Abstract
A comparative analysis of the lytic cycle of wild-type polyomavirus and middle T and small T defective mutants was carried out in the A2 genetic background. The results contrast with those obtained in comparisons between the hr-t type and their middle-T small-T-producing partners as previously described (20). The A2-derived mutants were found to share the maturation defect previously described for the hr-t mutants. However, their defect in DNA replication was more acute, resulting in a 5- to 100-fold decrease in the accumulation of viral genomes. Furthermore, their gene expression pattern was affected. A2-derived mutants displayed an early defect resulting in a 4- to 16-h delay in the expression of large T, and an alteration of the early-to-late transcriptional switch. In wild-type A2 infection, this switch is characterized by a large increase in the accumulation of early transcripts followed by late transcripts after the appearance of middle T and small T proteins and the onset of viral DNA replication (L. Chen and M. M. Fluck, J. Virol. 75: 8368–8379, 2001). In the mutant infection, increases in both classes of transcripts were delayed and reduced, but the effect on early transcripts was more pronounced. As has been described previously for the hr-t mutants (E. Goldman, J. Hattori, and T. Benjamin, Cell 13:505–513, 1979), the magnitude of these defects depended upon experimental conditions. Experiments using cytosine β-arabinofuranoside to reduce genome amplification suggest that the effect of middle T-small T on the transcriptional switch is not solely mediated by the effect of these protein(s) on increasing the number of templates. These data provide the first direct demonstration of an effect of middle T and/or small T in the viral transcription pattern during viral infection. The results agree with previous results obtained with plasmid reporters and with our understanding that the downstream targets of the middle T signaling pathway include three transcription factors that have binding sites in the enhancer domain that play a key regulatory role in the expression of the viral genes.
Most of the studies on the role of the middle T and small T proteins in the polyomavirus lytic infection have been carried out with the hr-t mutants, so named for their concomitant defects in host range and transformation. These mutants were selected for a differential growth pattern in normal versus polyomavirus-transformed host cells (2). Most mutants isolated using this strategy harbor an out-of-frame deletion in the large T intron (nucleotides [nt] 411 to 797) (10, 17, 25). Whether these deletions prevent the production of the middle T and small T mRNAs has not been tested. If not, the altered proteins have deletions and frameshifts that encompass the binding sites for the c-src family protein kinases or phosphatase 2A (PP2A) or both, as well as C termini that are either truncated or joined to the overlapping reading frames (5, 9, 22, 23). In addition, various sequence variations have been mapped among the hr-t mutants (10, 25). The availability of the hr-t mutants has been crucial for the discovery of the transforming function of polyomavirus (2) and many ensuing studies.
The lytic cycle defect of the hr-t mutants was analyzed by comparison with quasiisogenic “wild-type” strains with restored middle T-small T functions. These were obtained by marker rescue of the hr-t mutants by using sequences within the MspI fragment 4 from a wild-type strain (nt 399 to 1101) (17). Overall, these studies revealed no major differences in the early steps of infection, except for a small reduction in genome amplification (20, 50). The major growth defect was found to take place at a late stage of infection. While the production of capsid proteins and genomes appeared to be in the normal range, the yield of live virus was decreased by 2 orders of magnitude. This loss was due to a failure in maturation (20). It was correlated with the absence of phosphorylation of threonines 63 and 156 in the major VP1 viral capsid protein, which takes place before the encapsidation of the DNA (21, 34). Additionally, an essential serine (Ser-66) was shown to be an in vitro substrate for casein kinase II (CK II) phosphorylation. Although the exact capsid phosphorylation pathway has not been solved, it may be carried out by kinases, including CK II, that are activated by middle T signaling (35).
One of the first polyomavirus strains to be sequenced, the A2 strain is well characterized and one of the most widely used strains (19). It has been used in numerous studies of replication patterns in mice. Its tumor profile has been characterized as highly tumorigenic (14). An hr-t-like mutant with a deletion in MspI fragment 4 was created by site-directed mutagenesis by Lania et al. and called A185 (33). A direct comparison between A185 and the hr-t mutants lytic cycle was undertaken. The results demonstrate that, in the A2 background, the absence of middle T-small T proteins leads to multiple defects, which were not previously observed in hr-t mutants. In particular, the major transcriptional switch (described in reference 11) that coincides with the early-to-late transition and affects both early and late transcription is severely affected. In addition, the absence of middle T and/or small T leads to a large reduction in genome accumulation. These observations reveal a novel key role for middle T in the regulation of viral transcription and confirm the previously described defect in DNA replication (12).
MATERIALS AND METHODS
Virus, cells, and infections.
Polyomavirus wild-type strain A2 (WTA2) and the A185 middle T-small T defective mutant, derived from WTA2 by site-directed mutagenesis, have been characterized (19, 33). The hr-t mutant B2 harbors a 241-bp deletion in the early intron (nt 491 to 731, inclusive) and other sequence variations (25). An 11-bp deletion (nt 46 to 57) was repaired. Furthermore, a wild-type strain, WTB2, was reconstructed by replacing the BsrFI-BlpI fragment (nt 400 to 1079) with that of WTA2. A deletion identical to that of B2 was also generated into the A2 background by using primers with sequences flanking the deletion. This strain was called A2(−241).
Infections were carried out in cells arrested in the G0 phase of the cell cycle as follows. NIH 3T3 cells were plated at 3 × 104 cells per 60-mm culture plate in Dulbecco modified Eagle medium (Gibco-BRL) supplemented with 10% newborn calf serum (Gibco-BRL). After 3 days, when cells reached about 25% confluency, the calf serum supplement was lowered to 0.5% and cells were incubated for another 24-h period. The exit from the cell cycle into the G0 state was confirmed by fluorescence-activated cell sorting (FACS) analysis. The medium was removed and virus was added in 0.5 ml of 1 × phosphate-buffered saline (PBS) supplemented with 2% serum. A multiplicity of infection of 10 PFU was used. Prior to infection, stocks were tested to check whether the use of equal multiplicities resulted in equal levels of input genomes. When adjustments were necessary, the relative dilution factor was small (two- to three-fold) and could affect either parent. After 1 to 2 h, the unadsorbed virus was removed, and cells were washed once with 1× PBS and refed with medium supplemented with 10% newborn calf serum. The times given throughout (in hours postinfection [hpi]) also correspond to times post-release from G0. In all experiments, a sample was taken at between 4 and 12 hpi to ascertain the close equivalence in the level of “input” wild-type and mutant viral genomes.
For the experiments done in the presence of cytosine β-arabinofuranoside (AraC), the inhibitor was used at a concentration of 40 μg/ml and was added 30 min before the time shown to allow for DNA replication to stop (37).
Cell cycle analysis.
Cells were harvested at the times shown, washed twice, resuspended in 1 ml of cold 1× PBS containing 2% calf serum, and fixed by rapid injection into 10 volumes of ice-cold 80% ethanol. Cells were pelleted by centrifugation, washed in 1× PBS, and incubated in 300 μl of PI reagent (10 μg of propidium iodide per ml, 0.1% Triton X-100, 100 mM EDTA, and 10 μg of RNase A in 1× PBS [pH 7.4]) in the dark for at least 30 min at room temperature. The cell cycle stage of the cell population was determined by FACS (Becton Dickinson FACS Vantage) using Cell Quest. The analysis of the cell cycle was carried out using the Multiprime or Wincycle programs.
Protein analysis.
