Abstract
The expression of hepatitis B virus (HBV) genes is regulated by a number of transcription factors. One such factor, Sp1, has two binding sites in the core promoter and one in its upstream regulatory element, which is also known as the ENII enhancer. In this study, we have analyzed the effects of these three Sp1 binding sites on the expression of HBV genes. Our results indicate that both Sp1 binding sites in the core promoter are important for the transcription of the core RNA and the precore RNA. Moreover, while the downstream Sp1 site (the Sp1-1 site) in the core promoter did not affect the transcription of the S gene and the X gene, the upstream Sp1 site (the Sp1-2 site) in the core promoter was found to negatively regulate the transcription of the S gene and the X gene, as removal of the latter led to enhancement of transcription of these two genes. The Sp1 binding site in the ENII enhancer (the Sp1-3 site) positively regulates the expression of all of the HBV genes, as its removal by mutation suppressed the expression of all of the HBV genes. However, the suppressive effect of the Sp1-3 site mutation on the expression of the S gene and the X gene was abolished if the two Sp1 sites in the core promoter were also mutated. These results indicate that Sp1 can serve both as a positive regulator and as a negative regulator for the expression of HBV genes. This dual activity may be important for the differential regulation of HBV gene expression.
Hepatitis B virus (HBV) is an important human pathogen that can cause severe liver diseases, including acute and chronic hepatitis. This virus has a small, circular DNA genome with a size of about 3.2 kb. The viral genome carries four genes named the C, S, X, and P genes (Fig. 1). The C gene codes for the viral core protein that packages the viral genome and also for a related protein named precore protein. The precore protein is the precursor of the secreted e antigen, which may be important for the establishment of persistent infection following neonatal infection (for a review, see reference 20). The S gene codes for three co-carboxy-terminal envelope proteins named the pre-S-1, pre-S-2, and major S proteins. These S gene products are also known as surface antigens. The X gene codes for a transcriptional transactivator, and the P gene codes for the viral DNA polymerase, which is also a reverse transcriptase.
FIG. 1.
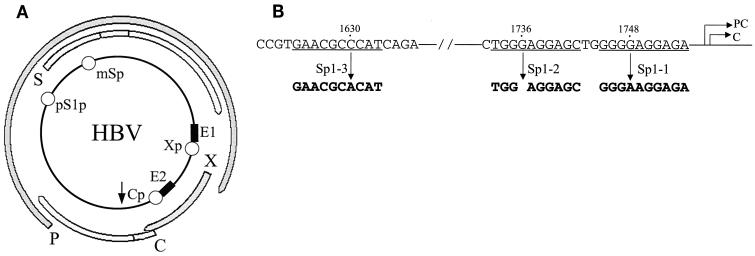
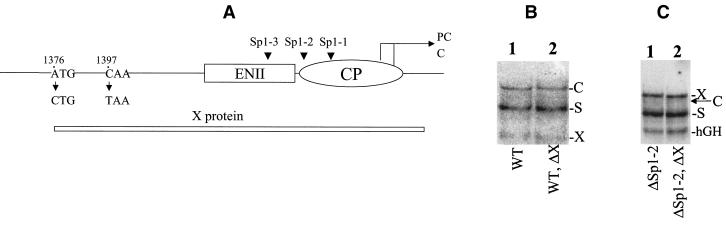
The HBV genome and Sp1 binding sites. (A) Schematic illustration of the HBV genome. P, S, X, and C, the four HBV genes. E1 and E2, ENI and ENII enhancers, respectively. Xp, Cp, pS1p, and mSp, X promoter, core promoter, pre-S-1 promoter, and major S promoter, respectively. The arrow indicates the unique poly(A) site. (B) The Sp1 binding sites in the core promoter and the ENII enhancer. The nucleotide numbers in the HBV genome are indicated. PC and C, transcription start sites of the precore RNA and the core RNA, respectively. The mutations introduced into the three Sp1 sites are also indicated.
The expression of the HBV genes is regulated by four different promoters and two enhancer elements (Fig. 1A) (34). The core promoter regulates the transcription of the precore RNA and the core RNA, the pre-S-1 promoter regulates the expression of the pre-S-1 RNA, the major S promoter regulates the transcription of the pre-S-2 RNA and the major S RNA, and the X promoter regulates the transcription of the X RNA (Fig. 1). The precore RNA and the core RNA are larger than the genome length. However, only the latter is used as the mRNA for the synthesis of the viral DNA polymerase (17, 21). The ENI enhancer partially overlaps the X promoter, and the ENII enhancer is located upstream of the core promoter (12, 28, 32, 33, 38). Both enhancers can upregulate the activities of all four HBV gene promoters (1, 33). Only one of the HBV DNA strands is coding, and therefore the transcription of the HBV genes is unidirectional. All of the HBV RNA transcripts terminate at the same poly(A) site in the viral genome (Fig. 1A). It has been demonstrated that cis-acting elements as well as the distance between the promoter and the poly(A) site play important roles in determining whether the poly(A) site should be used or bypassed for polyadenylation of the viral RNA (9, 13, 26). For example, as this site is located less than 200 bp from the core promoter, the C gene transcripts bypass this site the first time and become polyadenylated at this site only after they have circled around the genome once and encounter the site the second time. In contrast, this poly(A) site is located approximately 2 kbp away from the S gene promoters and is therefore used efficiently by the S gene transcripts for polyadenylation when the site is first encountered during transcription. The X promoter is located about 700 bp upstream of the poly(A) site, and therefore the X gene transcripts bypass this poly(A) site with an intermediate efficiency of approximately 50% (13). This leads to the production of two different X gene transcripts: one with a subgenomic size of 700 nucleotides (nt) and the other with a larger-than-genome size of 3.9 kb (13).
