Abstract
Cadherins constitute a family of cell-surface proteins that mediate intercellular adhesion through the association of protomers presented from juxtaposed cells. Differential cadherin expression leads to highly specific intercellular interactions in vivo. This cell–cell specificity is difficult to understand at the molecular level because individual cadherins within a given subfamily are highly similar to each other both in sequence and structure, and they dimerize with remarkably low binding affinities. Here, we provide a molecular model that accounts for these apparently contradictory observations. The model is based in part on the fact that cadherins bind to one another by “swapping” the N-terminal β-strands of their adhesive domains. An inherent feature of strand swapping (or, more generally, the domain swapping phenomenon) is that “closed” monomeric conformations act as competitive inhibitors of dimer formation, thus lowering affinities even when the dimer interface has the characteristics of high-affinity complexes. The model describes quantitatively how small affinity differences between low-affinity cadherin dimers are amplified by multiple cadherin interactions to establish large specificity effects at the cellular level. It is shown that cellular specificity would not be observed if cadherins bound with high affinities, thus emphasizing the crucial role of strand swapping in cell–cell adhesion. Numerical estimates demonstrate that the strength of cellular adhesion is extremely sensitive to the concentration of cadherins expressed at the cell surface. We suggest that the domain swapping mechanism is used by a variety of cell-adhesion proteins and that related mechanisms to control affinity and specificity are exploited in other systems.
Keywords: binding affinity, domain swapping, binding specificity, protein interfaces
Adhesive interactions between cadherin family members presented on cell surfaces is thought to provide a key driving force in the development of tissue architecture (1–3). Morphogenetic changes often correlate directly with changes in expression of individual cadherin family members, and genetic deletion of individual cadherins in vivo, or misexpression by transgenesis, interferes with tissue development in characteristic ways for each cadherin family member. Such data have been interpreted as evidence of the presence of highly specific homophilic interactions in the molecular recognition properties of individual cadherins. However, cell-sorting experiments in vitro often fail to reveal adhesive specificity in the binding behavior of cells expressing different cadherin types (4, 5). Moreover, analysis of cadherin sequences and structures do not clearly reveal why homodimer formation should be substantially preferred over the formation of heterodimers (6). Thus, cell–cell adhesion specificity is not simply correlated with molecular-binding specificity within the cadherin family. Indeed, it has been suggested that cellular binding specificity arises primarily from differences in overall cadherin cell surface concentration rather than from the identity of cadherins presented on cell surfaces (5, 7). In this article, we present a model for the function of cadherins on apposing cell surfaces that predicts cell–cell adhesive energies based on molecular binding affinities. The model describes how small affinity differences between individual cadherins can be translated into large affinity differences between cells expressing different cadherins. The model also demonstrates that the adhesive behavior of cells is highly sensitive to the cadherin concentration and, thus, offers an explanation of prior contradictory results that show adhesive specificity in some contexts, but not others, for the same cadherin pairs.
A central finding of the model is that small differences between the binding affinities of homodimers and heterodimers can be translated into large differences in intercellular adhesive strength, provided that dissociation constants are larger than the cadherin concentration at the cell surface, and provided that multiple interactions are used to amplify the affinity differences. We show that cadherin dimerization based on strand swapping, as revealed by crystallographic studies, imparts novel binding characteristics that are essential to this mechanism of generating specificity at the cellular level. The basis of the idea is that strand swapping, by its very nature, creates a situation in which a “closed” monomer form acts as a competitive inhibitor of dimer formation. Because of this competition, binding affinities are low even though the dimer interface has the characteristics of high-affinity protein–protein complexes.
In the next section, we summarize structural studies of cadherins and explain why it is difficult to understand the frequently observed preference of cadherins for homophilic dimerization. We then discuss how strand swapping lowers affinities for cadherin dimerization by describing the free-energy profile for the dimerization process with and without the energetic effects of strand swapping. Our model for adhesion at the cellular level is then presented and used to calculate the number of cadherin dimers formed between apposing cells as a function of cadherin surface density. The dependence of the number of dimers formed on dimerization affinities and cadherin concentration provides a simple framework for understanding the energetics of cadherin-mediated adhesion at the level of the cell. Last, we describe the relationship of strand swapping to alternate molecular mechanisms that function in related ways to achieve low affinity and high specificity in other systems.
