Abstract
Lafora disease (LD) is a fatal form of progressive myoclonus epilepsy caused by recessive mutations in either a gene encoding a dual-specificity phosphatase, known as laforin, or a recently identified gene encoding the protein known as malin. Here, we demonstrate that malin is a single subunit E3 ubiquitin (Ub) ligase and that its RING domain is necessary and sufficient to mediate ubiquitination. Additionally, malin interacts with and polyubiquitinates laforin, leading to its degradation. Missense mutations in malin that are present in LD patients abolish its ability to polyubiquitinate and signal the degradation of laforin. Our results demonstrate that laforin is a physiologic substrate of malin, and we propose possible models to explain how recessive mutations in either malin or laforin result in LD. Furthermore, these data distinguish malin as an E3 Ub ligase whose activity is necessary to prevent a neurodegenerative disease that involves formation of nonproteinacious inclusion bodies.
Keywords: progressive myoclonus epilepsy, phosphatase, neurodegenerative disease
The progressive myoclonus epilepsies (PMEs) share a characteristic triad of features: myoclonic seizures, tonic-clonic seizures, and progressive neurologic dysfunction. The five most common forms of PME, Unverricht–Lundborg disease, Lafora disease (LD), myoclonic epilepsy with ragged red fibers, the neuronal ceroid lipofuscinosis, and type I sialidosis, differ in the severity of their clinical features, molecular origins, pathogenesis, and prognosis (1). Although the mutated gene or genes are known and mouse models exist for each of the five most common PMEs, their molecular etiologies remain elusive.
LD (OMIM 254780) is an autosomal recessive progressive myoclonus epilepsy that was first described in 1911 (2–5). LD commonly presents in the first or second decade of life with epileptic seizures followed by progressive central nervous system degeneration beginning with myoclonic seizures, tonic-clonic seizures, focal occipital seizures, intellectual decline, severe motor and coordination deterioration, constant myoclonus, and, finally, death within 10 years of onset (4–7). A hallmark of the disease is the presence of periodic acid-Schiff-positive polyglucosan inclusion bodies, called Lafora bodies (LBs), located in the cytoplasm (5, 8). LBs are pathognomic of LD and may be the causative agent of LD neurodegeneration. The inclusion bodies appear to arise from an undefined defect or defects in glycogen metabolism (e.g., misregulated glycogen synthesis or degradation). LBs are found in most organs and are prominent in neuronal perikarya, the liver, and muscle (5, 9–11).
Two groups independently identified EPM2A (epilepsy of progressive myoclonus type 2 gene A) as a gene mutated in LD (12, 13). Mutations in EPM2A are responsible for ≈48% of LD cases (14). Of the remaining cases, ≈40% are the result of mutations in EPM2B (14, 15). To date, the molecular mechanisms underlying LD in patients harboring mutations in either EPM2A or EPM2B have not been defined.
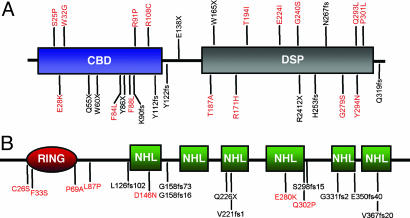
The human, mouse, and rat orthologs of EPM2A are 94% similar at the amino acid level (16). EPM2A encodes a protein product of 331 aa, named laforin, that contains the dual-specificity protein phosphatase (DSP) active site motif, HCXXGXXRS/T (Fig. 1A) (12, 13, 17, 18). Accordingly, recombinant laforin can hydrolyze phosphotyrosine and phosphoserine/threonine substrates in vitro (16, 19). The amino terminus of laforin contains a carbohydrate-binding domain (CBD) that targets it to subcellular sites of glycogen synthesis and promotes laforin's binding to glycogen in vitro and in vivo (Fig. 1 A) (19). Laforin preferentially binds LBs over glycogen in vitro and colocalizes with LBs in both a transgenic mouse model and in EPM2B mutated LD patient tissue (20, 21). Laforin also localizes to the endoplasmic reticulum and colocalizes with glycogen synthase (16, 20–22). Most of the reported missense mutations profoundly affect laforin's phosphatase activity and/or its ability to bind glycogen (19, 20, 23, 24). The exception is the G240S mutation, which has WT protein tyrosine phosphatase activity and binds glycogen but has reduced interaction with PTG/R5, one of the glycogen-targeting regulatory subunits of protein phosphatase 1 (24). Cumulatively, these data place laforin in the context of a multiprotein complex associated with intracellular glycogen particles and suggest that laforin is involved in the regulation of glycogen metabolism, either by promoting proper glycogen production and/or by removal of aberrant glycogen. However, the mechanism explaining laforin's role in the molecular etiology of LD is unknown.
Fig. 1.
A schematic of laforin and malin. LD mutations are shown for each protein, with missense mutations in red and other mutations in black. (A) Laforin contains a CBD and a DSP domain. (B) Malin contains a RING domain and six NHL repeats.
The second LD gene, EPM2B, encodes a 395-aa protein called malin (14). Malin contains a consensus RING-HC domain and six NHL domains (Fig. 1B) (14). RING domains are characteristic of one class of E3 ubiquitin (Ub) ligases (25, 26). Modification of proteins by Ub occurs by means of a three-step process in which Ub is activated by and transferred from the activating enzyme (E1) to a conjugating enzyme (E2) and finally to a substrate with the involvement of an Ub ligase (E3) (27). Ubiquitination of a protein most often leads to its degradation by the proteasome; alternatively, ubiquitination can change the activity, binding capacity, or localization of a protein (25, 26, 28, 29). NHL domains form a six-bladed β-propeller and are involved in protein–protein interactions, similar to WD40 repeats (30, 31). Although overexpressed malin colocalizes with laforin at the endoplasmic reticulum (14), currently no molecular mechanism connects malin with LD.
