Abstract
Many of the genes that comprise the vertebrate adaptive immune system are conserved across wide evolutionary time scales. Most notably, homologs of the mammalian MHC gene family have been found in virtually all jawed vertebrates, including sharks, bony fishes, reptiles, and birds. The CD1 family of antigen-presenting molecules are related to the MHC class I family but have evolved to bind and present lipid antigens to T cells. Here, we describe two highly divergent nonclassical MHC class I genes found in the chicken (Gallus gallus) that have sequence homology to the mammalian CD1 family of proteins. One of the chicken CD1 genes expresses a full-length transcript, whereas the other has multiple splice variants. Both Southern blot and single nucleotide polymorphism analysis indicates that chicken CD1 is relatively nonpolymorphic. Moreover, cross-hybridizing bands are present in other bird species, suggesting broad conservation in the avian class. Northern analysis of chicken tissue shows a high level of CD1 expression in the bursa and spleen. In addition, molecular modeling predicts that the potential antigen-binding pocket is probably hydrophobic, a universal characteristic of CD1 molecules. Genomic analysis indicates that the CD1 genes are located on chicken chromosome 16 and maps to within 200 kb of the chicken MHC B locus, suggesting that CD1 genes diverged from classical MHC genes while still linked to the major histocompatibility complex locus. The existence of CD1 genes in an avian species suggests that the origin of CD1 extends deep into the evolutionary history of terrestrial vertebrates.
Keywords: antigen presentation, comparative immunology/evolution, cell surface molecules
The essential elements that define the adaptive immune system emerged early in the course of vertebrate evolution. This finding is evidenced by the fact that virtually all jawed vertebrates share a conserved set of genes that have clear evolutionary homologs with the extensively characterized mammalian immune system (1). One of the most conserved parts of the adaptive cellular immune system are the genes that make up the major histocompatibility complex (MHC) locus. The MHCI genes encode highly polymorphic cell-surface glycoproteins that are expressed on all somatic cells, whereas MHCII has a more restricted expression pattern, being expressed primarily on professional antigen-presenting cells. It is well established that the MHCI and MHCII proteins have the capacity to bind and present pathogen-derived peptides to specific CD8+ and CD4+ T cells, respectively. The MHC gene family is therefore the core of the cell-mediated immune system's antigen-presenting function and is critical for host resistance to microbial infection.
The CD1 genes represent a third family of cell-surface proteins, in addition to MHCI and MHCII, which have the capacity to present antigens to T cells. The human CD1 gene family is composed of five nonpolymorphic genes (CD1A, -B, -C, -D, and -E) that are located in a small cluster on human chromosome 1 and are therefore unlinked to the MHC locus on chromosome 6 (2, 3). Although clearly related to the MHCI family of proteins, CD1 has evolved to bind and present lipid antigens to T cells in a manner analogous to the paradigm established for peptides and MHC (4). In addition, many of the CD1-restricted T cells recognize microbial lipids and glycolipids and have a proinflammatory phenotype with antimicrobial effector functions (5). These data suggest an important role for CD1 in the immune response to infection and other diseases (6).
Based on structural and sequence similarities, it is thought that CD1 and MHCI diverged from a common ancestral gene during the early evolution of vertebrates (7). One of our goals has been to investigate the origin of the CD1 genes by identifying homologs in more primitive animal species. These studies may allow a better appreciation of the role that CD1 has played in the evolution of the immune system. However, early ancestral or transitional forms of CD1 in extant nonmammalian vertebrate species have thus far remained elusive. Birds are generally accepted to be the living descendants of Theropod dinosaurs and, therefore, represent an ancient lineage of reptiles (8). Here, we describe two highly divergent nonclassical MHCI genes in the chicken (Gallus gallus) that share a common set of features typically found in the CD1 gene family. These genes reveal an ancient origin for CD1 and thus provide a unique opportunity for comparative analysis.
Materials and Methods
Animal Tissues. Tissue samples for Northern analysis and RT-PCR were obtained from Charles River SPAFAS (Wilmington, MA). Camperos chicken genomic DNA samples (starting from the left in fig. 3A in ref. 9) with previously described genotypes were used: 14 from the female parent line (f2, f3, f5, f6, f7, f8, f10, f11, f12, f13, f14, f15, f16, and f18) and three from the male parent line (m1, m2, and m3). Human genomic DNA was obtained from the MCF7 cell line. Other avian genomic DNA samples used for Fig. 3C were extracted as described in ref. 10 and are listed in Table 2, which is published on the PNAS web site.
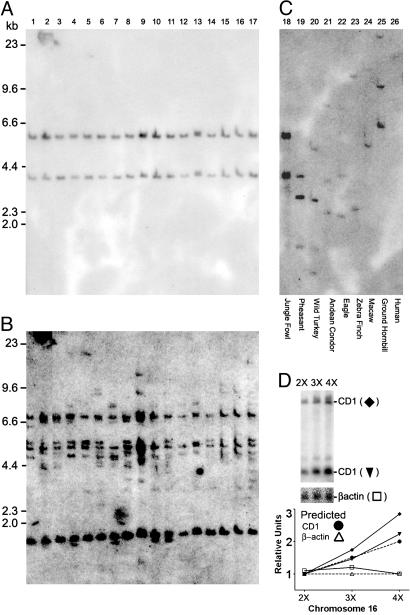
Fig. 3.
