Abstract
Trypanosoma brucei is a protozoan parasite that causes African sleeping sickness. T. brucei multiplies extracellularly in the bloodstream, relying on antigenic variation of a dense variant surface glycoprotein (VSG) coat to escape antibody-mediated lysis. We investigated the role of VSG in proliferation and pathogenicity by using inducible RNA interference to ablate VSG transcript down to 1-2% normal levels. Inhibiting VSG synthesis in vitro triggers a rapid and specific cell cycle checkpoint blocking cell division. Parasites arrest at a discrete precytokinesis stage with two full-length flagella and opposing flagellar pockets, without undergoing additional rounds of S phase and mitosis. A subset (<10%) of the stalled cells have internal flagella, indicating that the progenitors of these cells were already committed to cytokinesis when VSG restriction was sensed. Although there was no obvious VSG depletion in vitro after 24-h induction of VSG RNA interference, there was rapid clearance of these cells in vivo. We propose that a stringent block in VSG synthesis produces stalled trypanosomes with a minimally compromised VSG coat, which can be targeted by the immune system. Our data indicate that VSG protein or transcript is monitored during cell cycle progression in bloodstream-form T. brucei and describes precise precytokinesis cell cycle arrest. This checkpoint before cell division provides a link between the protective VSG coat and cell cycle progression and could function as a novel parasite safety mechanism, preventing extensive dilution of the protective VSG coat in the absence of VSG synthesis.
Keywords: antigenic variation, Trypanosoma brucei
African trypanosomes, including Trypanosoma brucei, are unicellular parasites transmitted by tsetse flies in sub-Saharan Africa, which are responsible for debilitating disease in humans and livestock. African sleeping sickness is resurgent, with an estimated 300,000-500,000 cases a year, and it is invariably fatal if untreated (1). T. brucei multiplies extracellularly in the bloodstream, exposing it to immune attack. Each bloodstream-form cell is coated with ≈107 variant surface glycoprotein (VSG) molecules of a given antigenic type, making up ≈10% of total cellular protein (2, 3). Eventually the host mounts an antibody response against a given VSG variant. However, as parasites can switch between hundreds of antigenically diverse VSG coats during the course of a chronic infection, trypanosomes expressing a new VSG variant can escape the host antibody response against the old one (4, 5). This highly sophisticated strategy of antigenic variation makes African trypanosomes a paradigm for antigenic variation in general.
Although a great deal is known about mechanisms mediating VSG switching, we know relatively little about the role of VSG itself in other aspects of immune evasion and pathogenicity. To investigate this, we inhibited VSG synthesis by performing inducible RNA interference (RNAi) both in vitro and in vivo. Inhibition of VSG synthesis in vitro triggers a rapid and specific precytokinesis arrest with cells blocked from undergoing additional rounds of S phase and mitosis. Blocking cell division prevents further dilution of the VSG coat, which could function as a safety mechanism preserving its integrity. Although the VSG coat appears superficially intact after induction of VSG RNAi for 24 h in vitro, induction of VSG RNAi in vivo results in rapid clearance of T. brucei in mice. We postulate that reduction of VSG transcript down to 1-2% normal levels results in stalled trypanosomes with minimally compromised VSG coats, which are nonetheless cleared by the immune system of the mouse. The extreme sensitivity of trypanosomes to reduced VSG synthesis in vivo makes this a potential drug target for trypanosomiasis.
Materials and Methods
Trypanosome Strains and Culture. VSG RNAi was performed in T. brucei 427 90-13 expressing VSG221 (6). These trypanosomes were transfected with the MC177VSG221 RNAi construct containing an 803-bp VSG221 target fragment (GenBank accession no. X56762, positions 122-925) inserted between the opposing T7 promoters of construct p2T7Ti-177 (7). To ensure that the transformants did not switch away from the VSG221 expression site (ES), the 221GP1 construct containing GFP and puromycin (8) was inserted immediately behind the 221 ES promoter, resulting in the T. brucei 221VG1.1 and 221VG2.1 cell lines. Alternatively, a blasticidin resistance gene was inserted behind the VSG221 ES promoter, resulting in T. brucei 221VB1.1 or 221VB2.1. T. brucei was cultured in vitro in HMI-9 medium (8) or in vivo in CBA/CA mice (Harlan UK, Bicester, U.K.). All results presented were comparable between the T. brucei 221VG1.1 and 221VG2.1 and the T. brucei VB1.1 and VB2.1 cell lines.
