Abstract
Dysfunctional mitochondria producing excessive ROS are the main factors that cause ovarian aging. Immp2l deficiency causes mitochondrial dysfunction and excessive ROS production, leading to ovarian aging, which is attributed to granulosa cell senescence. The pathway controlling mitochondrial proteostasis and mitochondrial homeostasis of the UPRmt and mitophagy are closely related with the ROS and cell senescence. Our results suggest that Immp2l knockout led to granulosa cell senescence, and enocyanin treatment alleviated Immp2l deficiency-induced granulosa cell senescence, which was accompanied by improvements in mitochondrial function and reduced ROS levels. Interestingly, redox-related protein modifications, including S-glutathionylation and S-nitrosylation, were markedly increased in Immp2l-knockout granulosa cells, and were markedly reduced by enocyanin treatment. Furthermore, STAT1 was significantly increased in Immp2l-knockout granulosa cells and reduced by enocyanin treatment. The co-IP results suggest that the expression of STAT1 was controlled by S-glutathionylation and S-nitrosylation, but not phosphorylation. The UPRmt was impaired in Immp2l-deficient granulosa cells, and unfolded and misfolded proteins aggregated in mitochondria. Then, the HIF1α/BNIP3-mediated mitophagy pathway was activated, but mitophagy was impaired due to the reduced fusion of mitophagosomes and lysosomes. The excessive aggregation of mitochondria increased ROS production, leading to senescence. Hence, Enocyanin treatment alleviated granulosa cell senescence through STAT1/ATF4-mediated UPRmt and STAT1/(ATF4)/HIF1α/BNIP3-mediated mitophagy.
Keywords: cell senescence, STAT1, UPRmt, mitophagy, enocyanin, HIF1α/BNIP3 pathway
1. Introduction
Female fertility is reduced or impaired by accelerated ovarian aging, and mitochondrial dysfunction is one of the hallmarks of aging. Mitochondrial dysfunction drives ovarian aging [1].
Mitochondrial dysfunction can induce cell senescence due to excessive ROS (Reactive Oxygen Species) production [2], and mitochondrial proteostasis and mitochondrial homeostasis are controlled by the mitochondrial quality control (MQC) system, which includes the mitochondrial unfolded protein response (UPRmt) and mitophagy. Mitophagy helps remove partially damaged mitochondria to ensure that mitochondria are healthy and functional, and the overactivation or inhibition of the UPRmt can even accelerate abnormal energy metabolism and mitochondrial dysfunction-induced cell senescence. Thus, an imbalance in MQC can accelerate cellular senescence and aging, and appropriate interventions in MQC can delay the aging process and extend the lifespan [3].
Signal transducer and activator of transcription 1 (STAT1) is a nuclear transcription factor that regulates genes involved in the cell cycle, cell survival and the immune response. Despite being activated downstream of common cytokines and growth factors, STAT1 plays antagonistic roles, and a disruption of this balance or phosphorylation levels shifts cells from survival to apoptotic cell death or from inflammatory to anti-inflammatory responses [4]. STAT proteins are latent in the cytoplasm of resting cells as monomers or unphosphorylated dimers until they are activated by growth factors and cytokines that bind to a specific cell receptor [4]. Aside from phosphorylation, STAT activity can be regulated by other posttranslational modifications, such as ubiquitination, acetylation, methylation, S-glutathionylation and ISGylation [4]; in particular, redox regulates STAT1, and redox-related protein posttranslational modifications, including S-glutathionylation [5] and S-nitrosylation [6], are closely related to posttranslational modifications of STAT1 [7]. In addition, STATs proteins, including STAT1, have been proven to localize in the mitochondria, and a deficiency of STAT1 protein in mitochondria is closely related with mitochondrial dysfunction, such as an imbalance of mitochondrial biogenesis and mitochondrial homeostasis. However, the STAT1 protein involved in maintaining the mitochondrial function is largely unknown, and whether UPRmt and mitophagy are regulated by STAT1 is unclear.
Our previous study suggested that ovarian aging in mice was caused by a deficiency in inner mitochondrial membrane peptidase 2-like (Immp2l), which is associated with the accelerated production of excessive ROS, then leading to granulosa cell senescence [8]. Whether the STAT1 protein was regulated by ROS-related posttranslational modifications, including S-glutathionylation and S-nitrosylation, and whether UPRmt and mitophagy regulated by STAT1 in Immp2l deficiency induced granulose cell senescence, is unknown.
Anthocyanidins, which are a class of water-soluble flavonoids, have been reported to possess several biological activities, including antioxidative, anti-inflammatory, and anticancer activities [9], and anthocyanidins can be extracted from different plant sources, such as fruits, flowers and vegetables; dietary sources of anthocyanidins include red and purple berries, grapes, apples, plums and cabbage [10]. Several studies have suggested that anthocyanidins play important roles in human health and that anthocyanidins are known for their therapeutic effects on many human diseases, such as cardiovascular and neurodegenerative diseases [10], obesity-induced inflammation [11], tension glaucoma [12], cancer [13], aging and cell senescence [14,15]. Enocyanin is a novel identified anthocyanidins, and it is extracted from grapes, but the role of enocyanin is largely unknown, especially in STAT1-regulated UPRmt, and that of mitophagy in Immp2l deficiency-induced granulose cell senescence is unclear.
Therefore, in this work, the role of STAT1-mediated UPRmt and mitophagy, as well as the role of enocyanin in Immp2l deficiency-induced granulosa cell senescence, were investigated. This study provides novel insights into and evidence of the role of STAT1 in maintaining mitochondrial function through UPRmt and mitophagy, as well as that of enocyanin in cell senescence, and reveals a novel target for alleviating mitochondrial dysfunction-induced granulosa cell senescence and ovarian aging.
2. Results
2.1. Impaired UPRmt and Mitophagy Caused by Immp2l Knockdown Induced Primary Granulosa Cell Senescence and Mitigated by Enocyanin
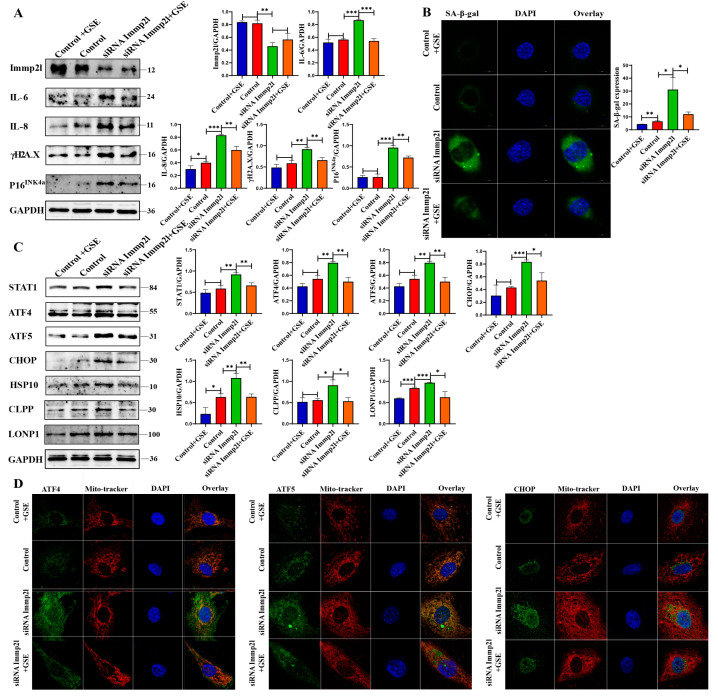
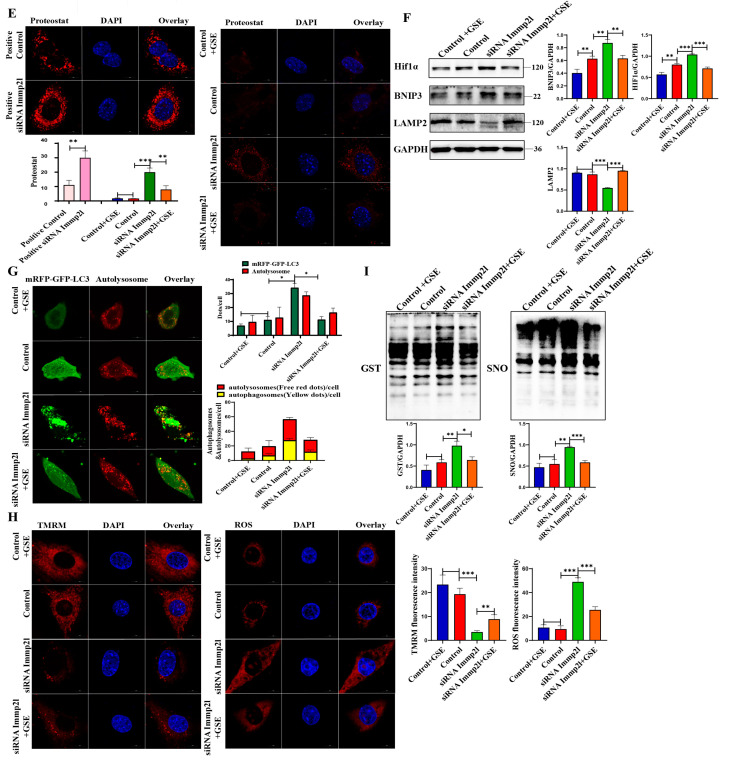
Immp2l-knockout mice were born, and limited granulosa cells were isolated from them in order to confirm the role of enocyanin in anti-aging. Primary granulosa cells were treated with enocyanin in vitro, and the results suggest that senescence was alleviated in enocyanin-treated Immp2l knockdown granulosa cells, which was accompanied by a significant reduction in the senescence markers IL-6, IL-8, γH2AX and P16INK4a (Figure 1A), which results were further supported by the staining of senescence-associated β-galactosidase (Figure 1B). In addition, the UPRmt and mitophagy were restored in enocyanin-treated Immp2l knockdown granulosa cells, which was accompanied by significant decreases in the STAT1 and UPRmt markers ATF4, ATF5, CHOP, HSP60, HSP10, CLPP and LONP1 (Figure 1C) and the mitophagy markers HIF1α and BNIP3, as well as significant increases in lysosome marker Lamp2 (Figure 1F). These results are further supported by results from the colocalization of ATF4, ATF5, CHOP and Mito-tracker (Figure 1D), as well as results from the detection of protein aggresomes (Figure 1E), autophagic flux (Figure 1G) and mitochondrial function (mitROS levels and MMP levels) (Figure 1H). Additionally, S-glutathionylation and S-nitrosylation were significantly decreased in enocyanin-treated Immp2l-knockdown granulosa cells (Figure 1I). Therefore, these results suggest that the impairment of UPRmt and mitophagy is closely related with the ROS-induced primary granulosa cell senescence in Immp2l-knockout primary granulosa cells, and improve UPRmt and mitophagy by enocyanin alleviated ROS level.