Many cellular protein factors that may regulate HBV gene expression have been identified. These factors include liver-enriched factors such as HNF1, HNF3, and C/EBP and ubiquitous factors such as Sp1, RFX1, NF-Y, and AP1 (4, 7, 15–19, 22, 24, 25, 30, 31, 35–37, 39, 40). In addition, members of the nuclear receptor superfamily such as HNF4, RXRα, PPARα, and COUP-TFs have also been found to regulate the expression of HBV genes (5, 6, 8, 11, 14, 23, 35). Despite extensive studies that have been conducted to study cis- and trans-acting factors that may regulate HBV gene expression, how these factors may interact with each other to allow differential expression of HBV genes remains largely unknown.
Sp1 is a ubiquitous transcription factor that binds to the GC-rich elements (3). Several Sp1 binding sites have been identified in the HBV genome, including one in the ENII enhancer, two in the core promoter, and four in the major S promoter (24, 25, 36, 40, 41). Previous studies indicate that the Sp1 sites in the core promoter are important for the core promoter activity and that those in the major S promoter are important for the major S promoter activity. However, those studies were conducted by using either subgenomic HBV DNA fragments or reporter DNA constructs (24, 25, 36, 40, 41). The possible effects of Sp1 on HBV gene expression in the context of the entire HBV genome were not studied. In this study, we have focused our attention on the two Sp1 sites in the core promoter and the Sp1 site in its upstream ENII enhancer and have examined their effects on HBV gene expression in the context of the entire HBV genome. Our results indicate that while these three Sp1 binding sites are important for the optimal activities of the core promoter, they have different effects on the activities of the S promoter and the X promoter.
MATERIALS AND METHODS
DNA plasmids.
pWTD contains the head-to-tail dimer of the wild-type HBV genome of the adw2 subtype (30) (Fig. 1). pHBVΔSp1-1 is identical to pWTD with the exception that it contains a G-to-A mutation at nt 1748 in the Sp1-1 site (Fig. 1). pHBVΔSp1-2 is identical to pWTD except that the G residue at nt 1736 in the Sp1-2 site has been deleted. pHBVΔSp1-3 is also identical to pWTD with the exception that it contains a C-to-A mutation at nt 1630 in the Sp1-3 site. pHBVΔSp1-1,2 contains both the G-to-A mutation at nt 1748 and a deletion of nt 1736. Similarly, pHBVΔSp1-1,2,3 contains all three nucleotide mutations in the Sp1-1, Sp1-2, and Sp1-3 sites. All of the mutants were created by PCR-based mutagenesis procedures as previously described (10). pXGH5 (Nichols Diagnostics) contains the human growth hormone (hGH) reporter under the control of the mouse metallothionein promoter.
Cell culture and DNA transfection.
Huh7 hepatoma cells were maintained in Dulbecco modified Eagle medium containing 10% fetal bovine serum. Cells were plated in a 10-cm-diameter petri dish the day before transfection and transfected when they were 80% confluent. Each plate of cells was transfected with 20 μg of DNA using the calcium phosphate precipitation method. In all cases, pXGH5 was included in the transfection procedures to serve as an internal control to monitor the transfection efficiency. Cells were lysed 48 h after transfection, and the RNA was isolated using our previously described procedures (14).
Northern blot analysis.
In general, 10 μg of total RNA was used for Northern blot analysis by our previous procedures (14). The linearized 3.2-kb HBV EcoRI DNA fragment was nick translated and labeled with 32P to serve as a probe. For the analysis of the 0.7-kb X RNA, the 550-bp BamHI-BglII HBV DNA fragment containing the X protein-coding sequence was used as the probe, as it increases the sensitivity of detection of the short X RNA (13).
EMSA.
The preparation of Huh7 nuclear extracts and procedures for electrophoretic mobility shift assay (EMSA) and supershift assay have been previously described (5, 14). The sequences of the oligonucleotide probes used for the EMSA are as follows: Sp1-1: wt 5′ GAGGAGCTGGGGGAGGAGATTA 3′
3′ CTCGACCCCCTCCTCTAATCCA 5′
mt 5′ GAGGAGCTGGGAGAGGAGATTA 3′
3′ CTCGACCCTCTCCTCTAATCCA 5′
Sp1-2: wt 5′ TGTTTAAGGACTGGGAGGAGCTGGGGG 3′
3′ AATTCCTGACCCTCCTCGACCCCCTCC 5′
mt 5′ TGTTTAAGGACTGG-AGGAGCTGGGGG 3′
3′ AATTCCTGACC-TCCTCGACCCCCTCC 5′
Sp1-3: wt 5′ ACCACCGTGAACGCCCATCAGATCCTG 3′
3′ TGGCACTTGCGGGTAGTCTAGGACGGG 5′
mt 5′ ACCACCGTGAACGCACATCAGATCCTG 3′
3′ TGGCACTTGCGTGTAGTCTAGGACGGG 5′
Primer extension analysis.