T. brucei Microscopy. Cells for cell cycle analysis by microscopy were fixed at an end concentration of 2% paraformaldehyde, allowed to settle on glass slides, washed, and mounted in Vectashield containing 40 ng·ml-1 DAPI (Vector Laboratories). For cell cycle analysis, an average of 1,200 cells was counted per time point. Cells for immunofluorescence were fixed comparably, but then reacted with an anti-VSG221 rabbit polyclonal antibody (gift of P. Borst's laboratory) and an anti-rabbit secondary antibody coupled to Alexa 488 (Molecular Probes). Microscopic analysis was performed with a Zeiss Axioplan 2 microscope. Images were obtained with a Roper Scientific (Trenton, NJ) Cool Snap HQ camera and analyzed with metamorph 6.20 imaging software (Universal Imaging, Downingtown, PA).
Endocytosis was followed by incubating cells in 10 μg·ml-1 of FITC-tomato lectin. Endocytosis ceases at 4°C. To probe for its activation, cells were transferred to 37°C for 4 min to allow internalization of the FITC-tomato lectin (9). Endocytosis experiments were performed with the T. brucei 221VB1.1 or 221VB2.1 transformants, which stalled in an equivalent fashion to the T. brucei 221VG1.1 and 221VG2.1 transformants after induction of VSG221 RNAi. Cells were prepared for scanning electron microscopy or transmission electron microscopy essentially as described (10, 11) except that the scanning electron microscopic cells were fixed in the culture medium.
T. brucei Analysis. RNA analysis was performed according to ref. 8. The VSG221 probe was the fragment used for VSG RNAi. Lysates for protein analysis were made by washing cells and then resuspending them at 109 cells per ml-1 in cold lysis buffer [50 mM Hepes, pH 7.5/1.5 mM MgCl2/1 mM EDTA/10% glycerol/1% Triton X-100/one mini protease inhibitor mixture tablet (Roche Diagnostics) per 10-ml mix]. After lysis and microcentrifuge centrifugation, the supernatants were analyzed further. Western blotting was performed by using standard protocols and antibodies against VSG221 (gift of P. Borst) or BiP (gift of Jay Bangs, University of Wisconsin, Madison). Flow cytometry was performed according to ref. 8. Cell volume measurements were carried out by using the Coulter size exclusion method on a CASY cell counter model TT (Schärfe, Reutlingen, Germany) according to the manufacturer's instructions.
Results and Discussion
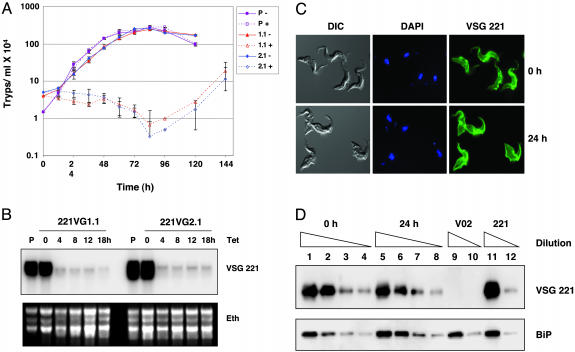
RNAi-Mediated Ablation of VSG Transcript Results in Rapid Growth Arrest in Vitro. To address the functional role of VSG, we specifically inhibited VSG synthesis by performing RNAi against the VSG expressed by our variant of the T. brucei 427 strain (VSG221) by using a tetracycline-inducible construct containing a fragment of VSG221 (6, 12). Transformants were obtained only by using an experimental strategy optimized for extremely low transcriptional read-through (7). Surprisingly, induction of VSG221 RNAi in vitro led to a very rapid inhibition of cell growth within one cell cycle (8 h) (Fig. 1A). This finding indicates that VSG has an essential function even in vitro in the absence of an immune system. VSG mRNA is one of the most abundant transcripts in T. brucei, yet after only 4-h induction of VSG RNAi it was reduced to 1-2% of normal levels (Fig. 1B). After tetracycline induction we did not detect dsRNA corresponding to VSG221, presumably because of very efficient processing by the RNAi machinery. After initial growth arrest, cells eventually grew out after ≈84 h. Analysis of 50 RNAi “escape” clones isolated in the absence of drug selection pressure on the active ES showed that 49 of 50 had switched to different VSG coats not recognized by the VSG221 RNAi rather than had inactivated the RNAi machinery (results not shown). As expected, if drug selection pressure on the active ES was maintained during induction of VSG RNAi, VSG switch variants were generated that had exclusively switched via DNA rearrangements (results not shown). These experiments argue for specificity of the VSG221 RNAi, as cells that express a new VSG escape the growth arrest. RNAi against a GFP gene inserted into the active VSG221 ES did not lead to growth defects (result not shown). Similarly, RNAi-mediated ablation of the different ESAG transcripts encoded within the VSG221 ES did not lead to a comparable growth arrest (result not shown), again arguing that the precytokinesis arrest that we observe is specific to VSG and that we are not merely disrupting a polycistronic transcription unit.
