Abstract
The prevalence of alternative splicing as a target for alterations leading to human genetic disorders makes it highly relevant for therapy. Here we have used in vitro splicing reactions with different splicing reporter constructs to screen 4,000 chemical compounds for their ability to selectively inhibit spliceosome assembly and splicing. We discovered indole derivatives as potent inhibitors of the splicing reaction. Importantly, compounds of this family specifically inhibit exonic splicing enhancer (ESE)-dependent splicing, because they interact directly and selectively with members of the serine-arginine-rich protein family. Treatment of cells expressing reporter constructs with ESE sequences demonstrated that selected indole derivatives mediate inhibition of ESE usage in vivo and prevent early splicing events required for HIV replication. This discovery opens the exciting possibility of a causal pharmacological treatment of aberrant splicing in human genetic disorders and development of new antiviral therapeutic approaches.
Keywords: splicing correction, exonic splicing enhancer, small chemicals, pathologic splicing
Removal of introns from newly transcribed RNA polymerase II precursors (pre-mRNA) during splicing not only is an essential step for the expression of most genes in higher eukaryotic cells but also constitutes an important mechanism for generation of protein diversity and regulation of gene expression (1, 2). It is estimated that >70% of human genes are subjected to alternative splicing, and it is not surprising that many point mutations causing human diseases are associated with aberrant splicing (3, 4).
Current models of constitutive and a fortiori alternative splicing suggest that splice site recognition is strongly modulated by the interaction of specific exonic and intronic pre-mRNA sequences with at least two classes of nonspliceosomal nuclear RNA-binding proteins: serine-arginine-rich (SR) proteins (5-7) and heterogeneous nuclear ribonucleoproteins (8-10). These proteins interact with spliceosomal components (5-7) and either activate or prevent the use of degenerate splice sites in their vicinity. Thus, binding of SR proteins to exonic splicing enhancers (ESE) through their RNA-recognition motif (RRM) promotes exon definition by recruiting constitutive factors via protein-protein interactions mediated by their arginine-serine-rich (RS) domain and prevents the action of nearby splicing silencers (4, 6, 11).
Mutations causing human diseases may affect splice sites as well as regulatory sequences leading to the production of defective proteins (4, 11). Thus, targeting either the mutated sequences or the factors that bind them may prove to be a valuable strategy to correct aberrant splicing. Recently, antisense strategies targeting ESE-dependent mechanisms have been used to induce skipping of exons containing nonsense mutations or, conversely, to restore exon inclusion by synthetic exon-specific effectors (bifunctional antisense peptide molecules or tailed antisense oligonucleotides) or spliceosome-mediated RNA trans-splicing (12-16).
As an alternative approach, small chemicals that target directly or indirectly splicing regulators could be used to inhibit and/or correct splicing. Over the past years, we have demonstrated that drugs that interfere with the kinase activity of topoisomerase I (topo I), and thereby with the phosphorylation status of SR proteins, prevent spliceosome assembly and modulate the splicing profile of several genes (17, 18). More importantly, topo I/kinase-mediated phosphorylation has been shown to be required for ESE-dependent splicing (19), implying that small molecules can achieve selective inhibition of splicing events. Here, we report the results of a large-scale screen for compounds that display selective inhibition of ESE-dependent splicing and their use to influence splicing efficiencies of target pre-mRNAs. We provide evidence that indole derivatives represent a recently discovered class of splicing inhibitors that have a selective action on SR proteins. These drugs will hopefully open new avenues for the development of therapeutic agents to correct splicing defects responsible for numerous human diseases or inhibit splicing events crucial for viral replication.
Materials and Methods
Chemical Library. The Institut Curie-Centre National de la Recherche Scientifique chemical library contains 6,720 molecules kept in 96-well microplates at a concentration of 10 mg/ml in DMSO. Extemporaneous dilutions were made with 10% DMSO. Microplates were kept at -20°C.
Recombinant Protein Purification Kinase Assays. Recombinant wild-type or truncated versions of SF2/ASF were produced and purified from Escherichia coli, as described (20). The internal deletion of the RS domain corresponding to the SF2/ASF Δ197-216 mutant was generated as described (21). Recombinant topo I and SF2/ASF were also produced and purified from baculovirus-infected Sf9 cells, as reported (21, 22). The kinase assays were performed with purified proteins as described (17, 20, 22).
Plasmid Constructs, Splicing, and Spliceosome Assembly Assays. The βglo-3S plasmid containing the β-globin cassette with a triplicate of a ASF/SF2 ESE (pSPHβ3S) has been described (23). βglo-3SF2 and βglo-SRp55 were obtained by replacing the ESEs of the βglo-3S plasmid by an AccI-BamHI fragment containing either three high-score ASF/SF2 ESE (5′-CACACGA CAGACGT CACACGA-3′) or one high-score SRp55 ESE (TGCGTC), as predicted by the ESE finder tool (24). The βGlo-3S-PDH construct used for in vitro splicing experiments was obtained as described (25).
In vitro transcription to obtain radiolabeled transcripts and splicing reactions were performed under standard conditions for 1 hour, as described (17), in the presence of 50 μM tested drug. Splicing products were analyzed by electrophoresis on denaturing 7% polyacrylamide gels and revealed by autoradiography.
Kinetics of appearance of splicing complexes were performed as described (17). Aliquots (5 μl) from the various reactions treated with heparin (2 mg/ml) were mixed with 1 μl of 97% glycerol/1% bromophenol blue and resolved directly on a 4% nondenaturing polyacrylamide gel (acrylamide/bis-acrylamide weight ratio of 80:1) in 50 mM Tris-glycine (pH 8.3).
