an almost spherical shape (28, 29). We used hydrophobic cluster analysis (HCA), a computer homology modeling program to predict the structural organization of VP5 based on its amino acid sequence. The prediction is essentially a two-dimensional helical representation of protein sequences, which combines the comparison of sequences and that of the protein secondary structures statistically centered on hydrophobic clusters (6, 21, 41, 42, 66). HCA analysis of VP5 demonstrated that the 526 residues are divided into two domains, an amino-terminal domain (aa 1 to 240) with the features of a coiled-coil structure and a carboxyl-terminal domain (aa 260 to 526) separated by a flexible alanine- and glycine-rich hinge region (Fig. 5A). Additional “coils” and “learncoils” programs were used to confirm the coiled coil regions of the molecule (3, 44). The coiled-coil organization of VP5 is very similar to the organization observed in the influenza A hemagglutinin (HA2), in the F proteins of paramyxoviruses, and in gp41 of human immunodeficiency virus type 1 (HIV-1) as deciphered through HCA analysis. Two amphipathic helices (helix 1 and helix 2) have also been identified in the first 40 residues at the amino terminus of VP5, followed by a strong stretch of hydrophobic residues. As shown in Fig. 5B, both helix 1 (aa 3 to 21) and helix 2 (aa 22 to 41) have a net-positive charge on their hydrophilic faces, which would allow them to bind to negatively charged phospholipids. These structural data support the notion that VP5 may be a membrane-destabilizing protein, consistent with a role in cell entry following virion internalization.
FIG. 5.
Diagram of VP5 configuration predicted from primary amino acid analysis. (A) Structural features and domains of VP5 identified using computer-assisted two-dimensional HCA analysis (6), as well as the “coils” and “learncoils” programs (3, 44). (B) Helical wheel representation of aa 1 to 19 and 22 to 41 of VP5. Each panel represents an α-helix viewed along the helix axis with the indicated amino acid residues. The VP5 sequence was searched for the presence of amphipathic structures initially using the program Moment of the GCG software package. The program Helical Wheel was then used to plot a helical wheel representation of the N-terminal amino acids of VP5 (18).
The amino terminus of VP5 is responsible for cytotoxicity.
To examine whether the predicted amphipathic helices of VP5 play a role in the observed cytotoxicity, a series of N- and C-terminal deletion mutants were constructed (Fig. 6A), and the mutant VP5 was expressed using the baculovirus expression system, either as VP5 sequences alone or, to facilitate purification, after fusion to GST. GST-tagged full-length VP5 was expressed at higher levels than was VP5 alone (see Fig. 1 and 6B), and progressive deletion of the N terminus (e.g., Δ42–526, Δ128–526, and Δ273–526) was found to increase the expression level still further (Fig 6B). Deletions from the C terminus failed to increase the amount of fusion protein produced after 48 h of infection, suggesting a sequence-specific effect and not simply the result of a reduced size of translated product (Fig 6B). Even when not fused to a GST tag, the deletion mutant Δ43–526 was expressed at a significant level compared to full-length VP5. All deletion constructions were capable of expression, however, since they could be detected by Western blot using an antiserum raised against the GST-VP5 fusion protein (Fig. 6C). However, for a fusion protein that consisted of only the first 39 amino acids of VP5 (see the arrow in Fig. 6C), the band was barely detectable by Western blot. This provides direct evidence of an inverse correlation between the presence of the extreme VP5 N terminus in unblocked form and the level of expression observed, a finding consistent with a role for the N terminus in membrane destabilization.
Destabilization of cellular plasma membrane by synthetic VP5 peptides encompassing the “amphipathic” helices.
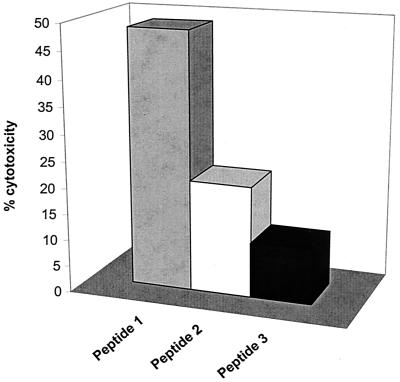
Amphipathic helices are a characteristic structural feature of many antimicrobial peptides that act on the plasma membrane. They are also found in the fusogenic proteins of enveloped viruses such as the hemagglutinin protein of influenza virus and the envelope glycoprotein (gp41) of HIV (1, 8, 48, 62, 63, 64, 65). Their amphipathic structure allows them to insert into membrane bilayers where they may form pores that destabilize membrane potential. To confirm the VP5 expression data and to ascertain if two amino-terminal “amphipathic” helices were sufficient on their own to trigger LDH release, we generated three peptides. Two of these peptides (aa 1 to 20 and 22 to 41) encompassed the two amphipathic helices and the third (aa 44 to 53) represented the hydrophobic residues immediately downstream. All three peptides were assessed for their effect on plasma membrane using LDH release as described above. Both peptides with amphipathic character caused substantial release of LDH, with peptide 1, the more basic of the two helices, exhibiting marginally higher activity than peptide 2 (Fig. 7), while the third peptide, although generally hydrophobic, failed to show any such effect.