Methods
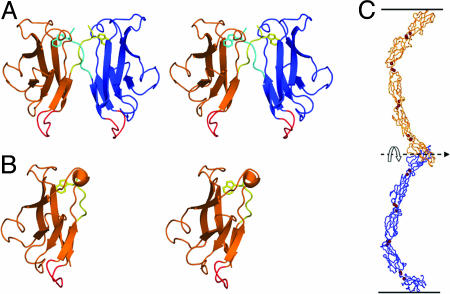
Homophilic vs. Heterophilic Interfaces: Affinity Differences Appear to Be Small. Crystallographic studies on a number of classical cadherins have provided detailed information about the nature of the dimer interface (8–10). Its characteristic feature is the swapping of the N-terminal β-strand in the EC1 domain (the A strand) so that the A strand of one monomer replaces the A strand of the other (Fig. 1A). A key element of this twofold symmetric interaction is the insertion of the side chain from Trp-2 of the A strand of one protomer into a pocket extending into the hydrophobic core of the adhesive partner (see Fig. 1 A). Mutations of Trp-2 or its acceptor pocket residues result in the loss of adhesive function for all cadherins that have been tested (11–13). Studies have also revealed structures for monomeric cadherins; notably, in one of these structures, the side chain of Trp-2 inserts into the core of its own rather than a partner protomer (Fig. 1B) (9, 14). Exchange of the A strand between two domains is characteristic of the general phenomenon of “3D domain swapping” that has been observed in many other protein complexes (15, 16). Consistent with the inherently symmetric nature of this binding mode, the EC1 domains of cadherin pairs that are presented from apposing cells bind in a parallel fashion even though the overall orientation of the two cadherins is antiparallel (Fig. 1C). The parallel orientation is crucial to achieve a true domain-swapped mode of interaction in which near-equivalent interfaces can be achieved in both the monomer and dimer states.
Fig. 1.
Structural models of C-cadherin. (A) Structure of the EC1 dimer of C-cadherin. The swapped A strands, including the conserved Trp-2 side-chain, are shown in yellow and cyan. The putative hinge loop is shown in red. (B) Structure of the E-cadherin monomer (PDB ID code 1O6S). The A-strand is shown in yellow with the Trp-2 side-chain facing the interior of its own protomer. The hinge loop is shown in red. (C) The crystal structure of the entire ectodomain of C-cadherin (8). Note that despite the fact that the two C termini point in opposite directions, as if toward apposing membranes, the “crescent” shape of the entire ectodomain orients the interacting EC1 domains in a parallel fashion in which the N termini are pointing in the same direction. This geometrical arrangement is necessary for domain swapping.
Strand swapping in cadherins was first observed in structures of the EC1 domain of N-cadherin (10) and was also observed in the only current structure of a complete ectodomain, that of C-cadherin (8). Most recently, a combined NMR and x-ray study of an EC1–EC2 fragment from E-cadherin (9) provided a biophysical characterization of the monomer–dimer transition. This work showed that, at low protein concentrations or in the absence of calcium, a monomer form is observed in which the A strand remains part of its own protomer, but as concentrations approach the millimolar range, a strand-swapped dimer is formed. Although the monomer form is not normally viewed as a biologically active conformation, we argue here that the monomer–dimer equilibrium is an essential property of the cadherin adhesion mechanism and is crucial for the evolution of highly specific homophilic interactions that characterize cadherin adhesion at the cellular level.
Fig. 2 shows a multiple sequence alignment of the EC1 domains from a number of type I cadherins. The interfacial residues in each dimer structure are shown in green. It is apparent in the figure that these residues are remarkably well conserved in all EC1 structures. With the exception of P-cadherin, the swapped A strand is essentially identical in all type I cadherins, although the polar residue at position 8 is somewhat variable. Structural analysis also fails to provide a clear indication of why homophilic dimerization might be preferred energetically over heterophilic dimerization. The backbones of C-, E-, and N-cadherin superimpose to within 1 Å and, when the molecular surfaces are compared, most surface points in the interface also superimpose to within 1 Å. Moreover, most of the key interactions are conserved in all type I cadherins, including that of the Trp-2 with the residues of the hydrophobic pocket into which it inserts, a pair of conserved salt bridges between Glu-89 and the partner N terminus, and a number of interfacial hydrogen bonds involving backbone groups.