Herein, we demonstrate that malin's RING domain possesses in vitro E3 Ub ligase activity. This activity depends on one of four E2 enzymes, UbcH2, UbcH5a, UbcH5c, or UbcH6. Using a yeast two-hybrid screen, we identified laforin as a binding partner of malin and present evidence that malin directly binds laforin in vitro and interacts with laforin in vivo. Additionally, we show that laforin is polyubiquitinated in a malin-dependent manner in vitro and in vivo and that this polyubiquitination leads to laforin degradation in vivo. LD mutations in malin abolish both laforin's polyubiquitination and its degradation. Thus, malin regulates the protein concentration of laforin by means of polyubiquitin-dependent degradation. We discuss how these results are in agreement with both clinical immunohistochemistry data and the genetics of LD.
Methods
Plasmids and Proteins. Plasmids encoding human malin were constructed by PCR using Open Biosystems clone CA450023 and a primer pair that amplified malin's coding region. This PCR product was inserted into pcDNA3.1NF (32), pET-GSTX (33), pET21a (Novagen), and pGBKT7 (Clontech). Mutations were introduced by using the QuikChange mutagenesis kit (Stratagene). HA3-Ub and HA3-Ubk-less were generous gifts from Vishva Dixit (Genetech, San Francisco). Hemagglutinin (HA)-Ub, HA-SUMO, pET21a laforin-His6, and pcDNA3.1NF-laforin are described in refs. 19 and 34.
Recombinant malin and laforin were purified from soluble bacterial lysates by using Ni-nitrilotriacetic acid (NTA) agarose or successive Ni-NTA agarose and glutathione-agarose affinity chromatography steps essentially as described in refs. 33 and 35; specific details are provided in Supporting Text, which is published as supporting information on the PNAS web site. WT ubiquitin, biotinylated Ub (biotin-Ub), human E1, and human E2 enzymes were purchased from Boston Biochem (Cambridge, MA).
Cell Culture and Transfection. Adenovirus-transformed human embryonic kidney (HEK) 293T cells were maintained at 37°C with 5% CO2 in DMEM (Invitrogen) supplemented with 10% FBS/50 units/ml penicillin/streptomycin/4 mM glutamine. Subconfluent cultures of HEK293T cells (2 × 106 cells per 100-mm dish) were transfected by using FuGENE transfection reagent (Roche Applied Science, Indianapolis) according to the manufacturer's protocol and were cultured for 24 h to allow protein expression.
Antibodies and Immunoprecipitations (IPs). IPs and denaturing IPs were essentially performed as described in refs. 36 and 37. Specific details are available in Supporting Text. Western blots were probed with one of the following antibodies: mouse anti-Ub (Covance, Richmond, CA), biotin-horseradish peroxidase (HRP), (Boston Biochem), mouse anti-His-HRP (Santa Cruz Biotechnology), mouse anti-myc-HRP (Roche Molecular Biochemicals), mouse anti-FLAG-HRP (Sigma), or rat anti-HA (Roche Molecular Biochemicals). Goat anti-rat-HRP (Roche Molecular Biochemicals), goat anti-mouse-HRP (Amersham Pharmacia), and rabbit antimouse (Amersham Pharmacia) secondary antibodies were used when needed. The HRP signal was detected by using SuperSignal West Pico or Femto (Pierce). Some blots were stripped with Restore buffer (Pierce) to reprobe with a second primary antibody.
Yeast Two-Hybrid Screen. The bait plasmid, pGBKT7-malin (pMG101), was constructed by subcloning full-length malin into pGBKT7 (Clontech). pGBKT7-RING (pMG106) contained malin's amino terminus through Asn-128, pGBKT7–6NHL contained malin's carboxyl terminus from Cys-78, pACTII-CBD contained laforin's amino terminus through Gly-120, and pACTII-DSP contained laforin's carboxyl terminus from Leu-161. All of the two-hybrid clones (including the pGBKT7-malin point mutants) expressed to similar degrees except for pGBKT7–6NHL, as measured by Western blotting. pGBKT7–6NHL expression was 2- to 3-fold lower. The yeast two-hybrid screen was performed per the manufacturer's instructions; specific details are in Supporting Text.
In Vitro Binding Assay. Empty Ni-NTA agarose (Qiagen, Valencia, CA) or Ni-NTA bound to 2 μg of bacterially expressed and purified laforin-His6 was incubated in buffer A (50 mM Tris, pH 8.0/300 mM NaCl/20 mM imidazole, pH 8.0/15% maltose/0.05% 2-mercaptoethanol/protease inhibitors) at 4°C for 2 h with 35S-labeled FLAG-malin that was made in vitro by using the TNT T7-coupled reticulocyte lysate system (Promega). The agarose was washed two times with buffer A and two times in buffer B (50 mM Tris, pH 8.0/600 mM NaCl/20 mM imidazole, pH 8.0/15% maltose/0.05% 2-mercaptoethanol/protease inhibitors). Proteins were eluted at 95°C in NuPage buffer, separated by SDS/PAGE, transferred to nitrocellulose, and analyzed to detect laforin-His6 and 35S-malin.
In Vitro Ubiquitination Assay. The in vitro ubiquitination assay was performed essentially as described in ref. 38; specific details are provided in Supporting Text. When laforin was used in this assay, 35S-labeled FLAG-laforin was made in vitro by using the TNT T7-coupled reticulocyte lysate system (Promega); it was then immunoprecipitated out of the reaction mixture, washed three times, and eluted into 50 μl of Ub assay buffer with 10 μg of FLAG peptide (Sigma).