The sequence of the antisense primer used for analyzing the C gene transcripts is 5′GGTGAGCAATGCTCAGGAGACTCTAAGG3′, which corresponds to nt 2051 to 2024 of the HBV genome. The antisense primer sequence used for analyzing the S gene transcripts is 5′CCATGTTCGTCACAGGGTCCCCAGTCCTCGCGGAGATTG3′, which corresponds to nt 123 to 161 of the HBV genome (30). The antisense primer sequence used for analyzing the hGH transcript is 5′GCCACTGCAGCTAGGTGAGCGTCC3′. The primer extension reaction was carried out as previously described (14).
RESULTS
Mutagenesis of the Sp1 sites in the ENII enhancer and C promoter.
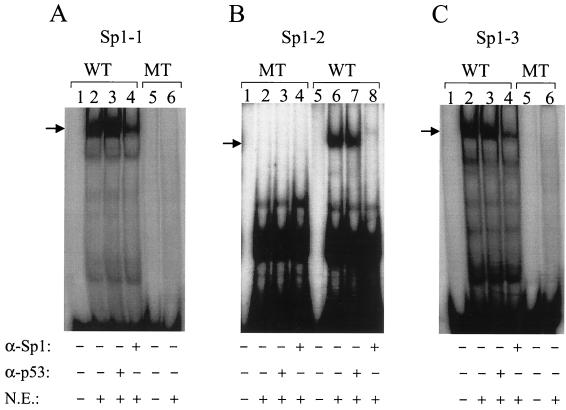
The three Sp1 sites located in the core promoter and the ENII enhancer are illustrated in Fig. 1B. Sp1-1 and Sp1-2 are located in the core promoter, and Sp1-3 is in the ENII enhancer. As shown in Fig. 2A, a synthetic DNA probe containing the Sp1-1 sequence could be bound by a protein factor in the nuclear extracts of Huh7 hepatoma cells to form a complex with a lower electrophoretic mobility on the EMSA gel. The signal of this complex was significantly reduced by the anti-Sp1 antibody but not by a control antibody, indicating that this complex was caused by the binding of Sp1 to the DNA probe. Similar results were obtained with the Sp1-2 DNA probe and the Sp1-3 DNA probe. These results confirm that the Sp1-1, Sp1-2, and Sp1-3 sites can indeed be bound by Sp1. In order to understand the functions of these Sp1 bindings sites in the expression of HBV genes, they were individually mutated to prevent the binding of Sp1 (Fig. 1). As shown in Fig. 2, the G-to-A mutation at nt 1748 prevented binding of Sp1 to the Sp1-1 DNA probe. Similarly, a single-nucleotide deletion at nt 1736 and a C-to-A mutation at nt 1630 (Fig. 1) also prevented binding of Sp1 to the Sp1-2 site and the Sp1-3 site, respectively. Head-to-tail dimers of the HBV genome containing these mutations were then constructed and transfected into Huh7 cells for gene expression studies.
FIG. 2.
EMSA analysis of the Sp1 binding sites. The preparation of Huh7 nuclear extracts (N.E.), the procedures for EMSA, and the sequences of the DNA probes used are described in Materials and Methods. The monoclonal anti-Sp1 antibody (α-Sp1) used for the supershift assay was purchased from Santa Cruz Biochemicals, and the control monoclonal anti-p53 antibody (α-p53) was purchased from Calbiochem. (A) Sp1-1 probe. (B) Sp1-2 probe. (C) Sp1-3 probe. WT, probe containing the wild-type sequence; MT, probe containing the mutated sequence. The arrows mark the locations of the Sp1-DNA complex.
Effects of Sp1-1 and Sp1-2 mutations on HBV gene expression.
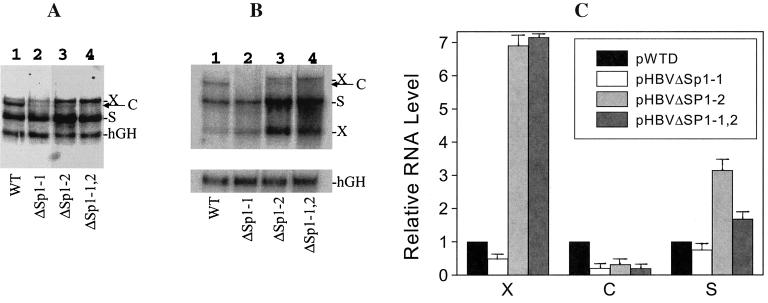
HBV RNAs expressed in Huh7 cells were extracted and analyzed by Northern blotting using the 32P-labeled 3.2-kb HBV genomic DNA fragment. As shown in Fig. 3A, the mutation introduced at the Sp1-1 binding site reduced the HBV C gene transcripts to an almost undetectable level. This mutation had little effect on the levels of the S RNA and the 3.9-kb X RNA. The mutation introduced at the Sp1-2 site also reduced the C gene transcripts to an almost undetectable level. Interestingly, while the Sp1-1 mutation had no apparent effects on the transcription of the S RNA and the 3.9-kb X RNA, the Sp1-2 mutation increased the level of the S RNA approximately threefold and the level of the 3.9-kb X RNA approximately sevenfold (Fig. 3A and C). The Sp1-1 and Sp1-2 double mutation also similarly reduced the C RNA level and increased the S RNA and the 3.9-kb X RNA levels (Fig. 3A and C). In all of our transfection experiments, we have included an hGH reporter plasmid for cotransfection to serve as an internal control to monitor the transfection efficiency (Fig. 3). The 32P-labeled hGH cDNA probe and the 3.2-kb HBV DNA probe were included in the hybridization buffer for Northern blotting. Unfortunately, the hGH internal control obscured the signal of the 0.7-kb X RNA, which migrated only slightly faster than the hGH mRNA on the gel. For this reason, an identical Northern blot experiment was repeated, and the hGH cDNA probe was used separately for the hybridization. In this experiment, the 32P-labeled 0.6-kb BamHI-BglII HBV DNA fragment containing sequences derived mostly from the X region was used as the probe. Based on our experience in the past, this X region-specific probe significantly enhanced the 0.7-kb X RNA signal (13). As shown in Fig. 3B, similar to the case for the 3.9-kb X RNA, the 0.7-kb X RNA level was increased six- to sevenfold by either the Sp1-2 single mutation or the Sp1-1 and Sp1-2 double mutation. This result indicates that the increase of the 3.9-kb X RNA level was not due to an increase of efficiency in bypassing the unique poly(A) site during RNA transcription but rather was most likely due to an increase of the X promoter activity.