Fig. 2.
Multiple sequence alignment of type I cadherins. Residues that are in the dimer interface are highlighted in green. Residues that may be involved in mediating specificity are shown in orange. The putative hinge loop residues are highlighted in pink with the conserved Gly-15 residue shown in red.
There are some subtype-specific differences at the periphery of the interface that potentially could affect specificity. For example, there is a hydrogen bond formed between Lys-8 and Gln-23 in C-cadherin that is not present in E- or N-cadherin, which have a Ser or Asn at position 8, respectively. Moreover, Asn-27 in C- and E-cadherin is replaced by Asp-27 in N-cadherin. It is also possible that long-range electrostatic interactions involving noninterfacial residues contribute to relative affinities. A detailed energetic analysis of affinity differences among closely related cadherins is beyond the scope of this article. However, it seems clear that the binding free-energy differences associated with homophilic and heterophilic complexes are quite small (and unlikely to be much greater than ≈1 kcal/mol, approximately a factor of 5 difference in affinities). How then is a high degree of adhesive selectivity achieved at the cellular level? We return to this question after discussing the molecular basis of low-affinity cadherin binding in the next section.
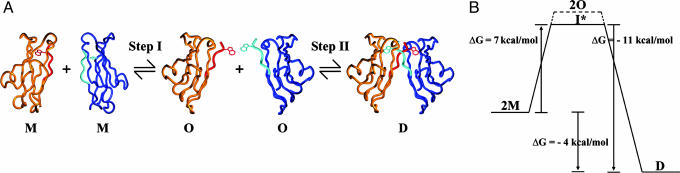
Strand Swapping as a Structural Basis of Low Affinity. In Fig. 3A, we define a hypothetical reaction in which two monomers (M) form a dimer (D). Step I involves a transition between the monomer and a hypothetical “open” state (O) in which the A strand has moved into the conformation that it will adopt in the dimer but the dimer itself has not yet formed. Step II corresponds to the association of two monomers, both in the open state, to form a dimer. O can be thought of as a high-energy reaction intermediate in which the attractive interactions between the A strand and its parent domain have been lost, but new interactions in the dimer state have not yet been made. Although it is unlikely that O corresponds to the true reaction intermediate, or set of intermediates (I*), we note that the energy of O is an upper limit to that of I* because the actual reaction will proceed via the lowest-energy activated state.
Fig. 3.
Putative cadherin binding reaction and conformational energy diagram. (A) In step I, two cadherin monomers with their A-strands in the “closed” state (M) undergo a conformational change whereby the A-strands assume the “open” state (O). In step II, the “open” cadherin protomers associate with each other to form a strand-swapped dimer (D). (B) Conformational free energies associated with each step of the binding reaction shown in A. Note that O is either equivalent or more positive in free energy than the true intermediate (I*).
Recent measurements on E-cadherin (9) allow us to assign approximate energies to various steps in the reaction. For EC1–EC2 constructs, the measured Kd of the reaction D→2M was found to be 0.7 mM, corresponding to a dimer formation free-energy difference of approximately -4 kcal/mol. In the presence of calcium, the activation energy for dimer formation, corresponding to the energy difference between I* and M, was estimated as 7 kcal/mol (9). Thus, the free-energy difference between D and I* is approximately -11 kcal/mol, as indicated in Fig. 3B. Below, we assume that I* may be represented by O, and we note that our main conclusions become stronger if we recognize that O is higher in energy than I*.