Figure 1.
Enocyanin delays Immp2l knockdown-induced primary granulosa cell senescence. (A) Western blot analysis of cell senescence marker molecules in Immp2l-knockdown granulosa cells treated with enocyanin. (B) Staining of senescence-associated β-galactosidase in Immp2l-knockdown granulosa cells treated with enocyanin, at least 50 different cells in different fields were analyzed, and the dots were blindly counted by three different individuals. (C) Western blot analysis of cell STAT1 and UPRmt molecules in Immp2l knockdown granulosa cells treated with enocyanin. (D) Detection of the colocalization of UPRmt molecules ATF4, ATF5, CHOP and Mito-tracker. (E) Detection of protein aggresomes with the PROTEOSTAT Aggresome Detection Kit in Immp2l-knockdown granulosa cells treated with enocyanin. (F) Western blot analysis of mitophagy molecules and lysosome membrane protein Lamp2 in Immp2l-knockdown granulosa cells treated with enocyanin. (G) Detection of autophagic flux with the mCherry-GFP-LC3 lentivirus in Immp2l-knockdown granulosa cells treated with enocyanin; at least 50 different cells in different fields were analyzed, and the dots were blindly counted by three different individuals. (H) Detection of mitROS and mitochondrial membrane potential in Immp2l-knockdown granulosa cells treated with enocyanin. (I) Western blot analysis of S-glutathionylation and S-nitrosylation in Immp2l-knockdown granulosa cells treated with enocyanin. (* p < 0.05; ** p < 0.01; *** p < 0.001), (Scale bar: 10 μm).
In addition, the ovaries from 28 days old mice were collected according to our previous study [8], and our results reveal that the three UPRmt core molecules ATF4, ATF5 and CHOP were all aggregated in the cytoplasm but not translocated into the nucleus in granulosa cells in the secondary follicle in Immp2l-knockout mice (Figure S1), and the UPRmt was impaired in senescent granulosa cells in secondary follicles in vivo.
2.2. Enocyanin Alleviates Granulosa Cell Senescence and Improves Oxidative Damage Induced by UPRmt and Mitophagy
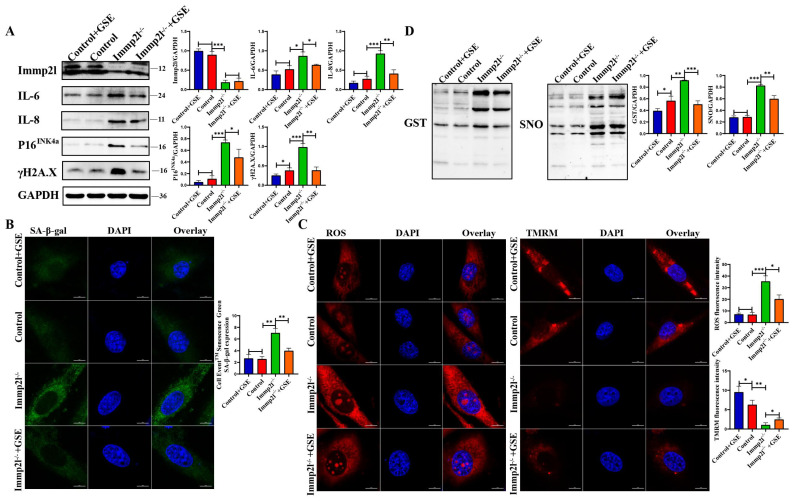
The KGN cells was employed to knock out Immp2l gene, and the Western blot results suggest that the knockout of the Immp2l gene (Immp2l−/−) in the human granulosa cell line KGN was successful, and the protein expression of the senescence markers IL-6, IL-8, γH2AX and P16INK4a was significantly increased in Immp2l−/− granulosa cells compared with the control (Immp2l+/+); therefore, granulosa cell senescence was triggered by Immp2l gene deficiency, and senescence was mitigated by enocyanin treatment, which was accompanied by a significant decrease in the senescence markers IL-6, IL-8, γH2AX and P16INK4a in enocyanin-treated Immp2l−/− granulosa cells compared with Immp2l−/− granulosa cells (Figure 2A). These results were further supported by the staining of senescence-associated β-galactosidase (Figure 2B). Immp2l deficiency causes granulosa cell senescence, and granulosa cell senescence is mitigated by enocyanin treatment. In addition, KGN cells shared the same traits with primary granulosa cells in ROS-induced granulosa cells.
Figure 2.
Enocyanin delays Immp2l deficiency-induced granulosa cell senescence. (A) Western blot analysis of cell senescence marker molecules in Immp2l-deficient granulosa cells treated with enocyanin. (B) Staining of senescence-associated β-galactosidase in Immp2l-deficient granulosa cells treated with enocyanin; at least 50 different cells in different fields were analyzed, and the dots were blindly counted by three different individuals. (C) Detection of mitROS and mitochondrial membrane potential in Immp2l-deficient granulosa cells treated with enocyanin. (D) Western blot analysis of S-glutathionylation and S-nitrosylation in Immp2l-deficient granulosa cells treated with enocyanin. (* p < 0.05; ** p < 0.01; *** p < 0.001), (Scale bar: 10 μm).
Cell senescence is closely related to mitochondrial dysfunction and ROS production [16]; therefore, mitochondrial function, including mitROS and mitochondrial membrane potential (MMP), was examined, and the results suggest that mitochondrial function was impaired, accompanied by significantly increased mitROS levels and decreased MMP in Immp2l−/− granulosa cells compared with the control. Enocyanin restored mitochondrial function, markedly decreased mitROS levels and increased MMP in enocyanin-treated Immp2l−/− granulosa cells compared with Immp2l−/− granulosa cells (Figure 2C). Then, S-glutathionylation and S-nitrosylation were detected in redox-related proteins, and the results suggest that S-glutathionylation (GST) and S-nitrosylation (SNO) levels were significantly increased and significantly decreased, respectively, by enocyanin treatment in Immp2l−/− granulosa cells (Figure 2D). Thus, mitochondrial dysfunction in granulosa cells induced by Immp2l deficiency was improved by enocyanin treatment.
2.3. STAT1 Was Regulated by GST and SNO but Not Phosphorylation
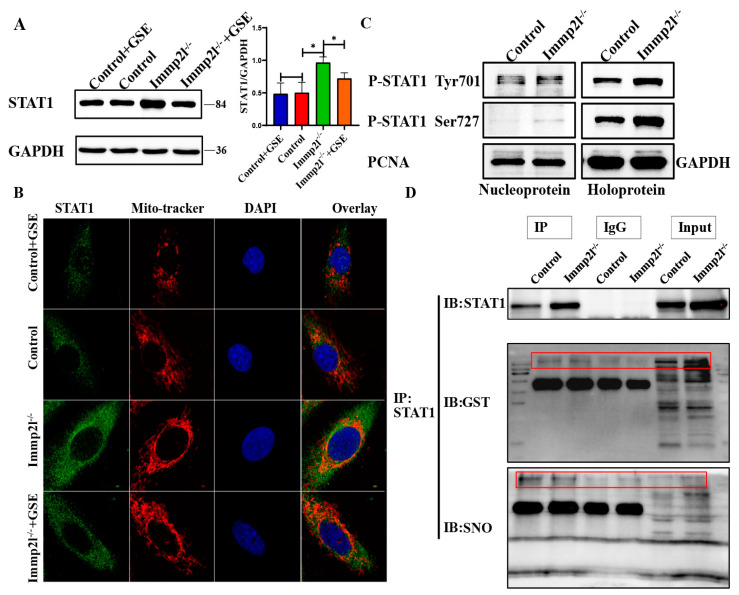
Previous studies suggested that ROS induces STAT1 [17], and our Western blot results suggest that STAT1 was markedly increased in Immp2l−/− granulosa cells compared with the control, while STAT1 was markedly reduced in enocyanin-treated Immp2l−/− granulosa cells compared with Immp2l−/− granulosa cells (Figure 3A). The STAT1 protein can be phosphorylated and translocate into the nucleus [18], and STAT1 is also regulated through transcriptional modification by S-glutathionylation and S-nitrosylation [5,6,7]. STAT1 was localized in the mitochondria and aggregated in the mitochondria of Immp2l-deficient granulosa cells, while the STAT1 protein’s accumulation in mitochondria was reduced by enocyanin treatment (Figure 3B). Consistently, phosphorylated STAT1 (Tyr701, Ser727) protein levels were not increased in the nucleus in Immp2l−/− granulosa cells (Figure 3C). Thus, the STAT1 protein was not posttranslationally modified by phosphorylation, and further Co-IP results show that the STAT1 protein was posttranslationally modified by S-glutathionylation and S-nitrosylation (Figure 3D).