Fig. 1.
Growth arrest after induction of VSG221 RNAi. (A) Growth curves of the T. brucei 90-13 parental line (P) or VSG221 RNAi cell lines 221VG1.1 (1.1) and 221VG2.1 (2.1) in the presence (+) or absence (-) of tetracycline to induce VSG RNAi. (B) Rapid ablation of VSG221 mRNA after VSG RNAi induction. RNA was isolated from the T. brucei 90-13 parental (P) line not containing the VSG221 RNAi construct or the 221VG1.1 or 221VG2.1 lines after tetracycline induction of VSG RNAi for the times indicated. The Northern blot was hybridized with a VSG221 probe, with an ethidium (Eth) stain of the gel indicated below. (C) Cells stalled after induction of VSG221 RNAi do not have an obviously depleted VSG coat. VSG RNAi was induced with tetracycline in T. brucei 221VB1.1 for 0 or 24 h. (Left) Cells were visualized by using differential interference contrast (DIC). (Center) DNA was stained with DAPI. (Right) The VSG221 coat was recognized by an anti-VSG221 primary antibody and a FITC-labeled secondary antibody. (D) Western blot analysis does not show significant depletion of the VSG221 coat in T. brucei 221VG1.1 cells after induction of VSG221 RNAi for 24 h. Protein lysate from T. brucei 221VG1.1 after VSG RNAi was induced for 0 or 24 h was analyzed from an equivalent of 1 × 105 cells (lanes 1 and 5) or diluted sequentially to the equivalent of 3.3 × 104 cells (lanes 2 and 6), 1.1 × 104 cells (lanes 3 and 7), or 3.7 × 103 cells (lanes 4 and 8). These lysates are compared with protein lysates from T. brucei expressing VSGV02 [T. brucei HNI(V02) (31)] or VSG221 (uninduced T. brucei 221VB1.1). Protein lysate was analyzed from the equivalent of 1 × 105 cells (lanes 9 and 11) or 1.1 × 104 cells (lanes 10 and 12). An antibody against BiP was used as a loading control (13).
Despite the radical reduction in VSG transcript after VSG RNAi, cells remained VSG221-positive with no observable decrease of VSG on the cell surface as monitored by immunofluorescence microscopy (Fig. 1C) or flow cytometric analysis (results not shown) with anti-VSG221 antibodies. In addition, there was no significant reduction in total VSG levels within 24 h of arrest as assayed by Western blot using dilutions of protein lysates and BiP as a loading control (Fig. 1D) (13). VSG has an extremely long half-life (33 ± 9 h) (14) and shows rapid lateral diffusion within the coat (15), presumably explaining why cells with reduced VSG synthesis remain viable for extensive periods in vitro. In some respects this longevity mimics stumpy-form trypanosomes, which are a nonreplicating form induced at high cell densities in a normal bloodstream infection and are able to survive for days despite having halted VSG synthesis (16, 17).
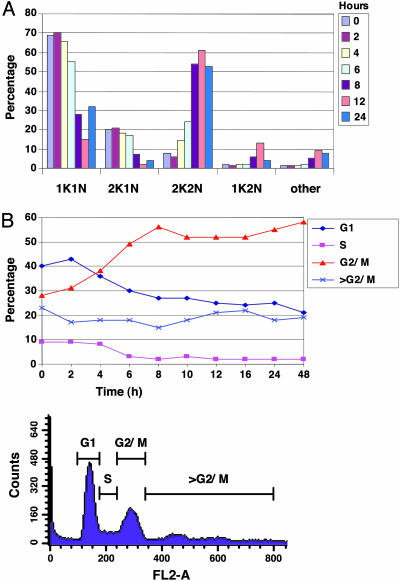
Cells Stalled After Inhibition of VSG Synthesis Arrest at a Precise Precytokinesis Stage. Given the very fast and effective growth arrest after the induction of VSG RNAi, we asked whether individual cells exhibited specific cell cycle phenotypes. DNA staining with DAPI allows analysis of T. brucei cell cycle stages. T. brucei has one nucleus (N) and kinetoplast (K) (mitochondrial genome), and post-S phase, the kinetoplast divides before the nucleus (18). After induction of VSG221 RNAi in vitro, cells rapidly accumulate at a 2K2N precytokinesis stage (Fig. 2A). This finding indicates that inhibiting VSG synthesis triggers a specific cell cycle checkpoint. Cell cycle arrest is highly effective, with no significant accumulation of multinucleated cells characteristic of successive rounds of S phase and mitosis. Flow cytometric analysis of propidium iodide-stained cells stalled after VSG RNAi indicated that these cells had not initiated additional rounds of S phase after induction of the precytokinesis cell cycle arrest (Fig. 2B). These results present an example of a precise precytokinesis arrest in trypanosomes. Such cell cycle arrest could operate to allow time for correction for fluctuations in VSG synthesis.