FIG. 7.
Membrane permeabilization of synthetic peptides encompassing the putative amphipathic helices. Three N terminal peptides were used for LDH release assay: (i) MGKIIKSLSRFGKKVGNALT (aa 1 to 20), (ii) NTAKKIYSTIGKAAERFAES (aa 22 to 41), and (iii) GAATIDGLVQGSVHSIITGE (aa 44 to 53). The peptides were dissolved in DMSO at a concentration of 10 μg/ml, and subjected to LDH release assay as described for Fig. 4. The cell cytotoxicity assay was conducted by adding various concentrations of each peptide to the 104 L929 cells. Readings (OD492) from duplicate wells of each concentration were used to calculate the percent cytotoxicity. The percent cytotoxicity was calculated according to a formula as described in Materials and Methods. Four different experiments were performed, any individual of which varied no more than 5%.
Purified VP5 is oligomeric and forms trimers in solution.
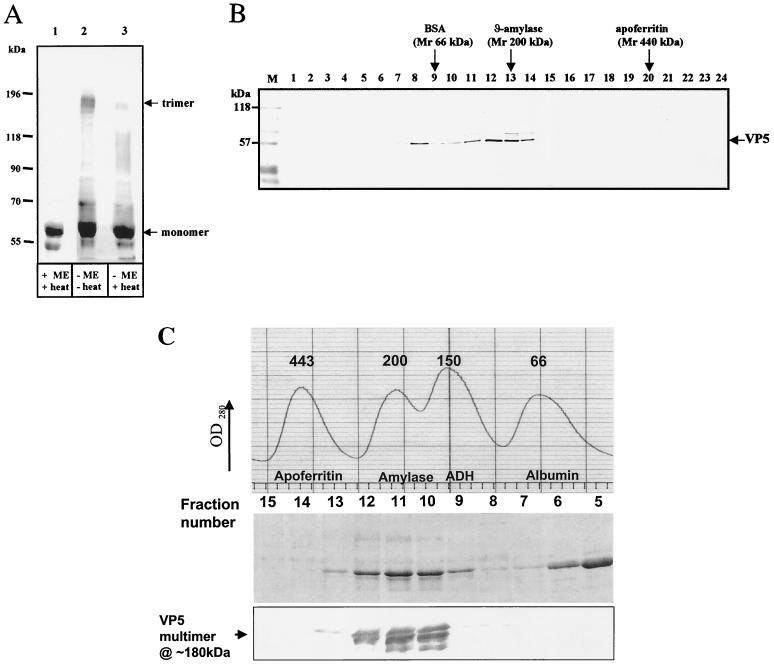
HCA analysis of VP5 strongly predicted a coiled-coil configuration downstream of the amphipathic helices. Coiled-coil motifs are found in a wide variety of viral and cellular proteins, including intermediate filaments, cell surface receptors, molecular motors, transcription factors, the hemagglutinin fusion protein of influenza virus, and HIV-1 gp41 (8, 62, 63). In most cases coiled coils serve as molecular connectors, allowing oligomerization of two or more protein molecules. Many of these proteins, including HA and gp41, form trimers in solution (8, 62, 63). Cryo-electron microscopy of BTV has suggested that BTV VP5 may also assemble into a trimer (28, 29). To provide biochemical support for this, VP5 purified from the GST carrier as described above was analyzed for its multimeric state by gel electrophoresis under reducing and nonreducing conditions by velocity gradient centrifugation and by gel filtration chromatography. When VP5 was dissociated by heat in the presence of β-mercaptoethanol, only a single band of full-length VP5 with an apparent molecular mass of ∼59 kDa was detected by SDS-PAGE (Fig. 8A, lane 1). In the absence of a reducing agent, however, an additional band with apparent molecular mass of ∼180 kDa was detected at a low level (Fig. 8A, lane 3), a finding consistent with the size of a VP5 trimer. The ∼180-kDa band was much more intense when the protein sample was not heated prior to application to the gel (Fig. 8A, lane 2). This suggests VP5 may exist as a trimer in solution, a small part of which, probably an artifact of the high VP5 concentration, is disulfide linked.