Spectroscopic Measurements. Fluorescence experiments were performed on a Fluorolog-II (Jobin Yvon, Longjumeau, France) spectrofluorometer at 25°C. The binding of the C76 indole derivative was monitored by quenching of the intrinsic fluorescence of the compound at 420 nm, upon excitation at 360 nm. The decrease in protein fluorescence was fitted to the appropriate form of the quadratic equation, as described (20, 22). The best fit was obtained with the Kd corresponding to maximal fluorescence quenching of the drug that binds SF2/ASF.
Ex Vivo Splicing Assays. HeLa cells (5 × 105 cells) were grown in RPMI medium 1640 (GIBCO/BRL), supplemented with 10% FCS on 3-cm-diameter dishes (Nunc) to 70-80% confluence. Transient transfections with splicing reporter constructs (1 μg) were performed with the LipofectAMINE Plus reagent (Invitrogen) according to the manufacturer's instructions.
Total cellular RNA was isolated from transfected HeLa cells 48 h posttransfection, and 3 μg of RNA was reversed-transcribed and PCR-amplified with forward primer BSS (5′-GGCTTGCTGAAGCGCGCACGGCAAGAGG-3′; nucleotides 700-727) and reverse primer SJ4.7A, which spans sites D4 and A7 (5′-TTGGGAGGTGGGTTGCTTTGATAGAG-3′; nucleotides 8369-8381 and 6032-6044) (26). To normalize the signals, GAPDH was used as an internal control of the PCR reactions as described (23). Amplification products were radiolabeled by performing a single round of PCR with the addition of 10 μCi (1 Ci = 37 GBq) of [α-32P]dCTP, and the products were analyzed by electrophoresis on 6% polyacrylamide 8 M urea gel, as described (26).
Results
Screen for Compounds That Inhibit Spliceosome Assembly. As an initial step toward the identification of compounds affecting splicing, we screened a large collection of known compounds, originating from the Centre National de la Recherche Scientifique-Institut Curie chemical library, for their ability to inhibit the kinase activity of topo I and thereby specific phosphorylation of SR proteins. In a primary screen, 10 μM of each compound was tested in duplicate against purified recombinant topo I/kinase in a standard kinase assay containing bacterially expressed recombinant SF2/ASF and [γ-32P]ATP. Of 2,500 compounds tested, 28 scored as potent inhibitors of the kinase activity of topo I. Several of the most efficient inhibitors of topo I/kinase activity were also tested for their ability to inhibit other purified SR protein kinases, namely SRPK1 and Clk-Sty, in the same assay. Although 10 μM drugs also inhibited Clk-Sty kinase, albeit to a lower extent than topo I, significant inhibition of SRPK1 required much higher drug concentrations (50 and 100 μM) (data not shown).
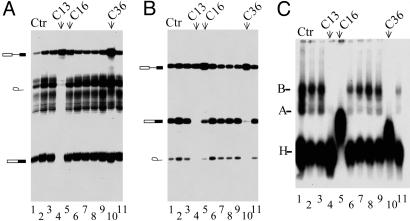
Given that topo I/kinase is required for ESE-dependent but not constitutive splicing (19), we tested the ability of the above compounds to selectively inhibit in vitro splicing of reporter pre-mRNAs. For this study, we used synthetic mRNA precursors derived from the adenovirus major late-transcription unit (Minx, Fig. 1A), which is a single-intron pre-mRNA not requiring ESE sequences in the second exon for efficient splicing. As shown in Fig. 1A, Minx pre-mRNA splicing was completely abolished in the presence of 50 μM of compound C13 (lane 4), whereas the other compounds, when used at the same concentration, have no effect (lanes 1-3 and 5-11). As expected, C13 also inhibited splicing of the βglo-3S pre-mRNA substrate, a β-globin derivative harboring three copies of a high-affinity ASF/SF2-binding site, as established by SELEX analysis (23) (Fig. 1B, lane 4) and whose splicing depends on these ESE sequences in the second exon. Interestingly, two compounds C16 and C36, which have structures related to C13 (Fig. 1 B, lanes 5 and 10, and D) also exhibited a strong inhibitory effect on βglo-3S but not on Minx pre-mRNA splicing, confirming that specific inhibitors of topo I/kinase-mediated phosphorylation may have some specificity toward ESE-dependent splicing and will interfere with spliceosome assembly.
Fig. 1.
Effect of selected drugs that inhibit topo I kinase activity on pre-mRNA splicing and spliceosome assembly. Fifty femtomol of 32P-labeled Minx, an adenovirus derivative (A), or βglo -3S, a β-globin derivative (B), pre-mRNAs were incubated in HeLa cell nuclear extracts under splicing conditions without (lanes 1) or in the presence of 50 μM selected drugs. The structure of the splicing products is depicted (Left). (C) Splicing complexes formed after 30-min incubation of 32P-labeled βglo-3S pre-mRNA in HeLa extracts alone (lane 1) or containing 50 μM (lanes 2-11) of the same drugs tested in A and B (lanes 2-11) were separated on a 4% nondenaturing polyacrylamide gel as described in Materials and Methods. Positions of splicing complexes H, A, and B are indicated. See Table 1 for chemical structure of compounds C13, C16, and C36, which have an effect on splicing and spliceosome assembly.