Results
Malin Functions as an E3 Ubiquitin Ligase in Vitro. Malin contains a RING-HC-type zinc finger and six NHL domains (14) (Fig. 1B). Based on subclassification of RING domains (39), malin falls into the RING-HCa family. The presence of a RING finger suggests that malin may function as an E3 Ub ligase; however, a recent study showed that only 60% of RING-HCa domain proteins exhibited in vitro E3 Ub ligase activity (39).
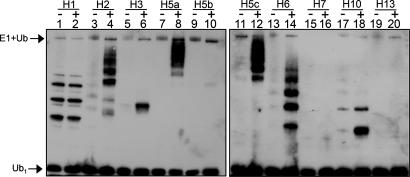
We therefore first tested whether malin possesses E3 ligase activity in an in vitro Ub assay (38, 40, 41). Previous work has demonstrated that E3 ligases can polyubiquitinate artificial protein substrates, often GST, in an in vitro reaction mixture (40). Recombinant, affinity-purified GST-malin-His6 was combined with a recombinant human E1 enzyme, 1 of 10 different recombinant human E2 enzymes, ATP, and biotin-Ub. Ubiquitinated species were monitored by Western blot analysis using avidin-HRP to detect biotin-Ub. Robust malin-dependent ubiquitination was consistently detected when one of four E2 enzymes was present: UbcH2, UbcH5a, UbcH5c, or UbcH6 (Fig. 2A, lanes 4, 8, 12, and 14). This ubiquitination was not dependent on the presence of malin's GST tag because malin-His6 also provided robust E3 ligase activity (data not shown). No ubiquitination was detected when bacterially purified GST was added to the reaction in place of GST-malin-His6 (Fig. 2A, compare lanes 3 and 4, 7 and 8, 11 and 12, and 13 and 14). The substrates ubiquitinated in this assay were largely the GST-E2 enzymes and not autoubiquitinated malin, because specific anti-E2 antibodies, and not an anti-malin antibody, detected the same laddered, high molecular mass bands (data not shown). Because RING domains are often sufficient to support ubiquitination, we tested both malin's RING and NHL domains for E3 activity. Indeed, ubiquitination depended on malin's RING domain but not on its NHL domains (Fig. 6A, which is published as supporting information on the PNAS web site). Additionally, ubiquitination was also dependent on the presence of all of the in vitro assay components (Fig. 6B). These results demonstrate that malin functions as an E2-dependent E3 Ub ligase in vitro.
Fig. 2.
Malin provides E3 Ub ligase activity in vitro. In vitro Ub assays were performed as described in Methods with E1 enzyme, 1 of 10 E2 enzymes, ATP, biotin-Ub, and recombinant GST-malin-H6 (+) or GST alone (-).
Malin Interacts with Laforin. To further understand malin's role in LD, we sought to identify malin's endogenous substrate(s). We performed a yeast two-hybrid screen using a human brain cDNA library and identified several clones that interacted with either malin or malin's NHL domains (data not shown). Plasmid rescue and sequencing identified three clones as EPM2A, the gene encoding laforin.
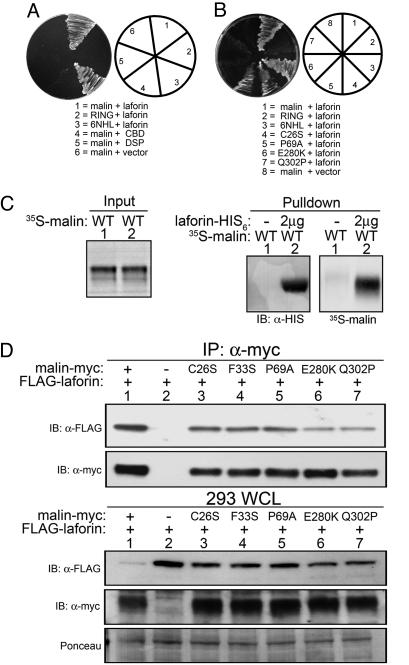
To map the domains responsible for the malin–laforin interaction, we generated two-hybrid clones containing malin's RING or NHL domain and tested the ability of these constructs to interact with laforin's CBD or DSP domain (Fig. 1). Malin's NHL domain interacted with laforin as strongly as full-length malin, whereas the RING domain did not interact with laforin (Fig. 3A). Alternatively, only full-length laforin interacted with malin (Fig. 3A). We then tested the effect of various LD missense mutations on the malin–laforin interaction. RING domain mutations (C26S and P69A) did not decrease the malin–laforin interaction, but NHL domain mutations (E280K and Q302P) abolished the interaction (Fig. 3B, compare 4 and 5 with 6 and 7). Thus, malin's six NHL domains likely function as a substrate-interacting motif, whereas malin's RING domain provides E3 Ub ligase activity. These data suggest that laforin could potentially serve as a substrate for malin. In addition, our results suggest that mutations in malin found in LD patients are likely to result in a loss of malin's ubiquitination activity by disrupting malin's substrate binding domain or its E2 binding domain.
Fig. 3.
Malin interacts with laforin. (A and B) Yeast cells were transformed with the various plasmid combinations, and the activation of the HIS and ADE reporters was assessed by growth on selective plates. (C) 35S-labeled, in vitro translated malin (35S-malin) was incubated with Ni-NTA agarose or Ni-NTA agarose bound to recombinant laforin-H6. The agarose was washed extensively, and bound proteins were analyzed by means of Western blotting with α-HIS and autoradiography for laforin and malin, respectively. Panels showing input and bound (pulldown) 35S-malin were exposed for different lengths of time. (D) HEK293T cells were cotransfected with 5 μg of malin-myc, empty vector, or mutant malin-myc and 5 μg of FLAG-laforin. WCLs were immunoblotted with α-myc and reprobed with α-FLAG. The nitrocellulose from the WCLs was stained with Ponceau S to monitor loading. Additionally, WCLs were subjected to IP with α-myc followed by α-myc immunoblotting and reprobed with α-FLAG.