Figure 3.
The translational modification of GST and SNO but not phosphorylation regulates STAT1. (A) Western blot analysis of STAT1 in Immp2l-deficient granulosa cells treated with enocyanin. (B) Colocalization of STAT1 and Mito-tracker in Immp2l-deficient granulosa cells treated with enocyanin. (C) Western blot analysis of phosphorylated STAT1 in Immp2l-deficient granulosa cells. (D) Co-IP confirmation of the interaction of STAT1 and S-glutathionylation and S-nitrosylation; at least 50 different cells in different fields were analyzed, and the dots were blindly counted by three different individuals. (* p < 0.05), (Scale bar: 10 μm).
2.4. Enocyanin Decrease Immp2l Deficiency-Induced UPRmt and Mitophagy
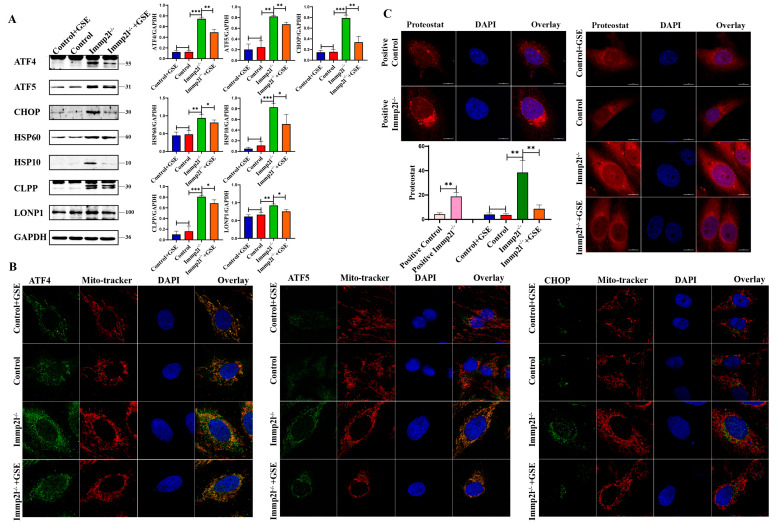
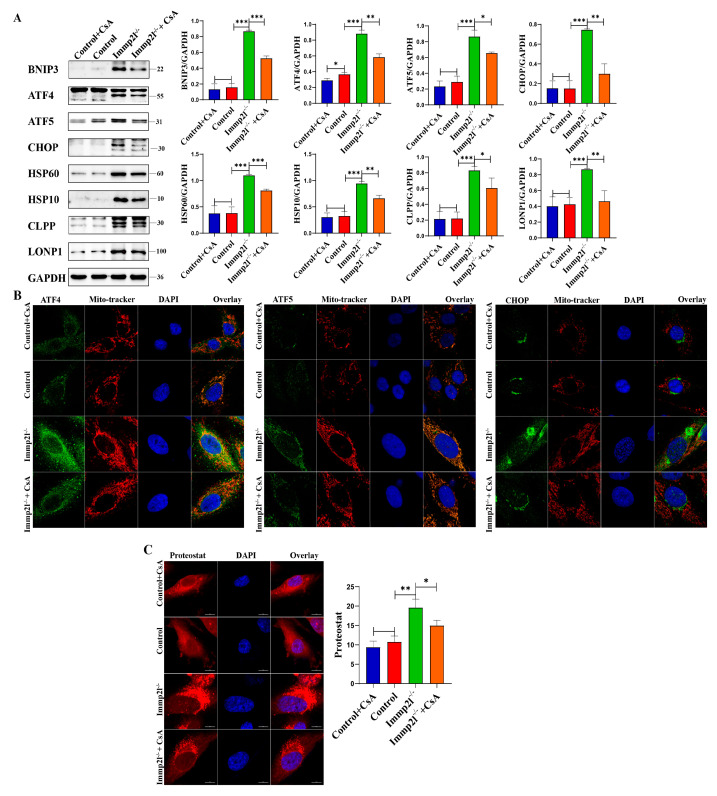
Mitochondrial proteostasis and mitochondrial homeostasis are maintained by the UPRmt [19,20] and mitophagy [21]. Mitochondrial dysfunction might be attributed to dysfunctional UPRmt and mitophagy. Our results reveal that the UPRmt marker proteins ATF4, ATF5, CHOP, HSP60, HSP10, CLPP and LONP1 were significantly increased (Figure 4A), but ATF4, ATF5 and CHOP did not translocate into the nucleus and accumulate in mitochondria (Figure 4B). Thus, UPRmt dysfunction occurred in Immp2l-knockout granulosa cells, and UPRmt impairment was alleviated by enocyanin treatment in Immp2l−/− granulosa cells accompanied with decreased UPRmt marker proteins and reduced accumulations of ATF4, ATF5 and CHOP proteins in mitochondria (Figure 4A,B). Indeed, mitochondrial proteostasis was impaired by Immp2l deficiency, and mitochondrial proteostasis was restored by enocyanin treatment. This result was further supported by the detection of protein aggresomes (Figure 4C).
Figure 4.
UPRmt pathway proteins were altered in Immp2l-deficient granulosa cells and regulated by enocyanin. (A) Western blot analysis of UPRmt marker molecules in Immp2l-deficient granulosa cells treated with enocyanin. (B) Colocalization of the UPRmt molecules ATF4, ATF5, CHOP and Mito-tracker in Immp2l-deficient granulosa cells treated with enocyanin. (C) Detection of protein aggresomes with the PROTEOSTAT Aggresome Detection Kit in Immp2l-deficient granulosa cells treated with enocyanin. (* p < 0.05; ** p < 0.01; *** p < 0.001) (scale bar: 10 μm).
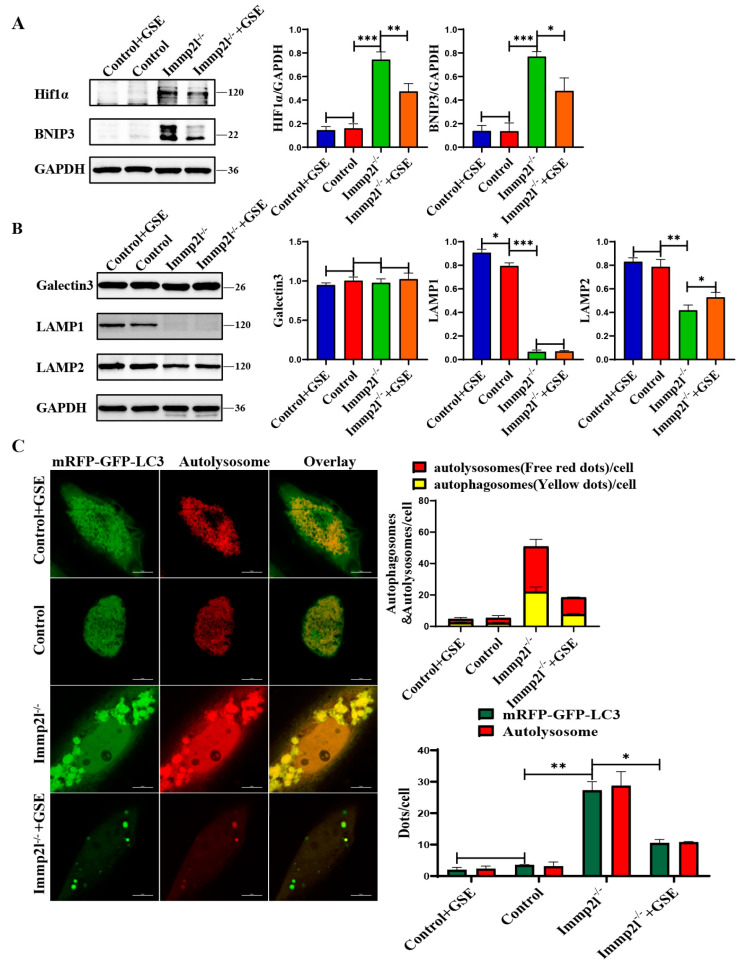
UPRmt impairment leads to unfolded and misfolded protein accumulation in mitochondria, resulting in mitochondrial dysfunction and aggregation in cells, and mitophagy is activated to clear aggregated mitochondria and proteins [22]. The HIF1α-BNIP3 mitophagy pathway plays an important role in maintaining mitochondrial homeostasis [23], and the HIF1α-BNIP3–mitophagy pathway is impaired in the ovarian granulosa cells of PCOS rats [24]. Thus, the role of the HIF1α-BNIP3-mitophagy pathway in Immp2l deficiency-induced senescence in granulosa cells was explored, and the results reveal that HIF1α and BNIP3 were significantly increased in Immp2l−/− granulosa cells and decreased by enocyanin treatment (Figure 5A). The lysosomal-associated membrane protein 2 (Lamp2) was significantly decreased in Immp2l−/− granulosa cells and increased by enocyanin treatment, while lysosomal-associated membrane protein 1 (Lamp1) and lysosome damage marker protein Galectin3 [25] were not altered by enocyanin treatment (Figure 5B). Therefore, the activated HIF1α/BNIP3-mediated mitophagy and markedly decreased Lamp2 levels indicate an imbalance in the mitophagy–lysosome degradation axis due to the impaired fusion of mitophagosomes and lysosomes, and restored the balance in the mitophagy–lysosome degradation axis attribute to increased Lamp2 levels in enocyanin-treated Immp2l−/− granulosa cells (Figure 5B). In addition, aggregated autophagosomes were markedly increased in Immp2l−/− granulosa cells and decreased by enocyanin treatment, as shown by mCherry-GFP-LC3 staining (Figure 5C). Thus, the UPRmt and HIF1α/BNIP3 mitophagy pathways were impaired in Immp2l−/− granulosa cells and subsequently restored by enocyanin.