Cells were lysed at various times with protein sample buffer (5% sodium dodecyl sulfate [SDS]; 0.03% bromophenol blue; 20% glycerol; 5% β-mercaptoethanol; 0.5 M Tris-HCl, pH 6.8) and boiled for 5 min. One-third aliquots of the lysates were electrophoresed in 10% polyacrylamide and electroblotted onto polyvinylidene difluoride membranes (Amersham). A polyclonal rat antitumor serum, harvested as ascites fluid, was used as the primary antibody. This antibody recognizes all three early proteins: the large, middle, and small T antigens. In addition, this serum reacts with a few cell proteins, which can serve as an internal loading control. Goat anti-rat horseradish peroxidase (HRP; Pierce) was used as the secondary antibody. Monoclonal antibodies directed against the amino terminus common to the large, middle, and small T proteins were generous gifts from B. Schaffhausen (PN116) and S. Dilworth (MAb762) (16a, 16b). A rabbit anticapsid antibody, a generous gift from R. L. Garcea, was used to for the detection of the VP1 capsid protein using goat anti-rabbit HRP as the second antibody.
Preparation and analysis of DNA.
Infected cells were lysed in 10 mM Tris-HCl–10 mM EDTA–0.2% SDS (pH 7.6) supplemented with 0.1 μg of proteinase K (Sigma) per ml. DNA was extracted with phenol-chloroform. DNA was digested with the restriction endonuclease EcoRI (Gibco-BRL), which has a single recognition site in the polyomavirus genome and linearizes it. Digested DNA was electrophoresed in 1% agarose, stained with ethidium bromide, and blotted onto nylon membranes (Amersham Pharmacia Biotech). The hybridization was carried out at 65°C in 1× Denhardt's solution–2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) by standard procedures with a 32P-radiolabeled probe containing the whole polyomavirus genomic DNA. Hybridization probes were labeled with [α-32P]dCTP (3,000 mCi/mmol; New England Nuclear) with a multiprime DNA labeling kit (Amersham). The hybridized blots were washed under stringent conditions. The blots were exposed to X-ray film (Kodak) for a few days at −70°C with an intensifying screen. To quantitate the level of viral genomes, membranes were scanned with a PhosphorImager (Molecular Dynamics).
Preparation and analysis of RNA.
Infected cells were lysed in Trizol solution (Gibco-BRL) at various times. Total RNA was extracted with chloroform and precipitated with isopropanol. For Northern blots. RNA samples were electrophoresed in a 1% agarose gel containing 2.2 M formaldehyde and transferred to a nylon membrane (Amersham).
The following probes were used to detect the six specific polyomavirus early and late mRNA species. To detect early transcripts, a pGEM1-based plasmid, pG3PyH4, was used that contains polyomavirus MspI fragment 4, cloned between the HindIII and EcoRI sites. The plasmid was cleaved with HindIII, and T7 RNA polymerase was used to synthesize the early-specific RNA probe. This probe spans the three overlapping early introns and can detect all three early mRNAs, i.e., the “19S” middle T and small T mRNAs and the “18S” large T mRNA. To score for late mRNA, sequences spanning nt 3918 to 2928 were inserted between the HindIII and BamHI sites of pSPT18 (Roche Molecular Biochemicals). The plasmid was cleaved with HindIII and T7 RNA polymerase was used to synthesize the late-specific RNA probe. Additionally, to simultaneously detect all transcripts and quantitate RNA, a double-stranded genomic DNA probe was used.
The strand-specific RNA probes were labeled with digoxigenin (DIG) with the kit from Roche Molecular Biochemicals, following the instructions of the manufacturer. Blots were prehybridized for 2 h at 68°C in 50% formamide–5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.02% SDS–0.1% N-laurylsarcosine–2% blocking reagent (Roche Molecular Biochemicals) and then hybridized overnight at 68°C in a hybridization mixture containing DIG-labeled RNA probe. The membranes were washed twice with 2× SSC and 0.1% SDS at room temperature for 15 min each time and twice with 0.5× SSC and 0.1% SDS at 68°C for 15 min each time. The membranes were treated with the blocking agent solution for 1 h and then with the anti-DIG-alkaline phosphatase, diluted 10,000-fold in blocking buffer for 30 min. After an extensive washing, the chemiluminescent phosphatase substrate detection reagent CSPD was applied for 1 min, and the membranes were exposed to X-ray films.
The 32P-substituted double-stranded genomic DNA probe was labeled as described above. Hybridization was performed at 42°C in 0.05 M sodium phosphate buffer (pH 7.0)–1 M NaCl–50% formamide–1% SDS–5% dextran sulfate–100 mg of salmon sperm DNA per ml. Membranes were washed and exposed to X-ray film for a few days at −70°C with an intensifying screen. Transcript levels were quantitated by counting with a PhosphorImager.
Plaque assays.
Intracellular (i.e., cell-associated) virus was obtained by disrupting cells after they were washed free of extracellular virus (i.e., virus already released into the medium). For the plaque assay, NIH 3T3 cells were grown to 80% confluence and infected with virus dilutions for 1 h at 37°C. Infected cells were refed with medium containing 5% calf serum and 0.9% agar, and incubated at 37°C for 7 to 9 days in the case of the wild type or 10 to 14 days in the case of the mutant. The plates were stained with neutral red. The plaques were counted after >4 h of incubation at 37°C. The difference in incubation times were used to compensate for the growth defect of the mutants, which results in a small plaque phenotype.
RESULTS
Physical characterization of the A185 viral strain.
The previously described hr-t mutants harbor deletions in the middle T-small T coding sequences that overlap with the large T intron (17). The A185 mutation was originally designed with the intention of reconstructing an hr-t-like mutant by site-specific mutagenesis in the well-characterized A2 background (33). The AvaI site at nt 659 located in the large T intron sequences was targeted, and the cleavage was extended by treatment with the Bal 31 exonuclease (33). Because the properties of the A185 mutant are so different from those of the previously described hr-t mutants, we initially sequenced key regions of its genome. The sequence of the regulatory region was confirmed to be identical to that of WTA2, and the deletion was found to encompass 71 nt (nt 646 to 717). Thus, similar to the case of most hr-t mutants, the deletion is out of frame (−1 frame) and should not interfere with the 5′ and 3′ splice sites for the three early mRNAs. The sequence for the PP2A binding site in small T and middle T is not affected (9, 23). The frameshift downstream of the deletion eliminates the middle T binding site for the tyrosine kinases of the c-Src family (5). If the splicing of the middle and small T introns were to take place normally, termination at an out-of-frame termination codon would yield a truncated protein of 156 amino acids with a J domain (48) and a PP2A binding site (9, 22). This protein fragment was not detected (see below).
To test whether the properties described for A185 are shared by other A2-derived early intron deletion mutants, we constructed a second mutant, with a 241-bp deletion (nt 491 to 731, inclusive) (+1 frameshift), identical to that of hr-t mutant B2 (10), extending into the PP2A binding domain. This exchange resulted in a mutant with the same phenotype as that of A185 for all properties tested. Most of the results described herein were obtained with mutant A185, except where noted.
Cell cycle analysis.
All experiments were carried out in G0-arrested cells as described in details in the accompanying article (11), following the protocol described in Materials and Methods. Briefly, NIH 3T3 cells were maintained in a subconfluent quiescent state for >24 h and then infected in parallel with WTA2 and A185 at the same “matched” multiplicity of infection (i.e., 10), as described in Materials and Methods. Infected cells were released from G0 at the time of infection by refeeding with serum-containing medium. In order to time the events in the lytic infection in relation to the host cell cycle progression, a cell cycle analysis was carried by FACS. No difference in progression of the infected cells though the cell cycle was observed between WTA2- and mutant-infected cells (data not shown). This was expected since the cell cycle of A2-infected cells does not differ from that of mock-infected cells (11).