Single subunit E3 ligases often interact directly with their substrates, whereas multisubunit E3 ligases require protein complexes to interact with substrate(s) (42, 43). To determine whether malin would behave as a single subunit E3 ligase, 35S-labeled in vitro translated malin was immunopurified from the reticulocyte extract and added to Ni-NTA agarose (control) or recombinant laforin-His6 immobilized on Ni-NTA agarose. Malin strongly associated with laforin without the aid of an adaptor protein, thus functioning like a single subunit E3 ligase (Fig. 3C).
To determine whether this interaction could be maintained in vivo, we coexpressed FLAG-tagged laforin and myc-tagged malin in HEK293T cells. Laforin coimmunoprecipitated with malin (Fig. 3D Upper, lane 1); it coimmunoprecipitated to a lesser degree with malin RING domain mutants (Fig. 3D Upper, compare lane 1 with lanes 3–5) and to a still lesser degree with malin NHL domain mutants (Fig. 3D Upper, compare lane 1 and lanes 6 and 7). Similar results were obtained by immunoprecipitating laforin and immunobloting for malin (data not shown). Although the NHL mutations decreased the malin–laforin interaction, they did not fully disrupt the interaction, as was observed by means of the yeast two-hybrid assay. Although we do not have a definite answer for these observations, the difference could result from a posttranslational modification or an additional interacting protein or proteins that may strengthen the interaction in mammalian cells. Strikingly, we noticed that there was much less total laforin in whole-cell lysate (WCL) when WT malin was present compared with empty vector or mutant malin (Fig. 3D Lower, top panel, compare lane 1 and lanes 2–7). The decreased overall amount of laforin in the WCL suggests that the malin–laforin interaction is quite robust. These data also show that laforin interacts with malin in vivo and suggest that malin is likely involved in regulating laforin protein concentrations.
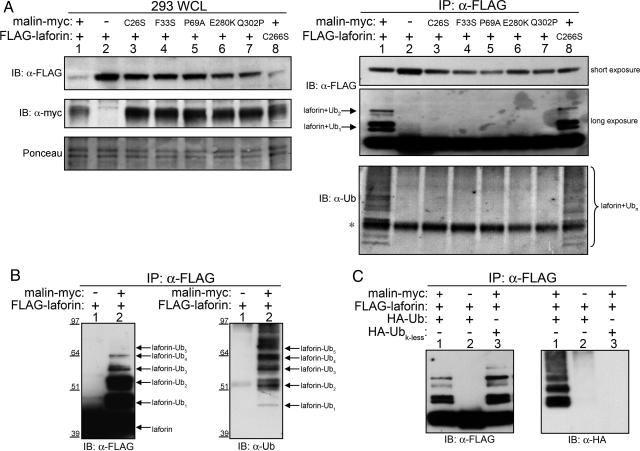
Laforin Is Ubiquitinated in a Malin-Dependent Manner. Because laforin levels are greatly decreased in cells expressing malin, we investigated whether malin might be responsible for regulating laforin levels by means of polyubiquitination and subsequent degradation. To determine whether malin was able to promote the ubiquitination of laforin in vivo, FLAG-tagged laforin was cotransfected with WT, mutant RING, or mutant NHL forms of malin in HEK293T cells. Of the three forms of malin, only WT malin reduced laforin protein levels (Fig. 4A Left Top, lanes 1 and 8 vs. lanes 2–7). Similarly, multiple Ub species coimmunoprecipitated with laforin only in cells coexpressing WT malin (Fig. 4A Right Bottom, lanes 1 and 8 vs. lanes 2–7). The Ub species that coimmunoprecipitated with laforin corresponded to ≈8-kDa increases in molecular mass (the molecular mass of ubiquitin) beginning at ≈47 kDa, ≈8 kDa larger than laforin. A longer exposure of the immunoprecipitated anti-FLAG blot revealed higher molecular mass versions of laforin when malin was present, again at ≈8-kDa intervals (Fig. 4A Right Middle). Disease mutations in either malin's RING or NHL domains abolished the reduction in laforin protein levels, the modified forms of laforin, and the coimmunoprecipitating Ub species (Fig. 4A). These data demonstrate that laforin is ubiquitinated and potentially targeted for degradation in a malin-dependent manner.
Fig. 4.
Laforin is polyubiquitinated in a WT malin-dependent manner. (A) HEK293T cells were cotransfected with 5 μg of malin-myc, empty vector, or mutant malin-myc and 5 μg of FLAG-laforin or mutant FLAG-laforin. WCLs were immunoblotted with α-FLAG and reprobed with α-myc. The nitrocellulose from the WCLs was stained with Ponceau S to monitor loading. Additionally, WCLs were subjected to IP with α-FLAG followed by immunoblotting with α-FLAG and reprobed with α-Ub. Asterisk marks a nonspecific band. (B) HEK293T cells were cotransfected with 3 μg of malin-myc or empty vector and 7 μg of FLAG-laforin. WCLs were subjected to IP with α-FLAG, immunoblotted with α-FLAG, and reprobed with α-Ub. (C) HEK293T cells were cotransfected with malin-myc, FLAG-laforin, HA-Ub, and HA-Ub lacking lysines (HA-Ubk-less) as indicated. WCLs were subjected to IP with α-FLAG, immunoblotted with α-FLAG, and reprobed with α-HA.
Phosphorylation often regulates ubiquitination, either positively or negatively (44–46). Because laforin is a phosphatase, we tested whether laforin's phosphatase activity was required for its ubiquitination. LaforinC266S lacks phosphatase activity (19), but it was still ubiquitinated and degraded in a malin-dependent manner (Fig. 4A Left and Right, lane 8). Thus, laforin's phosphatase activity is not required for its ubiquitination or degradation.