Figure 5.
Mitophagy pathway proteins are altered in Immp2l-deficient granulosa cells and regulated by enocyanin. (A) Western blot analysis of mitophagy marker molecules in Immp2l-deficient granulosa cells treated with enocyanin. (B) Western blot analysis of the lysosome marker molecules Lamp1, Lamp2 and Galectin3 in Immp2l-deficient granulosa cells treated with enocyanin. (C) Detection of autophagic flux with the mCherry-GFP-LC3 lentivirus in Immp2l-deficient granulosa cells treated with enocyanin; at least 50 different cells in different fields were analyzed, and the dots were blindly counted by three different individuals. (* p < 0.05; ** p < 0.01; *** p < 0.001) (scale bar: 10 μm).
2.5. UPRmt and Mitophagy Are Regulated by STAT1 Inhibition, Which Is Accompanied by the Alleviation of Granulosa Cell Senescence
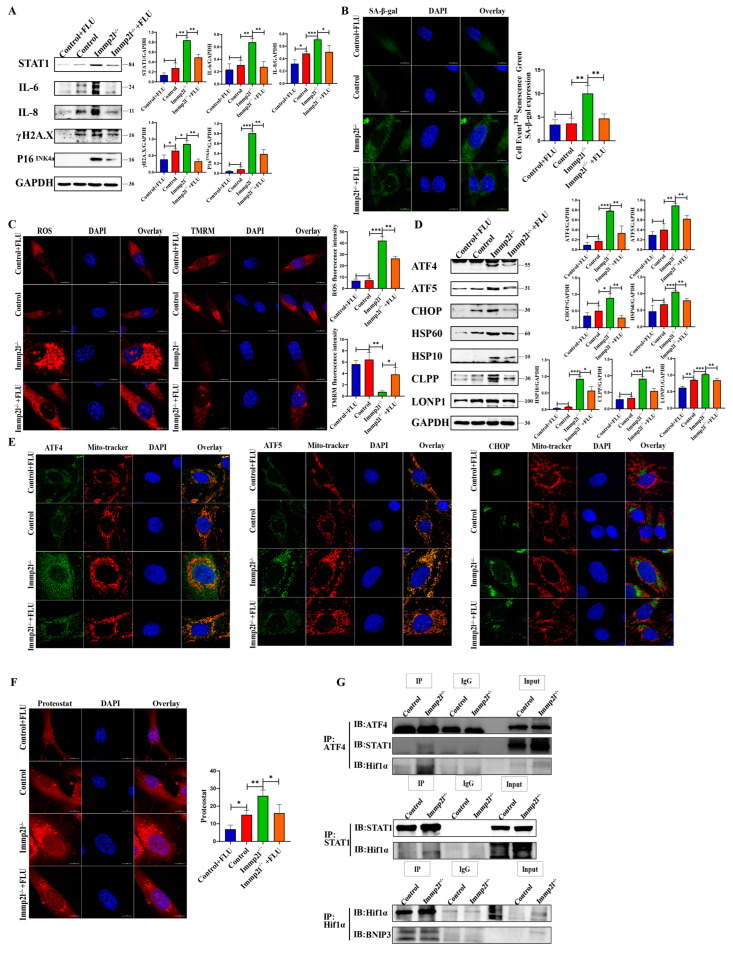
To explore the effect of STAT1 on granulosa cell senescence, the STAT1 inhibitors fludarabine (FLU) and siRNA were used to inhibit and knock down STAT1, respectively, and the results show that STAT1, IL-6, IL-8, γH2AX and P16INK4a were significantly decreased in STAT1-inhibited Immp2l−/− granulosa cells compared with Immp2l−/− granulosa cells (Figure 6A and Figure S2). These results are further supported by the results of the staining of senescence-associated β-galactosidase (Figure 6B). Inhibiting STAT1 restored mitochondrial function, markedly decreased mitROS levels and increased MMP in Immp2l−/− granulosa cells (Figure 6C). Therefore, inhibiting STAT1 alleviates granulosa cell senescence.
Figure 6.
The UPRmt and senescence proteins regulated by the STAT1 inhibitor fludarabine (FLU). (A) Western blot analysis of cell senescence marker molecules in Immp2l-deficient granulosa cells treated with the STAT1 inhibitor fludarabine (FLU). (B) Staining of senescence-associated β-galactosidase in Immp2l-deficient granulosa cells treated with the STAT1 inhibitor fludarabine (FLU); at least 50 different cells in different fields were analyzed, and the dots were blindly counted by three different individuals. (C) Detection of mitROS and mitochondrial membrane potential in Immp2l-deficient granulosa cells treated with the STAT1 inhibitor fludarabine (FLU); at least 50 different cells in different fields were analyzed, and the dots were blindly counted by three different individuals. (D) Western blot analysis of UPRmt marker molecules in Immp2l-deficient granulosa cells treated with the STAT1 inhibitor fludarabine (FLU). (E) Colocalization of the UPRmt marker molecules ATF4, ATF5, CHOP and Mito-tracker. (F) Detection of protein aggresome with the PROTEOSTAT Aggresome Detection Kit; at least 50 different cells in different fields were analyzed, and the dots were blindly counted by three different individuals. (G) Co-IP detection and analysis of the interaction of UPRmt ATF4 with STAT1, ATF4 and HIF1α, HIF1α and BNIP3, STAT1 and HIF1α. (* p < 0.05; ** p < 0.01; *** p < 0.001), (scale bar: 10 μm).
In addition, the UPRmt markers ATF4, ATF5, CHOP, HSP60, HSP10, CLPP and LONP1 were significantly decreased in STAT1-inhibited Immp2l−/− granulosa cells compared with Immp2l−/− granulosa cells (Figure 6D and Figure S3). The colocalization of ATF4, ATF5, CHOP and mitochondria (Figure 6E) with protein aggresomes (Figure 6F) was markedly decreased in STAT1-inhibited Immp2l−/− granulosa cells compared with Immp2l−/− granulosa cells. Furthermore, the co-IP results reveal that ATF4, which is a core molecule in the UPRmt, is regulated by STAT1 (Figure 6G). Therefore, the core molecule ATF4 in UPRmt is regulated by the STAT1.