Southern blot analysis of genomic DNA. (A) Purified genomic DNA from 17 individual Camperos chickens was digested with BglI. Hybridization was carried out with a chCD1-2 cDNA probe. Autoradiography shows two bands of 6.1 kb and 3.6 kb in all 17 birds, suggesting limited polymorphism of this gene in chickens. DNA molecular weight markers are indicated at left. (B) The blot used for A was reprobed with the F10 cDNA that hybridizes with MHC class I (B and Y) genes in chickens. Note the variability in the number and size of hybridizing bands between individual animals as compared with the uniform pattern in A. (C) Genomic DNA isolated from various bird species was digested with BglI and transferred to the same membrane as A and hybridized simultaneously with the chCD1-2 probe. The contrast of the autoradiogram was adjusted slightly to reveal weaker bands in lanes 18-25. No signal was detected in lane 26 (human). (D) Southern analysis of chCD1-2 gene in chickens with two, three, and four copies of GGA16. Equal amounts of genomic DNA from each animal were analyzed. Autoradiography shows increasing intensity of chCD1-2 probe hybridizing bands (♦ and ▾) as the copy number of GGA16 increases. The blot was rehybridized with a probe encoding β-actin cDNA (□). Densitometry of each band was normalized to the diploid (2X) bands for each the autoradiograms and plotted on the graph. Graph symbols are the same as for the autoradiogram. In addition, dashed lines represent the predicted intensities assuming localization of the chCD1-2 (•) on GGA16 and β-actin (▵) on GGA2.
Genes and Sequence Analysis. The non-human and non-mouse GenBank EST library database was searched with murine CD1d protein sequence (GenBank accession no. P11609) encompassing the α1-α2 domains. High scoring G. gallus candidates were re-screened against the same databases to extract other close matches and are listed in Table 3, which is published as supporting information on the PNAS web site. A candidate cDNA sequence (BG713169) that appeared in multiple EST libraries (designated K24) was chosen for more extensive analysis and coded for a putative protein we designated chCD1-2 (see Results and Discussion for nomenclature). Multiple sequence alignment and neighbor-joining trees were made with clustal-x and mega3 (11). The first draft of the red jungle fowl (G. gallus) genome sequence was used for genomic and single nucleotide polymorphism analysis (12, 13). Sequencing of two gaps in the chCD1 locus of G. gallus red jungle fowl genomic DNA (Table 2) was accomplished with primers 7 and 8 for chCD1-1 intron 2 and primers 9 and 10 for chCD1-2 intron 4, which are listed in Table 4, which is published as supporting information on the PNAS web site.
RT-PCR of chCD1-1 and chCD1-2 mRNA. Total RNA prepared from the bursa and spleen of Charles River SPAFAS chickens used for Northern analysis (see below) was used to generate cDNA with the Superscript RT-PCR kit by the oligo(dT) priming method according to the manufacturer's instructions (Invitrogen). All primer sequences are listed in Table 4. Bursa cDNA was used as template for PCR of full-length chCD1-1 and chCD1-2. Primers 1 and 2 were used for chCD1-1, and primers 3 and 4 were used for chCD1-2. Positive control primers were specific for chicken β2M (primers 11 and 12). Amplified DNA was analyzed on 1% agarose gel, and purified bands were ligated into the pCR4 TOPO-TA-cloning vector (Invitrogen) and sequenced.
Southern and Northern Hybridization. Southern blot and hybridizations were carried out by using standard techniques as described in ref. 14. This method was used in trisomy analysis and for mapping chCD1 locus to specific BACs. A detailed description of both Southern and Northern procedures are in Supporting Methods, which is published as supporting information on the PNAS web site.
Molecular Modeling and Structural Analysis. Molecular structure data for known CD1 and MHC proteins were obtained from the Protein Data Bank. Structural modeling of proteins and determination of the net hydropathy values is explained in detail in Supporting Methods.
Results and Discussion
Identification of Avian CD1 Genes. The CD1 cell-surface proteins are related to the MHCI family but have evolved to present lipid antigens to T cells rather than peptides. A more detailed understanding of the origin of CD1 would provide useful insights into the structural and functional evolution of this gene family. However, CD1 genes have thus far only been described in mammals. Therefore, we searched the available nonmammalian genomic databases for evidence of CD1-like proteins. Two genes with significant homology to mammalian CD1 were discovered in the chicken genome and EST databases. When aligned to the MHC and CD1 from other species, the chicken protein sequences were more similar to CD1 than to MHC sequences (Fig. 1A and Table 1). This relationship was maintained even when the protein sequence is parsed into individual domains (Table 1; Fig. 5, which is published as supporting information on the PNAS web site). We designated the chicken genes as chCD1-1 and chCD1-2 to denote their likely status as CD1 homologs.