To determine which step of the spliceosome assembly was affected by the drug, 32P-labeled βglo-3S pre-mRNA was incubated with HeLa nuclear extracts in the presence of 50 μM of each of the above compounds, and assembled ribonucleoprotein complexes were analyzed by native gel electrophoresis. As shown in Fig. 1C, the untreated control shows the characteristic pattern of spliceosome assembly pathway. Two heparin-resistant ATP-dependent complexes correspond to complexes A and B, in addition to a fast-migrating nonspecific complex H (lane 1). Treatment of HeLa extract with compounds C13, C16, and C36 completely abolished the formation of all splicing complexes, indicating that the drugs inhibit an early step in the spliceosome assembly pathway (lanes 4, 5, and 10). It is noteworthy that treatment of extracts with C16 and C36 induces the accumulation of a complex migrating slower than the H complex (lanes 5 and 10). Although further work is necessary to determine the composition of this complex, one can speculate that it might correspond to a stalled intermediate in the spliceosome assembly pathway that precedes the formation of complex A. As expected, however, inactive drugs did not affect spliceosome assembly (lanes 2, 3, 5-8, and 10), although they inhibit the kinase activity of topo I/kinase.
Because the latter findings indicated that spliceosome assembly can represent a more straightforward assay to screen for previously uncharacterized inhibitors of splicing, we decided to perform another screen based on this assay. During this secondary screen, we tested 1,500 small molecules, of which 25 scored positive. All these small molecules, including C13, C16, and C36 (Fig. 1D), have similar structures and belong to either pyrido-carbazoles, benzo-pyrido-indoles, or pyrido pyrrolo isoquinolines (all these compounds are referred to as indole derivatives).
Specific Interaction of the Drug with SF2/ASF. Pyrido-carbazole derivatives, like ellipticines, are DNA-intercalating agents endowed with antitumor activity (27). The finding that such drugs can inhibit kinase reactions without an apparent requirement for DNA suggests that either SF2/ASF and/or topo I are targets for these compounds. Therefore, the binding affinity of the drug to SF2/ASF or to topo I/kinase was determined by using the intrinsic fluorescence of some drugs. Compound C76 (Fig. 2C), which has the strongest fluorescence, was used for this experiment. Fig. 2A shows that increasing concentrations of SF2/ASF, but not topo I/kinase, induce a quenching of 80% of drug fluorescence, indicating that this drug binds to SF2/ASF rather than to topo I/kinase. The Kd value estimated from this analysis was 0.19 μM, which implies that the drug forms a stable complex with SF2/ASF.
Fig. 2.
Binding of compound C76 (see Table 1) to SF2/ASF. Fluorescence experiments were performed as described in Materials and Methods. (A) Curves of intrinsic fluorescence quenching after binding of the drug to full-length SF2/ASF (empty circles) but not topo I (filled circles). (B) Deletion of the RS domain (filled triangles) or part of the RS domain (filled squares) abolishes quenching of fluorescence, whereas unphosphorylated (empty circle) and phosphorylated (filled circles) forms of SF2/ASF induce quenching. Although several SF2/ASF RS domain deletion mutants were tested (197C, 207C, 215C, 223C, and Δ197-216 in ref. 20), none induced quenching of C76 fluorescence, and therefore only the result obtained by Δ197-216 is presented (filled squares). The estimated Kd values derived from this analysis are 0.19 μM for unphosphorylated and 0.4 μM for phosphorylated SF2/ASF.
To examine the potential requirement of different domain(s) of the SF2/ASF protein for drug binding, we used several SF2/ASF deletion mutants lacking either the entire C-terminal domain or only small regions of the RS domain (20). No binding was observed with mutants deleted of part or all of the RS domain, indicating that the integrity of the SF2/ASF structure is essential for efficient binding (Fig. 2B). This was confirmed by heat denaturation of full-length SF2/ASF and mutants lacking either the RRM or RS domain, which completely abolished binding. Subsequent renaturation led to 100% recovery of interaction with the full-length protein and ≈60% with the RS domain. Conversely, no significant recovery of interaction was observed with the RRM domain alone, indicating that the RS domain is the major drug-binding element (data not shown). Nevertheless, failure to recover complete binding with the RS domain alone indicates that the RRM domain also contributes to the overall structure required for drug binding. It is estimated from our analysis that at least three molecules of the drug bind to one molecule of SF2/ASF. The possibility of drug binding to contaminating nucleic acids was ruled out, because neither previous treatment of SF2/ASF preparation with DNase or RNases nor purification of SF2/ASF by cesium chloride affected binding.
Given that the RS domain is required for efficient phosphorylation and interaction between SF2/ASF and topo I/kinase, direct binding of the drug to this domain offers a plausible explanation for the inhibition of kinase activity (20). Interestingly, similar experiments performed with purified recombinant SF2/ASF produced in a baculovirus system where the phosphorylation of recombinant proteins is expected to take place demonstrated comparable binding activity (with an estimated Kd value of 0.4 μM) as unphosphorylated SF2/ASF expressed in E. coli (Fig. 2B), indicating that the phosphorylation of the RS domain has only a slight effect on binding.