Fig. 2.
Cells stall precytokinesis after the induction of VSG RNAi, but do not undergo reinitiation of S phase. (A) Induction of VSG221 RNAi results in T. brucei stalled before cytokinesis. T. brucei 221VG1.1 was induced with tetracycline for the times indicated. To determine in which stage of the cell cycle cells were stalled, cells were stained with DAPI to visualize the DNA. Cells containing the indicated number of nuclei (N) or kinetoplasts (K) are shown as a percentage of total. Comparable results were obtained with the T. brucei 221VG2.1 cell line. (B) Cells stalled precytokinesis after the induction of VSG221 RNAi do not undergo additional rounds of DNA replication. (Upper) T. brucei 221VB1.1 was induced with tetracycline for the times indicated. DNA was stained with propidium iodide and analyzed by flow cytometry according to ref. 8. Cells in a particular stage of the cell cycle (G1, S, G2/M, or >G2/M) are indicated as a percentage of total. (Lower) The gates used for quantitation of these different stages are shown.
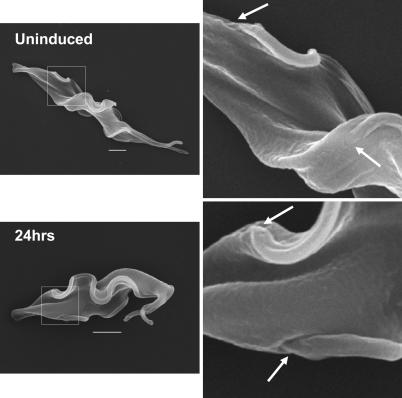
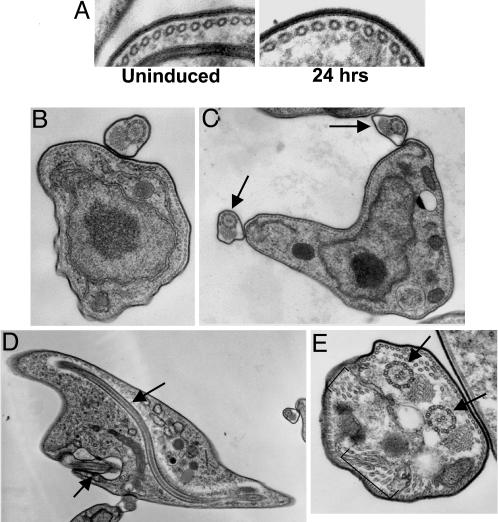
The trypanosome builds a new flagellum and flagellar pocket in its cell cycle, and flagella are instrumental in kinetoplast segregation. We used scanning electron microscopy to compare typical precytokinesis trypanosome stages in cultures that were uninduced or induced for VSG RNAi for 24 h (Fig. 3). Cells arrested after the induction of VSG RNAi had two full-length flagella, each with its own flagellar pocket at the base of the flagellum, but lacked a deep cleavage furrow (Fig. 3). In contrast to normal precytokinesis stages, cells stalled after VSG RNAi were characteristically shorter and broader (Fig. 3 and Fig. 7A, which is published as supporting information on the PNAS web site). The average volume of uninduced T. brucei VSG221 cells is ≈24 μm3 as measured by a Coulter counter, and this volume doubles in cultures induced for VSG RNAi for 12 h (Fig. 8, which is published as supporting information on the PNAS web site). As precytokinesis trypanosomes are expected to have double the volume of cells present in the modal peak of an uninduced culture, cells stalled after VSG RNAi are equivalent in volume to normal precytokinesis stages. The shorter and broader morphology of cells stalled after VSG RNAi could be a consequence of the trypanosomes attempting to minimize the surface area-to-volume ratio in the face of restricted VSG.
Fig. 3.
Scanning electron microscopy of trypanosomes stalled after VSG RNAi. Cells with two full-length flagella predominate in the T. brucei 221VG1.1 population after induction of VSG RNAi for 24 h. The boxed areas are expanded on the right. Arrows highlight the positioning of the flagellar pockets. (Scale bars: 2 μm.)