FIG. 8.
Identification of VP5 oligomers. (A) Western blot showing VP5 oligomers in reducing and nonreducing conditions. Purified soluble VP5 was analyzed by SDS–7% PAGE, blotted onto nitrocellulose paper, and probed with an anti-VP5 PAb. The VP5 were investigated in the presence (+ME) or absence (−ME) of 1% β-mercaptoethanol and with (+heat) or without (−heat) heating to 100°C for 4 min. The samples were resuspended in sample buffer containing 1% SDS, 15% glycerol, and 10 mM Tris-HCl (pH 6.8). The trimer (180 kDa) and monomer (∼59 kDa) are indicated. Additional smaller bands are the VP5 degraded products. (B) Western blot of purified VP5 after fractionation by glycerol gradient (20 to 40%) centrifugation. Lanes 1 to 24 represent fractions collected from the glycerol gradient; fraction 1 represents the top of the gradient, and fraction 24 represents the bottom. After SDS–10 PAGE, the gels were blotted onto nitrocellulose paper and probed with an anti-VP5 PAb. The positions of the standard size marker proteins of BSA (66 kDa), β-amylase (200 kDa), and apoferritin (440 kDa) are indicated at the top. The standard size markers were run in parallel in different tubes than the samples but in equivalent glycerol gradient fractions. The prestained markers on the blot are designated M. (C) Size exclusion chromatography of purified VP5. Recombinant VP5 expressed in insect cells was partially purified by anion-exchange and hydrophobic-interaction-exchange chromatography. VP5 was applied to a Superdex-200 column previously calibrated with a standard set of marker proteins (the profile is shown in the uppermost panel). VP5 fractions 5 to 13 covering the molecular mass range from 66 to 400 kDa were analyzed by SDS–10% PAGE (middle panel), and fractions 8 to 13 only were analyzed by Western blot with polyclonal VP5 antiserum (lower panel). The identity of the high-molecular-mass VP5 species as the putative trimer is indicated.
Further evidence for the solution structure of VP5 was obtained by sedimentation through glycerol gradients, followed by fractionation and analysis by SDS-PAGE and Western blot. Two peaks of VP5 were detected in fraction numbers 6 to 8 and 11 to 14 (Fig. 8B), coinciding with marker BSA (66 kDa) and β-amylase (200 kDa), respectively. This profile would be consistent with VP5 existing as a mixture of monomeric and trimeric forms in solution.
Additional evidence for the oligomeric state of VP5 was obtained by gel filtration chromatography of part-purified VP5 preparations on a Superdex-200 column. The column was calibrated using a set of protein molecular mass size standards that included apoferritin (443 kDa), amylase (200 kDa), alcohol dehydrogenase (150 kDa), and BSA (66 kDa) covering the range of molecular masses expected of oligomeric VP5 structures. Purified VP5, examined under the same buffer conditions, was identified by SDS-PAGE of the elution fractions in fractions 5 and 6 (∼57 kDa) and as a broad band eluting in fractions 10 to 12 (the 180- to 210-kDa range of the gel) (Fig. 8C, middle panel). No higher-molecular-weight species were observed. Since the VP5 preparation used for gel filtration showed monomeric and oligomeric proteins, the presence of VP5 was confirmed by Western blot of fractions 8 to 14 using a polyvalent BTV serum (Fig. 8C, lower panel).
Thus, by three independent measures of the oligomeric state, the migration of VP5 was consistent with it being predominantly a trimer in solution.