To determine the specificity of malin's ligase activity, we also tested whether SUMO, a small Ub-like modifier, could be attached to laforin in a malin-dependent manner. Only HA-Ub coimmunoprecipitated with laforin in a malin-dependent manner (Fig. 7 Top, which is published as supporting information on the PNAS web site). In addition, laforin was still modified in cells overexpressing malin and HA-SUMO by the endogenous Ub (Fig. 7 Bottom), showing that SUMO is not transferred to laforin.
Because mono-/di-ubiquitination and polyubiquitination signal very different fates for an ubiquitinated protein, we wanted to determine the ubiquitination status of laforin. To determine whether laforin had multiple Ubs covalently attached to it, we coexpressed FLAG-laforin and malin-myc in HEK293T cells. IP with anti-FLAG antibody and immunoblotting with anti-FLAG and anti-Ub detected up to six forms of laforin, including five modified forms that comigrated with Ub (Fig. 4B). Additionally, the Ub ladder coimmunoprecipitated with laforin after lysing cells in buffer containing 2% SDS and denaturing proteins by heating the lysate at 95°C for 10 min (data not shown). This treatment should break all noncovalent protein–protein interactions. These results demonstrate that laforin is covalently modified with multiple Ub molecules in the presence of malin.
Multiple Ub attachment could occur by means of two different mechanisms: multiple single Ubs attached to multiple lysines (multiubiquitination) or a chain of Ubs attached to a single lysine (polyubiquitination). These two types of ubiquitination trigger different outcomes for a target protein. To determine whether laforin was multiubiquitinated or polyubiquitinated, we expressed FLAG-laforin, malin-myc, and either HA-Ub or HA-Ub-lacking lysines (HA-Ubk-less) in HEK293T cells. The Ub lacking all lysines is unable to form polyubiquitin chains and thus can only be efficiently involved in multiubiquitination. Laforin was ubiquitinated by HA-Ub but not by HA-Ubk-less (Fig. 4C), demonstrating that laforin is polyubiquitinated in a malin-dependent manner.
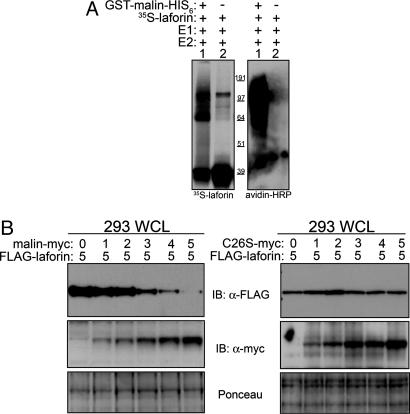
Malin Polyubiquitinates Laforin and Promotes Its Degradation. To determine whether laforin serves as a substrate for malin in vitro,we again used the same in vitro ubiquitination assay previously described but supplemented the reactions with immunopurified 35S-labeled in vitro translated laforin. After performing the reaction at 37°C, the 35S-laforin was immunoprecipitated out of the reaction to eliminate all other ubiquitinated proteins in the mixture (specifically, the GST-E2s). Ubiquitination of 35S-laforin was evaluated by Western blot analysis of biotin-Ub and autoradiography. High molecular mass laforin species that comigrated with biotin-Ub were seen only in the presence of bacterially expressed GST-malin-His6 and all other reaction components (Fig. 5A). These data provide direct evidence that malin induces polyubiquitination of laforin in vitro and that malin does not need additional cellular components to polyubiquitinate laforin.
Fig. 5.
Malin polyubiquitinates laforin and promotes its degradation. (A) In vitro ubiquitination assays were performed as described in Fig. 2 but were supplemented with 35S-labeled, in vitro translated laforin (35S-laforin). 35S-laforin ubiquitination was monitored by autoradiography, and biotin-Ub was detected with avidin-HRP. (B) HEK293T cells were cotransfected with increasing micrograms (0–5) of malin-myc or mutant malin-myc (C26S-myc), 5 μg of FLAG-laforin, and enough empty vector to make the total transfection amount 10 μg. WCLs were immunoblotted with α-FLAG and reprobed with α-myc. The nitrocellulose was stained with Ponceau S to monitor loading.
To prove that laforin polyubiquitination triggers its degradation, we coexpressed FLAG-laforin with either malin-myc or malinC26S-myc in HEK293T cells. MalinC26S is a LD mutation that disrupts malin's RING domain and decreases malin's in vitro E3 activity (data not shown), likely by disrupting malin's interaction with E2 enzymes. Laforin protein levels were decreased in a malin dose-dependent manner (Fig. 5B Left). Alternatively, laforin protein levels were unchanged with increasing amounts of malinC26S-myc (Fig. 5B Right). Therefore, the malin-dependent reduction in laforin levels is not due to a transcription/translation artifact but to the presence of WT malin. Together with our in vitro data, these results show that laforin is polyubiquitinated by malin and targeted for degradation.
Discussion
In this study, we show that malin is a single-subunit, E2-dependent E3 Ub ligase and that it interacts with laforin both in vitro and in vivo. Missense mutations in malin's NHL domains found in LD patients diminish this interaction. Furthermore, malin polyubiquitinates laforin, and this polyubiquitination promotes laforin's degradation. Mutations in malin found in LD patients abolish both laforin's polyubiquitination and its degradation. Thus, malin regulates laforin protein concentrations by means of polyubiquitin-dependent degradation.