A typical feature of trypanosome-arrested precytokinesis by VSG RNAi was the reduced distance between the two kinetoplasts and nuclei (Fig. 7 B and C). This distance is characteristic of the bloodstream-form T. brucei “phase 5” stage of the cell cycle immediately before cell division (19). Trypanosomes stalled precytokinesis by VSG RNAi typically had parallel flagella and opposed flagellar pockets (Fig. 3), again suggesting arrest at a precise precytokinesis point. This configuration could also be found in the uninduced population, but very infrequently, indicating that VSG RNAi has resulted in a cell cycle block, resulting in the accumulation of a normally transient stage in the cell cycle.
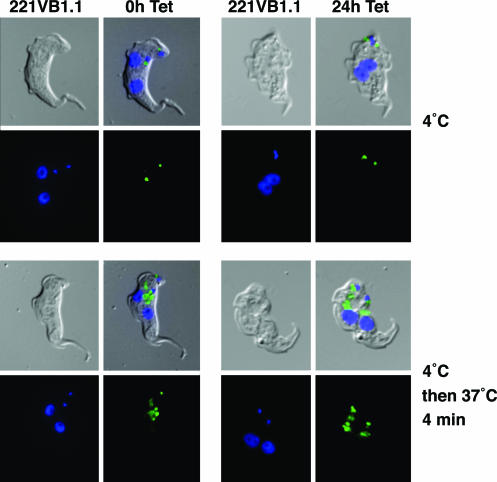
Endocytosis Is Unimpaired in Arrested Cells. Bloodstream-form T. brucei has one of the highest rates of endocytosis measured, with the entire pool of surface VSG recycled every 12 min (20, 21). This very high rate of endocytosis, which protects the trypanosome by removing anti-VSG antibody complexes, is restricted to the flagellar pocket (21). We therefore asked whether both flagellar pockets were functional for endocytosis in these trypanosomes arrested by VSG221 RNAi. FITC-tomato lectin binds flagellar pocket glycoproteins and can be used as a probe for the endocytic pathway (9). Endocytosis stops at 4°C (22), and incubation of stalled T. brucei cells with FITC-tomato lectin at 4°C labeled the two flagellar pockets, with a slightly smaller signal in the new flagellar pocket as in the controls (Fig. 4). Endocytosis into the cell reactivates upon subsequent incubation at 37°C (22), resulting in tracks of lectin-labeled vesicles extending from the flagellar pockets, again as in controls. Therefore, both flagellar pockets can be functionally active in these arrested cells.
Fig. 4.
VSG221 RNAi results in stalled trypanosomes that can undergo endocytosis from two functional flagellar pockets. (Left) T. brucei 221VB1.1 was incubated with FITC-tomato lectin at 4°C or a subsequent incubation at 37°C to activate endocytosis. (Right) T. brucei 221VB1.1 was first induced with tetracyline for 24 h before the FITC-tomato lectin incubation. Trypanosomes are shown as differential interference contrast images, with FITC-tomato lectin in green and DNA in blue (DAPI stain). (Magnification: ×1,000.)
VSG RNAi Results in Cells with Internal Flagella. The VSG coat was originally defined by electron microscopy as an electron-dense layer coating bloodstream-form T. brucei (23). We confirmed the apparent integrity of the VSG coat on the many stalled precytokinesis cells in the VSG RNAi-induced population by using this approach. After 24 h of VSG RNAi induction, the VSG coat on cross sections of stalled cells looked comparable to controls (Fig. 5A). We further investigated the subcellular architecture of the VSG RNAi cells by using electron microscopy. The majority of noninduced T. brucei had one external flagellum (Fig. 5B). After induction of VSG RNAi, cells accumulated with two external flagella (indicated by arrows in Fig. 5C). Further, we saw no disruption of the subpellicular microtubule corset after 24 h of VSG RNAi, and both flagella were associated with the normal cytoplasmic structures of the flagellum attachment zone (Fig. 5C). We did not detect systematic changes in endomembrane organization. In a subset (<10%) of the VSG RNAi-induced population, trypanosomes were seen with one external and one internal flagellar axoneme and associated paraflagellar rod (Fig. 5D). We propose that VSG RNAi is so effective that the majority of the cells implement the precytokinesis checkpoint within one cell cycle (Fig. 9A, which is published as supporting information on the PNAS web site). However, cells committed to cytokinesis upon induction are likely to divide before the RNAi is fully effective (Fig. 9B). After cell division, these trypanosomes will be experiencing a major reduction in VSG coat synthesis some 4 h later when they initiate new flagellum formation. We suggest that their new basal body fails to make a productive connection with the cell surface, or there is insufficient VSG-coated surface membrane, and hence the new flagellum is extended internally in the cytoplasm (Fig. 9B). This phenomenon reveals that although surface membrane/coat and subpellicular microtubule cytoskeleton construction is normally coordinated, there is no dependency relationship between the two. Cytoskeletal assembly can sometimes continue extensively in the face of this VSG restriction, such that occasionally at later stages cells exhibit cytoplasmically located microtubule bundles along with multiple internal flagella (Fig. 5E).