Selected ESE-Dependent Splicing Inhibition by Indole Derivatives. The RS domain is responsible for specific protein-protein interactions between RS domain-containing proteins required for constitutive and alternative splicing (5), and it has recently been shown to be also involved in RNA-protein interactions (28). This domain is found not only in the so-called family of SR proteins involved in alternative splicing but also in constitutive splicing factors like the U1-snRNP-specific protein U1-70K and the splicing factor U2AF. If we assume that these domains are the general targets for indole derivatives, we could expect to find among them two categories of drugs: (i) those binding general splicing factors and constituting nonspecific inhibitors of all splicing events and (ii) those targeting one particular SR protein and therefore inhibiting splicing events depending on this protein and its cognate regulatory sequences. The second category is obviously the most likely to yield drugs with therapeutic potential, because they would be expected to be less toxic for normal tissues.
Among the 6,720 compounds of the chemical library used in the initial screen, 220 indole derivatives were subsequently screened for selective inhibition of ESE-dependent splicing events. In a first step, all these drugs were tested for inhibition in an in vitro splicing assay by using a βglo-3S substrate carrying an SF2/ASF-dependent ESE. Drugs scoring positive (inhibitors) and negative were separately tested in a second assay based on a βglo-SRp55 substrate in which SF2/ASF high-affinity sequences have been substituted by an optimal binding site for SRp55 (Fig. 3E). In Fig. 3A, which represents the result of one particular set of drug tested on βglo-3S, compounds C6-10 (lanes 15, 16, 18, 19, and 21), C26-28 (lanes 10, 11, and 14), and C78 (lane 13) demonstrate a very strong inhibitory effect. Although several compounds inhibited splicing of both βglo-3S and βglo-SRp55, some were specific for each substrate (see Tables 1 and 2, which are published as supporting information on the PNAS web site). As shown in a sample screening gel on βglo-SRp55 of drugs scoring negative on βglo-3S, C22 constitutes a good example of a drug that severely impedes splicing of βglo-SRp55 (Fig. 3B, lane 9) but has no effect on βglo-3S. Conversely, several compounds, like C81-82, C38, C85, and C30, which inhibit βglo-3S, have only a minor effect on βglo-SRp55 (Fig. 3C, lanes 2, 3, 12, 20, and 21, respectively). It is also noteworthy that some inhibitors of βglo-3S splicing, like C80, C79, and C78, displayed a significant increase of βglo-SRp55 pre-mRNA splicing when compared with the control (Fig. 3C, lanes 5, 9, and 17, respectively). Such drugs could be good candidates for interfering with an inhibitory effect mediated by some SR or SR-like proteins.
Fig. 3.
Effect of indole derivatives on splicing of substrates containing different ESE. (A) One set of indole derivatives tested for their ability to inhibit βglo-3S pre-mRNA splicing is shown. The compounds showing very strong effects on splicing (lanes 10, 11, 13-16, 18, 19, and 21) are indicated on the top with arrows. Indole derivatives that inhibit (C) or not (B) βglo-3S splicing pre-mRNA were tested in splicing reactions with βglo-SRp55 pre-mRNA. The drugs that have selective action on βglo-SRp55 (B, lane 9) or on βglo-3S (C, lane 2, 3, 5, 9, 12, 17, 20, and 21) pre-mRNAs are shown by arrows on the top. (D) Dose-dependent inhibition of βglo-SRp55 (Left), βglo-3S (Center), or βglo-3SF2 (Right) pre-mRNA splicing with 200 μM (lane 2), 100 μM (lane 3), 50 μM (lane 4), 10 μM (lane 5), and 1 μM (lane 6) compound C22.(E) Structures of the different ESE-containing transcripts.
To confirm the specificity of the selected drugs, 5-fold dilution series were performed to determine the concentration required to inhibit 50% of the splicing reaction (ID50) on both βglo-SRp55 and βglo-3S substrates. As an example, C22 inhibits both reactions in a dose-dependent manner (Fig. 3D, compare lanes 2-6 in Left and Center), but the inhibitory effect is much higher for βglo-SRp55 (ID50 ≈ 1 μM) than for βglo-3S (ID50 ≈ 25 μM). The specificity of the inhibitory effect of the drug was further confirmed by using another β-globin derivative that harbors three copies of a high-affinity ASF/SF2-binding site different from that used in βglo-3S (Fig. 3E). This substrate demonstrated the same inhibition behavior as βglo-3S (Fig. 3D, lanes 2-6, compare Center and Right), confirming the idea that the major target for inhibition is the SR protein itself rather than the ESE that it binds.