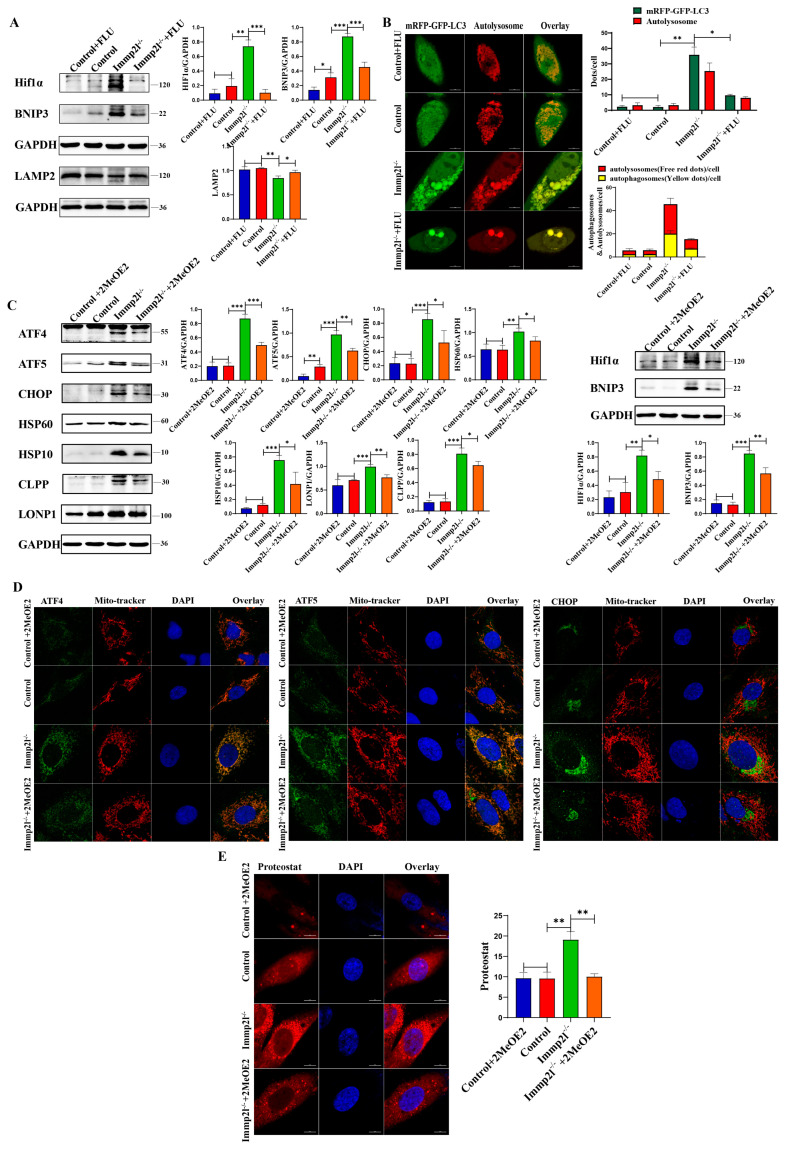
The mitophagy markers HIF1α and BNIP3 were significantly decreased, but the lysosomal molecule Lamp2 was significantly increased, in STAT1-inhibited Immp2l−/− granulosa cells compared with Immp2l−/− granulosa cells (Figure 7A and Figure S4). These results were further confirmed by detecting autophagic flux, while the impairment of the autophagic flux was detected in Immp2l−/− granulosa cells due to the impaired fusion of mitophagosomes and lysosomes, with the restoration of the autophagic flux attributed to the increased Lamp2 levels in STAT1-inhibited Immp2l−/− granulosa cells (Figure 7B). HIF1α was reduced by the inhibitor 2-methoxyestradiol (2-MeOE2), and the mitophagy marker BNIP3 and UPRmt markers ATF4, ATF5, CHOP, HSP60, HSP10, CLPP and LONP1 were significantly decreased in 2-MeOE2-treated Immp2l−/− granulosa cells (Figure 7C), which was accompanied by decreases in ATF4, ATF5, and CHOP aggregates in mitochondria in 2-MeOE2-treated Immp2l−/− granulosa cells (Figure 7D), as well as markedly reduced protein aggresomes in 2-MeOE2-treated Immp2l−/− granulosa cells (Figure 7E). Mitophagy was reduced by the inhibitor cyclosporin A (CsA), which significantly decreased the mitophagy marker BNIP3. The UPRmt markers ATF4, ATF5, CHOP, HSP60, HSP10, CLPP and LONP1 were significantly decreased by CsA treatment in Immp2l−/− granulosa cells (Figure 8A), which was accompanied by decreases in ATF4, ATF5, and CHOP aggregation in mitochondria induced by CsA treatment in Immp2l−/− granulosa cells (Figure 8B), as well as markedly reduced protein aggresomes in CsA-treated Immp2l−/− granulosa cells (Figure 8C). Thus, mitophagy was regulated by STAT1 via the STAT1/HIF1α/BNIP3 pathway or STAT1/ATF4/HIF1α/BNIP3 pathway. The UPRmt was also regulated by HIF1α/BNIP3-mediated mitophagy.
Figure 7.
Mitophagy and lysosomal proteins regulated by the STAT1 inhibitor fludarabine (FLU). (A) Western blot analysis of mitophagy and lysosome molecule in Immp2l-deficient granulosa cells treated by STAT1 inhibitor fludarabine (FLU). (B) Detection of autophagic flux with the mCherry-GFP-LC3 lentivirus in Immp2l-deficient granulosa cells treated by STAT1 inhibitor fludarabine (FLU); at least 50 different cells in different fields were analyzed, and the dots were blindly counted by three different individuals. (C) Western blot analysis of UPRmt and mitophagy marker molecules in Immp2l-deficient granulosa cells treated by HIF1α inhibitor 2-Methoxyestradiol (2-MeOE2). (D) Detection of the colocalization of UPRmt molecules ATF4, ATF5, CHOP and Mito-tracker. (E) Detection of protein aggresome with PROTEOSTAT Aggresome Detection Kit in Immp2l-deficient granulosa cells treated by HIF1α inhibitor 2-Methoxyestradiol (2-MeOE2); at least 50 different cells in different fields were analyzed, and the dots were blindly counted by three different individuals. (* p < 0.05; ** p < 0.01; *** p < 0.001), (Scale bar: 10 μm).
Figure 8.
UPRmt regulated by mitophagy inhibitor cyclosporin A (CsA) in Immp2l-deficient granulosa cells. (A) Western blot analysis of UPRmt marker molecules in Immp2l-deficient granulosa cells treated by mitophagy inhibitor cyclosporin A (CsA). (B) Detection of the colocalization of UPRmt molecules ATF4, ATF5, CHOP and Mito-tracker. (C) Detection of protein aggresome with PROTEOSTAT Aggresome Detection Kit in Immp2l-deficient granulosa cells treated by mitophagy inhibitor cyclosporin A (CsA); at least 50 different cells in different fields were analyzed, and the dots were blindly counted by three different individuals. (* p < 0.05; ** p < 0.01; *** p < 0.001), (scale bar: 10 μm).
3. Discussion
Mitochondria are the multifunctional powerhouse of cells, including oocytes and granulosa cells. Compared to nuclear DNA, mtDNA is vulnerable to oxidative damage, then tends to mutations [26] and protein misfolding [27]. Mitochondrial dysfunction is one of the hallmarks of aging, and mitochondrial DNA (mtDNA) copy number and function decline with age in various tissues. There is increasing evidence to suggest that mitochondrial dysfunction drives aging, including ovarian aging.
Accelerated ovarian aging reduces female fertility and even causes infertility. Ovarian aging is triggered by several factors [28], such as mitochondrial dysfunction, excessive ROS production, endoplasmic reticulum stress, and autophagy. Under physiological conditions, ovarian function is controlled by mitochondrial homeostasis [19], and ovarian aging is triggered by mitochondrial dysfunction. Therefore, mitochondrial dysfunction and ROS are considered to be important in initiating ovarian aging and cell senescence.
As a nuclear gene encoding a mitochondrial inner membrane protein, Immp2l plays important roles in several biological processes. Behavior changed in the Immp2l knockdown or knockout mice, which is linked with autism [29]. The behavioral changes in Immp2l knockdown or knockout mice are associated with an antioxidant-like phenotype, but ROS levels were not increased and were significantly lowered in Immp2l knockout or knockout mice [30,31]. The Immp2l knockout mice had significantly less lean mass and overall body weight compared with wildtype littermates [30]. Immp2l plays an important role in maintaining ovarian function, and the deficiency of the Immp2l gene in mice causes infertility due to ovarian aging, which is caused by the cessation of ovarian follicle development in the secondary stage, as well as ovulation disorder [32]. Our previous study revealed that ovarian aging associated with Immp2l deficiency was mainly caused by granulosa cell senescence, and granulosa cell senescence and ovarian aging were delayed by treatment with the antioxidant melatonin [8]. Therefore, inhibiting the senescence of granulosa cells is the best choice for inhibiting aging in the ovary. Antioxidant treatment might be the best method to prevent Immp2l deficiency-caused age-dependent degeneration [33].
As an anthocyanidin, enocyanin might play important antioxidant and anti-inflammation roles. Indeed, granulosa cell senescence caused by Immp2l deficiency was alleviated by enocyanin treatment, which was accompanied by inhibitions of the senescence markers γH2AX, P16INK4a, IL-6 and IL-8. IL-6 and IL-8 are inflammation-related senescence markers and senescence-associated cytokines that induce self- and cross-reinforced senescence [34,35]. Therefore, enocyanin inhibits inflammation-related granulosa cell senescence. Cell senescence is triggered by excessive ROS production, and Immp2l deficiency increases ROS production. Our results reveal that ROS levels were reduced and MMP was increased in enocyanin-treated Immp2l−/− cells compared with Immp2l−/− cells, and enocyanin mitigated granulosa cell senescence, which was attributed to the reduction in ROS levels and improvements in mitochondrial function. In addition, the enocyanin belongs to flavonoids, and the flavonoids act as a potential redox balance mediator [36], improving the redox balance [37]. Redox regulates ROS production [38]. Therefore, enocyanin might recover the redox balance and mitochondrial function in Immp2l-deficient granulosa cells, and then reduce ROS production.
Mitochondria are the main source of ROS [39], and mitochondrial function is impaired by Immp2l deficiency, which might be attributed to an imbalance in mitochondrial proteostasis and mitochondrial homeostasis. Indeed, mitochondrial proteostasis was impaired in Immp2l-deficient granulosa cells due to the impaired UPRmt, leading to more unfolded and misfolded proteins aggregating in mitochondria. Then, the mitophagy/lysosome degradation system was activated to maintain mitochondrial proteostasis and mitochondrial homeostasis by clearing protein aggregates and organelles, such as mitochondria (i.e., mitophagy) [40]. In Immp2l-deficient cells, HIF1α-BNIP3-mediated mitophagy is activated to clear protein aggregates and organelles, but impairs mitophagosome–lysosome fusion, which is accompanied by autophagosome accumulation in senescent granulosa cells due to reduced Lamp1 and Lamp2 protein levels [24,41,42]. The HIF1α-BNIP3–mitophagy pathway is impaired in the ovarian granulosa cells of PCOS rats [24], and restoring the balance of the HIF1α-BNIP3–mitophagy pathway might be a novel therapeutic strategy for PCOS. Therefore, restoring the balance of HIF1α-BNIP3–mitophagy and lysosomes might be a novel mechanism by which enocyanin alleviates granulosa cell senescence. Indeed, mitophagy controls the mitochondrial quality through removing impaired/dysfunctional mitochondria to maintain redox homeostasis and sustain cell viability [43]. Our results suggest that the balance of HIF1α-BNIP3–mitophagy and lysosomes was restored by enocyanin treatment, which was accompanied by a marked decrease in cell senescence markers and ROS. Thus, enocyanin delays granulosa cell senescence by maintaining the balance of the HIF1α-BNIP3–mitophagy pathway and redox homeostasis.