DISCUSSION
An increasing body of evidence suggests that the ability of viruses to modify membrane permeability and induce lysis is mediated by a single viral gene product and does not require the assembly of viral particle (7). Expression of proteins by heterologous expression systems, such as the baculovirus system, can provide sufficient protein for an examination of the intrinsic membrane permeability properties of individual viral proteins. We have used this approach to prepare tagged and nontagged forms of the BTV VP5 protein. N-terminally GST-tagged VP5 fusion protein was expressed at higher levels than untagged VP5, suggesting that masking the amino terminus allows higher levels of protein to stably accumulate. VP5 alone, in contrast, has been shown to be cytotoxic and easily degraded following expression (45, 47; also unpublished results). The GST-VP5 expression level was sufficient to enable purification to homogeneity and further characterization of its biological function. Purified protein was able to bind to mammalian cells in monolayers, whereas data obtained by the Huismans et al. (31) reported that BTV particles stripped of VP2 and with VP5 exposed did not bind to BHK-21 cells in suspension. Differences in cell culture conditions or the forms of VP5 used in our study and that used by Huismans et al. may be responsible for the apparent contrast in cell binding activity. However, purified protein was not internalized, suggesting that it is unlikely to be involved in receptor-mediated endocytosis, a function that has been recently mapped to the 110-kDa VP2 protein, the larger protein of the outer capsid (26). The soluble purified VP5 was also found to form trimers, a finding consistent with the predictions of cryoelectron microscopy and image analysis (28, 29). Of note, several other viral fusion proteins are found as trimers, e.g., the influenza virus hemagglutinin (65) and the fusogenic proteins of togaviruses, rhabdoviruses, and some retroviruses (8, 11, 62, 63).
VP5 intrinsic cytotoxicity was investigated using a LDH release assay, which showed that purified protein causes cellular permeabilization. The disruption of membrane integrity was observed to be greater for C6/36 insect cells than for mammalian cells, possibly due to the different lipid composition of each. LDH release by purified protein did not require acidic pH, suggesting that the acidic environment of endosome through which the virion enters the cell may be required only for the removal of VP2 and not for the activity of VP5. In support of this hypothesis, Huismans et al. (32) showed that VP2 could be completely removed from BTV in vitro at pH 5.0 to leave VP5 exposed. The solubilized and trypsin-cleaved outer glycoprotein of rotavirus, VP7, also induces permeabilization of cell membrane vesicles (9).
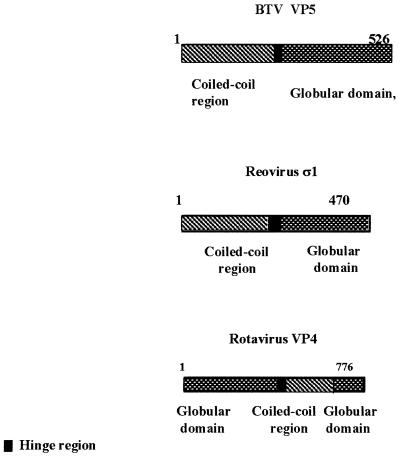
Secondary amino acid sequence analysis of VP5 revealed that the protein has structural features consistent with a role in virus penetration of the cell. The amino terminus is high in helical content, with a strongly predicted coiled-coil structure connected to a globular domain in the carboxyl half of the protein. Three-dimensional reconstructions of cryo-electron microscopy studies at low resolution have also predicted one of the outer capsid proteins to have an overall globular structure (24, 25). Two “amphipathic” helices were identified at the beginning of the coiled-coil domain, a finding consistent with a role in insertion into the lipid membrane bilayer. For reovirus, an analogous function has been assigned to the predicted two amphipathic helices (aa 534 to 551 and aa 591 to 604) of the μ1 protein (43). Similar motifs have been also identified in the VP4 protein of rotavirus (13). Interestingly, both μ1 and VP4 are the hemagglutinin and virus attachment proteins for cellular receptors, a situation unlike that for BTV, in which the VP2 protein, not VP5, possesses these activities. However, there are clearly some structural features and domain organization that are shared by all three proteins. It is noteworthy that the position of the domains of rotavirus VP4 is inverted in relation to the BTV VP5 (Fig. 9).
FIG. 9.
Overall organization of primary structures of outer capsid proteins of BTV, rotavirus, and reovirus. The overall structural organization of VP5 shows the similarity to the organization of the rotavirus outer capsid protein, VP4, and the reovirus outercapsid protein, ς1. Note that all three proteins consist of coiled-coil and globular domains.
To examine the predicted amphipathic helices for direct membrane permeabilization activity, each sequence was synthesized in vitro and tested using the LDH release assay. The peptide representing the first amphipathic helix of VP5 produced the highest levels of LDH release, with significant activity also shown by the second VP5 α-helix. A third peptide synthesized from sequences downstream of the two amphipathic helices showed no LDH release effect. Similarly, data obtained from the deletion mutation analysis confirmed that the presence of the two “amphipathic” helices at the N terminus correlated with cell cytotoxicity and membrane damage. These data are consistent with a model of VP5 function in which amphipathic helices at the amino terminus of a coiled-coil domain of VP5 are unmasked following low-pH removal of VP2 and are responsible for the membrane destabilization that is required for normal virus cell entry.