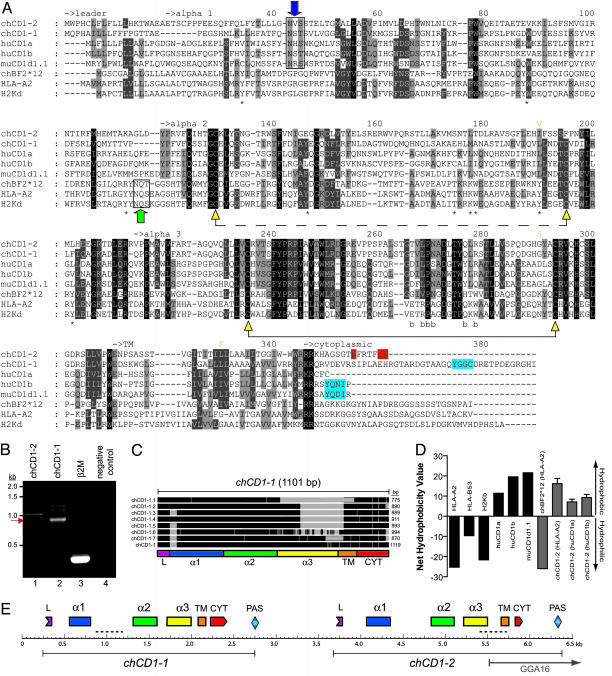
Fig. 1.
Molecular comparison, characterization, and mapping of chCD1 genes and protein sequences. (A) Alignment of the chicken chCD1-2, chCD1-1, human CD1a, human CD1b, mouse CD1d, chBF2*12 (chicken MHCI), human HLA-A2, and mouse H2Kd. Note that chCD1-1 is a hypothetical sequence based on conceptual splicing of genomic DNA. Black boxes, identical or conserved substitutions at all residues at that position; dark gray, 80% conserved; light gray, 60% conserved. Intramolecular disulfide bonds (connected yellow triangles) are indicated. Note that the α2 disulfide bond (connected dashed line) is not present in the human CD1a crystal structure and cannot form in chCD1-2. MHCI peptide anchor residues (*) and residues that interact with β2M (b) are also indicated. A putative dileucine motif (DXXXXLI) at the carboxyl terminus of chCD1-2 protein is highlighted (red), and tyrosine motifs in chCD1-1, human CD1b, and mouse CD1d are in blue. Conserved N-linked NX(S/T) glycosylation site motifs of MHCI and CD1 is indicated by filled green and blue arrows, respectively. Three nucleotide polymorphisms that result in amino acid changes from the red jungle fowl chCD1-2 are indicated in light green above the K24 clone chCD1-2 sequence. (B) PCR of bursa cDNA with primers specific for chCD1-2 (lane 1), chCD1-1 (lane 2), β2M (lane 3), and no template control (lane 4). The red arrow indicates a dominant chCD1-1 product at 900 bp. (C) Schematic diagram showing cloned chCD1-1 cDNA fragments. Sequenced PCR products from B (lane 2) were aligned with the hypothetical full-length chCD1-1 (Lower). Identical residues are in black, and missing DNA from the sequenced clones are indicated in light gray. (D) Calculation of hydrophobicity (NHV) of the α1 helix lining residues of the antigen binding pocket by using known MHCI and CD1 crystal structure data (black bars). The chicken BF2*12 (MHCI) and chCD1-2 protein sequences were modeled on HLA-A2 or human CD1 crystal structures (see Supporting Methods) and the net hydropathy values calculated (gray bars). The modeled protein sequence is listed for each bar with the template crystal structure in parentheses. These data suggest that the putative chCD1-2 antigen binding pocket forms a hydrophobic surface.(E) Map of the G. gallus CD1 locus based on public genome database with minor gaps filled in by this study (dashed lines). The fragment of the chCD1 locus currently mapped to GGA16 in the public database is indicated. PAS, putative polyA sites.
Table 1. Percent identity of chCD1-2 to MHC and CD1.