FIG. 3.
(A and B) Northern blot analysis of the HBV RNAs. Huh7 cells transfected with pWTD (lanes 1), pHBVΔSp1-1 (lanes 2), pHBVΔSp1-2 (lanes 3), and pHBVΔSp1-1,2 (lanes 4) genomic DNA constructs were lysed 48 h after transfection for RNA isolation. (A) Northern blot analysis using the 32P-labeled 3.2-kb HBV DNA probe and the hGH cDNA probe together. X, C, and S mark the locations of the 3.9-kb X RNA (13), the C gene transcripts, and the S gene transcripts, respectively. The location of the hGH RNA internal control is also indicated. (B) Northern blot analysis using the 32P-labeled HBV X region-specific DNA probe and the hGH cDNA probe separately. X, C, and S mark the locations of the X, C, and S gene transcripts, respectively. (C) Relative RNA levels expressed by various HBV DNA constructs. X, C, and S indicate the 3.9-kb X gene transcript, the C gene transcripts, and the S gene transcripts, respectively. The RNA bands shown in panel A were measured with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) and normalized against the hGH RNA internal control. The X, C, and S RNA levels expressed by the wild-type genome, pWTD, were arbitrarily set as 1. Error bars indicate standard deviations.
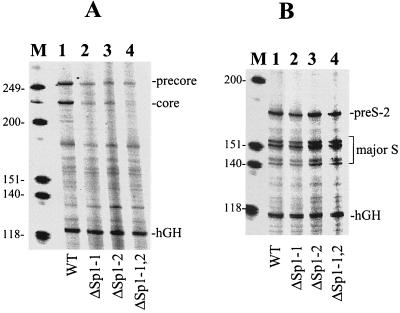
Since the resolution of the Northern blot gel is not high enough to resolve precore and core RNAs, we also conducted primer extension analysis to analyze the effects of Sp1-1 and Sp1-2 mutations on the transcription of these two RNAs. As shown in Fig. 4A, the mutation of the Sp1-1 site significantly reduced both precore and core RNA levels, which is consistent with the Northern blot results. Similarly, the Sp1-2 mutation also significantly reduced both precore RNA and core RNA levels, which is again consistent with the Northern blot result. The Sp1-1 and Sp1-2 double mutation appeared to have an additive effect and further reduced the precore RNA and core RNA levels. Note that although the wild-type HBV genome expressed the precore RNA and core RNA at similar levels, the Sp1-1 and Sp1-2 double mutation reduced the core RNA to an almost undetectable level without totally abolishing the expression of the precore RNA (Fig. 4A). This result indicates that the double mutation had a greater effect on expression of the core RNA than on that of the precore RNA. Similar primer extension experiments were also conducted to analyze the S gene transcripts. As shown in Fig. 4B, five major S gene transcripts could be detected. These bands were consistent with those previously reported (19, 29); the uppermost band represented the pre-S-2 RNA, and the four lower bands were the major S RNAs (19, 29). In agreement with the Northern blot results, the Sp1-1 mutation had no effect on the pre-S-2 RNA and major S RNA levels, but the Sp1-2 single mutation or the Sp1-1 and Sp1-2 double mutation increased both pre-S-2 and major S RNA levels approximately twofold (Fig. 4B).
FIG. 4.
Primer extension analysis of HBV RNAs. HBV RNA isolated from Huh7 cells was analyzed by primer extension as described in Materials and Methods. (A) Primer extension analysis of the C gene transcripts. The locations of the precore RNA, core RNA, and hGH RNA are indicated. (B) Primer extension analysis of the S gene transcripts. The locations of the pre-S-2 RNA, major S RNA, and hGH RNA are indicated. Lanes 1, cells transfected with pWTD; lanes 2, cells transfected with pHBVΔSp1-1; lanes 3, cells transfected with pHBVΔSp1-2; lanes 4, cells transfected with pHBVΔSp1-1,2. Lanes M, DNA markers and their sizes (in kilodaltons).
Lack of effects of the X protein on HBV gene expression in Huh7 cells.