Amphipathic helices have been implicated in the membrane-binding activities of a variety of proteins, including toxins, viral fusion proteins, and many pore-forming proteins (7). For example, in influenza virus the fusion peptides in the hemagglutinins show a propensity to form amphipathic helices (5, 64). Similarly, the fusion peptides of other enveloped viruses (e.g., respiratory syncytial virus, HIV-1, Newcastle disease virus, Sindbis virus, etc.) also form sided helices, with most of the hydrophobic amino acids falling on one face of the helix and most of the apolar residues, including the majority of the alanine and glycine residues, falling on the opposite face of the helix (see reviews in references 5 and 64). BLAST searches for proteins with sequences similar to those defining the two amphipathic helices of VP5 identified cecropin, an antibacterial peptide toxic to bacterial cells and which causes permeabilization (49, 54). Interestingly, peptides representing the first amphipathic helix of VP5, the most cytotoxic, exhibited the highest sequence homology (55%) with the cecropin peptide, while the second helix showed less homology (33%).
A coiled-coil domain in VP5 separates the amphipathic helices from a hinge sequence that connects it to the globular body of the protein. It is tempting to speculate that the coiled-coil is the region responsible for the VP5 trimerization and that a conformational change occurs following VP2 removal to enable the penetration reaction. In this regard, the mechanism of this nonenveloped virus entry into the cell may resemble the more well characterized fusion mechanisms of hemagglutinin (62, 65) and HIV (8, 48) in which the coiled-coil has been likened to an unsprung spring mechanism that drives the fusion peptide into the target bilayer. For HIV, peptides that mimic the coiled coil have been shown to inhibit fusion by the membrane glycoprotein (35). It is conceivable that peptide mimics of the VP5 coiled-coil sequence might be used to verify a similar mechanism for BTV and to provide an opportunity for therapy that would be much less susceptible to the strain variation that hinders effective vaccine control of the disease (for a review, see reference 57).
ACKNOWLEDGMENTS
We are grateful to I. Callebaut (Universite Pierre et Marie Curie, Paris, France) for generating the VP5 structural data by using the HCA Programme and to Katalina Di Gleria, Molecular Medicine Department, University of Oxford.
This work was partially funded by BBSRC and the National Institute of Allergy and Infectious Diseases (5 R01-AI26879). S. Hassan was supported by a studentship from the Public Services Department of Malaysia.
REFERENCES
- 1.Baker K A, Dutch R E, Lamb R A, Jardetzky T S. Structural basis for paramyxovirus-mediated membrane fusion. Mol Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 2.Basak A K, Stuart D I, Roy P. Preliminary crystallographic study of bluetongue virus capsid protein, VP7. J Mol Biol. 1992;228:687–689. doi: 10.1016/0022-2836(92)90850-j. [DOI] [PubMed] [Google Scholar]
- 3.Berger B, Singh M. An iterative method for improved protein structural motif recognition. J Comp Biol. 1997;4:261–273. doi: 10.1089/cmb.1997.4.261. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Bradshaw J P, Duff K C, Gilchrist P J, Saxena A M. Neutron diffraction studies of amphipathic helices in phospholipid bilayers. Basic Life Sci. 1996;64:191–202. doi: 10.1007/978-1-4615-5847-7_17. [DOI] [PubMed] [Google Scholar]
- 6.Callebaut I, Labesse G, Durand P, Poupon A, Canard L, Chomilier J, Henrissat B, Mornon J P. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell Mol Life Sci. 1997;53:621–645. doi: 10.1007/s000180050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrasco L. Entry of animal viruses and macromolecules into cells. FEBS Lett. 1994;350:151–154. doi: 10.1016/0014-5793(94)00780-2. [DOI] [PubMed] [Google Scholar]