Interestingly, the STAT1 protein, which is closely related to oxidative stress and ROS, was markedly increased in Immp2l-knockdown granulosa cells, and was mostly regulated by the S-glutathionylation [5,6] and S-nitrosylation [7] of redox-related proteins. Our results suggest that STAT1 did not translocate into the nucleus and accumulate in mitochondria; it is not controlled by phosphorylation, but is controlled by the posttranslational modifications S-glutathionylation and S-nitrosylation. Furthermore, S-glutathionylation and S-nitrosylation were regulated by enocyanin treatment. Therefore, the STAT1 protein was regulated by enocyanin, which controlled S-glutathionylation and S-nitrosylation. The modification sites for S-glutathionylation and S-nitrosylation on STAT1 need to be explored in further studies. Additionally, S-glutathionylation and S-nitrosylation are redox-mediated modifications that regulate protein function [44]. Immp2l deficiency causes imbalances in mitochondria-associated redox status [45], and redox regulates the STAT1 [4].
In addition, UPRmt and mitophagy were regulated by STAT1 inhibition in Immp2l-deficient granulosa cells.
ATF4 is a core molecule in UPRmt [20]; UPRmt is initiated by ATF4, and ATF4 is downstream of STAT1 [46]. ATF4 was downregulated by inhibiting STAT1 in the context of Immp2l deficiency-induced granulosa cell senescence, and the co-IP results reveal that STAT1 targeted the ATF4 protein. Thus, Immp2l deficiency impairs the UPRmt via STAT1/ATF4, and enocyanin improves UPRmt by regulating the STAT1/ATF4 pathway.
Additionally, ATF4 targets HIF1α [47,48]. The co-IP results reveal that ATF4 targeted HIF1α, STAT1 targeted HIF1α, and HIF1α targeted BNIP3. Thus, Immp2l deficiency activates mitophagy via STAT1/ATF4/HIF1α/BNIP3 or STAT1/HIF1α/BNIP3, and enocyanin improves mitophagy by regulating the STAT1/ATF4/HIF1α/BNIP3 or STAT1/HIF1α/BNIP3 pathway (Figure 9). Inhibiting HIF1α with 2MeOE2 and inhibiting mitophagy with CsA further confirmed that the HIF1α/BNIP3 pathway was activated in Immp2l deficiency-induced granulosa cell senescence, and that HIF1α/BNIP3 was regulated by STAT1. Conversely, the UPRmt was regulated by HIF1α/BNIP3-mediated mitophagy.
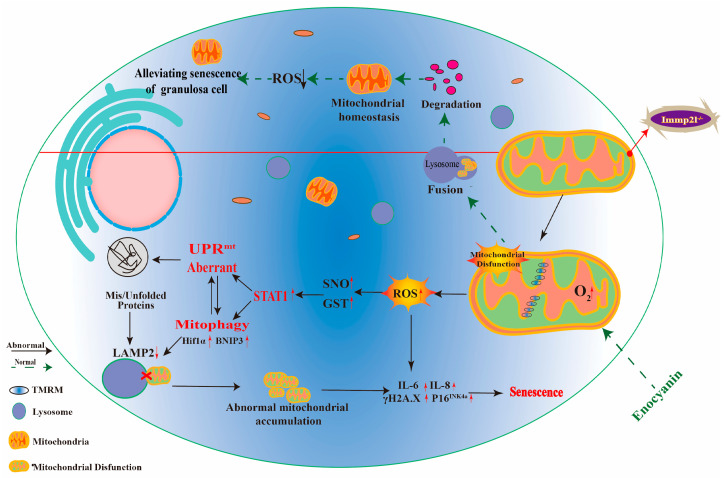
Figure 9.
The pathway by which enocyanin alleviates Immp2l deficiency-induced granulosa cell senescence. Under normal physiological conditions, mitochondrial proteostasis and mitochondrial homeostasis in granulosa cells are maintained by the UPRmt and mitophagy. However, Immp2l deficiency in granulosa cells impairs the UPRmt and mitophagy, leading to excessive unfolded and misfolded protein aggregation in mitochondria, dysfunctional mitochondria aggregation in cells, and excessive aggregated dysfunctional mitochondria producing excessive ROS, thereby triggering granulosa cell senescence. Enocyanin treatment alleviates granulosa cell senescence and improves the function of UPRmt and mitophagy through STAT1/ATF4-mediated UPRmt and STAT1/(ATF4)/HIF1α/BNIP3-mediated mitophagy, restoring mitochondrial proteostasis and mitochondrial homeostasis in granulosa cells and reducing ROS production.
Overall, enocyanin delays granulosa cell senescence induced by Immp2l deficiency through STAT1-mediated mitophagy and UPRmt, and STAT1 is regulated by enocyanin-controlled posttranslational modifications of S-glutathionylation and S-nitrosylation (Figure 9).
4. Materials and Methods
4.1. Mice and Cells
Immp2l gene-knockout mice were purchased from Cyagen Biosciences Inc. (Suzhou, China). The 21- to 26-day-old female mice were purchased and bred in the Laboratory Animal Center at Ningxia Medical University and were maintained at 24 ± 2 °C in a light-controlled room (12 h light:12 h darkness) with free access to food and water. The animal procedures strictly followed the US guidelines of the National Institutes of Health and were approved by the Institutional Animal Care and Use Committee at Ningxia Medical University (2021-Z011). All animals were operated on under sodium pentobarbital anesthesia, and all efforts were made to minimize pain and discomfort.
Primary granulosa cells from mice were isolated and cultured in vitro according to our previous studies [8]. The human granulosa tumor cell line KGN maintains most physiological activities of immature granulosa cells, including steroidogenesis, such as AMH, and apoptosis [49,50,51]; therefore, it is a very widely used and acceptable cell line for exploring granulosa cell function. Correspondingly, our previous study suggested that Immp2l gene deficiency causes ovarian follicle developmental arrest in the secondary stage, attributable to granulosa cell senescence [8]. Therefore, KGNs are optimal cells to explore the role of Immp2l in granulosa cell. KGN cells were purchased from Cyagen, the Immp2l gene was knocked out in the KGN cell line via the CRISPR/Cas9 method by Cyagen (Suzhou, China), and the procedure of knocking out Immp2l in the KGN cell line is briefly described in the Supplemental Materials (Figures S5–S7), while the siRNA sequence of STAT1 and Immp2l is shown in Table S1. The granulosa cells were treated with 10 mM enocyanin (HY-114336, MCE, grape-skin extract, GSE) according to previous studies [52,53] with minor modifications; after 24 h, the treated granulosa cells were collected for further analysis.
4.2. Western Blot Analysis
Western blot analysis was performed according to our previous study [8]. The information on antibodies is shown in Table S2. GAPDH was used as a reference protein, and the protein bands were analyzed using ImageJ software (V1.8.0.112).
4.3. Coimmunoprecipitation
The coimmunoprecipitation (co-IP) procedure was performed according to our previous study [8]. The antibodies used for co-IP were the same as those described for Western blotting.
4.4. Immunofluorescence Analysis
The immunofluorescence procedure was performed as described in our previous study [8]. The primary antibodies used for Western blotting were used for these experiments, and goat anti-rabbit FITC and goat anti-mouse TRITC secondary antibodies were purchased from ZSGB-BIO (Beijing, China). MitoSOX™ Red mitochondrial superoxide indicator (M36008) was purchased from Invitrogen (Waltham, MA, USA). TMRM (I34361) was purchased from Thermo Fisher (Waltham, MA, USA). HBAD-mRFP-GFP-LC3 (HBAD-1006) was purchased from HANBIO (Shanghai, China). The procedure of staining with the fluorescent probe was performed according to the manufacturer’s instructions. The images were obtained with confocal laser scanning microscopy (CLSM), and the fluorescence intensity was analyzed using ImageJ software.
4.5. Mitochondrial Membrane Potential and Mitochondrial Superoxide Assays
Mitochondrial membrane potential (MMP) was determined in cells for ex vivo analysis using the potentiometric probe TMRM (Molecular Probes, Life Technologies, Carlsbad, CA, USA). After surface-staining, the cells were incubated with TMRM (100 nM) for 15 min at 37 °C and photographed with a confocal laser scanning microscope. Images were obtained from different parts of the cells and analyzed using ImageJ software.
Mitochondrial superoxide levels were measured in cells and were incubated with MitoSOX Red dye (5 μm; Thermo Fisher) for 15 min at 37 °C according to the manufacturer’s protocol, and images were obtained by confocal laser scanning microscopy (CLSM). Images were obtained from different parts of the cells and analyzed using ImageJ software.