| Protein | Species | α1 | α2 | α3 | α1-2 | α123 |
|---|---|---|---|---|---|---|
| chCD1-1 | Chicken | 22 | 23 | 97 | 23 | 48 |
| huCD1a | Human | 11 | 25 | 34 | 16 | 24 |
| huCD1b | Human | 17 | 23 | 35 | 19 | 25 |
| muCD1d1.1 | Mouse | 12 | 21 | 35 | 17 | 23 |
| chBF2*12 | Chicken | 4 | 13 | 38 | 8 | 18 |
| HLA-A2 | Human | 7 | 13 | 27 | 9 | 15 |
| H2Kd | Mouse | 7 | 13 | 26 | 9 | 15 |
Conceptual splicing and translation of the chCD1-1 genomic DNA sequence revealed the theoretical possibility of a full-length ORF with CD1 sequence homology. However, a full-length EST for chCD1-1 was not present in the public databases and only appeared in two clones from a single bursa library (Table 3). In contrast, the chCD1-2 sequence appeared in multiple libraries and full-length EST clones (Table 3). One EST clone designated K24 (BG713169) was completely sequenced and used as the reference clone for chCD1-2. Primers were made to the 5′ and 3′ ends of the ORF of chCD1-1 (primer 1 and 2) and chCD1-2 (primer 3 and 4) and used to PCR amplify bursa-derived cDNA. Amplification of chCD1-1 revealed multiple products ranging in size from 700 to 1,300 bp with a prominent band of 900 bp (Fig. 1B, lane 2). We extracted the DNA fragments, derived clones, and sequenced the inserts. Fig. 1C shows a schematic summary of the aligned chCD1-1 sequence inserts from a random selection of seven clones. These sequence inserts reveal multiple deletions in the chCD1-1 cDNA sequences. As shown in Fig. 1B (lane 2, arrow), the most intense band is ≈900 bp, which is consistent with the size of the predominant 872-bp splice variant (Fig. 1C, clones chCD1-1.2, -1.3, -1.4, and -1.5). Variations in the size of CD1 mRNA transcripts have been described (15-17). Some of these variants also lack the transmembrane domain and result in soluble forms of CD1 (15, 17, 18). The biological significance of these variants is unclear, but they may be a mechanism for post-transcriptional regulation.
In contrast to the chCD1-1 gene described above, a single band was obtained for the chCD1-2 PCR reaction, which sequencing confirmed as a full-length 1044-bp ORF identical to the original chCD1-2 K24 EST sequence (AY375530). A similar result to Fig. 1B was obtained for both chCD1 genes by using spleen cDNA (data not shown). Subsequent molecular analysis focused on the chCD1-2 gene because we were unable to obtain a full-length chCD1-1. In summary, two distinct CD1 genes are expressed from the chicken genome with one of these genes (chCD1-1) expressing multiple splice variants.
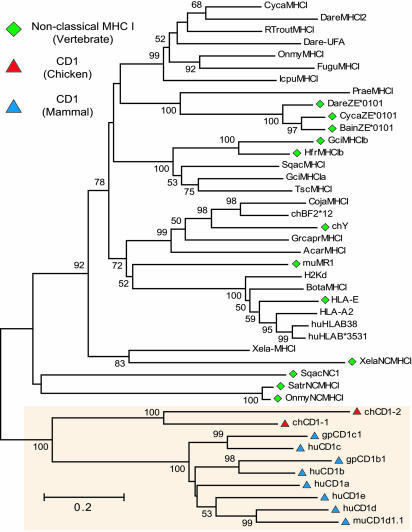
Relationship of Chicken CD1 to Vertebrate MHC and CD1 Genes. Phylogenetic analysis of the mammalian CD1 protein sequences shows that these proteins form a homology cluster that is distinct from the MHC (3, 19). Therefore, we examined the relationship of the putative chicken CD1 proteins to a broad array of MHC and CD1 proteins from various species (Fig. 2). Similar to the initial analysis in Table 1, there is significant homology between the chicken CD1 protein sequences and the mammalian CD1 cluster. However, there is a greater degree of divergence between the avian and mammal CD1 branches as compared with the differences between individual mammal CD1 proteins. This divergence is likely a result of the large elapsed time (≈310 million years) since birds and mammals last shared a common ancestor (20). We also performed an identical phylogenetic analysis of the individual domains (Fig. 6, which is published as supporting information on the PNAS web site). In each case, the affinity of the chicken CD1 sequences for the mammalian CD1 cluster was maintained. In addition, none of the previously identified nonclassical MHCI proteins from sharks, teleosts, or amphibians exhibited significant homology to the chCD1 proteins or to the overall CD1 cluster (Fig. 2). We also used the chCD1 sequences to search for homologs from nonmammalian vertebrate species. However, blast analysis of the available nonmammalian databases found no other clear homologs for the chCD1 genes. Taken together, these data support the hypothesis that the chicken and mammal CD1 genes are homologs that likely share a common ancestor.
Fig. 2.
Relationship of chCD1 proteins to other members of the MHC class I family. A neighbor-joining tree based on the alignment of the α1-3 domains of the MHCI family of protein sequences including classical, nonclassical, and CD1 sequences from multiple vertebrate species. A complete list of the taxa with accession numbers used can be found in Table 5, which is published as supporting information on the PNAS web site. The chCD1 proteins group with the mammalian CD1 in a distinct cluster (shaded region). Bootstrap values are indicated as a percentage of 1,000 iterations. Values <50% are not shown. The scale bar is the number of substitutions per position.