- 8.Chan F D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 9.Charpilienne A, Abad M J, Michelangeli F, Alvarado F, Vasseur M, Cohen J, Ruiz M C. Solubilized and cleaved VP7, the outer glycoprotein of rotavirus, induces permeabilization of cell membrane vesicles. J Gen Virol. 1997;78:1367–1371. doi: 10.1099/0022-1317-78-6-1367. [DOI] [PubMed] [Google Scholar]
- 10.Chow M B R, Hogle J M. The role of conformational transitions in poliovirus pathogenesis. In: Chiu W, Garcea R, Burnett R, editors. Structural biology of viruses. New York, N.Y: Oxford University Press; 1997. pp. 157–186. [Google Scholar]
- 11.Crise B, Ruusala A, Zagouras P, Shaw A, Rose J K. Oligomerization of glycolipid-anchored and soluble forms of the vesicular stomatitis virus glycoprotein. J Virol. 1989;63:5328–5333. doi: 10.1128/jvi.63.12.5328-5333.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Decker T, Lohmann-Matthes M L. A quick and simple method for the quantitation of lactate dehydrogenase release in measurements of cellular cytotoxicity and tumor necrosis factor (TNF) activity. J Immunol Methods. 1988;115:61–69. doi: 10.1016/0022-1759(88)90310-9. [DOI] [PubMed] [Google Scholar]
- 13.Denisova E, Dowling W, LaMonica R, Shaw R, Scarlata S, Ruggeri F, Mackow E R. Rotavirus capsid protein VP5* permeabilizes membranes. J Virol. 1999;73:3147–3153. doi: 10.1128/jvi.73.4.3147-3153.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowling W, Denisova E, LaMonica R, Mackow E R. Selective membrane permeabilization by the rotavirus VP5* protein is abrogated by mutations in an internal hydrophobic domain. J Virol. 2000;74:6368–6376. doi: 10.1128/jvi.74.14.6368-6376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eaton B T, Crameri G S. The site of bluetongue virus attachment to glycophorins from a number of animal erythrocytes. J Gen Virol. 1989;70:3347–3353. doi: 10.1099/0022-1317-70-12-3347. [DOI] [PubMed] [Google Scholar]
- 16.Eaton B T, Hyatt A D, Brookes S M. The replication of bluetongue virus. Curr Top Microbiol Immunol. 1990;162:89–118. doi: 10.1007/978-3-642-75247-6_4. [DOI] [PubMed] [Google Scholar]
- 17.Estes M K. Rotaviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1995. pp. 1625–1655. [Google Scholar]
- 18.Finer-Moore J, Stroud R M. Amphipathic analysis and possible formation of the ion channel in an acetylcholine receptor. Proc Natl Acad Sci USA. 1984;81:155–159. doi: 10.1073/pnas.81.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.French T J, Roy P. Synthesis of bluetongue virus (BTV) corelike particles by a recombinant baculovirus expressing the two major structural core proteins of BTV. J Virol. 1990;64:1530–1536. doi: 10.1128/jvi.64.4.1530-1536.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fricks C E, Hogle J M. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J Virol. 1990;64:1934–1945. doi: 10.1128/jvi.64.5.1934-1945.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaboriaud C, Bissery V, Benchetrit T, Mornon J-P. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987;224:149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert J M, Greenberg H B. Virus-like particle-induced fusion from without in tissue culture cells: role of outer-layer proteins VP4 and VP7. J Virol. 1997;71:4555–63. doi: 10.1128/jvi.71.6.4555-4563.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorman B M, Taylor J, Walker P J. Orbiviruses. In: Joklik W K, editor. The reoviridae. New York, N.Y: Plenum Press; 1983. pp. 287–357. [Google Scholar]
- 24.Graber U F, Willets M, Webster P, Helenius A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell. 1993;75:477–486. doi: 10.1016/0092-8674(93)90382-z. [DOI] [PubMed] [Google Scholar]
- 25.Grubman M J, Appleton J A, de Letchworth G J. Identification of bluetongue virus type 17 genome segments coding for polypeptides associated with virus neutralization and intergroup reactivity. Virology. 1983;131:355–366. doi: 10.1016/0042-6822(83)90503-2. [DOI] [PubMed] [Google Scholar]
- 26.Hassan S H, Roy P. Expression and functional characterization of bluetongue virus VP2 protein: role in cell entry. J Virol. 1999;73:9832–9842. doi: 10.1128/jvi.73.12.9832-9842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hewat E A, Booth T F, Loudon P T, Roy P. Three-dimensional reconstruction of baculovirus expressed bluetongue virus core-like particles by cryo-electron microscopy. Virology. 1992;189:10–20. doi: 10.1016/0042-6822(92)90676-g. [DOI] [PubMed] [Google Scholar]