We first consider the reaction 2O→D, which can be thought of as a standard binding reaction in the absence of swapping (i.e., if no low-energy monomer state were available). Using the C-cadherin structure as a basis of our analysis, the dimer interface buries ≈1,840 Å2 (920 Å2 per protomer) of accessible area, of which 51% is nonpolar. N- and E-cadherins were found to have dimer interfaces of comparable sizes. E-cadherin is the only cadherin for which both monomeric and dimeric crystal structures are available. For E-cadherin, the surface area buried in the process O→D is very close to that buried in the reaction O→M. Based on the energy diagram shown in Fig. 3B, the binding free energy of the 2O→D reaction is at least -11 kcal/mol, corresponding to a ≈10-nM affinity. It is evident from the figure that the effect of swapping is to reduce the binding free energy from approximately -11 (10 nM) to approximately -4 kcal/mol (0.7 mM). Thus, strand swapping makes it possible for an interface that has the structural characteristics of a nanomolar binding reaction to have a weak millimolar binding affinity.
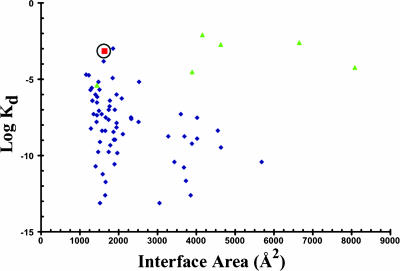
A number of studies have characterized the physical and chemical properties of protein–protein interfaces in some detail, and the results of the analysis have been compared with measured dissociation constants (17, 18). The data are replotted in Fig. 4, in which, for consistency, the buried accessible areas have been recalculated with our own algorithm (19), and some additional values of Kd have been taken from the literature. The interfaces in the figure vary in size between 500 and 8,000 Å2, and the Kd values vary by over 11 orders of magnitude. For interfaces with buried areas >1,500 Å2, almost all of the Kd values are <10 μM, with many of the dimeric complexes exhibiting subnanomolar affinities. Note that the lower limit for the hypothetical 2O→D binding reaction, of -11 kcal/mol (a ≈10-nM binding affinity) is consistent with the range of affinities observed for many of the complexes that bury approximately the same amount of surface area as buried in the cadherin dimer. However, as shown strikingly in Fig. 4, classical cadherins and several other dimeric complexes are highly unusual in that they have relatively large interfacial surface areas and extremely low binding affinities. Indeed, it is remarkable that, with few exceptions, all of the complexes in the figure that have affinities of 0.1 mM or weaker have interfaces that involve domain swapping.
Fig. 4.
Plot of buried accessible areas vs. Kd constants for various dimers. Blue diamonds correspond to data points taken from refs. 17 and 18, although surface areas are replotted as indicated in the text. Green triangles correspond to domain-swap dimers (33–38). The circled red square denotes the E-cadherin dimer. Note that the domain swapped dimers tend to have low affinities even when they have large buried surface areas.
The reason that domain swapping is associated with weak affinities is that the same interface is formed by the swapped domain in both the monomeric and dimeric molecules, as pointed out by Eisenberg and coworkers (15, 16). The existence in the monomer of a docking site for the swapped strand that is essentially identical to the docking site on the partner domain in the dimer leads to a competition between monomeric and dimeric species and, hence, lowers free energies of complex formation. This competition enables the formation of low-affinity dimers that have the characteristics of protein–protein complexes, which are highly specific in the sense that they bind tightly and exhibit well formed interdomain interfaces. Thus, the swapping mechanism enables cadherins to be highly specific with respect to noncadherins but, as described above, not with respect to other family members. Also, the conversion from monomer to dimer occurs on the order of seconds (9, 20). It is essential for the function of cadherins for domain swapping to be fast, and in this sense, the phenomenon differs from other domain-swapping events that have been described, such as the diphtheria toxin oligomerization process (15).
The source of the small energetic difference between the monomers and dimers may reside in part in the “hinge loop” (21), which in cadherins corresponds to the AB loop that links the swapped A strand to the rest of the EC1 domain (see Figs. 1 and 2) Comparison of the monomer and dimer conformations of E-cadherin reveals that the largest changes in ϕ, ϕ angles involve residues 14–16, especially Gly-15. The AB loop contains two conserved calcium-binding residues (Glu-11 and Asn-12). Because calcium binding is known to promote cadherin dimerization (14, 22, 23), direct Ca+2 interaction with the hinge loop may regulate the monomer–dimer equilibrium. Moreover, some of the residues in this loop (in particular, Gly-15 and Pro-18) are highly conserved in different cadherin subfamilies, suggesting that strand swapping mediated by conformational changes in the hinge loop is a conserved mechanism used by different cadherins.