Analysis of the Chicken Genome. In humans and other mammals, the CD1 and MHC loci are located on separate chromosomes (2, 3). This separation is thought to have arisen from a primordial genome duplication event (reviewed in refs. 1 and 21). To examine this feature in chickens, we searched the recently completed G. gallus genome sequence to determine the chromosomal location of the chCD1 genes (12). However, the current genome assembly is incomplete with only ≈700 bp of the 3′ end of the chCD1-2 gene mapped to the GGA16 chromosome (Fig. 1E). Using the available contig data, we sequenced across two discreet gaps to generate a complete chCD1 locus, thereby linking the entire chicken CD1 locus to the mapped GGA16 data (Fig. 1E). This chicken microchromosome contains the two known MHC (B and Y) loci of chickens (22, 23). We also performed a trisomy analysis (see Supporting Methods) as an independent method of mapping the CD1 locus to GGA16 (Fig. 3D). This method further supports localization of the chCD1 locus on GGA16. Taken together, these data strongly support the location of the chicken CD1 locus on the GGA16 microchromosome.
To determine whether chCD1 is located near the chicken classical MHC B gene cluster or near the second genetically independent MHC Y gene cluster, we screened 12 BAC clones previously isolated with an MHCII probe and then assigned these clones to either the B or Y locus based on Southern hybridizations with B specific and Y specific probes. Hybridization with the K24 chCD1-2 probe revealed a 3.6-kb BglI fragment in one B locus BAC indicating that the chCD1 locus is within 200 kb of the B locus MHCII genes (Fig. 7, which is published as supporting information on the PNAS web site). Taken together, these data are consistent with linkage of the chCD1 to the MHC B locus and supports the evolution of an ancestral CD1 gene before its separation from the MHC. This linkage may be a primordial feature that has been preserved in birds and is in contrast to the mammalian CD1 genes that reside on separate chromosomes from the MHC.
The CD1 and MHC loci in humans have been proposed to be parts of the “MHC paralogous group” (reviewed in ref. 1). Duplication and subsequent diversification of a primordial vertebrate genome has been proposed to account for the large-scale paralogy observed between chromosome 1 (CD1) and chromosome 6 (MHC) in humans (24, 25). Our findings raise some questions about this model with respect to CD1 because the chicken genome data supports the emergence of CD1 before its complete separation from the MHC to another chromosome. One possibility that preserves the paralogy hypothesis is that CD1 diverged from the MHCI genes in a recent common ancestor just before the synapsid-diapsid split as suggested by Hughes (26). In this scenario, the CD1 genes remained linked to the MHC locus in the diapsid lineage (reptiles and birds), whereas large-scale translocation of the CD1 locus to a another chromosome occurred in the synapsid lineage (mammals) after the bird-mammal split. Supporting this model is the observation that none of the three completed teleost genomes have obvious CD1 homologs. However, this apparent absence may be a derived feature related to the fragmentation of the MHCI and MHCII loci in teleosts (27).
Conserved Structural Features in the Avian CD1 Proteins. The chCD1 gene sequences provide a unique opportunity to examine specific features that have been conserved in both the mammal and bird CD1 proteins since their divergence from a common ancestor. Previous studies have noted a highly conserved glycosylation site (NX(S/T)) present in the loop (Fig. 1A, green arrow) at the distal end of the classical MHCI α1 helix (28). This site is found in the same position in all classical MHC class I proteins and is involved in calnexin binding (29). Interestingly, a conserved glycosylation site is present in all known CD1 protein sequences at position 44 (Fig. 1A, blue arrow). Examination of the CD1 crystal structures shows that this position is in spatial proximity to the MHCI glycosylation site but is shifted to the adjacent β-sheet loop (data not shown). Importantly, both chCD1 proteins have the putative glycosylation site common to the CD1 family of proteins and not the classical MHCI (Fig. 1A). The conserved position of this motif in all known CD1 proteins suggests an important function, possibly also for calnexin binding (30). It is possible that steric constraints imparted by the structural changes to the antigen-binding pocket of CD1 necessitated a shift in the position of this glycosylation site to preserve its function.
Each of the CD1 isoforms in humans has a distinct pattern of intracellular traffic (31). This feature is thought to facilitate a broad surveillance of the cell's intracellular environment for possible antigenic lipids (3). Interestingly, the last two amino acids of the chCD1-2 protein are Leu and Ile, with an Asp five residues up from the carboxyl terminus (Fig. 1A). Together, these residues form a putative dileucine-based sorting motif that would allow binding of AP-2 for internalization from the cell surface to recycling endosomes (32). These data suggest that chCD1-2 may have an intracellular localization that is distinct from MHCI and may resemble those of the nonprimate CD1a isoforms that also have dileucine motifs and have cytoplasmic tail lengths similar to chCD1-2.
The most distinctive structural feature of CD1 proteins is the deep antigen binding pocket which is lined predominantly by nonpolar amino acids for binding hydrophobic lipid acyl chains (33). We calculated the overall hydrophobicity of part of the antigen binding surface of known MHC I and CD1 structures (see Supporting Methods; Fig. 8, which is published as supporting information on the PNAS web site). Fig. 1D shows that the MHCI proteins exhibit a charged surface, whereas CD1 proteins have a net hydrophobic surface. We then modeled the structure of the chCD1-2 protein sequence by using MHCI or CD1 structures. Fig. 1D shows that in each case, the chCD1-2 models exhibited a positive net hydropathy value indicative of a hydrophobic surface and consistent with a CD1 antigen pocket.