Fig. 5.
Transmission electron microscopy of cells with two external flagella predominating after VSG221 RNAi induction. (A) No evidence was seen for a compromised VSG coat after 24-h induction of VSG221 RNAi. (B and C) Cross sections of T. brucei noninduced (B) or after VSG RNAi induction for 24 h (external flagella indicated with arrows) (C). (D) After induction of VSG RNAi, an internal nonmembrane bound flagellum is observed in a subset of the cells (flagella indicated with arrows). (E) Cells with internal flagella (arrows) and misplaced subpellicular microtubules (brackets) after 60-h VSG221 RNAi. (Magnifications: ×63,000 (A), ×13,000 (B), ×10,000 (C), ×4,000 (D), and ×16,000 (E).
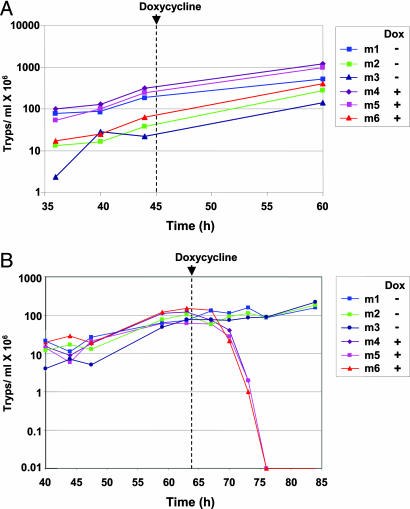
VSG Is Essential in Vivo. Finally, we addressed the role of the VSG coat in pathogenicity by establishing infections in mice (Fig. 6). Growth of the parental T. brucei 90-13 strain lacking the VSG221 RNAi construct was not affected by addition of the tetracycline analogue doxycycline to the drinking water (Fig. 6A) (24). We next infected six mice with T. brucei 221VG1.1 and induced VSG221 RNAi by using doxycycline in half of the mice 64 h after inoculation. Although there was no significant reduction of the VSG221 coat detectable in vitro after VSG RNAi, induction of VSG RNAi in vivo resulted in rapid clearance of the infection within 12 h. This finding argues that even minor, and experimentally not easily detectable, damaging of the VSG coat is fatal for the parasite, and that a fully uncompromised VSG coat is essential for successful infection. We postulate that in the absence of VSG synthesis, even in the presence of a cell cycle arrest, chinks form in the protective layer that can be exploited by an immunocompetent mouse. These results were comparable with different T. brucei VSG221 RNAi transformants and mouse strains (results not shown).
Fig. 6.
Clearance of T. brucei in mice after induction of VSG221 RNAi. Mice were infected with either the parental T. brucei 90-13 cell line not containing the VSG221 RNAi construct (A) or with T. brucei 221VG1.1 (B), and densities were monitored at the times indicated. Doxycycline (Dox) was added to the water bottles of mice 4-6 (m4, m5, and m6) at the times indicated by an arrow. This procedure resulted in induction of VSG RNAi in the T. brucei 221VG1.1 cell line.
The rapid clearance of parasites must be caused by recognition by the immune system or a mouse lytic factor, rather than a consequence of the cell growth arrest. The stumpy forms of T. brucei are naturally occurring nondividing forms that are arrested in their cell cycle during an infection. However, stumpy-form trypanosomes possess an intact VSG coat, and although nonproliferative, have a longevity between 48 and 72 h (25). Therefore, cell cycle arrest on its own would not lead to the dramatic clearance kinetics that we observe. Stumpy-form T. brucei are fundamentally different from the slender bloodstream form and have been shown to be more resistant to various acidic and proteolytic stresses (26). We cannot exclude that this arrested lifecycle stage is therefore more resistant to mouse lytic factors than our stalled slender-form T. brucei cells.
The kinetics of clearance were not affected if VSG RNAi was induced at the earlier time point of 40 h rather than >60 h after infection (result not shown). As antibody titers against VSG221 would be expected to increase during the course of the infection, the unchanged kinetics of clearance indicate that another component of the mouse immune response instead of antibody-mediated lysis is involved in clearance. One possibility is that there is increased complement deposition on the surface of trypanosomes stalled after the induction of VSG RNAi, which could result in increased clearance of these trypanosomes by macrophages. This scenario, however, still remains to be tested. At the moment we cannot exclude that the trypanosomes stalled by VSG RNAi are cleared by a mouse lytic factor that is not part of the complement system.