Samples were taken at various times to assay viral transcripts, proteins, and genomes, as well as live virus. Most experiments were stopped at 48 hpi, when cytopathic effects became visible, prior to reaching the maximal live-virus titer. Various aspects of these analyses were repeated in multiple experiments, and all conclusions are based on at least two experiments. The data from one extensive experiment are shown in Fig. 1, 2, 7, and 8. Some of the results for the wild type in that experiment were shown in the accompanying study (11) and are represented for comparison. As summarized in the discussion, biological variations were obtained between experiments carried out under “identical” conditions, in the extent of the defect of the middle T-small T defective mutant. Among the experiments described herein, the one demonstrating the weakest middle T-small T defect is that presented in Fig. 1, 2, 7, and 8.
FIG. 1.
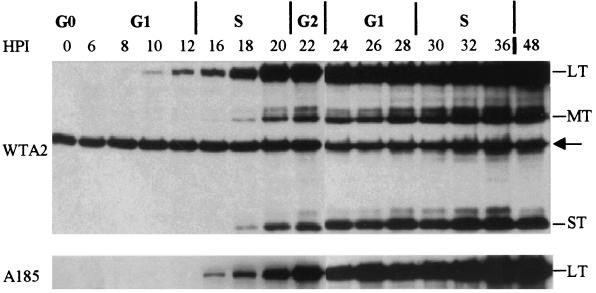
Analysis of the expression of the early protein(s). Cells were infected with WTA2 and mutant A185, as described in Materials and Methods. Proteins were extracted at the times shown on top of the lanes (HPI = hpi) and processed as described in Materials and Methods. The first lane represents an uninfected control. The Western blot was probed with a polyclonal antiserum directed against all three early polyomavirus proteins. This antiserum also detects a cellular protein (arrow) which serves as a loading control. All three proteins produced in the wild-type infection are identified on the right: large (LT), middle (MT), and small (ST) T antigens. In the case of A185, only the large T protein is produced and the figure was cropped. The phase of the cell cycle, for the majority of the infected cells, at the time of harvest is shown on the top of the figure.
FIG. 2.
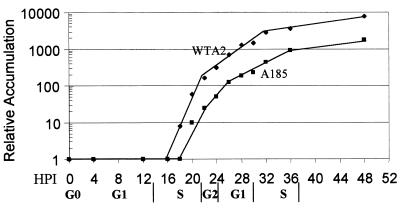
Viral genome amplification. Cells were infected as described in the legend to Fig. 1, total DNA was extracted at the times shown, digested with EcoRI which linearizes the viral genome, processed for Southern blotting, and hybridized to a 32P-labeled genomic probe as described in Materials and Methods. Hybridized counts were determined with a PhosphorImager and corrected for dilution, and the increase relative to the input was graphed in function of time postinfection (HPI = hpi). The phase of the cell cycle, for the majority of the infected cells at the time of harvest is shown below.
FIG. 7.
Late proteins. Cells were infected with WTA2 or A185, proteins extracted at the times shown and processed as described in the legend to Fig. 3. Samples from uninfected or A2- or A185-infected cells were collected at the times shown (HPI = hpi) and analyzed by Western blotting using a rabbit antibody directed against the VP1 capsid protein.
FIG. 8.
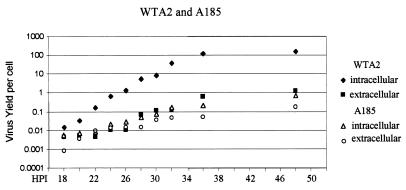
Live virus production. The production of live virus in infections by WTA2 or mutant A185 was assayed by plaque assays as described in Materials and Methods at the times shown (HPI = hpi). Both the cell-associated virus and the virus released in the medium were assayed.
Experiments carried out in parallel with or without serum stimulation demonstrated a profound early defect in A185 infection in the absence of serum. No or very low levels of large T protein were expressed, and the infected cells remained arrested in G0. A further characterization of this defect, which is not seen in the case of hr-t mutants, will be presented elsewhere. All data presented in this report were obtained in cells that were serum stimulated at the end of the adsorption period.
Expression of early proteins.
Since the transcript levels remain very low throughout the early phase of infection (below the detection level by Northern blot analysis) (see the related study [11] and Fig. 4 below), the assay of early proteins represents a convenient measure of gene expression. A kinetic analysis of their expression was carried out in serum-released NIH 3T3 cells infected in G0 in parallel with WTA2 and A185, as described in Materials and Methods. Protein samples were extracted at the times shown and analyzed by Western blotting using a polyclonal antibody that detects large T, middle T, and small T proteins. The data are shown in Fig. 1. All three early proteins were observed in wild-type infection, while the mutant produced only large T. No other protein fragment was detected. Tests with 2 monoclonal antibodies (PN116 and MAb762) directed toward the amino-terminal protein domain common to large, middle, and small T proteins also failed to detect additional protein fragments. A 4- to 6-h delay was observed in large T protein detection when we compared the A185 mutant to the wild-type, and lower levels were observed throughout the infection (Fig. 1) or longer (not shown). Therefore, the delay in large T detection is likely to be a consequence of lower expression. Examination of large T expression by immunofluorescence demonstrated that the staining of the mutant infected cells was less bright than that of wild-type-expressing cells (data not shown). This early defect is not observed with hr-t mutants (see Fig. 3). It is not due to a lower input genome dosage, as shown in Fig. 2, nor, apparently, to a defect in virus entry. Indeed, the genomes of WTA2 and A185 were shown to reach the nucleus with similar kinetics and to induce similar levels of receptor-mediated signaling (data not shown). Furthermore, both populations of wild-type- and mutant-infected cells expressed large T in close to 100% of the cells by 32 hpi. As described above and as summarized in the Discussion, biological variations were observed between experiments. In other experiments, a more pronounced decrease in the level of large T protein was observed in mutant infection. However, these differences compared to the wild type were observed with two different synchronization protocols, as well as when exponentially growing cells were used.
FIG. 4.
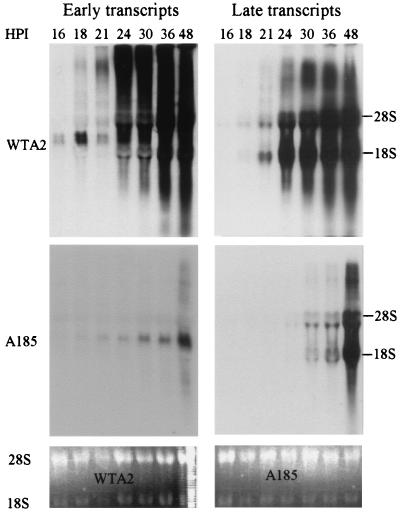
Early-to-late transcriptional switch. Cells were infected with WTA2 or mutant A185 as described in the legend to Fig. 1. Samples were harvested at the times shown (HPI = hpi). Total RNA was extracted and electrophoresed as described in Materials and Methods. The ethidium bromide staining of the 28S and 18S rRNA bands is shown (bottom two gels). Note that the 21-hpi wild-type sample was underloaded. The blots were hybridized with a DIG-substituted RNA probe detecting the early transcripts (left side), stripped, and rehybridized with a probe for the late transcripts (right side). The early-transcript-specific probe (nt to 399 to 1101) detects all early RNAs. The late-transcript-specific probe (nt 3918 to 2928) detects all late RNAs. WTA2, top two gels; A185, middle two gels.
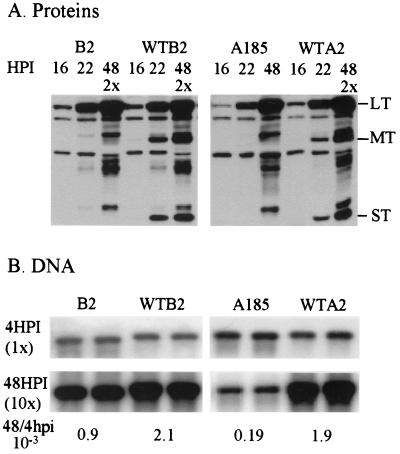
FIG. 3.