Polymorphism of Chicken CD1. Compared with the classical MHCI genes, the CD1 and most other nonclassical MHCI genes have limited sequence variation between individuals (34). To address the polymorphism of CD1 in birds, we performed Southern blot analysis by using a chCD1-2 probe and DNA from 17 genetically diverse Camperos breed chickens (9). Fig. 3A shows two bands of 6.1 and 3.6 kb that appear in all of the individuals tested. To compare this pattern with MHC, the blot in Fig. 3A was stripped and reprobed with F10 cDNA that hybridizes to both B and Y genes in the chicken (35). As expected, the banding pattern of the autoradiogram in Fig. 3B varies significantly between individual birds reflecting the increased polymorphism of the classical MHCI genes relative to CD1. These data suggest that the chCD1-2 gene and its surrounding 5′ and 3′ untranslated regions are of limited genetic diversity, which is consistent with the data for human CD1.
We also compared the chCD1-2 K24 cDNA sequence (broiler) with the red jungle fowl genome derived chCD1-2 sequence (12). Alignment of the predicted chCD1-2 cDNA from red jungle fowl with the chCD1-2 K24 cDNA revealed seven single nucleotide polymorphisms within the ORF. Only three of these single nucleotide polymorphisms resulted in conserved amino acid substitutions to the chCD1-1 protein sequence with one each in the α2, α3, and transmembrane domains (Fig. 1A). We also compared the single nucleotide polymorphism rate of chCD1-2 with rates obtained from the recently completed G. gallus genomewide variation map (13). These data are shown in Table 6, which is published as supporting information on the PNAS web site, and also indicate a low rate of polymorphism in the chCD1-2 gene relative to the overall rate of the GGA16 chromosome.
CD1 Genes in Other Avian Species. The CD1 genes appear to be ubiquitous in mammals, although there is a wide variation in the precise number of CD1 genes and specific isoforms that are present in a particular mammalian species (36). Therefore, we performed Southern hybridization with DNA from a random sample of wild bird species by using radiolabeled chCD1-2 cDNA as the probe. All of the avian species tested exhibited two to four cross-hybridizing bands of varying molecular weights (Fig. 3C, lanes 18-25). Taken together, these data indicate that the CD1 genes are conserved across a range of avian species and may be widespread in birds.
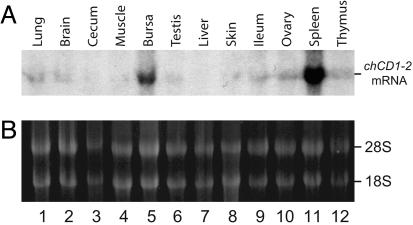
Differential Expression of CD1 in Chicken Tissues. The classical MHCI genes are expressed on all somatic cells in the body. In contrast, CD1 proteins are expressed predominantly on professional antigen-presenting cells (dendritic cells and B cells) and, therefore, have a more restricted expression pattern. Northern blot analysis shows the highest level of chCD1-2 mRNA in the bursa and spleen (Fig. 4A, lanes 5 and 11). Normalization of the hybridizing bands in the autoradiograph with the 28S rRNA band (Fig. 4B) also revealed weak expression in the thymus and ovary (Fig. 9, which is published as supporting information on the PNAS web site). These bands were more apparent on longer exposure autoradiograms (data not shown). Thus, the chCD1-2 gene has a pattern of expression that is similar to mammalian CD1 and is restricted primarily to the bird lymphoid organs.
Fig. 4.
Tissue expression of chCD1-2. Total RNA isolated from various chicken lymphoid and nonlymphoid tissues was analyzed by Northern blot. (A) Autoradiogram of chCD1-2 cross-hybridizing bands. Strong signals are present in the bursa (lane 5) and spleen (lane 11). Densitometry (Fig. 9) also revealed weak but detectable signals in the thymus and ovary. (B) Ethidium stained total RNA.
Conclusion. The presence of CD1 homologs in both birds and mammals implies that CD1 was present in a reptilian ancestor common to both groups. The split of this common ancestor into two distinct lineages represents one of the major milestones in vertebrate evolution: the synapsid-diapsid split. After this separation, mammals evolved from the Synapsida lineage, whereas reptiles were derived from the Diapsida (20). It is generally accepted that modern Aves arose from within the Archosaura group of Diapsid reptiles, more specifically the Theropod lineage of bipedal predatory dinosaurs (8). Fossil evidence strongly supports the divergence of the synapsid and diapsid lineages at ≈310 million years ago in the late Permian era (20). Thus, the origin of the CD1 gene family extends deep into the early evolution of tetrapods, well before the emergence of the first true mammals.