4.6. Detection of Senescent Cells
The Invitrogen™ CellEvent™ Senescence Green Detection Kit (Invitrogen, Waltham, MA, USA) and Senescence β-Galactosidase Staining Kit (KeyGEN, Nanjing, China) were used to stain the senescent cells, and the fluorescence intensity was analyzed by ImageJ software.
4.7. Protein Aggregation Assay
KGN cells were seeded at a density of 2 × 105/well in 12-well cell culture plates (Corning, Corning, NY, USA) containing glass coverslips, and aggregated proteins were detected using the PROTEOSTAT Aggresome Detection Kit (ENZ-51035; Enzo Life Sciences, Farmingdale, NY, USA) according to the manufacturer’s instructions. Briefly, cells were seeded in 12-well culture plates, fixed with 4% formaldehyde at 25 °C for 30 min and permeabilized with 0.5% Triton X-100 and 3 mM EDTA (pH 8.0) on ice for 30 min. Subsequently, the cells were stained with a dual detection reagent containing Hoechst 33258 (nuclear staining) and PROTEOSTAT dye reagent and then incubated at 25 °C for 30 min. Aggregated protein imaging was performed using confocal laser scanning microscopy (CLSM). Images were obtained from different parts of the cells and analyzed using ImageJ software.
4.8. Statistical Analysis
All experiments were repeated at least thrice in triplicates each time. The data are presented as the mean ± SEM. Data from more than two groups were analyzed with One-way ANOVA followed by Fisher’s least significant difference test, and data from two groups were analyzed with t tests using SPSS software (version 21.0; SPSS, Inc., Chicago, IL, USA). Differences were considered significant at p < 0.05.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms252011122/s1.
Author Contributions
Conceptualization, Y.Y.; methodology, X.Q., P.P., S.C., Y.M. and J.Y.; validation, X.Q.; formal analysis, X.Q.; investigation, X.Q., P.P., S.C., Y.M. and J.Y.; resources, H.G., X.P. and Y.Y.; data curation, X.Q., P.P. and S.C.; writing—original draft preparation, Y.Y.; writing—review and editing, Y.Y.; visualization, X.Q.; supervision, Y.Y.; project administration, Y.Y.; funding acquisition, Y.Y. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This work was approved by the Institutional Animal Care and Use Committee at Ningxia Medical University (2021-Z011).
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, and further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This article was supported by Key research and development program of Ningxia (2022BEG03145); the Natural Science Foundation of Ningxia (Grant number: 2021AAC03133).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Chiang J.L., Shukla P., Pagidas K., Ahmed N.S., Karri S., Gunn D.D., Hurd W.W., Singh K.K. Mitochondria in Ovarian Aging and Reproductive Longevity. Ageing Res. Rev. 2020;63:101168. doi: 10.1016/j.arr.2020.101168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sachdeva K., Do D.C., Zhang Y., Hu X., Chen J., Gao P. Environmental Exposures and Asthma Development: Autophagy, Mitophagy, and Cellular Senescence. Front. Immunol. 2019;10:2787. doi: 10.3389/fimmu.2019.02787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miwa S., Kashyap S., Chini E., von Zglinicki T. Mitochondrial dysfunction in cell senescence and aging. J. Clin. Investig. 2022;132:2787. doi: 10.1172/JCI158447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butturini E., Carcereri de Prati A., Mariotto S. Redox Regulation of STAT1 and STAT3 Signaling. Int. J. Mol. Sci. 2020;21:7034. doi: 10.3390/ijms21197034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butturini E., Boriero D., Carcereri de Prati A., Mariotto S. STAT1 drives M1 microglia activation and neuroinflammation under hypoxia. Arch. Biochem. Biophys. 2019;669:22–30. doi: 10.1016/j.abb.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 6.Matsui R., Ferran B., Oh A., Croteau D., Shao D., Han J., Pimentel D.R., Bachschmid M.M. Redox Regulation via Glutaredoxin-1 and Protein S-Glutathionylation. Antioxid. Redox Signal. 2020;32:677–700. doi: 10.1089/ars.2019.7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalinina E., Novichkova M. Glutathione in Protein Redox Modulation through S-Glutathionylation and S-Nitrosylation. Molecules. 2021;26:435. doi: 10.3390/molecules26020435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Q., Gu L., Lin Q., Ma Y., Liu C., Pei X., Li P.A., Yang Y. The Immp2l Mutation Causes Ovarian Aging Through ROS-Wnt/β-Catenin-Estrogen Pathway: Preventive Effect of Melatonin. Endocrinology. 2020;161:bqaa119. doi: 10.1210/endocr/bqaa119. [DOI] [PubMed] [Google Scholar]
- 9.Chen B.H., Stephen Inbaraj B. Nanoemulsion and Nanoliposome Based Strategies for Improving Anthocyanin Stability and Bioavailability. Nutrients. 2019;11:1052. doi: 10.3390/nu11051052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattioli R., Francioso A., Mosca L., Silva P. Anthocyanins: A Comprehensive Review of Their Chemical Properties and Health Effects on Cardiovascular and Neurodegenerative Diseases. Molecules. 2020;25:3809. doi: 10.3390/molecules25173809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ngamsamer C., Sirivarasai J., Sutjarit N. The Benefits of Anthocyanins against Obesity-Induced Inflammation. Biomolecules. 2022;12:852. doi: 10.3390/biom12060852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nomi Y., Iwasaki-Kurashige K., Matsumoto H. Therapeutic Effects of Anthocyanins for Vision and Eye Health. Molecules. 2019;24:3311. doi: 10.3390/molecules24183311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J., Xu B., Sun J., Jiang X., Bai W. Anthocyanin supplement as a dietary strategy in cancer prevention and management: A comprehensive review. Crit. Rev. Food Sci. Nutr. 2022;62:7242–7254. doi: 10.1080/10408398.2021.1913092. [DOI] [PubMed] [Google Scholar]
- 14.Hair R., Sakaki J.R., Chun O.K. Anthocyanins, Microbiome and Health Benefits in Aging. Molecules. 2021;26:537. doi: 10.3390/molecules26030537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong Y., Wu X., Han L., Bian J., He C., El-Omar E., Gong L., Wang M. The Potential Roles of Dietary Anthocyanins in Inhibiting Vascular Endothelial Cell Senescence and Preventing Cardiovascular Diseases. Nutrients. 2022;14:2836. doi: 10.3390/nu14142836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren X., Li B., Xu C., Zhuang H., Lei T., Jiang F., Zhou P. High expression of Piezo1 induces senescence in chondrocytes through calcium ions accumulation. Biochem. Biophys. Res. Commun. 2022;607:138–145. doi: 10.1016/j.bbrc.2022.03.119. [DOI] [PubMed] [Google Scholar]
- 17.Rendra E., Riabov V., Mossel D.M., Sevastyanova T., Harmsen M.C., Kzhyshkowska J. Reactive oxygen species (ROS) in macrophage activation and function in diabetes. Immunobiology. 2019;224:242–253. doi: 10.1016/j.imbio.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 18.Swiatek-Machado K., Kaminska B. STAT Signaling in Glioma Cells. Adv. Exp. Med. Biol. 2020;1202:203–222. doi: 10.1007/978-3-030-30651-9_10. [DOI] [PubMed] [Google Scholar]
- 19.Tian Y., Liu X., Pei X., Gao H., Pan P., Yang Y. Mechanism of Mitochondrial Homeostasis Controlling Ovarian Physiology. Endocrinology. 2022;164:bqac189. doi: 10.1210/endocr/bqac189. [DOI] [PubMed] [Google Scholar]
- 20.Gu L.F., Chen J.Q., Lin Q.Y., Yang Y.Z. Roles of mitochondrial unfolded protein response in mammalian stem cells. World J. Stem Cells. 2021;13:737–752. doi: 10.4252/wjsc.v13.i7.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liou Y.H., Personnaz J., Jacobi D., Knudsen N.H., Chalom M.M., Starost K.A., Nnah I.C., Lee C.H. Hepatic Fis1 regulates mitochondrial integrated stress response and improves metabolic homeostasis. JCI Insight. 2022;7:e150041. doi: 10.1172/jci.insight.150041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin Q., Chen J., Gu L., Dan X., Zhang C., Yang Y. New insights into mitophagy and stem cells. Stem Cell Res. Ther. 2021;12:452. doi: 10.1186/s13287-021-02520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu Z.J., Wang Z.Y., Xu L., Chen X.H., Li X.X., Liao W.T., Ma H.K., Jiang M.D., Xu T.T., Xu J., et al. HIF-1α-BNIP3-mediated mitophagy in tubular cells protects against renal ischemia/reperfusion injury. Redox Biol. 2020;36:101671. doi: 10.1016/j.redox.2020.101671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F., Han J., Wang X., Liu Y., Zhang Z. Roles of HIF-1α/BNIP3 mediated mitophagy in mitochondrial dysfunction of letrozole-induced PCOS rats. J. Mol. Histol. 2022;53:833–842. doi: 10.1007/s10735-022-10096-4. [DOI] [PubMed] [Google Scholar]