Multiple Weak Interactions Enhance Selectivity if Concentrations Are Low. To explain how specificity within the cadherin family is achieved at the cellular level, we describe a simple model for adhesion mediated by two cadherin subtypes i and j presented on apposing cell surfaces, I and J, respectively. (Note that i and j can be the same subtype, whereas I and J always denote different cells.) The model provides a theoretical basis for linking the molecular binding behavior of cadherins with the adhesive properties of cadherin-expressing cells. We assume that a local chemical equilibrium is rapidly established when two cells come into contact (i.e., that cadherins have enough time to diffuse within and into the contact region and form dimers). For simplicity, we ignore the formation of more complicated structures, such as intercellular junctions, which takes place on a longer time scale.
The adhesive energy, ΔG(I, J), between two cells, I and J, can be written as follows:
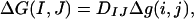 |
[1] |
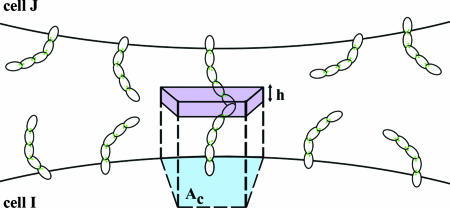
where DIJ is the number of dimers formed between two cells and Δg(i, j) is the binding free energy of the monomer–dimer reaction (see above). A value of DIJ can be approximated by calculating the relative concentrations of monomers and dimers in an interface in which the total number of monomers is known. We use a Kd (0.7 mM) determined from the NMR measurements of Haussinger et al. (22) to estimate the dimerization probability of EC1 domains from cadherins on apposing cell surfaces based on the following simple model. We assume that cadherins are freely diffusible on the cell surface, with 2D surface densities ρ(I) = N(I)/A(I), ρ(J) = N(J)/A(J), where N denotes the number of cadherins per cell and A is the surface area of a cell. To relate the 2D densities to 3D concentrations, C(I) and C(J), we identify an “interfacial shell” of volume V = Ach containing the EC1 domains of cadherins from both cells, with Ac denoting the average surface area associated with one cadherin monomer. The shell thickness h may be identified with the amplitude of EC1 fluctuations normal to the cell surface, which are limited because of their membrane attachment through EC2–EC5 (see Fig. 5). We assume that the EC1 domains diffuse within this shell and are able to dimerize as if they were in a bulk solution (This is clearly an approximation because there are restrictions in rotational motion for cadherins anchored on the cell surface.) For the concentrations of EC1 domains, we now have C(I) = ρ(I)/h, C(J) = ρ(J)/h.
Fig. 5.
Model for adhesion mediated by cadherins presented on apposing cell surfaces, I and J. Cadherin monomers in random orientations on the cell surfaces are shown as crescent-shaped structures. The blue patch denotes the average surface area Ac, occupied by each monomer on the cell surface. The “interfacial shell,” with thickness h, containing the interfacial EC1 domain is shown in light purple.
The equilibrium constant for the dimer dissociation reaction, Kd, is given by the following:
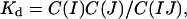 |
[2] |
where C(IJ) is the concentration of dimers. In the next section, we calculate values of C(I, J) and DIJ for two cells, each having a diameter of 10 μm and each presenting 25,000 cadherins on the cell surface (80 cadherins per μm2). This value is at the lower end of the range used by Foty and Steinberg in their studies of transfected cells (24). To estimate h, we note that cadherins may undergo bending fluctuations and, thus, assume a range of angles, implying a range of distances between the EC1 domains and the cell surface. A cryoelectron microscopy study (25) revealed a minimum intermembrane distance of ≈150 Å, or ≈75 Å per cadherin monomer. Based on the crystal structure, the maximal extension of a cadherin dimer is 385 Å (8) so that each cadherin can maximally extend up to ≈195 Å from a cell surface. Assuming that the EC1 domain of a free cadherin monomer will be located ≈75–195 Å from the cell surface, we obtain h ≈ 120 Å.