Both the Sp1-1 site and the Sp1-2 site reside in the X protein-coding region. The nt 1748 G-to-A mutation in the Sp1-1 site is a silent mutation for the X protein and thus should not affect its expression. Our attempts to find a silent mutation for the X protein in the Sp1-2 site that would abolish the binding of Sp1 have been unsuccessful (data not shown). For that reason, the G residue at nt 1736 in the Sp1-2 site was deleted. This deletion created a frameshift in the X protein sequence, which might be responsible for the observed increase of the S gene and X gene levels. To rule out this possibility, two new mutations were introduced into the X gene coding sequence. The first mutation converted nt 1376 from A to C. This mutation removed the initiation codon of the X protein-coding sequence (Fig. 5A). The second mutation is a C-to-T mutation at nt 1397. This mutation created a premature termination codon in the X protein sequence. These two mutations would abolish the expression of the 16.5-kDa X protein. As shown in Fig. 5B, in agreement with a previous report (2), these two mutations, when introduced into the HBV genome, had no apparent effect on HBV gene expression in Huh7 cells. These two mutations were then introduced into pHBVΔSp1-2, the HBV genomic construct that carried the Sp1-2 mutation, to abolish the expression of the frameshifted X protein. As shown in Fig. 5C, pHBVΔSp1-2 with and without the two additional mutations that abolished the frameshifted X protein expression produced indistinguishable viral RNA phenotypes (Fig. 5C). These results indicate that the enhancement of the S gene and X gene expression by the Sp1-2 mutation was not due to the frameshifted X protein.
FIG. 5.
Northern blot analysis of HBV RNA expressed by the X-negative HBV DNA constructs. (A) Schematic illustration of the mutations introduced into the X protein-coding sequence. The locations of the three Sp1 binding sites are indicated by arrowheads. ENII, ENII enhancer; CP, C promoter; X protein, X protein-coding sequence; PC and C, transcription start sites of precore and core RNAs, respectively. (B) HBV RNA expressed in Huh7 cells by pWTD without (lane 1) or with (lane 2) the mutations that abolished the expression of the 16.5-kDa X protein. C, S, and X mark the locations of the C RNA, the S RNA, and the 0.7-kb X RNA, respectively. (C) HBV RNA expressed in Huh7 cells by pHBVΔSp1-2 without (lane 1) or with (lane 2) the additional mutations in the X protein-coding sequence. X, C, S, and hGH mark the locations of the 3.9-kb X RNA, the C RNA, the S RNA, and the hGH RNA, respectively. In both panels B and C, the 3.2-kb HBV genomic DNA was used as the probe.
Role of the Sp1 binding site in the ENII enhancer.
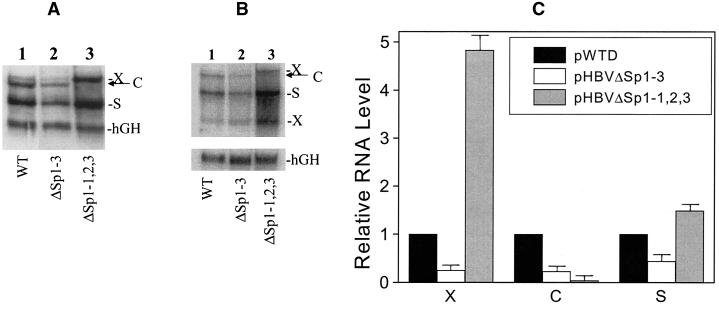
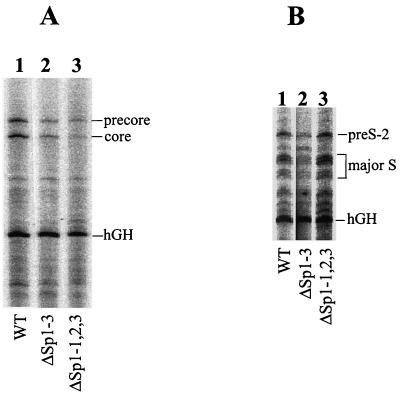
As mentioned above (Fig. 1), upstream from the core promoter is another Sp1 binding site, which we named Sp1-3. This site is located in the ENII enhancer. To investigate the possible role of this Sp1 binding site in HBV gene expression, a C-to-A mutation at nt 1630 was introduced into this site. This nucleotide mutation created a silent mutation in the X protein-coding sequence. As discussed above, this mutation abolished the binding of Sp1 (Fig. 2C). The mutation was introduced into the HBV genome, which was then transfected into Huh7 cells. As shown in Fig. 6, the removal of the Sp1-3 binding site resulted in an approximately two- to threefold reduction of the levels of all of the HBV RNAs (Fig. 6C). This result suggests that the optimal activity of the ENII enhancer is dependent on Sp1, and hence abolishing Sp1 binding to this enhancer element results in the suppression of all of the HBV promoter activities. When the Sp1 binding sites in the ENII enhancer and the core promoter were mutated simultaneously, the C RNA level was further reduced, but interestingly, the S RNA and 3.9-kb X RNA levels were increased by nearly twofold and fivefold, respectively (Fig. 6A and C). The 0.7-kb X RNA level also showed a similar increase (Fig. 6B). The Northern blot results were again verified by primer extension experiments. As shown in Fig. 7A, the mutation of the Sp1-3 site reduced precore and core RNA levels, and the mutations of all three Sp1 sites further reduced the expression levels of these two RNAs, with a greater effect on the core RNA. In contrast, while the mutation of the Sp1-3 site reduced the pre-S-2 RNA and major S RNA levels, the mutation of all three Sp1 sites slightly increased the levels of pre-S-2 and major S RNAs. These results indicate that the Sp1-3 site in the ENII enhancer is a positive regulator for the expression of all of the HBV genes, and its mutation reduces the expression levels of all of the HBV genes. The suppressive effect of this mutation on the expression of S and X genes, however, can be compensated for and further enhanced by the mutation of the downstream Sp1 sites in the core promoter.