- 25.Arhzaouy K., Papadopoulos C., Schulze N., Pittman S.K., Meyer H., Weihl C.C. VCP maintains lysosomal homeostasis and TFEB activity in differentiated skeletal muscle. Autophagy. 2019;15:1082–1099. doi: 10.1080/15548627.2019.1569933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S.R., Kim N., Noh Y.H., Xu Z., Ko K.S., Rhee B.D., Han J. Mitochondrial DNA, mitochondrial dysfunction, and cardiac manifestations. Front. Biosci. 2017;22:1177–1194. doi: 10.2741/4541. [DOI] [PubMed] [Google Scholar]
- 27.Pickles S., Vigié P., Youle R.J. Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr. Biol. 2018;28:R170–R185. doi: 10.1016/j.cub.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y., Cheung H.H., Zhang C., Wu J., Chan W.Y. Melatonin as Potential Targets for Delaying Ovarian Aging. Curr. Drug Targets. 2019;20:16–28. doi: 10.2174/1389450119666180828144843. [DOI] [PubMed] [Google Scholar]
- 29.Leung B.K., Merlin S., Walker A.K., Lawther A.J., Paxinos G., Eapen V., Clarke R., Balleine B.W., Furlong T.M. Immp2l knockdown in male mice increases stimulus-driven instrumental behaviour but does not alter goal-directed learning or neuron density in cortico-striatal circuits in a model of Tourette syndrome and autism spectrum disorder. Behav. Brain Res. 2023;452:114610. doi: 10.1016/j.bbr.2023.114610. [DOI] [PubMed] [Google Scholar]
- 30.Clarke R.A., Govindaraju H., Beretta M., Olzomer E., Lawther A.J., Walker A.K., Fang Z., Eapen V., Hyams T.C., Killingsworth M., et al. Immp2l Enhances the Structure and Function of Mitochondrial Gpd2 Dehydrogenase. Int. J. Mol. Sci. 2024;25:990. doi: 10.3390/ijms25020990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawther A.J., Zieba J., Fang Z., Furlong T.M., Conn I., Govindaraju H., Choong L.L.Y., Turner N., Siddiqui K.S., Bridge W., et al. Antioxidant Behavioural Phenotype in the Immp2l Gene Knock-Out Mouse. Genes. 2023;14:1717. doi: 10.3390/genes14091717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu B., Poirier C., Gaspar T., Gratzke C., Harrison W., Busija D., Matzuk M.M., Andersson K.E., Overbeek P.A., Bishop C.E. A mutation in the inner mitochondrial membrane peptidase 2-like gene (Immp2l) affects mitochondrial function and impairs fertility in mice. Biol. Reprod. 2008;78:601–610. doi: 10.1095/biolreprod.107.065987. [DOI] [PubMed] [Google Scholar]
- 33.Liu C., Li X., Lu B. The Immp2l mutation causes age-dependent degeneration of cerebellar granule neurons prevented by antioxidant treatment. Aging Cell. 2016;15:167–176. doi: 10.1111/acel.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ortiz-Montero P., Londoño-Vallejo A., Vernot J.P. Senescence-associated IL-6 and IL-8 cytokines induce a self- and cross-reinforced senescence/inflammatory milieu strengthening tumorigenic capabilities in the MCF-7 breast cancer cell line. Cell Commun. Signal. 2017;15:17. doi: 10.1186/s12964-017-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng Y., Wu S., Ke H., Peng S., Hu C. Secretion of IL-6 and IL-8 in the senescence of bone marrow mesenchymal stem cells is regulated by autophagy via FoxO3a. Exp. Gerontol. 2023;172:112062. doi: 10.1016/j.exger.2022.112062. [DOI] [PubMed] [Google Scholar]
- 36.Chen L., Shi M., Lv C., Song Y., Wu Y., Liu S., Zheng Z., Lu X., Qin S. Dihydromyricetin Acts as a Potential Redox Balance Mediator in Cancer Chemoprevention. Mediat. Inflamm. 2021;2021:6692579. doi: 10.1155/2021/6692579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gentile D., Fornai M., Pellegrini C., Colucci R., Blandizzi C., Antonioli L. Dietary flavonoids as a potential intervention to improve redox balance in obesity and related co-morbidities: A review. Nutr. Res. Rev. 2018;31:239–247. doi: 10.1017/S0954422418000082. [DOI] [PubMed] [Google Scholar]
- 38.Pecchillo Cimmino T., Ammendola R., Cattaneo F., Esposito G. NOX Dependent ROS Generation and Cell Metabolism. Int. J. Mol. Sci. 2023;24:2086. doi: 10.3390/ijms24032086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Battaglia A.M., Chirillo R., Aversa I., Sacco A., Costanzo F., Biamonte F. Ferroptosis and Cancer: Mitochondria Meet the “Iron Maiden” Cell Death. Cells. 2020;9:1505. doi: 10.3390/cells9061505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Triolo M., Oliveira A.N., Kumari R., Hood D.A. The influence of age, sex, and exercise on autophagy, mitophagy, and lysosome biogenesis in skeletal muscle. Skelet. Muscle. 2022;12:13. doi: 10.1186/s13395-022-00296-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y., Zhao D., Peng W., Xue P., Jiang X., Chen S., Gao H., Wang X., Feng S. Atmospheric PM2.5 blocking up autophagic flux in HUVECs via inhibiting Sntaxin-17 and LAMP2. Ecotoxicol. Environ. Saf. 2021;208:111450. doi: 10.1016/j.ecoenv.2020.111450. [DOI] [PubMed] [Google Scholar]
- 42.Shi W., Lu D., Wu C., Li M., Ding Z., Li Y., Chen B., Lin X., Su W., Shao X., et al. Coibamide A kills cancer cells through inhibiting autophagy. Biochem. Biophys. Res. Commun. 2021;547:52–58. doi: 10.1016/j.bbrc.2021.01.112. [DOI] [PubMed] [Google Scholar]
- 43.Nakashima M., Suga N., Ikeda Y., Yoshikawa S., Matsuda S. Inspiring Tactics with the Improvement of Mitophagy and Redox Balance for the Development of Innovative Treatment against Polycystic Kidney Disease. Biomolecules. 2024;14:207. doi: 10.3390/biom14020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halloran M., Parakh S., Atkin J.D. The role of s-nitrosylation and s-glutathionylation of protein disulphide isomerase in protein misfolding and neurodegeneration. Int. J. Cell Biol. 2013;2013:797914. doi: 10.1155/2013/797914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan L., Zhai L., Qian L., Huang D., Ding Y., Xiang H., Liu X., Thompson J.W., Liu J., He Y.H., et al. Switching off IMMP2L signaling drives senescence via simultaneous metabolic alteration and blockage of cell death. Cell Res. 2018;28:625–643. doi: 10.1038/s41422-018-0043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu T.S., Tan C.T., Chang C.C., Lin B.R., Lai W.T., Chen S.T., Kuo M.Y., Rau C.L., Jaw F.S., Chang H.H. B-cell lymphoma/leukemia 10 promotes oral cancer progression through STAT1/ATF4/S100P signaling pathway. Oncogene. 2015;34:1207–1219. doi: 10.1038/onc.2014.43. [DOI] [PubMed] [Google Scholar]
- 47.Zhu K., Jiao H., Li S., Cao H., Galson D.L., Zhao Z., Zhao X., Lai Y., Fan J., Im H.J., et al. ATF4 promotes bone angiogenesis by increasing VEGF expression and release in the bone environment. J. Bone Miner. Res. 2013;28:1870–1884. doi: 10.1002/jbmr.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chee N.T., Carriere C.H., Miller Z., Welford S., Brothers S.P. Activating transcription factor 4 regulates hypoxia inducible factor 1α in chronic hypoxia in pancreatic cancer cells. Oncol. Rep. 2023;49:14. doi: 10.3892/or.2022.8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishi Y., Yanase T., Mu Y., Oba K., Ichino I., Saito M., Nomura M., Mukasa C., Okabe T., Goto K., et al. Establishment and characterization of a steroidogenic human granulosa-like tumor cell line, KGN, that expresses functional follicle-stimulating hormone receptor. Endocrinology. 2001;142:437–445. doi: 10.1210/endo.142.1.7862. [DOI] [PubMed] [Google Scholar]
- 50.Xu L., Wang W., Zhang X., Ke H., Qin Y., You L., Li W., Lu G., Chan W.Y., Leung P.C.K., et al. Palmitic acid causes insulin resistance in granulosa cells via activation of JNK. J. Mol. Endocrinol. 2019;62:197–206. doi: 10.1530/JME-18-0214. [DOI] [PubMed] [Google Scholar]
- 51.Roy S., Gandra D., Seger C., Biswas A., Kushnir V.A., Gleicher N., Kumar T.R., Sen A. Oocyte-Derived Factors (GDF9 and BMP15) and FSH Regulate AMH Expression Via Modulation of H3K27AC in Granulosa Cells. Endocrinology. 2018;159:3433–3445. doi: 10.1210/en.2018-00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiang Y., Lai F., He G., Li Y., Yang L., Shen W., Huo H., Zhu J., Dai H., Zhang Y. Alleviation of Rosup-induced oxidative stress in porcine granulosa cells by anthocyanins from red-fleshed apples. PLoS ONE. 2017;12:e0184033. doi: 10.1371/journal.pone.0184033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang S., Li X., Liu R., Hu J., Li Y., Sun J., Bai W. Malvidin-3-O-Glucoside Ameliorates Cadmium-Mediated Cell Dysfunction in the Estradiol Generation of Human Granulosa Cells. Nutrients. 2023;15:753. doi: 10.3390/nu15030753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, and further inquiries can be directed to the corresponding authors.