Comparisons of middle T-small T defective mutants in different genetic backgrounds. Cells were infected with equal multiplicities of the hr-t mutant B2, its wild-type version WTB2, A185, and WTA2. Proteins and DNA were analyzed as described in the legends to Fig. 1 and 2. Protein samples for B2, WTB2, and WTA2 collected at 48 hpi were diluted twofold. The DNA samples collected at 48 hpi were diluted 10-fold. The counts in the bands were determined with a PhosphorImager, and the ratio (10−3) of the output at 48 hpi over the input (4 hpi) is given under the lanes. HPI = hpi.
We have also tested the same pair of mutants (A2 and A185), in NIH 3T3 clone 7, a subline from C. Scherr's laboratory with a regulated cyclin D1. The level of WTA2 proteins was lower than that observed in the subline used in the present report. In this case, the defect of the A185 mutant was very severe, since no early viral proteins were detected (data not shown).
Viral DNA replication.
The accumulation of viral genomes was assayed by Southern blotting as described in Materials and Methods in the same experiment as that for which the proteins are shown in Fig. 1. Total DNA was isolated at the times shown and analyzed by Southern blotting using a probe for polyomavirus genomic DNA, and the band signal was quantitated by using a PhosphorImager. The ratio of output to input was calculated, and the resulting curves are shown in Fig. 2. The levels of input genomes (4-hpi samples) for WTA2 and mutant A185 were equivalent. In the WTA2 infection, an increase in the level of genomes became detectable at between 16 and 18 hpi; in the case of the mutant, detection of an increase was delayed by approximately 2 h. In addition to this delay, a substantial reduction in genome accumulation was observed. Qualitatively similar results were seen in many different experiments. However, the overall reduction in genome accumulation varied from experiment to experiment by between a factor of 5 and a factor of 100. This variation is further discussed below. In the experiments described here, the reduction was 5-fold in the experiment described in Fig. 2 (and in Fig. 1, 7, and 8), 10-fold for that in Fig. 3, 74-fold for that in Fig. 4, and 20-fold for that in Fig. 6. Qualitatively similar results were obtained using different synchronization protocols, exponentially growing cells, and other cell lines, as summarized in the Discussion. A reduction in A185 genome amplification relative to WTA2 was also observed in the infection of semipermissive FR3T3 rat cells (data not shown).
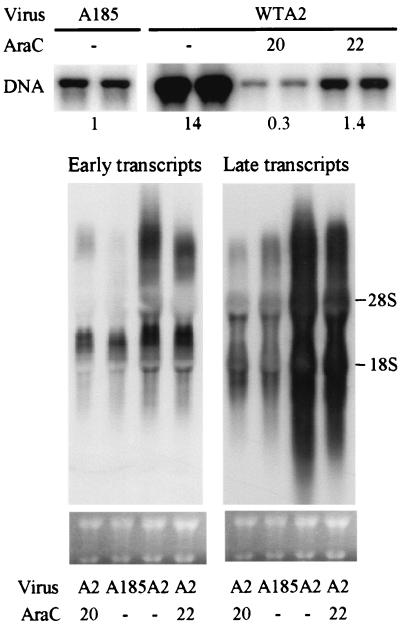
FIG. 6.
Comparisons of transcripts and genome levels in A185-infected and AraC-treated A2-infected cells. Cells were infected with WTA2 or A185 as described in the legend to Fig. 1. A2-infected cells were treated with AraC at 20 or 22 hpi or were not treated. All samples were collected at 26 hpi. DNA was extracted and analyzed as described in the legend to Fig. 2. Duplicate samples are shown. The level of hybridized counts in the bands relative to the level for A185 taken as 1 is shown. Transcripts were analyzed as described in the legend to Fig. 4. Duplicate samples were analyzed either for the levels of early transcripts (left) or for the levels of late transcripts (right). The ethidium bromide staining pattern of the rRNAs is shown.
The defect in DNA replication did not stem from the lower level of large T expressed in the A185 infection, since infection of cells stably producing a high level of large T protein did not increase the replication potential of the mutant (data not shown).
Comparisons of the properties of middle T-small T defective mutants in different genetic backgrounds.
As mentioned in the introduction, the analysis of hr-t mutants did not reveal a defect in early protein expression and detected a mild defect in DNA replication (20, 50). These properties were compared directly between A2-derived and hr-t middle T-small T defective mutants. For this purpose, the B2 hr-t mutant was chosen, and a quasi-isogenic wild type, WTB2, was reconstructed by repairing the 241-bp intron deletion, as described in Materials and Methods. The results of a direct comparison of the expression of early proteins and DNA replication patterns in infections with WTA2 and A185, on the one hand, and with WTB2 and B2, on the other, are shown in Fig. 3. Lower levels of large T expression in A185 compared to A2 infection was evident at 16 and 22 hpi, while no difference could be seen in the comparison of B2 with WTB2. Analysis of genome levels showed a 10-fold reduction in genome amplification (ratio of output at 48 hpi/input) in the comparison of A185 with A2, while a 2.3-fold difference was seen between B2 and WTB2, in agreement with previous results (20, 50).
The defect in genome amplification in the A2 background was tested with a second mutant, A2(−241), in which a deletion identical to that of B2 was introduced (see Materials and Methods). In a direct comparison, the output/input ratio of the levels of genomes at 48 hpi was 4 × 103 for WTA2, 3 × 102 for A185, and 2.5 × 102 to 4 × 102 for the four independent stocks of the 241-bp deletion mutant (data not shown).
Transcriptional control of the early-to-late transition.
Various experiments demonstrated that the level of transcripts was also diminished in A185- versus A2-infected cells (data not shown), including a previously published study (12). In the accompanying study (11), we reported that a major transcriptional switch takes place at the transition between the early-to-late phase of infection. The pattern of transcripts in A185 mutant-infected cells was examined by Northern blotting during the period encompassing the switch and compared to that obtained with WTA2-infected cells (Fig. 4). In infection with WTA2, the early transcripts were detected from 16 hpi on. As reported in the accompanying study (11), their levels began to increase around 18 hpi, shortly following the onset of viral DNA replication, and continued to rise until the end of the experiment. As noted in the legend to Fig. 4, the 21-hpi sample was underloaded. The increase in early transcripts was monitored by the detection of late transcripts from 18 hpi on. Giant oligomeric transcripts were observed at from 18 to 21 hpi among the early transcripts and from 21 to 24 hpi among the late transcripts. These aspects of polyomavirus transcription are discussed in detail elsewhere (11). The absence of the middle T-small T proteins severely altered the patterns of early and late transcription. A large decrease in the levels of early transcripts was observed. The giant transcripts were essentially absent. The induction of the late transcripts was also affected: these became detectable with a delay of 6 to 9 h, and their levels remained lower until very late times. However, giant late transcripts were synthesized at late times postinfection. In contrast, although delayed by 2 h as described above, genome amplification did take place. In the experiment shown in Fig. 4, the level of genomes in the A2 and A185 input samples was equal, and the amplification of the wild-type genome surpassed that of A185 by 74-fold at 36 hpi.
Coupling between transcription and DNA replication.