FIG. 6.
(A and B) Northern blot analysis of HBV RNAs expressed by the HBV genomic construct carrying the Sp1-3 mutation. Huh7 cells transfected by the HBV DNA constructs were lysed 48 h after transfection for RNA isolation. The HBV RNA was then analyzed by Northern blotting using the HBV DNA probe and the hGH cDNA probe either together (A) or separately (B). Lanes 1, Huh7 cells transfected by pWTD; lanes 2, Huh7 cells transfected by pHBVΔSp1-3; lanes 3, Huh7 cells transfected by pHBVΔSp1-1,2,3. The locations of the X, C, S, and hGH RNAs are marked. (C) Relative RNA levels quantified with a PhosphorImager. The gel shown in panel A was used for measurements based on the procedures described in the legend to Fig. 3C. X, C, and S indicate the 3.9-kb X gene transcript, the C gene transcripts, and the S gene transcripts, respectively. Error bars indicate standard deviations.
FIG. 7.
Primer extension analysis of HBV RNA expressed from the HBV DNA constructs carrying the Sp1-3 mutation. (A) Primer extension analysis of the C gene transcripts. The locations of the precore RNA, the core RNA, and the hGH RNA are marked. (B) Primer extension analysis of the S gene transcripts. The locations of the pre-S-2 RNA, the major S RNA, and the hGH RNA are marked. Lanes 1, Huh7 cells transfected by the wild-type HBV DNA; lanes 2, Huh7 cells transfected by the HBV DNA genome carrying the Sp1-3 mutation; lanes 3, Huh7 cells transfected by the HBV DNA genome carrying the mutations of all three Sp1 sites.
DISCUSSION
The HBV DNA genome is circular and contains four transcription units. Due to its compact genome size, these four transcription units extensively overlap. Significant progress has been made during the past 15 years toward identifying cis- and trans-acting factors that may regulate the activities of these four transcription units. One of the trans-acting factors is the ubiquitous transcription factor Sp1. Sp1 has two binding sites in the HBV core promoter and one in its upstream ENII enhancer (36, 40, 41). Our results demonstrate that both Sp1 sites in the core promoter are important for the transcription of the precore RNA and the core RNA, although their effects on the core RNA appear to be more prominent (Fig. 4A). These results are consistent with a report by Yu and Mertz (36), who found that Sp1 had a greater effect on core RNA transcription than on precore RNA transcription in their in vitro transcription experiments.
A surprising finding of ours is that the removal of the Sp1-2 site resulted in the enhancement of transcription of the S gene and the X gene. The enhancement for the S gene ranged from two- to threefold, and that for the X gene ranged from five- to sevenfold, in different experiments. Due to the low expression level of the pre-S-1 RNA, the possible effect of the Sp1-2 site on the transcription of this RNA was not investigated. Previous studies have shown that when the four HBV promoters were analyzed individually, the X promoter was found to display a very strong transcriptional activity (1, 12). However, in the context of the entire HBV genome, this promoter is relatively weak (e.g., see Fig. 3B, 5B, and 6B). The results shown in this report indicate that the suppression of the X promoter activity in the entire HBV genome could be at least partially attributed to the Sp1-2 site.
The enhancement of S gene and X gene transcription by the mutation of the Sp1-2 site cannot be simply due to suppression of the core promoter activity, as the mutation of the Sp1-1 site, which similarly suppressed the core promoter activity, did not enhance the transcription of the S and X genes. How Sp1 that binds to the Sp1-2 site suppresses the transcription of the S and X genes remains unclear. It does not appear likely that this binding or mutation affects the RNA stability, as Sp1 is a DNA binding protein and the mutation is located in a region not known to be involved in the regulation of RNA stability. It is perhaps more likely that this Sp1 interacts with other transcription factors binding to the S promoter and the X promoter to suppress their activities. Alternatively, since the Sp1-2 site is located within the transcription units of the S and X genes, Sp1 binding to this site may suppress the process of elongation of S and X gene transcripts. In either case, it is rather intriguing that Sp1 binding to the adjacent Sp1-1 site fails to do so. It will be interesting to determine whether this is due to the subtle difference between the Sp1-1 and Sp1-2 sequences (Fig. 1) or to their respective locations on the core promoter. Experiments are being conducted to resolve these issues. Our results are reminiscent of recent findings which indicate that Sp1 can serve both as a positive regulator and as a negative regulator for gene expression (27, 39).
The Sp1-3 site in the ENII enhancer is apparently important for the transcription of all of the HBV genes, as the removal of this site by mutagenesis led to the suppression of expression of all of the HBV genes (Fig. 6). If the mutation of this site is also accompanied by mutations of the Sp1-1 and Sp1-2 sites, then the expression level of the C RNAs is further reduced (Fig. 6 and 7). However, in this case, the S RNA and X RNA levels are increased rather than reduced (Fig. 6 and 7). This result indicates that the negative activity of the Sp1 factor binding to the Sp1-2 site likely plays a more prominent role than the positive activity of the Sp1 factor binding to the Sp1-3 site in the regulation of S gene and X gene expression.
In summary, in this report we demonstrate that the two Sp1 binding sites in the core promoter and the Sp1 site in the ENII enhancer are required for optimal activities of the core promoter, and while the Sp1-2 site is a positive regulator of the core promoter, it is a negative regulator of the major S promoter and the X promoter. The dual activities of Sp1 may be important for the differential regulation of HBV gene expression during natural HBV infection.