Because malin is a single subunit E3 ligase, malin's function could be abrogated by means of mutations that disrupt its ability to bind either an E2 enzyme or its substrate(s). This assertion is supported by the location of missense mutations within the malin protein in LD patients. The mutations fall in malin's RING and NHL domains. The only missense mutation not located in one of these domains is L87P, and crystal structures of other E2–E3 interactions suggest that this residue is likely involved in malin's ability to bind an E2 enzyme (47–49). These data, together with our findings, strongly suggest that loss of malin's E3 activity is the impetus for the onset of LD in patients with EPM2B mutations.
There is indirect evidence suggesting that laforin is involved in proper glycogen anabolism. Laforin binds R5/PTG, a regulatory subunit of protein phosphatase 1 that is a critical positive regulator of glycogen synthesis (24, 50). R5/PTG directly binds laforin, glycogen synthase, phosphorylase, and phosphorylase kinase, acting as a molecular scaffold by assembling the glycogen machinery (24, 50, 51). Additionally, these proteins are likely to compete for binding to the PTG scaffold (24, 52). Therefore, laforin could promote proper glycogen anabolism by dephosphorylating a necessary component of glycogen synthesis and/or by competing for binding on R5/PTG.
Alternatively, recent studies suggest that laforin may act as a regulator of aberrant glycogen to inhibit its accumulation. This model postulates that laforin recognizes LBs, by means of its CBD, and initiates mechanisms to inhibit LB formation and/or promote LB destruction through its phosphatase domain (20, 21). Multiple groups have shown that laforin preferentially binds LBs or LB-like material over glycogen both in vitro and in vivo (20, 21, 53). Thus, it is possible that laforin's role in suppressing LD begins after LB formation by inhibiting the formation, or by clearance and/or degradation, of LBs.
Interestingly, Chan et al. (21) reported that they were unable to detect endogenous laforin in tissue from non-LD patients by using multiple polyclonal α-laforin antibodies, but they could detect endogenous laforin in tissue from LD patients carrying mutations in malin (21). These clinical results mirror our biochemical findings, which demonstrate that malin promotes laforin degradation. Collectively, these clinical and biochemical data demonstrate that malin regulates the concentration of laforin in situ by means of Ub-dependent degradation and that loss-of-function malin mutations cause laforin levels to increase. However, if laforin degradation is malin's only LD-linked function, then these results appear to be in conflict with the genetics of LD, because recessive mutations in either laforin or malin cause LD. We envision two scenarios that explain these data. The models are valid if malin and laforin are involved in proper glycogen anabolism or aberrant glycogen catabolism (e.g., LB degradation/clearance).
Malin could promote the degradation of another protein, protein X, and it is the accumulation of protein X that causes LD when malin is mutated. Malin's regulation of laforin concentrations would be a secondary regulatory function of malin, and increases in laforin concentrations would not be the cause of LD in patients with mutations in malin. Because phosphorylation often regulates ubiquitination, protein X could be dephosphorylated by laforin, and this dephosphorylation event would trigger protein X to be ubiquitinated by malin and degraded. Once protein X is degraded, glycogen metabolism could progress properly. This model proposes that laforin and malin function on the same substrate (protein X) and promote its degradation.
Alternatively, laforin could be both an activator and repressor of proper glycogen metabolism. In this model, laforin's CBD would function to localize laforin and allow laforin to dephosphorylate substrate X; after dephosphorylation, the next event, either proper glycogen formation or inhibition of aberrant glycogen accumulation, would not occur until laforin is polyubiquitinated and degraded. In this model, laforin is analogous to previously postulated “kamikaze” or “single-shot rifle” transcriptional activators (54, 55). The activators function to assemble the necessary reagents for transcription, but their degradation is required to enable RNA polymerase to leave the promoter and transcribe downstream sequence. We propose that a similar mechanism could be the cause of LD in patients with mutations in malin. Patients with mutations in malin would accumulate laforin and develop LD because destruction of laforin is necessary for glycogen metabolism to proceed. Patients with mutations in laforin would develop LD because laforin would be unable to dephosphorylate a necessary substrate in glycogen metabolism.
Our finding that malin polyubiquitinates laforin and promotes its degradation, coupled with the clinical immunohistochemistry data from affected and unaffected patient tissue (21), suggests that malin's regulation of laforin protein concentrations could be malin's primary LD-linked function. Thus, increased levels of laforin would be the molecular etiology of LD in patients with mutations in malin. However, both models should be explored in detail to determine which provides the best molecular explanation of LD.
Supplementary Material
Acknowledgments
We thank Vishva Dixit for various Ub plasmids, Mike Begley for assistance with the in vitro binding assay, and Dr. Gregory S. Taylor and David J. Pagliarini for insightful discussions. This work was supported by National Institutes of Health (NIH)/National Cancer Institute Grant T32 CA09523 (to M.S.G.), NIH Grant 18024 (to J.E.D.), and the Walther Cancer Institute (J.E.D.).
Author contributions: M.S.G. and J.E.D. designed research; M.S.G. and C.A.W. performed research; M.S.G. and C.A.W. contributed new reagents/analytic tools; M.S.G., C.A.W., and J.E.D. analyzed data; and M.S.G. wrote the paper.
Abbreviations: LD, Lafora disease; LB, Lafora body; HA, hemagglutinin; IP, immunoprecipitation; CBD, carbohydrate-binding domain; DSP, dual-specificity protein phosphatase; NTA, nitrilotriacetic acid; HEK, human embryonic kidney; HRP, horseradish peroxidase; Ub, ubiquitin; biotin-Ub, biotinylated Ub; WCL, whole-cell lysate.