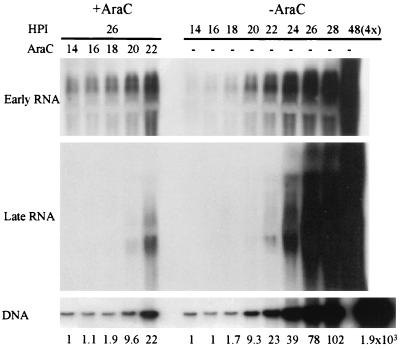
Since the absence of middle T-small T results in a decrease in the accumulation of genomes which can be severe, the possibility that part or all the defect in the early-to-late transcriptional switch of the A185 strain is a consequence of a reduction in DNA replication was considered. Therefore, the dependence of transcription on DNA replication in WTA2 infection was examined, using AraC as an inhibitor of DNA chain elongation (37). AraC was added at various times at between 14 and 22 hpi (as described in Materials and Methods), while cells were in S phase. All AraC-treated samples were collected at 26 hpi. The FACS analysis of treated cells shows that the addition of the inhibitor caused cells to arrest with the DNA content reached at the time of AraC addition (data not shown). Treated samples harvested at 26 hpi contained the same levels of genomes as those harvested at the time of AraC addition, verifying AraC inhibition of viral DNA synthesis. In the untreated cells, an increase in the level of viral genomes was observed at from 16 hpi on, with a 1.7-fold increase occurring between 16 and 18 hpi and a further 5.4-fold increase occurring between 18 and 20 hpi (Fig. 5, DNA). The level of transcripts was followed by Northern blotting. In the case of the early genes, no or low levels of transcripts were detected in untreated samples at 14, 16, and 18 hpi, in agreement with data in Fig. 4 and in the accompanying study (11). In contrast, in cells treated with AraC at 14, 16, and 18 hpi transcripts were clearly detected. Thus, transcripts were synthesized between the time of addition of the inhibitor and the time of sample collection (26 hpi). This increase in transcript levels took place in the absence of (AraC at 14 and 16 hpi) or with very low (1.9-fold; AraC at 18 hpi) detectable genome amplification prior to AraC addition. The level of these transcripts was almost equivalent to that seen in the untreated 20-hpi sample, which had undergone a 9.3-fold genome amplification. Very similar results were seen for the large T protein levels, that is, the levels of large T in the samples treated at 14, 16, or 18 hpi were very similar to those in the untreated 20-hpi sample (data not shown). Thus, an increase in early transcript levels took place in the absence of or with very low detectable DNA amplification. Nevertheless, the inhibition in replication did lead to a reduction in transcript levels. The results for the late transcripts contrasted with those for the early transcripts. In this case, neither transcripts nor VP1 protein (not shown) was detected in samples treated before 20 hpi. Late transcripts were observed in the sample treated at 20 hpi/ and in the sample treated at 22 hpi their level was clearly more abundant than in the untreated 22-hpi sample, while the degree of genome amplification was nearly identical (22-fold versus 23-fold). VP1 protein was detected when transcripts were seen, i.e., as long as AraC was added after 18 hpi (data not shown).
FIG. 5.
Coupling between transcription and DNA replication. Cells were infected as described in the legend to Fig. 2 and either treated with AraC at 14, 16, 18, 20, or 22 hpi (+AraC) or left untreated (−AraC). AraC-treated cells were all collected at 26 hpi. Untreated cells were sampled at 2-h intervals at between 14 and 28 hpi and processed for analysis. Times (HPI = hpi) shown above the figure refer to the time of sampling. Untreated and treated samples were processed for the analysis of early as well as late transcripts and DNA as described in Materials and Methods and in the legends of Fig. 2 and 4. The increase in viral genome levels over the input assayed at 14 hpi is given under the figure.
A more direct comparison of the levels of transcripts and genomes between A2-versus A185-infected cells is shown in Fig. 6. Cells were infected with equal multiplicities of the two viral strains, using the standard protocol. Wild-type-infected cells were treated with AraC at 20 or 22 hpi or were not treated. All samples were harvested at 26 hpi. The relative levels of genomes was determined by phosphorimaging of the blots, and the numbers are shown underneath the lanes. The level of genomes was set at 1 for the A185 case. In the untreated wild-type-infected cells the relative genome level was 14, representing a 20-fold difference in DNA amplification between the wild type and the mutant at 26 hpi. The relative level of genomes in the wild-type-infected cells treated with AraC at 22 hpi was very similar to that of the untreated-mutant-infected cells (1.4 versus 1.0). However, the levels of either early or late transcripts in the wild-type-infected AraC-treated cells were higher than the corresponding transcripts in the mutant-infected cells.
Expression of the VP1 major capsid protein.
The expression of the major late capsid protein VP1 was compared in mutant and wild-type infections by Western blotting using a rabbit antibody directed against VP1 (Fig. 7). As expected from the late transcript pattern, the level of the VP1 protein was also decreased in the mutant relative to the wild-type infection. Similar results were obtained when VP1 expression was examined by immunofluorescence (data not shown). As is the case for the detection of wild-type and mutant early proteins (and as described in detail in reference 11), the VP1 protein synthesized in infections with the A185 mutant could be detected by Western blotting 2 h prior to the detection of its mRNA by Northern blots. This reduction was quantitated by dilution and estimated to be less than 10-fold.
Production of live virus.
The maturation process of the A185 mutant was compared to that of the wild type. Live virus particles were quantitated by plaque assays in extracts from the infected cell monolayer (intracellular virus) and from their supernatants (extracellular virus) harvested at the times shown in Fig. 8. A 100-fold decrease in live virus levels was observed. A decrease of fivefold could be expected from the decrease in genome levels in the same experiment (Fig. 2). Similarly, a decrease in VP1 expression was also observed. However, the decrease in yield was 20 times larger than the decrease in genome level, suggesting a defect in encapsidation. Such a defect has been documented in detail for the middle T-small T mutants in the hr-t background (20, 21, 34, 35). This defect is also observed in the production of mutant virus stocks, which typically reach titers at least 10-fold lower than those of wild-type virus. Not surprisingly, the A2-derived middle T-small T defective viruses produce plaques that are smaller and delayed compared to those of WTA2.
DISCUSSION
The results presented here demonstrate that the loss of middle T and/or small T function in the A2 genetic background results in the development of phenotypes that were not observed in the case of comparisons between the hr-t mutants and their corresponding wild types. The most extensive study was carried out in NIH 3T3 cells synchronized by serum deprivation in a subconfluent state. However, various phenotypes (mostly the difference in the level of early protein expression and in viral genome accumulation) were confirmed by using different conditions. These include (i) NIH 3T3 cells, synchronized by contact inhibition and serum deprivation, trypsinized and replated 4 h prior to infection; (ii) exponentially growing NIH 3T3; (iii) a different subline of NIH 3T3 named clone 7; and (iv) the semipermissive FR3T3 rat cell line. Decreases similar to those reported above, or more dramatic ones (NIHcl7), in early proteins and/or genome levels or both were observed in all cases. This suggests that the differences described are inherent of the viral strains rather than specific to the conditions used. The effect of the absence of middle T-small T in the A2 genetic background was also observed with another deletion (−241) with a larger deletion and a different frameshift. This suggests that, in the A2 background, the observed differences are not specific to the A185 mutant.
The absence of middle T-small T results in an early defect in gene expression.
One altered phenotype of the A2-derived middle T-small T mutants is manifested very early in the infection and results in a delay in large T protein expression (range, 4 to 12 h) when the mutant is compared to the wild type. Preliminary data suggest that this defect is not related to the early entry and decapsidation steps, since the mutant chromatin becomes accessible to nucleases as rapidly as does wild-type chromatin. The delay may be a reflection of differences between mutant and wild-type chromatin modification (43) that result in a lower level of expression of the mutant. This defect is first seen prior to the time when expression of middle T-small T proteins can be detected in infection with the wild-type virus. Thus, it appears not to be a consequence of the absence of middle T-small T in the current infection. This defect is particularly severe in NIHcl7. Further preliminary experiments have shown that the defect is most severe if the cells are infected in the absence of serum, in which case essentially no viral proteins are expressed and the infection is aborted. This is in contrast to infection with wild type, which is almost insensitive to the presence of serum. This defect is under further investigation.
Role of middle T-small T in the activation of the early-to-late transcriptional switch.