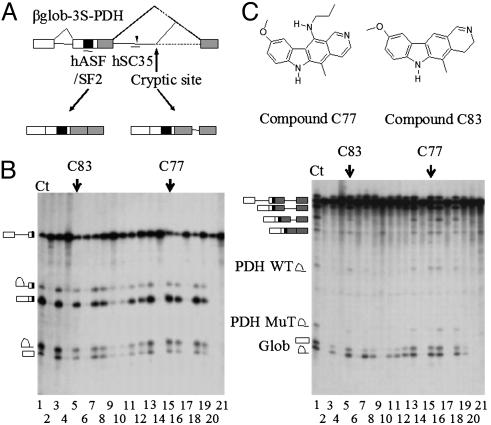
Inhibition of Aberrant Splicing by Indole Derivatives. SR protein function in ESE recognition is largely nonredundant, and multiple classes of ESE consensus motifs have been described (11). Mutations affecting these sequences have been shown to lead to exon skipping, because they abrogate or significantly reduce binding of specific SR proteins (11). To test whether some indole derivatives could differentially alter splicing events depending on distinct classes of ESE within the same substrate, we used a chimeric three-exon model substrate (βglo-3S-PDH) in which sequences of exon 7, intron 7 containing a G to A substitution at +26, and exon 8 of the E1α pyruvate dehydrogenase (PDH) gene (29) were inserted downstream of the βglo-3S sequences (Fig. 4A). The G to A change at position 26 generates an ESE that serves as a binding site for SC35 and thereby activates a cryptic 5′ splice site 20 nucleotides downstream of the mutation within the same intron (ref. 25 and Fig. 4A). The splicing products of this chimeric transcript include: species resulting from the splicing of the βglo-3S mono-intronic β-globin substrate (i.e., exon 1 and first-intron lariat), species specific for the PDH part (i.e., the two lariat introns and lariat introns-exon 8 resulting from splicing at the wild-type and cryptic 5′ splice sites) as well as intermediate and final products containing β-globin and PDH sequences. In the control sample, the final product containing three exons and 45 nucleotides from the PDH intron is predominant, whereas the three-exon product missing the latter intron sequences is less abundant (Fig. 4B Right, lane 1). All drugs that inhibit splicing of βglo-3S have a dramatic effect on the splicing of βglo-3S-PDH, because they block the formation of all products (Fig. 4B, compare Left and Right, lanes 2, 4, 6, 9, 10, 14, 17, 20, and 21), indicating that, in vitro, the splicing of the β-globin intron proceeds before that of PDH. Interestingly, however, drugs C77 and C83 (Fig. 4C), which have no effect on the βglo-3S transcript, preferentially inhibit the formation of the three-exon product with additional intron sequences, whereas the appearance of globin-derived products and the three-exon product alone is not affected (Fig. 4B Right, lanes 5 and 15). Accordingly, drug treatment also reduces significantly the production of the short intron derived from aberrant splicing but not that from authentic splicing of PDH sequences. These results are consistent with the idea that indole derivatives may mediate selective inhibition of splicing, depending on the SR protein involved. Thus, in the case of the βglo-3S-PDH transcript, the splicing event mediated by SC35, but not that depending on SF2/ASF is preferentially inhibited by drugs C77 and C83.
Fig. 4.
Ability of selected indole derivatives to inhibit aberrant but not normal splicing. (A) Schematic representation of βglo-3S-PDH artificial transcript and expected splicing products. (B) The same set of indole derivatives was tested for their ability to inhibit βglo-3S (Left) and βglo-3S-PDH (Right). C77 (lane 15) and C83 (lane 5) have no effect on βglo-3S, whereas they prevent aberrant but not wild-type splicing of βglo-3S-PDH. (C) Chemical structure of compounds C77 and C83.
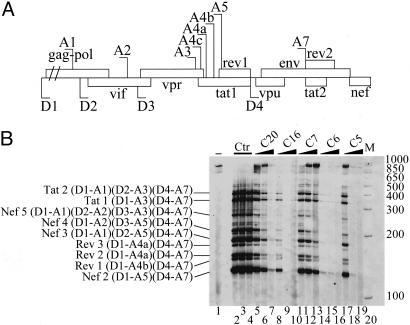
Indole Derivatives Inhibit HIV-1 RNA Splicing. To determine the effectiveness of indole derivatives in vivo, we used HIV-1 pre-mRNA splicing as a model substrate regulated by alternative splicing. Indeed, to express key viral proteins, HIV-1 uses a combination of several alternative 5′ and 3′ splice sites to generate >40 different mRNAs from its singly transcribed genomic pre-mRNA. The choice of these alternative splice sites is strongly influenced and regulated by interactions of specific HIV pre-mRNA sequences with SR proteins (30). The HIV-1 transcript therefore constitutes an ideal natural substrate to test the efficacy of our drugs on alternative splicing. As a first step, we used chronically infected promonocytic U1 cell lines (31), which produce large quantities of HIV-1 mRNAs after stimulation with phorbol myristate acetate (PMA). RT-PCR and p24 antigen enzyme-linked immunosorbent assays after treatment of these PMA-treated cell lines with various indole derivatives revealed that several drugs completely prevent HIV-1 production (data not shown). Because in vitro assays have limitations in the size of substrates they can splice, they are inadequate to test the effects of selected drugs on specific splicing events occurring on an RNA containing all HIV-1 splicing sites. We therefore transfected HeLa cells with a pΔPSP plasmid containing the HIV-1 proviral genome deleted between nucleotides 1511 and 4550. Despite being deleted of most gag-pol sequences, the resulting miniHIV-1 transcript yields the same amount of spliced subgenomic products as the authentic HIV-1 pre-mRNA (31, 32). After transfection and treatment with the various drugs, the splicing products of this transcript were analyzed by RT-PCR (32) and normalized to neomycin mRNA for transfection efficiency and GAPDH mRNA for total mRNA content. As previously observed in chronically infected cells, treatment of transfected HeLa cells with 5 μM of drugs C20, C16, C6, or C5 strongly impedes the production of spliced products (Fig. 5B, compare control lanes 2-4 with lanes 7, 10, 16, and 19, respectively). Interestingly, compound C7 does not completely abolish the production of splicing products but rather leads to changes in the splicing profile of HIV-1 RNA. Indeed, drug C7 induced preferential accumulation of larger transcripts with concomitant depletion of the smaller ones (lane 13). Similar splicing alterations were also observed with C5 (lane 17) and C20 (lanes 5 and 6) when they were used at lower concentrations. This dose-dependent alteration of splicing strongly indicates that drugs are inhibiting the use of several weak 3′ splice sites located upstream of key regulatory viral proteins like Tat, Rev, Vpu, Env, and Nef whose expression critically depends upon the activity of one or several SR proteins such as SF2/ASF and SC35. In this context, the C6 compound is one of the most potent drugs, exhibiting a very strong inhibitory effect at the lowest concentration used in this study (1 μM). Control RT-PCR reactions revealed that drug treatment alters neither the splicing profiles of CD44, SC35, Clk/Sty, or BclX genes nor their global level of expression, thereby indicating that splicing is not globally affected by drug treatment (see Fig. 6, which is published as supporting information on the PNAS web site). Treatment of cells for at least 3 days with 5 μM drug each day did not further affect cell viability or the splicing profile of tested genes (data not shown). Altogether, these data indicate that modulation of HIV-1 alternative splicing can be achieved by indole derivatives and open interesting avenues concerning novel pharmacological therapeutic approaches for AIDS treatment.