Cell Cycle Arrest in T. brucei. Why are the trypanosomes that are stalled after VSG RNAi cleared in vivo despite the presence of a cell cycle checkpoint preventing significant depletion of the VSG coat? Inhibiting VSG synthesis triggers a cell cycle checkpoint, which could have evolved to absorb less drastic fluctuations in VSG synthesis than are occurring in our VSG RNAi experiments. In our experiments VSG RNAi resulted in an ablation of VSG transcript to <1-2% normal levels, presumably more disruption than can be absorbed without small but lethal chinks appearing in the coat that can be exploited by the immune system of the mouse.
Tsetse-form trypanosomes lacking the major surface molecule procyclin are viable and proliferate in vitro (27). In contrast, we show here that bloodstream-form T. brucei is acutely sensitive of the amount of its major surface molecule VSG, or of the VSG mRNA itself, as ablation of VSG transcript results in a rapid and specific precytokinesis block. Earlier studies have shown that disruption of the mitotic cyclin gene CYC6 in bloodstream-form T. brucei resulted in a cell cycle arrest before cell division, but this arrest was not precise, as cells reinitiated S phase and accumulated multiple kinetoplast DNA (28). Similarly, disruption of GPI8 encoding the catalytic subunit of a complex that adds glycosylphosphatidylinositol (GPI) anchors onto nascent polypeptides resulted in growth-arrested cells that had undergone repeated rounds of S phase and mitosis, resulting in the accumulation of “monster” cells with multiple nuclei, kinetoplasts, and flagella (29). This GPI8 disruption would prevent all GPI-anchored proteins (including VSG) from reaching the cell surface. In contrast, we find that our cells arrest at a precise stage of precytokinesis without undergoing further rounds of DNA replication. The difference between the VSG RNAi phenotype and those presented for the GPI8 knockout argues for the presence of a specific cytokinesis checkpoint sensing VSG synthesis rather than VSG addition to the surface.
It is intriguing that the correlation between cessation of VSG synthesis and cell cycle arrest revealed in our VSG RNAi cells is also seen in the nonreplicating stumpy-form parasites characteristic of a natural infection. In both cases, progression through the cell cycle could be linked to monitoring of VSG synthesis. This precytokinesis checkpoint has many hallmarks of the Cdc 14/Flp1/SIN checkpoint in yeast and other eukaryotes (30). Although we have not excluded that it is the VSG transcript that is being “sensed,” we find it more likely that this precytokinesis checkpoint is triggered in the absence of VSG production, which would otherwise eventually lead to a disastrous dilution of the protective VSG coat. This process would provide a safety mechanism protecting the integrity of the VSG coat, which we show is vital for survival and pathogenicity in the mammalian host.
Supplementary Material
Acknowledgments
We thank Jay Bangs and Kevin Tyler for discussions; Bill Wickstead, Michael Ginger, Niall Aitcheson, Stephen Terry, and Suzanne Talbot for comments on the manuscript; Catarina Gadelha for help with data analysis; Peter Warne for experimental assistance; George Cross (The Rockefeller University, New York) and Mark Field (University of Cambridge, Cambridge, U.K.) for the T. brucei 90-13 cell line; Bill Wickstead (University of Oxford) for the MC177p2T7Ti RNAi construct; and Jay Bangs for generous amounts of antibodies. G.R. is a Wellcome Senior Fellow in the Basic Biomedical Sciences. K.G. is a Wellcome Principal Research Fellow. This work was funded by the Wellcome Trust.
Author contributions: K.G. and G.R. designed research; K.S., S.V., J.M., K.H., and G.R. performed research; K.S., S.V., J.M., K.H., K.G., and G.R. analyzed data; and K.G. and G.R. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: VSG, variant surface glycoprotein; RNAi, RNA interference; ES, expression site.
References
- 1.World Health Organization (2001) African Trypanosomiasis or Sleeping Sickness, Fact Sheet 259 (W.H.O., Geneva).