The most striking novel defect of the A2-derived middle T-small T defective mutants uncovered in these experiments takes place at the early-to-late transcriptional switch. In the accompanying study (11), we describe this major change in the transcriptional program at the transition between the early and late phases of infection. At the switch time, a rapid and large increase is observed in the levels of early and, 2 to 4 h later, late transcripts. The present results show that, in the absence of middle T-small T, this activation is both delayed and reduced. An extreme example is shown in Fig. 4. In this case, the detection of the early transcripts was delayed 5 h and showed a dramatic reduction, while the late transcripts were also delayed (approximately 6 h), although their level was less severely reduced.
The transcriptional switch defect is not likely to be solely due to a decrease in transcription templates and thus an indirect effect of the reduction in genome amplification. Indeed, the addition of an inhibitor of DNA synthesis (AraC) in wild-type-infected cells demonstrates that a level of genome amplification lower than that observed in mutant-infected cells allows for the synthesis of higher levels of transcripts than is observed in infections with the middle T-small T defective mutants. Thus, we conclude that middle T and/or small T play a crucial role in mediating the early-to-late transcriptional switch. Further implications of the results observed following AraC treatment are discussed below.
Although the data described here represent the first demonstration of an effect of middle T-small T on the transcription of the viral genome during viral infection, the results could be expected. Middle T signaling is known to activate transcription factors, namely, AP1/PEA1, PEA3/ets/elk, and c/EBP (31, 44, 45, 51, 52, 53) that have binding sites in the polyomavirus enhancer (38, 49, 51, 52, 53). It has been well documented that these sites play a crucial role in the regulation of both early and late gene expression, as well as in DNA replication (3, 7, 13, 27, 28, 39, 40, 49, 55). The present data also agree with previous results obtained with transfected plasmid reporters. An enhancement of transcription by middle T has been demonstrated for reporter genes that are controlled by the PEA1 and/or PEA3 sites and either the early or the late promoter elements (29, 55). In the case of the plasmid reporters, the evidence suggests that middle T rather than small T is the major contributing factor (55). The present experiments did not distinguish between middle T and small T function. Although the downstream targets of the signaling pathways of these two proteins appear to at least partially overlap, the activation of the transcription factors listed above has been specifically demonstrated in the case of middle T. However, while awaiting further clarification with mutants that are defective in middle T but not small T, this report is written in terms of middle T and/or small T effects.
The method of analysis used here is not suitable to determine whether the effect of middle T and/or small T in the early-to-late transcriptional switch is effected at the transcriptional or the posttranscriptional level. The documented activation of transcription factors by middle T suggests that at least part of the activation may be transcriptional. This notion is supported by the observation that cellular genes, such as JunB, c-fos, and transin, that are under the control of the same transcription factors (6, 30, 46) are also transcriptionally activated shortly after the early-to-late viral transcriptional switch (12, 56; data not shown). These cellular genes are not induced in the absence of middle T-small T (preliminary data). These effects of middle T and/or small T on viral and cellular genes take place with a substantial delay from the time of protein detection. They appear to coincide with the appearance of novel phosphorylated proteins coimmunoprecipitated in the middle kinase assay (see reference 11). Whether some effects of the presence of middle T and/or small T are exerted at earlier times was not determined in the present experiments nor, to our knowledge, in previous experiments in the literature. Further experiments are under way to clarify these points.
Activation of the late promoter in the absence of middle T-small T.
Despite the parallelism in the control of early and late transcription, differences were noted as reported and reviewed elsewhere (11). Additional differences emerged in the comparison of these transcripts in the wild-type and mutant infections. In a condition where the absence of middle T-small T resulted in a severe reduction in enhancement of early transcription (Fig. 4), the induction of late transcripts did take place, albeit with a long delay and at a reduced level. One possible mechanism for this late transcription induction could be large T mediated, as has been amply documented in the case of simian virus 40 (SV40) (see references 1 and 47 for a review). At present, the results for polyomavirus are unclear. An effect of large T on late-promoter induction has been reported previously in the case of an Ori defective plasmid reporter that contained the whole enhancer and half of the early region (nt 5022 to 1587). This effect required sequences located between nt 5055 and 5182 (29). However, no large T effect was obtained with a very similar reporter containing the same control sequences (55). Other possible mechanisms could be replication linked. It has been suggested that the effect of a repressor of late transcription is diluted by genome amplification (8, 36). This reasoning has also been applied to the putative occlusion of the late Inr TATA-less initiation site by binding of TFIID on the early TATA box (55).
A differential effect on early versus late transcripts synthesis was also observed following inhibition of DNA replication by AraC, an inhibitor of elongation. As previously described for both SV40 and polyomavirus (8, 37; see references 1 and 47 for reviews), AraC, added prior to the detection of increases in genome levels, totally abolished the synthesis of late transcripts and proteins. In contrast, the levels of early transcripts continued to increase, albeit to lower levels than without inhibitor.
The absence of middle T-small T results in a large reduction in genome amplification.
The level of viral genomes accumulated during an infection with the A2-derived middle T-small T defective mutants is considerably lower than that obtained in WTA2 infection (with a 5- to 100-fold decrease). This defect in DNA replication of A185 has been noted previously (12). A modest defect in DNA replication of hr-t mutants has also been reported (20, 50) and is confirmed here. We have further characterized the replication defect of A2-derived mutants and demonstrated that an increase in the level of viral genomes is observed when middle T, but not large T or small T, expression vectors are transfected along with the A185 infection. Using Origin-enhancer containing plasmid reporters, we have also shown that middle T has a more important role than small T in the stimulation of viral DNA replication (unpublished data).
We have hypothesized that the role of middle T and/or small T in the enhancement of DNA replication, like their role in transcription discussed above, is mediated by transcription factors that are among the downstream targets of the middle T signaling pathways (12). These include AP1/PEA1, PEA3, and c/EBP, which have binding sites in the polyomavirus enhancer. These sites are known to play an important role in viral DNA replication as well as transcription (13, 15, 38, 41, 42, 50). In the latter case, the effect of activating c-jun and c-fos on polyomavirus origin-dependent DNA replication has been directly demonstrated (42).
Maturation defect.
As reviewed in the introduction, the major defect reported so far in the lytic cycle of middle T-small T defective hr-t mutants takes place at a maturation step (20, 21, 34, 35). This defect was also observed in the present study. In the experiment described in Fig. 1, 2, 7, and 8, a 5-fold reduction in genome amplification (Fig. 2) was observed, as well as a modest decrease in the VP1 capsid protein level (also about 10-fold) (Fig. 7). In contrast, the reduction in live virus level at 48 hpi was at least 200-fold (Fig. 8). Thus, the A2-derived middle T-small T mutants also display a failure to mature.
Biological variations in the defects of middle T-small T mutants.
A variability in the severity of the various defects of A2-derived middle T-small T mutants (delay in early protein expression, reduction in genome amplification and defect in the early-to-late transcriptional switch) was observed between experiments carried out in “identical” conditions. Similar variations have been observed previously in the case of the hr-t mutants. In that case, variations in the yield of live virus particles were seen, presumably reflecting changes in the degree of the maturation defect (24). The term “permissivity” was used to describe the ability of the host cells to bypass the defect of hr-t mutants (24). Permissivity reflects a yet-undefined cellular state. This state appears to be modulated by unknown serum factors, which vary from batch to batch. It is likely that serum batches vary in their content of factors which modulate a cellular signaling pathway that overlaps with that of middle T-small T. Changes are also linked to the number of passages of the host cells in tissue culture and, in turn, these may be related to changes in growth conditions.
Conclusions.
The comparative analysis of the lytic cycle of wild type and middle T-small T defective mutants in the A2 background has uncovered defects that were either not seen or showed a lesser degree of severity when middle T-small T defective hr-t mutants were first examined. The hr-t mutants appear to bypass these problems, by the action of at least one “compensatory” mutation (to be published elsewhere). This mutation appears to be responsible for the “dominant lethal” phenotype attributed to the hr-t mutants and their multiple probably related phenotypes: a high level of gene expression of early and late proteins and of genome replication, competition for expression, and replication in mixed infections with other strains (18).