- 28.Hewat E A, Booth T F, Roy P. Structure of bluetongue virus particles by cryoelectron microscopy. J Struct Biol. 1992;109:61–69. doi: 10.1016/1047-8477(92)90068-l. [DOI] [PubMed] [Google Scholar]
- 29.Hewat E A, Booth T F, Roy P. Structure of Bluetongue virus-like particles by cryo-electron microscopy. J Struct Biol. 1994;109:61–69. doi: 10.1016/1047-8477(92)90068-l. [DOI] [PubMed] [Google Scholar]
- 30.Huismans H, Erasmus B J. Identification of the serotype-specific and group-specific antigens of bluetongue virus. Onderstepoort J Vet Res. 1981;48:51–58. [PubMed] [Google Scholar]
- 31.Huismans H, van der Walt N T, Cloete M, Erasmus B J. The biochemical and immunological characterization of bluetongue virus outer capsid polypeptides. In: Compans R W, Bishop D, editors. Double-stranded RNA viruses. New York, N.Y: Elsevier; 1983. pp. 165–172. [Google Scholar]
- 32.Huismans H, van Dijk A A, Els H J. Uncoating of parental bluetongue virus to core and subcore particles in infected L cells. Virology. 1987;157:180–188. doi: 10.1016/0042-6822(87)90327-8. [DOI] [PubMed] [Google Scholar]
- 33.Hussy P, Schmid G, Mous J, Jacobsen H. Purification and in vitro-phospholabeling of secretory envelope proteins E1 and E2 of hepatitis C virus expressed in insect cells. Virus Res. 1996;45:45–57. doi: 10.1016/0168-1702(96)01365-2. [DOI] [PubMed] [Google Scholar]
- 34.Kahlon J, Sugiyama K, Roy P. Molecular basis of bluetongue virus neutralization. J Virol. 1983;48:627–632. doi: 10.1128/jvi.48.3.627-632.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kilby J M, Hopkins S, Venetta T M, DiMassimo B, Cloud G A, Lee J Y, Alldredge L, Hunter E, Lambert D, Bolognesi D, Matthews T, Johnson M R, Nowak M A, Shaw G M, Saag M S. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 36.King L A, Possee R D, editors. The Baculovirus expression system: a laboratory guide. London, England: Chapman and Hall; 1992. [Google Scholar]
- 37.Kirkegaard K. Mutations in VP1 of poliovirus specifically affect both encapsidation and release of viral RNA. J Virol. 1990;64:195–206. doi: 10.1128/jvi.64.1.195-206.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korzeniewski C, Callewaert D M. An enzyme-release assay for natural cytotoxicity. J Immunol Methods. 1983;64:313–320. doi: 10.1016/0022-1759(83)90438-6. [DOI] [PubMed] [Google Scholar]
- 39.Lama J, Carrasco L. Mutations in the hydrophobic domain of poliovirus protein 3AB abrogate its permeabilizing activity. FEBS Lett. 1995;367:5–11. doi: 10.1016/0014-5793(95)00523-c. [DOI] [PubMed] [Google Scholar]
- 40.Lee W M, Monroe S S, Rueckert R R. Role of maturation cleavage in infectivity of picornaviruses: activation of an infectosome. J Virol. 1993;67:2110–2122. doi: 10.1128/jvi.67.4.2110-2122.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lemesle-Varloot L, Henrissat B, Gaboriaud C, Bissery V, Morgat A, Mornon J-P. Hydrophobic cluster analysis: procedures to derive structural and functional information from 2-D representation of protein sequences. Biochimie. 1990;72:555–574. doi: 10.1016/0300-9084(90)90120-6. [DOI] [PubMed] [Google Scholar]
- 42.Lim V I. Algorithms for prediction of alpha-helical and beta-structural regions in globular proteins. J Mol Biol. 1974;88:873–894. doi: 10.1016/0022-2836(74)90405-7. [DOI] [PubMed] [Google Scholar]
- 43.Lucia-Jandris P, Hooper J W, Fields B N. Reovirus M2 gene is associated with chromium release from mouse L cells. J Virol. 1993;57:5339–5345. doi: 10.1128/jvi.67.9.5339-5345.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 45.Marshall J J, Roy P. High-level expression of the two outer capsid proteins of bluetongue virus serotype 10: their relationship with the neutralization of virus infection. Virus Res. 1990;15:189–95. doi: 10.1016/0168-1702(90)90027-9. [DOI] [PubMed] [Google Scholar]