- 2.Cross, G. A. (1975) Parasitology 71, 393-417. [DOI] [PubMed] [Google Scholar]
- 3.Bohme, U. & Cross, G. A. (2002) J. Cell Sci. 115, 805-816. [DOI] [PubMed] [Google Scholar]
- 4.Barry, J. D. & McCulloch, R. (2001) Adv. Parasitol. 49, 1-70. [DOI] [PubMed] [Google Scholar]
- 5.Pays, E., Vanhamme, L. & Perez-Morga, D. (2004) Curr. Opin. Microbiol. 7, 369-374. [DOI] [PubMed] [Google Scholar]
- 6.Wirtz, E., Leal, S., Ochatt, C. & Cross, G. A. (1999) Mol. Biochem. Parasitol. 99, 89-101. [DOI] [PubMed] [Google Scholar]
- 7.Wickstead, B., Ersfeld, K. & Gull, K. (2002) Mol. Biochem. Parasitol. 125, 211-216. [DOI] [PubMed] [Google Scholar]
- 8.Sheader, K., te Vruchte, D. & Rudenko, G. (2004) J. Biol. Chem. 279, 13363-13374. [DOI] [PubMed] [Google Scholar]
- 9.Nolan, D. P., Geuskens, M. & Pays, E. (1999) Curr. Biol. 9, 1169-1172. [DOI] [PubMed] [Google Scholar]
- 10.Sherwin, T. & Gull, K. (1989) Philos. Trans. R. Soc. London B 323, 573-588. [DOI] [PubMed] [Google Scholar]
- 11.Moreira-Leite, F. F., Sherwin, T., Kohl, L. & Gull, K. (2001) Science 294, 610-612. [DOI] [PubMed] [Google Scholar]
- 12.LaCount, D. J., Barrett, B. & Donelson, J. E. (2002) J. Biol. Chem. 277, 17580-17588. [DOI] [PubMed] [Google Scholar]
- 13.Bangs, J. D., Uyetake, L., Brickman, M. J., Balber, A. E. & Boothroyd, J. C. (1993) J. Cell Sci. 105, 1101-1113. [DOI] [PubMed] [Google Scholar]
- 14.Seyfang, A., Mecke, D. & Duszenko, M. (1990) J. Protozool. 37, 546-552. [DOI] [PubMed] [Google Scholar]
- 15.Bulow, R., Overath, P. & Davoust, J. (1988) Biochemistry 27, 2384-2388. [DOI] [PubMed] [Google Scholar]
- 16.Amiguet-Vercher, A., Perez-Morga, D., Pays, A., Poelvoorde, P., Van Xong, H., Tebabi, P., Vanhamme, L. & Pays, E. (2004) Mol. Microbiol. 51, 1577-1588. [DOI] [PubMed] [Google Scholar]
- 17.Vasella, E., Reuner, B., Yutzy, B. & Boshart, M. (1997) J. Cell Sci. 110, 2661-2671. [DOI] [PubMed] [Google Scholar]
- 18.Woodward, R. & Gull, K. (1990) J. Cell Sci. 95, 49-57. [DOI] [PubMed] [Google Scholar]
- 19.Tyler, K. M., Matthews, K. R. & Gull, K. (2001) Protist 152, 367-378. [DOI] [PubMed] [Google Scholar]
- 20.Engstler, M., Thilo, L., Weise, F., Grunfelder, C. G., Schwarz, H., Boshart, M. & Overath, P. (2004) J. Cell Sci. 117, 1105-1115. [DOI] [PubMed] [Google Scholar]
- 21.Overath, P. & Engstler, M. (2004) Mol. Microbiol. 53, 735-744. [DOI] [PubMed] [Google Scholar]
- 22.Brickman, M. J., Cook, J. M. & Balber, A. E. (1995) J. Cell Sci. 108, 3611-3621. [DOI] [PubMed] [Google Scholar]
- 23.Vickerman, K. (1978) Nature 273, 613-617. [DOI] [PubMed] [Google Scholar]
- 24.Krieger, S., Schwarz, W., Ariyanayagam, M. R., Fairlamb, A. H., Krauth-Siegel, R. L. & Clayton, C. (2000) Mol. Microbiol. 35, 542-552. [DOI] [PubMed] [Google Scholar]
- 25.Turner, C. M. R., Aslam, N. & Dye, C. (1995) Parasitology 111, 289-300. [DOI] [PubMed] [Google Scholar]
- 26.Nolan, D. P., Rolin, S., Rodriguez, J. R., Van Den Abbeele, J. & Pays, E. (2000) Eur. J. Biochem. 267, 18-27. [DOI] [PubMed] [Google Scholar]
- 27.Vasella, E., Butikofer, P., Engstler, M., Jelk, J. & Roditi, I. (2003) Mol. Biol. Cell 14, 1308-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammarton, T. C., Clark, J., Douglas, F., Boshart, M. & Mottram, J. C. (2003) J. Biol. Chem. 278, 22877-22886. [DOI] [PubMed] [Google Scholar]
- 29.Lillico, S., Field, M. C., Blundell, P., Coombs, G. H. & Mottram, J. C. (2003) Mol. Biol. Cell 14, 1182-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simanis, V. (2003) J. Cell Sci. 116, 4263-4275. [DOI] [PubMed] [Google Scholar]
- 31.Rudenko, G., Chaves, I., Dirks-Mulder, A. & Borst, P. (1998) Mol. Biochem. Parasitol. 95, 97-109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.