The present data imply a role for middle T and/or small T in the activation of the early-to-late transcriptional switch and the enhancement of viral DNA replication. Both events take place at the early-to-late transition. Both are likely to be effected by downstream targets of the middle T-small T signaling cascade. At least three transcription factors with binding sites in the enhancer—AP1/PEA1, PEA3/etc, and a member of the c/EBP family—may be implicated. The effects of these factors in the viral transcription and replication process are likely to be mechanistically linked and to involve modifications of the viral chromatin.
ACKNOWLEDGMENTS
Louis King is gratefully acknowledged for help with the FACS analysis, and we thank the following colleagues for generous gifts of antibodies and cells: R. Garcea for the anti-VP1 antibody, B. Schaffhausen and S. Dilworth for the monoclonal antibodies against early proteins, and C. Scherr for NIH 3T3 clone 7 cells.
This work was supported by grant R01-CA29270 from the National Cancer Institute.
REFERENCES
- 1.Acheson N H. Lytic cycle of SV40 and polyoma virus. In: Tooze J, editor. DNA tumor virus. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1981. pp. 125–204. [Google Scholar]
- 2.Benjamin T L. Host range mutants of polyoma virus. Proc Natl Acad USA. 1970;67:394–399. doi: 10.1073/pnas.67.1.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourachot B, Yaniv M, Herbomel P. Control elements situated downstream of the major transcriptional start are sufficient for highly efficient polyomavirus late transcription. J Virol. 1989;63:2567–2577. doi: 10.1128/jvi.63.6.2567-2577.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brabant F, Acheson N H. RNA footprint mapping of RNA polymerase II molecules stalled in the intergenic region of polyomavirus DNA. J Virol. 1995;69:4423–4430. doi: 10.1128/jvi.69.7.4423-4430.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brewster C E, Glover H R, Dilworth S M. pp60c-src binding to polyomavirus middle T-antigen (MT) requires residues 185 to 210 of the MT sequence. J Virol. 1997;71:5512–5520. doi: 10.1128/jvi.71.7.5512-5520.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buttice G, Duterque-Coquillaud M, Basuyaux J P, Carrere S, Kurkinen M, Stehelin D. Erg, an Ets-family member, differentially regulates human collagenase 1 (MMP1) and stromelysin 1 (MMP3) gene expression by physically interacting with the Fos/Jun complex. Oncogene. 1996;13:2297–2306. [PubMed] [Google Scholar]
- 7.Cahill K B, Carmichael G G. Deletion analysis of the polyomavirus late promoter: evidence for both positive and negative elements in the absence of early proteins. J Virol. 1989;63:3634–3642. doi: 10.1128/jvi.63.9.3634-3642.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cahil K B, Roome A J, Carmichael G C. Replication-dependent transactivation of the polyoma late promoter. J Virol. 1990;64:992–1101. doi: 10.1128/jvi.64.3.992-1001.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell K S, Auger K R, Hemmings B A, Roberts T M, Pallas D C. Identification of regions in polyomavirus middle T and small t antigens important for association with protein phosphatase 2A. J Virol. 1995;69:3721–3728. doi: 10.1128/jvi.69.6.3721-3728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carmichael G G, Benjamin T L. Identification of DNA sequence changes leading to loss of transforming ability in polyoma virus. J Biol Chem. 1980;255:230–235. [PubMed] [Google Scholar]
- 11.Chen L, Fluck M M. Kinetic analysis of the steps of the polyomavirus lytic cycle. J Virol. 2001;75:8368–8379. doi: 10.1128/JVI.75.18.8368-8379.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen M C, Redenius D, Ozati-Ashtiani F, Fluck M M. Enhancer-mediated role for polyomavirus middle T/small T in DNA Replication. J Virol. 1995;69:326–333. doi: 10.1128/jvi.69.1.326-333.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dailey L, Basilico C. Sequences in the polyomavirus DNA regulatory region involved in viral DNA replication and early gene expression. J Virol. 1985;54:739–749. doi: 10.1128/jvi.54.3.739-749.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawe C J, Freund R, Mandel G, Ballmer-Hofer K, Talmage D A, Benjamin T L. Variations in polyoma virus genotype in relation to tumor induction in mice: characterization of wild-type strains with widely differing tumor profiles. Am J Pathol. 1987;127:243–261. [PMC free article] [PubMed] [Google Scholar]
- 15.de Villiers J, Schaffner W, Tyndall C, Lupton S, Kamen R. Polyoma virus DNA replication requires an enhancer. Nature. 1984;312:242–246. doi: 10.1038/312242a0. [DOI] [PubMed] [Google Scholar]
- 16a.Dilworth S M, Griffin B E. Monoclonal antibodies against polyoma virus tumor antigens. Proc Natl Acad Sci USA. 1982;79:1059–1063. doi: 10.1073/pnas.79.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16b.Dilworth S M, Horner V P. Novel monoclonal antibodies that differentiate between the binding of pp60c-src or protein phosphatase 2A by polyomavirus middle T antigen. J Virol. 1993;67:2235–2244. doi: 10.1128/jvi.67.4.2235-2244.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feunteun J, Sompayrac L, Fluck M M, Benjamin T L. Localization of gene functions in polyoma virus DNA. Proc Natl Acad Sci USA. 1976;73:4169–4173. doi: 10.1073/pnas.73.11.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fluck M M, Staneloni R J, Benjamin T L. Hr-t and Ts-a: two early gene functions of polyoma virus. Virology. 1977;77:510–624. doi: 10.1016/0042-6822(77)90486-x. [DOI] [PubMed] [Google Scholar]
- 19.Fried M, Griffin B E, Lund E, Robberson L. Polyoma DNA: a study of wild type, mutant and defective DNAs. Cold Spring Harbor Symp Quant Biol. 1974;39:45–52. doi: 10.1101/sqb.1974.039.01.009. [DOI] [PubMed] [Google Scholar]
- 20.Garcea R L, Benjamin T L. Host range transforming gene of polyoma virus plays a role in virus assembly. Proc Natl Acad Sci USA. 1983;80:3613–3617. doi: 10.1073/pnas.80.12.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcea R L, Ballmer-Hofer K, Benjamin T L. Virion assembly of polyomavirus hr-t mutants: underphosphorylation of the major capsid proteins VP1 before viral DNA encapsidation. J Virol. 1985;54:311–316. doi: 10.1128/jvi.54.2.311-316.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glenn G M, Eckhart W. Mutation of a cysteine residue in polyomavirus middle T antigen abolishes interactions with protein phosphatase 2A, pp60c-src, and phosphatidylinositol 3-kinase, activation of c-fos expression, and cellular transformation. J Virol. 1993;67:1945–5192. doi: 10.1128/jvi.67.4.1945-1952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glenn G M, Eckhart W J. Amino-terminal regions of polyomavirus middle T antigen are required for interactions with protein phosphate 2A. J Virol. 1995;69:3729–3736. doi: 10.1128/jvi.69.6.3729-3736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldman E, Hattori J, Benjamin T. Cellular and C-type viral factors in infections by polyoma virus hr-t mutants. Virology. 1979;95:373–384. doi: 10.1016/0042-6822(79)90492-6. [DOI] [PubMed] [Google Scholar]
- 25.Hattori J, Carmichael G G, Benjamin T L. DNA sequence alterations in hr-t deletion mutants of polyoma virus. Cell. 1979;13:505–513. doi: 10.1016/0092-8674(79)90025-4. [DOI] [PubMed] [Google Scholar]