ACKNOWLEDGMENTS
We thank Jinah Choi for help with the preparation of some of the figures and Jinah Choi and T. S. Benedict Yen for critical reading of the manuscript.
This work was supported by a research grant from the National Institutes of Health.
REFERENCES
- 1.Antonucci T K, Rutter W J. Hepatitis B virus (HBV) promoters are regulated by the HBV enhancer in a tissue-specific manner. J Virol. 1989;63:579–583. doi: 10.1128/jvi.63.2.579-583.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blum H E, Zhang Z S, Galun E, von Weizsacker F, Garner B, Liang T J, Wands J R. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J Virol. 1992;66:1223–1227. doi: 10.1128/jvi.66.2.1223-1227.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briggs M R, Kadonaga J T, Bell S P, Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986;234:47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- 4.Buckwold V E, Chen M, Ou J H. Interaction of transcription factors RFX1 and MIBP1 with the gamma motif of the negative regulatory element of the hepatitis B virus core promoter. Virology. 1997;227:515–518. doi: 10.1006/viro.1996.8360. [DOI] [PubMed] [Google Scholar]
- 5.Buckwold V E, Xu Z, Chen M, Yen T S, Ou J H. Effects of a naturally occurring mutation in the hepatitis B virus basal core promoter on precore gene expression and viral replication. J Virol. 1996;70:5845–5851. doi: 10.1128/jvi.70.9.5845-5851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckwold V E, Xu Z, Yen T S, Ou J H. Effects of a frequent double-nucleotide basal core promoter mutation and its putative single-nucleotide precursor mutations on hepatitis B virus gene expression and replication. J Gen Virol. 1997;78:2055–2065. doi: 10.1099/0022-1317-78-8-2055. [DOI] [PubMed] [Google Scholar]
- 7.Chen M, Hieng S, Qian X, Costa R, Ou J H. Regulation of hepatitis B virus ENI enhancer activity by hepatocyte-enriched transcription factor HNF3. Virology. 1994;205:127–132. doi: 10.1006/viro.1994.1627. [DOI] [PubMed] [Google Scholar]
- 8.Chen M, Ou J H. Cell type-dependent regulation of the activity of the negative regulatory element of the hepatitis B virus core promoter. Virology. 1995;214:198–206. doi: 10.1006/viro.1995.9940. [DOI] [PubMed] [Google Scholar]
- 9.Cherrington J, Russnak R, Ganem D. Upstream sequences and cap proximity in the regulation of polyadenylation in ground squirrel hepatitis virus. J Virol. 1992;66:7589–7596. doi: 10.1128/jvi.66.12.7589-7596.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge L, Rudolph P. Simultaneous introduction of multiple mutations using overlap extension PCR. BioTechniques. 1997;22:28–30. doi: 10.2144/97221bm03. [DOI] [PubMed] [Google Scholar]
- 11.Guidotti L G, Eggers C M, Raney A K, Chi S Y, Peters J M, Gonzalez F J, McLachlan A. In vivo regulation of hepatitis B virus replication by peroxisome proliferators. J Virol. 1999;73:10377–10386. doi: 10.1128/jvi.73.12.10377-10386.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo W T, Bell K D, Ou J H. Characterization of the hepatitis B virus EnhI enhancer and X promoter complex. J Virol. 1991;65:6686–6692. doi: 10.1128/jvi.65.12.6686-6692.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo W T, Wang J, Tam G, Yen T S, Ou J S. Leaky transcription termination produces larger and smaller than genome size hepatitis B virus X gene transcripts. Virology. 1991;181:630–636. doi: 10.1016/0042-6822(91)90896-j. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Buckwold V E, Hon M W, Ou J H. Mechanism of suppression of hepatitis B virus precore RNA transcription by a frequent double mutation. J Virol. 1999;73:1239–1244. doi: 10.1128/jvi.73.2.1239-1244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li M, Xie Y, Wu X, Kong Y, Wang Y. HNF3 binds and activates the second enhancer, ENII, of hepatitis B virus. Virology. 1995;214:371–378. doi: 10.1006/viro.1995.0046. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Cabrera M, Letovsky J, Hu K Q, Siddiqui A. Transcriptional factor C/EBP binds to and transactivates the enhancer element II of the hepatitis B virus. Virology. 1991;183:825–829. doi: 10.1016/0042-6822(91)91019-d. [DOI] [PubMed] [Google Scholar]
- 17.Nassal M, Junker-Niepmann M, Schaller H. Translational inactivation of RNA function: discrimination against a subset of genomic transcripts during HBV nucleocapsid assembly. Cell. 1990;63:1357–1363. doi: 10.1016/0092-8674(90)90431-d. [DOI] [PubMed] [Google Scholar]
- 18.Ori A, Shaul Y. Hepatitis B virus enhancer binds and is activated by the hepatocyte nuclear factor 3. Virology. 1995;207:98–106. doi: 10.1006/viro.1995.1055. [DOI] [PubMed] [Google Scholar]
- 19.Ou J, Rutter W J. Hybrid hepatitis B virus-host transcripts in a human hepatoma cell Proc. Natl Acad Sci USA. 1985;82:83–87. doi: 10.1073/pnas.82.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ou J H. Molecular biology of hepatitis B virus e antigen. J Gastroenterol Hepatol. 1997;12:S178–S187. doi: 10.1111/j.1440-1746.1997.tb00499.x. [DOI] [PubMed] [Google Scholar]