Fig. 5.
Selective inhibition of HIV-1 RNA splicing ex vivo by indole derivatives. (A) Schematic representation of HIV-1 genome. The 5′ splice sites (D1-D4) and 3′ splice sites (A1-A7) are indicated. The various ORFs are boxed. (B) HeLa cells transfected with the pΔPSP construct were either untreated (lane 2) or were treated with 0.003% DMSO (lane 3); 0.015% DMSO (lane 4); 1, 2.5, or 5 μm of compound C20 (lanes 5-7, respectively); C16 (lanes 8-10, respectively); C7 (lanes 11-13, respectively); C6 (lanes 14-16, respectively); or C5 (lanes 17-19, respectively). Multiple spliced products of HIV-1 RNA were amplified by RT-PCR by using the oligonucleotide primers BSS and SJ4.7A. The PCR products were analyzed by polyacrylamide gel electrophoresis after normalization with GAPDH (see Experimental Procedures). Nomenclature of the RT-PCR products on the left is according to ref. 26. Size markers (in base pairs) are shown on the right (lane 20). RT-PCR from untransfected HeLa cells (lane 1).
Discussion
A large number of genetic diseases are caused by defects in the proper processing of primary transcripts, and changes in alternative splicing are the basis for multiple human pathologies including cancer, viral infections, inflammatory responses, and neurological defects. Identification of molecules capable of correcting and/or inhibiting pathological splicing events is therefore an important issue for future therapeutic approaches. In this study, we have demonstrated that screening of a chemical library with splicing substrates that depend on ESE sequences resulted in the identification of potential inhibitors of alternative splicing. Furthermore, several indole derivatives proved highly selective for the inhibition of specific splicing events mediated by different classes of ESE sequences. Significantly, some drugs, like compounds C77 and C83, retained a strict selectivity for one specific ESE even in substrates containing multiple enhancer sequences.
Despite the use of simple splicing substrates for screening, the selected drugs were also capable of inhibiting splicing events in vivo with good specificity. Several indole derivatives (like C5, C6, C7, C16, and C20) were shown to be potent inhibitors of HIV-1 RNA production in cells chronically infected with the virus. Furthermore, because these compounds were also effective in preventing viral RNA splicing in cells transfected with the pΔPSP plasmid, it is unlikely that the drug interferes with other processes involved in viral RNA synthesis, such as reverse transcription and/or integration of the proviral DNA, as suggested (33). Because HIV-1, like other human viruses, uses alternative splicing to produce the large number of proteins required for its multiplication, inhibition of splicing appears a likely explanation for the inhibition of viral production by chronically infected cells. In agreement with this, drug treatment of HeLa cells transfected with the pΔPSP plasmid altered the splicing pattern of viral RNA in a dose-dependent manner. Such alterations of regulated splicing may well be a key step accounting for the remarkable antiviral activities exhibited by indole derivatives in cell culture systems (33).
Although the exact mechanism responsible for the selective action of the indole derivatives remains to be elucidated, it most likely involves a direct interaction with target SR proteins. That both the RS and RRM domains contribute to SR protein activity in ESE-dependent splicing is in keeping with our binding experiments, which show that the affinity for isolated SR domains is lower than for the entire protein. We can therefore surmise that the RRM domains provide an additional level of specificity with respect to drug binding. Alternatively, drug binding could be influenced by the overall composition of the activator complex assembled on a given ESE. Both hypotheses are consistent with the ability of the RS domain to mediate both RNA-protein and protein-protein interactions.
Along this line, these drugs might alter posttranslational modifications and/or interaction of SR proteins with specific and/or constitutive splicing factors. Consistent with this hypothesis, several indole derivatives prevent phosphorylation of the RS domain by topo I/kinase and, to a lesser extent, by Clk-Sty kinase, a modification shown to be required for ESE-dependent splicing (19). It is therefore possible that modulation of the phosphorylation status of specific SR proteins could be relevant to the drug effect. Because this modification may alter not only the splicing activity of SR proteins but also their cellular localization, indole derivatives may have a pleiotropic effect on multiple functions of SR proteins, i.e., mRNA export (34) and translation (35), and may therefore have a general impact on the assembly of specific mRNPs.
Aside from antisense molecules designed to block or activate specific splicing events and the use of vectors either overexpressing specific splicing factors or mediating transsplicing (16) to bypass the effects of mutations, several small chemical compounds have been shown to counteract splicing alterations, such as that of the SMN2 gene. The first, butyrate, was shown to restore the splicing pattern of SMN2 mRNA in cultured cells of SMA patients (36). Butyrate is well known to have a low toxicity, but its extremely short half-life in human serum (6 min) makes it inadequate for therapeutic purposes. Other drugs, such as sodium vanadate and aclarubicin, have also been shown to correct SMN2 splicing (16), but their side effects and toxicity preclude their use for extended therapeutic regimens. More recently, valproic acid, a widely used antiepileptic drug with low toxicity and rare long-term adverse side effects, was shown to increase the expression level of the functional SMN2 protein (37).