References
- 1.Lehesjoki, A. E. (2002) Adv. Neurol. 89, 193-197. [PubMed] [Google Scholar]
- 2.Van Heycop Ten Ham, M. W. (1975) in Handbook of Clinical Neurology, eds. Vinken, P. J. & Bruyn, G. W. (North–Holland, Amsterdam), Vol. 15, pp. 382-422. [Google Scholar]
- 3.Berkovic, S. F., Andermann, F., Carpenter, S. & Wolfe, L. S. (1986) N. Engl. J. Med. 315, 296-305. [DOI] [PubMed] [Google Scholar]
- 4.Minassian, B. A. (2002) Adv. Neurol. 89, 199-210. [PubMed] [Google Scholar]
- 5.Lafora, G. A. B. G. (1911) Z. Ges. Neurol. Psychiatr. 6, 1-14. [Google Scholar]
- 6.Berkovic, S. F., So, N. K. & Andermann, F. (1991) J. Clin. Neurophysiol. 8, 261-274. [PubMed] [Google Scholar]
- 7.Berkovic, S. F., Cochius, J., Andermann, E. & Andermann, F. (1993) Epilepsia 34, Suppl. 3, S19-S30. [DOI] [PubMed] [Google Scholar]
- 8.Collins, G. H., Cowden, R. R. & Nevis, A. H. (1968) Arch. Pathol. 86, 239-254. [PubMed] [Google Scholar]
- 9.Harriman, D. G., Millar, J. H. & Stevenson, A. C. (1955) Brain 78, 325-349. [DOI] [PubMed] [Google Scholar]
- 10.Schwarz, G. A. & Yanoff, M. (1965) Arch. Neurol. 12, 172-188. [DOI] [PubMed] [Google Scholar]
- 11.Carpenter, S. & Karpati, G. (1981) Neurology 31, 1564-1568. [DOI] [PubMed] [Google Scholar]
- 12.Minassian, B. A., Lee, J. R., Herbrick, J. A., Huizenga, J., Soder, S., Mungall, A. J., Dunham, I., Gardner, R., Fong, C. Y., Carpenter, S., et al. (1998) Nat. Genet. 20, 171-174. [DOI] [PubMed] [Google Scholar]
- 13.Serratosa, J. M., Gomez-Garre, P., Gallardo, M. E., Anta, B., de Bernabe, D. B., Lindhout, D., Augustijn, P. B., Tassinari, C. A., Malafosse, R. M., Topcu, M., et al. (1999) Hum. Mol. Genet. 8, 345-352. [DOI] [PubMed] [Google Scholar]
- 14.Chan, E. M., Young, E. J., Ianzano, L., Munteanu, I., Zhao, X., Christopoulos, C. C., Avanzini, G., Elia, M., Ackerley, C. A., Jovic, N. J., et al. (2003) Nat. Genet. 35, 125-127. [DOI] [PubMed] [Google Scholar]
- 15.Chan, E. M., Omer, S., Ahmed, M., Bridges, L. R., Bennett, C., Scherer, S. W. & Minassian, B. A. (2004) Neurology 63, 565-567. [DOI] [PubMed] [Google Scholar]
- 16.Ganesh, S., Agarwala, K. L., Ueda, K., Akagi, T., Shoda, K., Usui, T., Hashikawa, T., Osada, H., Delgado-Escueta, A. V. & Yamakawa, K. (2000) Hum. Mol. Genet. 9, 2251-2261. [DOI] [PubMed] [Google Scholar]
- 17.Yuvaniyama, J., Denu, J. M., Dixon, J. E. & Saper, M. A. (1996) Science 272, 1328-1331. [DOI] [PubMed] [Google Scholar]
- 18.Maehama, T. & Dixon, J. E. (1998) J. Biol. Chem. 273, 13375-13378. [DOI] [PubMed] [Google Scholar]
- 19.Wang, J., Stuckey, J. A., Wishart, M. J. & Dixon, J. E. (2002) J. Biol. Chem. 277, 2377-2380. [DOI] [PubMed] [Google Scholar]
- 20.Ganesh, S., Tsurutani, N., Suzuki, T., Hoshii, Y., Ishihara, T., Delgado-Escueta, A. V. & Yamakawa, K. (2004) Biochem. Biophys. Res. Commun. 313, 1101-1109. [DOI] [PubMed] [Google Scholar]
- 21.Chan, E. M., Ackerley, C. A., Lohi, H., Ianzano, L., Cortez, M. A., Shannon, P., Scherer, S. W. & Minassian, B. A. (2004) Hum. Mol. Genet. 13, 1117-1129. [DOI] [PubMed] [Google Scholar]
- 22.Minassian, B. A., Andrade, D. M., Ianzano, L., Young, E. J., Chan, E., Ackerley, C. A. & Scherer, S. W. (2001) Ann. Neurol. 49, 271-275. [PubMed] [Google Scholar]
- 23.Ianzano, L., Young, E. J., Zhao, X. C., Chan, E. M., Rodriguez, M. T., Torrado, M. V., Scherer, S. W. & Minassian, B. A. (2004) Hum. Mutat. 23, 170-176. [DOI] [PubMed] [Google Scholar]
- 24.Fernandez-Sanchez, M. E., Criado-Garcia, O., Heath, K. E., Garcia-Fojeda, B., Medrano-Fernandez, I., Gomez-Garre, P., Sanz, P., Serratosa, J. M. & Rodriguez de Cordoba, S. (2003) Hum. Mol. Genet. 12, 3161-3171. [DOI] [PubMed] [Google Scholar]
- 25.Pickart, C. M. (2001) Annu. Rev. Biochem. 70, 503-533. [DOI] [PubMed] [Google Scholar]
- 26.Hershko, A. & Ciechanover, A. (1998) Annu. Rev. Biochem. 67, 425-479. [DOI] [PubMed] [Google Scholar]
- 27.Hershko, A., Heller, H., Elias, S. & Ciechanover, A. (1983) J. Biol. Chem. 258, 8206-8214. [PubMed] [Google Scholar]
- 28.Sun, L. & Chen, Z. J. (2004) Curr. Opin. Cell Biol. 16, 119-126. [DOI] [PubMed] [Google Scholar]