- 46.Martinez-Torrecuadrada J L, Langeveld J P, Vento A, Sanz A, Dalsgaard K, Hamilton D O, Meloen R H, Casal J I. Antigenic profile of African horse sickness virus serotype 4 VP5 and identification of a neutralizing epitope shared with bluetongue virus and epizootic hemorrhagic disease virus. Virology. 1999;257:449–459. doi: 10.1006/viro.1999.9680. [DOI] [PubMed] [Google Scholar]
- 47.Martyn J C, Gould A R, Yu M. Expression of the outer capsid proteins VP2 and VP5 of bluetongue virus in Saccharomyces cerevisiae. Virus Res. 1994;33:11–25. doi: 10.1016/0168-1702(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 48.Matthews J M, Young T F, Tucker S P, Mackay J P. The core of the respiratory syncytial virus fusion protein is a trimeric coiled coil. J Virol. 2000;74:5911–5920. doi: 10.1128/jvi.74.13.5911-5920.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merrifield R B, Vizioli L D, Boman H G. Synthesis of the antibacterial peptide cecropin A (1–33) Biochemistry. 1982;21:5020–5031. doi: 10.1021/bi00263a028. [DOI] [PubMed] [Google Scholar]
- 50.Nemerow G R, Stewart P L. Role of alpha integrins in adenovirus cell entry and gene delivery. Microbiol Mol Biol Rev. 1999;63:725–734. doi: 10.1128/mmbr.63.3.725-734.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nibert M, Schiff L, Fields B N. Reoviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1557–1596. [Google Scholar]
- 52.Nibert M L, Fields B N. A carboxy-terminal fragment of protein μ1/μ1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J Virol. 1992;66:6408–6418. doi: 10.1128/jvi.66.11.6408-6418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prasad B V, Yamaguchi S, Roy P. Three-dimensional structure of single-shelled bluetongue virus. J Virol. 1992;66:2135–2142. doi: 10.1128/jvi.66.4.2135-2142.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Putsep R A, Branden C I, Boman H G, Normark S. Antibacterial peptide from H. pylori. Nature. 1999;398:671–672. doi: 10.1038/19439. [DOI] [PubMed] [Google Scholar]
- 55.Rossman M G, Bella J, Kolatkaer P R, He Y, Wimmer E, Kuhn R J, Baker T S. Cell recognition and entry by rhino- and enteroviruses. Virology. 2000;269:239–247. doi: 10.1006/viro.2000.0258. [DOI] [PubMed] [Google Scholar]
- 56.Roy P. Orbivirus structure and assembly. Virology. 1996;216:1–11. doi: 10.1006/viro.1996.0028. [DOI] [PubMed] [Google Scholar]
- 57.Roy P. Orbiviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1995. pp. 1709–1734. [Google Scholar]
- 58.Roy P, Urakawa T, van Dijk A A, Erasmus B J. Recombinant virus vaccine for bluetongue disease in sheep. J Virol. 1990;64:1998–2003. doi: 10.1128/jvi.64.5.1998-2003.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rueckert R R. Picornaviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 2nd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1990. pp. 507–548. [Google Scholar]
- 60.Ruiz M C, Abad M J, Charpilienne A, Cohen J, Michelangeli F. Cell lines susceptible to infection are permeabilized by cleaved and solubilized outer layer proteins of rotavirus. J Gen Virol. 1997;78:2883–2893. doi: 10.1099/0022-1317-78-11-2883. [DOI] [PubMed] [Google Scholar]
- 61.Verwoerd D W, Els H J, De Villiers E M, Huismans H. Structure of the bluetongue virus capsid. J Virol. 1972;10:783–794. doi: 10.1128/jvi.10.4.783-794.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weissenhorn W, Dessen A, Calder L J, Harrison S C, Skehel J J, Wiley D C. Structural basis for membrane fusion by enveloped viruses. Mol Membr Biol. 1999;16:3–9. doi: 10.1080/096876899294706. [DOI] [PubMed] [Google Scholar]
- 63.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 64.White J M. Viral and cellular membrane fusion proteins. Annu Rev Physiol. 1990;52:675–697. doi: 10.1146/annurev.ph.52.030190.003331. [DOI] [PubMed] [Google Scholar]
- 65.Wiley D C, Skehel J J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 66.Woodcock S, Mornon J P, Henrissat B. Detection of secondary structure elements in proteins by hydrophobic cluster analysis. Protein Eng. 1992;5:629–635. doi: 10.1093/protein/5.7.629. [DOI] [PubMed] [Google Scholar]