The main disadvantage of these molecules is their broad mechanism of action, not restricted to pre-mRNA splicing, which makes them less specific. Both butyrate and valproic acid are known to act as histone deacetylase inhibitors enhancing transcription of some genes, among which are splicing factors of the SR family, whereas aclarubicin seems to increase the expression of transcription factors (38). Last, sodium vanadate has been reported to inhibit ATPase, alkaline, and tyrosine, as well as multiple other phosphatases, and its likely targets are SR proteins, snRNPs, and hnPNPs (39). This remark about the lack of specificity also stands for drugs that interfere with the kinase activity of DNA topo I. Indeed, we have shown that drugs such as NB506 can inhibit ESE-dependent splicing in general by decreasing the phosphorylation level of SR proteins (17).
Because some of the compounds reported here can specifically inhibit a subset and possibly even a single member of the SR protein family, they can be expected to have an acceptable toxicity and may therefore represent significant progress toward the development of clinically usable drugs. Along this line, it must be pointed out that, although the indole derivatives of the Institut Curie library were initially designed for use in cancer chemotherapy, most of those selected in our in vitro splicing inhibition screens had not been previously considered good candidates for cancer therapy because of their low cytotoxicity, which was at that time considered an essential prerequisite.
Additional studies will now be needed to confirm in vivo the potency and lack of deleterious side effects of indole derivatives in animal models of genetic diseases. In this context, it is encouraging that ellipticine and its derivatives used for clinical purposes have resulted in high efficiencies against several types of cancer with rather limited toxic side effects (40). The discovery that some indole derivatives target individual SR proteins will definitely open exciting perspectives of causal therapies, not only for genetic diseases resulting from aberrant splicing but also for cancer or viral infections where splicing regulators are essential for the pathological process.
Supplementary Material
Acknowledgments
We thank C. Dupon and V. Ferreira for skillful technical assistance, Dr. Ch. Branlant (Université Henri Poincaré, Nancy, France) for the kind gift of the pΔPSP plasmid, and Dr. M. Benkirane (Institut de Génétique Humaine, Montpellier, France) for providing us with the U1 cell lines. This work was supported by the GIS-Institut des Maladies Rares and by grants from the Association pour la Recherche sur le Cancer, the Agence Nationale pour la Valorisation des Activités de Recherche (ANVAR), and the Action Incitative Concertée “Molécules et Cibles Thérapeutiques.” M.G., W.F., and S.D. were supported by a graduate fellowship from the Ministère Délégué à la Recherche et aux Technologies. N.B. and L.Z. are recipients of ANVAR and Ligue Contre le Cancer (Comité de l'Herault) fellowships, respectively.
Author contributions: J.T. designed research; J.S., N.B., S.M., S.D., L.Z., M.G., W.F., G.D., and P.J. performed research; J.S., C.R., D.D., and C.H.N. contributed new reagents/analytic tools; J.T. analyzed data; and J.T. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ESE, exonic splicing enhancers; SR, serine-arginine-rich; RS, arginine-serine-rich; topo I, topoisomerase I; RRM, RNA-recognition motif.
References
- 1.Maniatis, T. & Tasic, B. (2002) Nature 418, 236-243. [DOI] [PubMed] [Google Scholar]
- 2.Kriventseva, E. V., Koch, I., Apweiler, R., Vingron, M., Bork, P., Gelfand, M. S. & Sunyaev, S. (2003) Trends Genet. 19, 124-128. [DOI] [PubMed] [Google Scholar]
- 3.Johnson, J. M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P. M., Armour, C. D., Santos, R., Schadt, E. E., Stoughton, R. & Shoemaker, D. D. (2003) Science 302, 2141-2144. [DOI] [PubMed] [Google Scholar]
- 4.Faustino, N. A. & Cooper, T. A. (2003) Genes Dev. 17, 419-437. [DOI] [PubMed] [Google Scholar]
- 5.Graveley, B. R. (2000) RNA 6, 1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manley, J. L. & Tacke, R. (1996) Genes Dev. 10, 1569-1579. [DOI] [PubMed] [Google Scholar]
- 7.Caceres, J. F., Misteli, T., Screaton, G. R., Spector, D. L. & Krainer, A. R. (1997) J. Cell Biol. 138, 225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayeda, A., Munroe, S. H., Caceres, J. F. & Krainer, A. R. (1994) EMBO J. 13, 5483-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayeda, A. & Krainer, A. R. (1992) Cell 68, 365-375. [DOI] [PubMed] [Google Scholar]