- 21.Ou J H, Bao H, Shih C, Tahara S M. Preferred translation of human hepatitis B virus polymerase from core protein- but not from precore protein-specific transcript. J Virol. 1990;64:4578–4581. doi: 10.1128/jvi.64.9.4578-4581.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pei D Q, Shih C H. Transcriptional activation and repression by cellular DNA-binding protein C/EBP. J Virol. 1990;64:1517–1522. doi: 10.1128/jvi.64.4.1517-1522.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raney A K, Johnson J L, Palmer C N, McLachlan A. Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J Virol. 1997;71:1058–1071. doi: 10.1128/jvi.71.2.1058-1071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raney A K, Le H B, McLachlan A. Regulation of transcription from the hepatitis B virus major surface antigen promoter by the Sp1 transcription factor. J Virol. 1992;66:6912–6921. doi: 10.1128/jvi.66.12.6912-6921.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raney A K, Milich D R, Easton A J, McLachlan A. Differentiation-specific transcriptional regulation of the hepatitis B virus large surface antigen gene in human hepatoma cell lines. J Virol. 1990;64:2360–2368. doi: 10.1128/jvi.64.5.2360-2368.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russnak R, Ganem D. Sequences 5′ to the polyadenylation signal mediate differential poly(A) site use in hepatitis B viruses. Genes Dev. 1990;4:764–776. doi: 10.1101/gad.4.5.764. [DOI] [PubMed] [Google Scholar]
- 27.Sasahara R M, Takahashi C, Noda M. Involvement of the Sp1 site in ras-mediated downregulation of the RECK metastasis suppressor gene. Biochem Biophys Res Commun. 1999;264:668–675. doi: 10.1006/bbrc.1999.1552. [DOI] [PubMed] [Google Scholar]
- 28.Shaul Y, Rutter W J, Laub O. A human hepatitis B viral enhancer element. EMBO J. 1985;4:427–430. doi: 10.1002/j.1460-2075.1985.tb03646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Standring D N, Rutter W J, Varmus H E, Ganem D. Transcription of the hepatitis B surface antigen gene in cultured murine cells initiates within the presurface region. J Virol. 1984;50:563–571. doi: 10.1128/jvi.50.2.563-571.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valenzuela P, Quriroga M, Zaldivar J, Gray P, Rutter W J. The nucleotide sequence of the hepatitis B viral genome and the identification of the major genes. In: Fields B N, Jaenisch R, Fox C F, editors. Animal virus genetics. New York, N.Y: Academic Press; 1980. pp. 57–70. [Google Scholar]
- 31.Wang W X, Li M, Wu X, Wang Y, Li Z P. HNF1 is critical for the liver-specific function of HBV enhancer II. Res Virol. 1998;149:99–108. doi: 10.1016/s0923-2516(98)80085-x. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Chen P, Wu X, Sun A L, Wang H, Zhu Y A, Li Z P. A new enhancer element, ENII, identified in the X gene of hepatitis B virus. J Virol. 1990;64:3977–3981. doi: 10.1128/jvi.64.8.3977-3981.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yee J K. A liver-specific enhancer in the core promoter region of human hepatitis B virus. Science. 1989;246:658–661. doi: 10.1126/science.2554495. [DOI] [PubMed] [Google Scholar]
- 34.Yen T S B. Regulation of hepatitis B virus gene expression. Semin Virol. 1993;4:33–42. [Google Scholar]
- 35.Yu X, Mertz J E. Differential regulation of the pre-C and pregenomic promoters of human hepatitis B virus by members of the nuclear receptor superfamily. J Virol. 1997;71:9366–9374. doi: 10.1128/jvi.71.12.9366-9374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu X, Mertz J E. Promoters for synthesis of the pre-C and pregenomic mRNAs of human hepatitis B virus are genetically distinct and differentially regulated. J Virol. 1996;70:8719–8726. doi: 10.1128/jvi.70.12.8719-8726.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuh C H, Ting L P. C/EBP-like proteins binding to the functional box-alpha and box-beta of the second enhancer of hepatitis B virus. Mol Cell Biol. 1991;11:5044–5052. doi: 10.1128/mcb.11.10.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuh C H, Ting L P. The genome of hepatitis B virus contains a second enhancer: cooperation of two elements within this enhancer is required for its function. J Virol. 1990;64:4281–4287. doi: 10.1128/jvi.64.9.4281-4287.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaid A, Li R, Luciakova K, Barath P, Nery S, Nelson B D. On the role of the general transcription factor Sp1 in the activation and repression of diverse mammalian oxidative phosphorylation genes J. Bioenerg Biomembr. 1999;31:129–135. doi: 10.1023/a:1005499727732. [DOI] [PubMed] [Google Scholar]
- 40.Zhang P, McLachlan A. Differentiation-specific transcriptional regulation of the hepatitis B virus nucleocapsid gene in human hepatoma cell lines. Virology. 1994;202:430–440. doi: 10.1006/viro.1994.1359. [DOI] [PubMed] [Google Scholar]
- 41.Zhang P, Raney A K, McLachlan A. Characterization of functional Sp1 transcription factor binding sites in the hepatitis B virus nucleocapsid promoter. J Virol. 1993;67:1472–1481. doi: 10.1128/jvi.67.3.1472-1481.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]