The mapping of CD1 to the MHC in birds has important evolutionary implications for the emergence of CD1 in vertebrates. The linkage of the chicken CD1 and MHC to within 200 kb suggests that the CD1 genes originated from a primordial MHCI gene, probably through gene duplication and neofunctionalization. One possibility to explain the extant organization of CD1 and MHC in vertebrates is that the CD1 genes first evolved in a common ancestor close to, but before, the bird-mammal split as described above. The implication of this model is that CD1 evolved well after the emergence of the MHC, possibly in an early terrestrial tetrapod. Interestingly, preliminary molecular clock analysis of the chCD1 protein sequences strongly supports this model (C.C.D., unpublished data). Further analysis of lower vertebrate genome data may reveal more transitional forms of these genes, although initial attempts to find these transitional forms have not been successful. Alternatively, if CD1 is as ancient as the MHC, then genome duplication in an early vertebrate resulted in two paralogous chromosomes, each with the linked MHC-CD1 locus as predicted by the paralogy hypothesis (1, 7, 37). In this scenario, the synapsid lineage deleted MHC genes from one chromosome and the CD1 from the paralogous chromosome, whereas an entire paralogous chromosome was deleted in birds leaving a single linked MHC-CD1 locus. Completion of the G. gallus genome assembly and additional lower vertebrates should help clarify these models.
The role of CD1 in host defense, together with the conservation of CD1 homologs in birds, suggests that these molecules may be an important part of the avian immune system. A functional comparison between the avian and mammalian CD1 systems may provide additional insights into how and why this unique gene family evolved.
Supplementary Material
Acknowledgments
We thank Joan Burnside (Delaware Biotechnology Institute, Newark, DE) for supplying the chicken EST clones and tissue samples, Gabriela Iglesias and Instituto Nacional de Tecnología Agropecuaria (Pergamino, Argentina) for generously providing the Camperos DNA samples, Elwood Briles for providing avian DNA samples, Charles Auffray for providing the F10 probe, Mary Delany for discussing the genetic history of the chicken breeds, Dirk Zajonc for discussing the CD1 structure, and Jonathan Higgins for reviewing of the manuscript. M.M.M. was supported by National Cancer Institute Grant SR21CA105425 and U.S. Department of Agriculture Grant 2002-35205-11628.
Author contributions: M.M.M., C.W., E.P., R.D.C., R.M.G., S.Y.L., D.C.B., H.L.F., M.B.B., and C.C.D. designed research; M.M.M., C.W., E.P., R.D.C., R.M.G., S.Y.L., D.C.B., M.T., C.R.-M., H.L.F., and C.C.D. performed research; M.M.M., R.D.C., R.M.G., S.Y.L., and C.C.D. contributed new reagents/analytic tools; M.M.M., C.W., E.P., R.D.C., R.M.G., S.Y.L., D.C.B., H.L.F., M.B.B., and C.C.D. analyzed data; and C.C.D. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: MHC, major histocompatibility complex.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [AY375530 (chCD1-2 complete cDNA sequence), AY874074-AY874080 (chCD1-1 splice variants 1-7), AY939845 and AY939846 (additional G. gallus genomic DNA sequences for chCD1-1 intron 2 and chCD1-2 intron 4), and DQ007238 and DQ007237 (probes to map chicken MHCII and C4 genes)].
See Commentary on page 8399.
References
- 1.Flajnik, M. F. & Kasahara, M. (2001) Immunity 15, 351-362. [DOI] [PubMed] [Google Scholar]
- 2.Calabi, F. & Milstein, C. (1986) Nature 323, 540-543. [DOI] [PubMed] [Google Scholar]
- 3.Dascher, C. C. & Brenner, M. B. (2003) Trends Immunol. 24, 412-418. [DOI] [PubMed] [Google Scholar]
- 4.Brigl, M. & Brenner, M. B. (2004) Annu. Rev. Immunol. 22, 817-890. [DOI] [PubMed] [Google Scholar]
- 5.Vincent, M. S., Gumperz, J. E. & Brenner, M. B. (2003) Nat. Immunol. Rev. 4, 517-523. [DOI] [PubMed] [Google Scholar]
- 6.Dascher, C. C. & Brenner, M. B. (2003) in Host Response Mechanisms in Infectious Disease, ed. Herwald, H. (Karger, Basel), Vol. 10, pp. 164-182. [Google Scholar]
- 7.Martin, L. H., Calabi, F. & Milstein, C. (1986) Proc. Natl. Acad. Sci. USA 83, 9154-9158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sereno, P. C. (1999) Science 284, 2137-2147. [DOI] [PubMed] [Google Scholar]