- 10.Chabot, B., LeBel, C., Hutchison, S., Nasim, F. H. & Simard, M. J. (2003) Prog. Mol. Subcell. Biol. 31, 59-88. [DOI] [PubMed] [Google Scholar]
- 11.Cartegni, L., Chew, S. L. & Krainer, A. R. (2002) Nat. Rev. Genet. 3, 285-298. [DOI] [PubMed] [Google Scholar]
- 12.Cartegni, L. & Krainer, A. R. (2003) Nat. Struct. Biol. 10, 120-125. [DOI] [PubMed] [Google Scholar]
- 13.Skordis, L. A., Dunckley, M. G., Yue, B., Eperon, I. C. & Muntoni, F. (2003) Proc. Natl. Acad. Sci. USA 100, 4114-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunckley, M. G., Manoharan, M., Villiet, P., Eperon, I. C. & Dickson, G. (1998) Hum. Mol. Genet. 7, 1083-1090. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Blanco, M. A. (2003) J. Clin. Invest. 112, 474-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Blanco, M. A., Baraniak, A. P. & Lasda, E. L. (2004) Nat. Biotechnol. 22, 535-546. [DOI] [PubMed] [Google Scholar]
- 17.Pilch, B., Allemand, E., Facompre, M., Bailly, C., Riou, J. F., Soret, J. & Tazi, J. (2001) Cancer Res. 61, 6876-6884. [PubMed] [Google Scholar]
- 18.Labourier, E., Riou, J. F., Prudhomme, M., Carrasco, C., Bailly, C. & Tazi, J. (1999) Cancer Res. 59, 52-55. [PubMed] [Google Scholar]
- 19.Soret, J., Gabut, M., Dupon, C., Kohlhagen, G., Stevenin, J., Pommier, Y. & Tazi, J. (2003) Cancer Res. 63, 8203-8211. [PubMed] [Google Scholar]
- 20.Labourier, E., Rossi, F., Gallouzi, I. E., Allemand, E., Divita, G. & Tazi, J. (1998) Nucleic Acids Res. 26, 2955-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allemand, E., Gattoni, R., Bourbon, H. M., Stevenin, J., Caceres, J. F., Soret, J. & Tazi, J. (2001) Mol. Cell. Biol. 21, 1345-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossi, F., Labourier, E., Gallouzi, I. E., Derancourt, J., Allemand, E., Divita, G. & Tazi, J. (1998) Nucleic Acids Res. 26, 2963-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labourier, E., Allemand, E., Brand, S., Fostier, M., Tazi, J. & Bourbon, H. M. (1999) Nucleic Acids Res. 27, 2377-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cartegni, L., Wang, J., Zhu, Z., Zhang, M. Q. & Krainer, A. R. (2003) Nucleic Acids Res. 31, 3568-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gabut, M., Miné M., Marsac, C., Brivet, M., Tazi, J. & Soret, J. (2005) Mol. Cell. Biol. 25, 3286-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacquenet, S., Mereau, A., Bilodeau, P. S., Damier, L., Stoltzfus, C. M. & Branlant, C. (2001) J. Biol. Chem. 276, 40464-40475. [DOI] [PubMed] [Google Scholar]
- 27.Alvarez, M. & Joule, J. A. (2001) Alkaloids Chem. Biol. 57, 235-272. [DOI] [PubMed] [Google Scholar]
- 28.Shen, H. & Green, M. R. (2004) Mol. Cell 16, 363-373. [DOI] [PubMed] [Google Scholar]
- 29.Mine, M., Brivet, M., Touati, G., Grabowski, P., Abitbol, M. & Marsac, C. (2003) J. Biol. Chem. 278, 11768-11772. [DOI] [PubMed] [Google Scholar]
- 30.Ropers, D., Ayadi, L., Gattoni, R., Jacquenet, S., Damier, L., Branlant, C. & Stevenin, J. (2004) J. Biol. Chem. 279, 29963-29973. [DOI] [PubMed] [Google Scholar]
- 31.Clouse, K. A., Powell, D., Washington, I., Poli, G., Strebel, K., Farrar, W., Barstad, P., Kovacs, J., Fauci, A. S. & Folks, T. M. (1989) J. Immunol. 142, 431-438. [PubMed] [Google Scholar]
- 32.Bilodeau, P. S., Domsic, J. K. & Stoltzfus, C. M. (1999) J. Virol. 73, 9764-9772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ducrocq, C., Wendling, F., Tourbez-Perrin, M., Rivalle, C., Tambourin, P., Pochon, F., Bisagni, E. & Chermann, J. C. (1980) J. Med. Chem. 23, 1212-1216. [DOI] [PubMed] [Google Scholar]
- 34.Huang, Y., Gattoni, R., Stevenin, J. & Steitz, J. A. (2003) Mol. Cell 11, 837-843. [DOI] [PubMed] [Google Scholar]
- 35.Sanford, J. R., Gray, N. K., Beckmann, K. & Caceres, J. F. (2004) Genes Dev. 18, 755-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang, J. G., Hsieh-Li, H. M., Jong, Y. J., Wang, N. M., Tsai, C. H. & Li, H. (2001) Proc. Natl. Acad. Sci. USA 98, 9808-9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brichta, L., Hofmann, Y., Hahnen, E., Siebzehnrubl, F. A., Raschke, H., Blumcke, I., Eyupoglu, I. Y. & Wirth, B. (2003) Hum. Mol. Genet. 12, 2481-2489. [DOI] [PubMed] [Google Scholar]
- 38.Andreassi, C., Jarecki, J., Zhou, J., Coovert, D. D., Monani, U. R., Chen, X., Whitney, M., Pollok, B., Zhang, M., Androphy, E., et al. (2001) Hum. Mol. Genet. 10, 2841-2849. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, M. L., Lorson, C. L., Androphy, E. J. & Zhou, J. (2001) Gene Ther. 8, 1532-1538. [DOI] [PubMed] [Google Scholar]
- 40.Stiborova, M., Bieler, C. A., Wiessler, M. & Frei, E. (2001) Biochem. Pharmacol. 62, 1675-1684. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.