- 26.Hyde-DeRuyscher R, Carmichael G G. Polyomavirus early-late switch is not regulated at the level of transcription initiation and is associated with changes in RNA processing. Proc Natl Acad Sci USA. 1988;85:8993–8997. doi: 10.1073/pnas.85.23.8993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jat P, Novak U, Cowie A, Tyndall C, Kamen R. DNA sequences required for specific and efficient initiation of transcription at the polyomavirus early promoter. Mol Cell Biol. 1982;2:737–751. doi: 10.1128/mcb.2.7.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kern F G, Dailey L, Basilico C. Common regulatory elements control gene expression from polyoma early and late promoters in cells transformed by chimeric plasmids. Mol Cell Biol. 1985;5:2070–2079. doi: 10.1128/mcb.5.8.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kern F G, Pellegrini S, Cowie A, Basilico C. Regulation of polyomavirus late promoter activity by viral early proteins. J Virol. 1986;60:275–285. doi: 10.1128/jvi.60.1.275-285.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerr L D, Holt J T, Matrisian L M. Growth factors regulate transin gene expression by c-fos-dependent and c-fos-independent pathways. Science. 1988;242:1424–1427. doi: 10.1126/science.2462278. [DOI] [PubMed] [Google Scholar]
- 31.Kowenz-Leutz E, Twamley G, Ansieau S, Leutz A. Novel mechanism of C/EBP beta (NF-M) transcriptional control: activation through derepression. Genes Dev. 1994;15:2781–2791. doi: 10.1101/gad.8.22.2781. [DOI] [PubMed] [Google Scholar]
- 32.Kumar M, Carmichael G G. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc Natl Acad Sci USA. 1997;15:3542–3547. doi: 10.1073/pnas.94.8.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lania L, Griffiths M, Cooke B, Ito Y, Fried M. Untransformed rat cells containing free and integrated DNA of a polyoma nontransforming (hr-t) mutant. Cell. 1979;18:793–802. doi: 10.1016/0092-8674(79)90132-6. [DOI] [PubMed] [Google Scholar]
- 34.Li M, Garcea R L. Identification of the threonine phosphorylation sites on the polyomavirus major capsid protein VP1: relationship to the activity of middle T antigen. J Virol. 1994;68:320–327. doi: 10.1128/jvi.68.1.320-327.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li M, Lyon M K, Garcea R L. In vitro phosphorylation of the polyomavirus major capsid protein VP1 on serine 66 by casein kinase II. J Biol Chem. 1995;270:26006–26011. doi: 10.1074/jbc.270.43.26006. [DOI] [PubMed] [Google Scholar]
- 36.Liu Z, Carmichael G G. Polyoma virus early-late switch: regulation of late RNA accumulation by DNA replication. Proc Natl Acad Sci USA. 1993;90:8494–8498. doi: 10.1073/pnas.90.18.8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manteuil S, Girard M. Inhibitors of DNA synthesis: their influence on replication and transcription of simian virus 40 DNA. Virology. 1974;60:438–454. doi: 10.1016/0042-6822(74)90338-9. [DOI] [PubMed] [Google Scholar]
- 38.Martin M E, Piette J, Yaniv M, Tang W J, Folk W R. Activation of the polyomavirus enhancer by a murine activator protein 1 (AP1) homolog and two contiguous proteins. Proc Natl Acad Sci USA. 1988;85:5838–5843. doi: 10.1073/pnas.85.16.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mueller C R, Mes-Masson A M, Bouvier M, Hassell J A. Location of sequences in polyomavirus DNA that are required for early gene expression in vivo and in vitro. Mol Cell Biol. 1984;4:2594–2609. doi: 10.1128/mcb.4.12.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mueller C R, Muller W J, Hassell J A. The polyomavirus enhancer comprises multiple functional elements. J Virol. 1988;62:1667–1678. doi: 10.1128/jvi.62.5.1667-1678.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muller W J, Dufort D, Hassell J A. Multiple subelements within the polyomavirus enhancer function synergistically to activate DNA replication. Mol Cell Biol. 1988;8:5000–5015. doi: 10.1128/mcb.8.11.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murakami Y, Satake M, Yamaguchi-Twai Y, Sakai M, Muramatsu M, Ito Y. The nuclear protooncogenes c-jun and c-fos as regulators of DNA replication. Proc Natl Acad Sci USA. 1991;88:3947–3951. doi: 10.1073/pnas.88.9.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaffhausen B S, Benjamin T L. Deficiency in histone acetylation in nontransforming host range mutants of polyoma virus. Proc Natl Acad Sci USA. 1976;73:1092–1096. doi: 10.1073/pnas.73.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schonthal A, Srinivas S, Eckhart W. Induction of c-jun proto-oncogene expression and transcription factor AP-1 activity by the polyoma virus middle-sized tumor antigen. Proc Natl Acad Sci USA. 1992;89:4972–4976. doi: 10.1073/pnas.89.11.4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srinivas S, Schonthal A, Eckhart W. Polyomavirus middle-sized tumor antigen modulates c-Jun phosphorylation and transcriptional activity. Proc Natl Acad Sci USA. 1994;91:10064–10068. doi: 10.1073/pnas.91.21.10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986;46:567–574. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- 47.Salzman N P, Natarajan V, Selzer G B. Transcription of SV40 and polyoma virus and its regulation. In: Salzman N P, editor. The viruses. New York, N.Y: Plenum Press, Inc; 1986. [Google Scholar]
- 48.Sheng Q, Denis D, Ratnofsky M, Roberts T M, DeCaprio J A, Schaffhausen B. The DnaJ domain of polyomavirus large T antigen is required to regulate Rb family tumor suppressor function. J Virol. 1997;71:9410–9416. doi: 10.1128/jvi.71.12.9410-9416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang WJ, Berger S L, Triezenberg S J, Folk W R. Nucleotides in the polyomavirus enhancer that control viral transcription and DNA replication. Mol Cell Biol. 1987;7:1681–1690. doi: 10.1128/mcb.7.5.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turler H, Salomon C. Small and middle T antigens contribute to lytic and abortive polyomavirus infection. J Virol. 1985;53:579–586. doi: 10.1128/jvi.53.2.579-586.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wasylyk C, Imler J L, Wasylyk B. Transforming but not immortalizing oncogenes activate the transcription factor PEA1. EMBO J. 1988;7:2475–2483. doi: 10.1002/j.1460-2075.1988.tb03094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wasylyk C, Flores P, Gutman A, Wasylyk B. PEA3 is a nuclear target for transcription activation by non-nuclear oncogenes. EMBO J. 1989;8:3371–3378. doi: 10.1002/j.1460-2075.1989.tb08500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wasylyk B, Wasylyk C, Flores P, Begue A, Leprince D, Stehelin D. The c-ets proto-oncogenes encode transcription factors that cooperate with c-fos and c-jun for transcriptional activation. Nature. 1990;315:191–193. doi: 10.1038/346191a0. [DOI] [PubMed] [Google Scholar]
- 54.Wiley S R, Kraus R J, Zuo F, Murray E E, Loritz K, Mertz J E. SV40 early-to-late switch involves titration of cellular transcriptional repressors. Genes Dev. 1993;7:2206–2219. doi: 10.1101/gad.7.11.2206. [DOI] [PubMed] [Google Scholar]
- 55.Yoo W, Martin M E, Folk W R. PEA1 and PEA3 enhancer elements are primary components of the polyomavirus late transcription initiator element. J Virol. 1991;65:5391–5400. doi: 10.1128/jvi.65.10.5391-5400.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zullo J, Stiles C D, Garcea R L. Regulation of c-myc and c-fos mRNA levels by polyomavirus: distinct roles for the capsid protein VP1 and the viral early proteins. Proc Natl Acad Sci USA. 1987;84:1210–1214. doi: 10.1073/pnas.84.5.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]