- 9.Iglesias, G. M., Soria, L. A., Goto, R. M., Jar, A. M., Miquel, M. C., Lopez, O. J. & Miller, M. M. (2003) Anim. Genet. 34, 88-95. [DOI] [PubMed] [Google Scholar]
- 10.Goto, R. M., Miyada, C. G., Young, S., Wallace, R. B., Abplanalp, H., Bloom, S. E., Briles, W. E. & Miller, M. M. (1988) Immunogenetics 27, 102-109. [DOI] [PubMed] [Google Scholar]
- 11.Kumar, S., Tamura, K. & Nei, M. (2004) Brief. Bioinformatics 5, 150-163. [DOI] [PubMed] [Google Scholar]
- 12.Hillier, L. W., Miller, W., Birney, E., Warren, W., Hardison, R. C., Ponting, C. P., Bork, P., Burt, D. W., Groenen, M. A., Delany, M. E., et al. (2004) Nature 432, 695-716.15592404 [Google Scholar]
- 13.Ka-Shu Wong, G., Liu, B., Wang, J., Zhang, Y., Yang, X., Zhang, Z., Meng, Q., Zhou, J., Li, D., Zhang, J., et al. (2004) Nature 432, 717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller, M. M., Goto, R., Bernot, A., Zoorob, R., Auffray, C., Bumstead, N. & Briles, W. E. (1994) Proc. Natl. Acad. Sci. USA 91, 4397-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kojo, S., Adachi, Y., Tsutsumi, A. & Sumida, T. (2000) Biochem. Biophys. Res. Commun. 276, 107-111. [DOI] [PubMed] [Google Scholar]
- 16.Angenieux, C., Salamero, J., Fricker, D., Cazenave, J. P., Goud, B., Hanau, D. & de La Salle, H. (2000) J. Biol. Chem. 275, 37757-37764. [DOI] [PubMed] [Google Scholar]
- 17.Woolfson, A. & Milstein, C. (1994) Proc. Natl. Acad. Sci. USA 91, 6683-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kojo, S., Tsutsumi, A., Goto, D. & Sumida, T. (2003) J. Rheumatol. 30, 2524-2528. [PubMed] [Google Scholar]
- 19.Porcelli, S. A. (1995) Adv. Immunol. 59, 1-98. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, S. & Hedges, S. B. (1998) Nature 392, 917-920. [DOI] [PubMed] [Google Scholar]
- 21.Ohno, S. (1999) Semin. Cell Dev. Biol. 10, 517-522. [DOI] [PubMed] [Google Scholar]
- 22.Bloom, S. E. & Bacon, L. D. (1985) J. Hered. 76, 146-154. [PubMed] [Google Scholar]
- 23.Miller, M. M., Bacon, L. D., Hala, K., Hunt, H. D., Ewald, S. J., Kaufman, J., Zoorob, R. & Briles, W. E. (2004) Immunogenetics 56, 261-279. [DOI] [PubMed] [Google Scholar]
- 24.Kasahara, M., Kandil, E., Salter-Cid, L. & Flajnik, M. F. (1996) Res. Immunol. 147, 278-284; discussion 284-285. [DOI] [PubMed] [Google Scholar]
- 25.Kasahara, M. (1999) Immunogenetics 50, 134-145. [DOI] [PubMed] [Google Scholar]
- 26.Hughes, A. L. (1991) Mol. Biol. Evol. 8, 185-201. [DOI] [PubMed] [Google Scholar]
- 27.Ohta, Y., Okamura, K., McKinney, E. C., Bartl, S., Hashimoto, K. & Flajnik, M. F. (2000) Proc. Natl. Acad. Sci. USA 97, 4712-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barber, L. D., Patel, T. P., Percival, L., Gumperz, J. E., Lanier, L. L., Phillips, J. H., Bigge, J. C., Wormwald, M. R., Parekh, R. B. & Parham, P. (1996) J. Immunol. 156, 3275-3284. [PubMed] [Google Scholar]
- 29.Parham, P. (1996) Trends Biochem. Sci. 21, 427-433. [DOI] [PubMed] [Google Scholar]
- 30.Sugita, M., Porcelli, S. A. & Brenner, M. B. (1997) J. Immunol. 159, 2358-2365. [PubMed] [Google Scholar]
- 31.Moody, D. B. & Porcelli, S. A. (2003) Nat. Rev. Immunol. 3, 11-22. [DOI] [PubMed] [Google Scholar]
- 32.Pond, L., Kuhn, L. A., Teyton, L., Schutze, M. P., Tainer, J. A., Jackson, M. R. & Peterson, P. A. (1995) J. Biol. Chem. 270, 19989-19997. [DOI] [PubMed] [Google Scholar]
- 33.Zeng, Z., Castano, A. R., Segelke, B. W., Stura, E. A., Peterson, P. A. & Wilson, I. A. (1997) Science 277, 339-345. [DOI] [PubMed] [Google Scholar]
- 34.Han, M., Hannick, L. I., DiBrino, M. & Robinson, M. A. (1999) Tissue Antigens 54, 122-127. [DOI] [PubMed] [Google Scholar]
- 35.Guillemot, F., Billault, A., Pourquie, O., Behar, G., Chausse, A. M., Zoorob, R., Kreibich, G. & Auffray, C. (1988) EMBO J. 7, 2775-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dascher, C. C., Hiromatsu, K., Naylor, J. W., Brauer, P. P., Brown, K. A., Storey, J. R., Behar, S. M., Kawasaki, E. S., Porcelli, S. A., Brenner, M. B., et al. (1999) J. Immunol. 163, 5478-5488. [PubMed] [Google Scholar]
- 37.Kasahara, M., Nakaya, J., Satta, Y. & Takahata, N. (1997) Trends Genet. 13, 90-92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.