- 29.Aguilar, R. C. & Wendland, B. (2003) Curr. Opin. Cell Biol. 15, 184-190. [DOI] [PubMed] [Google Scholar]
- 30.Edwards, T. A., Wilkinson, B. D., Wharton, R. P. & Aggarwal, A. K. (2003) Genes Dev. 17, 2508-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slack, F. J. & Ruvkun, G. (1998) Trends Biochem. Sci. 23, 474-475. [DOI] [PubMed] [Google Scholar]
- 32.Taylor, G. S., Maehama, T. & Dixon, J. E. (2000) Proc. Natl. Acad. Sci. USA 97, 8910-8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor, G. S., Liu, Y., Baskerville, C. & Charbonneau, H. (1997) J. Biol. Chem. 272, 24054-24063. [DOI] [PubMed] [Google Scholar]
- 34.Orth, K., Xu, Z., Mudgett, M. B., Bao, Z. Q., Palmer, L. E., Bliska, J. B., Mangel, W. F., Staskawicz, B. & Dixon, J. E. (2000) Science 290, 1594-1597. [DOI] [PubMed] [Google Scholar]
- 35.Maehama, T., Taylor, G. S., Slama, J. T. & Dixon, J. E. (2000) Anal. Biochem. 279, 248-250. [DOI] [PubMed] [Google Scholar]
- 36.Abbott, D. W., Wilkins, A., Asara, J. M. & Cantley, L. C. (2004) Curr. Biol. 14, 2217-2227. [DOI] [PubMed] [Google Scholar]
- 37.Kotaja, N., Karvonen, U., Janne, O. A. & Palvimo, J. J. (2002) Mol. Cell. Biol. 22, 5222-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leng, R. P., Lin, Y., Ma, W., Wu, H., Lemmers, B., Chung, S., Parant, J. M., Lozano, G., Hakem, R. & Benchimol, S. (2003) Cell 112, 779-791. [DOI] [PubMed] [Google Scholar]
- 39.Stone, S. L., Hauksdottir, H., Troy, A., Herschleb, J., Kraft, E. & Callis, J. (2005) Plant Physiol. 137, 13-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorick, K. L., Jensen, J. P., Fang, S., Ong, A. M., Hatakeyama, S. & Weissman, A. M. (1999) Proc. Natl. Acad. Sci. USA 96, 11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joazeiro, C. A., Wing, S. S., Huang, H., Leverson, J. D., Hunter, T. & Liu, Y. C. (1999) Science 286, 309-312. [DOI] [PubMed] [Google Scholar]
- 42.Borden, K. L. (2000) J. Mol. Biol. 295, 1103-1112. [DOI] [PubMed] [Google Scholar]
- 43.Jackson, P. K., Eldridge, A. G., Freed, E., Furstenthal, L., Hsu, J. Y., Kaiser, B. K. & Reimann, J. D. (2000) Trends Cell Biol. 10, 429-439. [DOI] [PubMed] [Google Scholar]
- 44.Yaron, A., Hatzubai, A., Davis, M., Lavon, I., Amit, S., Manning, A. M., Andersen, J. S., Mann, M., Mercurio, F. & Ben-Neriah, Y. (1998) Nature 396, 590-594. [DOI] [PubMed] [Google Scholar]
- 45.Skowyra, D., Craig, K. L., Tyers, M., Elledge, S. J. & Harper, J. W. (1997) Cell 91, 209-219. [DOI] [PubMed] [Google Scholar]
- 46.Feldman, R. M., Correll, C. C., Kaplan, K. B. & Deshaies, R. J. (1997) Cell 91, 221-230. [DOI] [PubMed] [Google Scholar]
- 47.Zheng, N., Wang, P., Jeffrey, P. D. & Pavletich, N. P. (2000) Cell 102, 533-539. [DOI] [PubMed] [Google Scholar]
- 48.Albert, T. K., Hanzawa, H., Legtenberg, Y. I., de Ruwe, M. J., van den Heuvel, F. A., Collart, M. A., Boelens, R. & Timmers, H. T. (2002) EMBO J. 21, 355-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng, N., Schulman, B. A., Song, L., Miller, J. J., Jeffrey, P. D., Wang, P., Chu, C., Koepp, D. M., Elledge, S. J., Pagano, M., et al. (2002) Nature 416, 703-709. [DOI] [PubMed] [Google Scholar]
- 50.Printen, J. A., Brady, M. J. & Saltiel, A. R. (1997) Science 275, 1475-1478. [DOI] [PubMed] [Google Scholar]
- 51.Fong, N. M., Jensen, T. C., Shah, A. S., Parekh, N. N., Saltiel, A. R. & Brady, M. J. (2000) J. Biol. Chem. 275, 35034-35039. [DOI] [PubMed] [Google Scholar]
- 52.Brady, M. J. & Saltiel, A. R. (2001) Recent Prog. Horm. Res. 56, 157-173. [DOI] [PubMed] [Google Scholar]
- 53.Liu, A. W., Delgado-Escueta, A. V., Serratosa, J. M., Alonso, M. E., Medina, M. T., Gee, M. N., Cordova, S., Zhao, H. Z., Spellman, J. M. & Peek, J. R. (1995) Am. J. Hum. Genet. 57, 368-381. [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas, D. & Tyers, M. (2000) Curr. Biol. 10, R341-R343. [DOI] [PubMed] [Google Scholar]
- 55.Lipford, J. R. & Deshaies, R. J. (2003) Nat. Cell Biol. 5, 845-850. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.