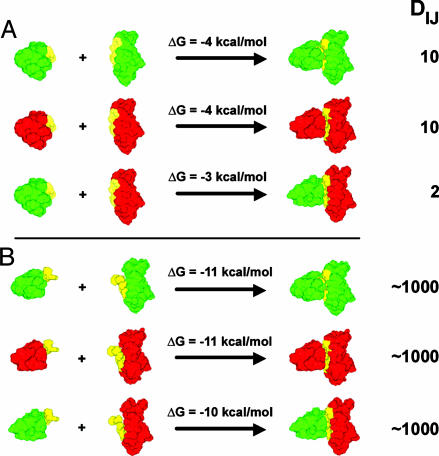
By using the values of ρ and h given above, C(I) and C(J) are found to be ≈10 μM, which is well below the binding affinity of 0.7 mM for the cadherin dimerization reaction (9). C(IJ) is calculated from Eq. 2 to be ≈0.1 μM, so that approximately one dimer is formed per 100 monomers. To obtain a rough estimate of the number of monomers in the contact region between two cells, we note that a single perfectly spherical cell could contact 12 other identical cells. Thus, at most, ≈8% of the surface of each cell is available to interact with another cell, although the actual contact region must be <8%. We arbitrarily assume that the contact region between any two cells is half of this value, or 4% of the cell surface. Thus, for a cell containing 25,000 monomers, a rough estimate of the number of monomers available to contact a complementary cell surface is ≈1,000, but only ≈10 of these will form dimers. Given the uncertainties in the numbers, this estimate could clearly change in either direction, but the essential point for the purpose of our discussion is that the number of dimers involved in the initial contact between two cells must be quite small in absolute terms. For illustrative purposes, we summarize some of the numbers used in this discussion in Fig. 6A. Red and green indicate two different cadherins (i.e., E and N), which can form homodimers or heterodimers. The reactants correspond to the closed state of each monomer.
Fig. 6.
Illustrative example of the effects of domain swapping on the extent of dimer formation. Green and red indicate the EC1 domains of two different cadherin family members. The A-strand is shown in yellow. DIJ, the number of dimers, is calculated as described in the text. (A) The reactions involve domain swapping so that the two interacting monomers are in the closed state when they are not bound. (B) The dimerization reactions depicted involve two monomers that are in the open state so that no domain swapping occurs. Note that at physiological cadherin concentrations differential binding is shown only in the domain-swapped case (in A).
If we assume that binding free energy associated with heterophilic adhesion is slightly weaker (by 1 kcal/mol; see above) than that for homophilic adhesion, DIJ will be reduced by a factor of ≈5, corresponding to only approximately two dimers per pair of cells (Fig. 6A). By using a value of Δg(i, j) = -4 kcal/mol for homophilic dimerization and -3 kcal/mol for heterophilic dimerization, Eq. 1 predicts that cell–cell adhesion free energies, ΔG(I, J), are -40 and -6 kcal/mol for homophilic and heterophilic adhesion, respectively. The difference in the two values is so large that, under equilibrium conditions, heterophilic cell–cell adhesion would never be observed. The example shows clearly how even small affinity differences between members of the same cadherin subfamily can be translated into large equilibrium specificity differences if enough contacts are made. However, the strength of adhesive forces upon initial contact has kinetic consequences as well because if cells are in contact for enough time, additional cadherins will diffuse into the contact region and will eventually form junctions. Depending on the conditions of a particular in vitro experiment (i.e., shear forces, flow rates, etc.) cells may remain together long enough to strengthen their interactions or may move apart. It is essential then that specificity is manifest at a kinetic level so that the wrong cells will move apart before they are stuck together. When only a few dimers are formed per cell pair, cells are unlikely to adhere strongly. However, the situation changes dramatically if affinities are large or if cadherin densities are high.
If no swapping occurred to lower binding affinities, and cadherins formed homodimers with nanomolar affinities, a 1-kcal/mol reduction in the binding free energy would yield heterodimers that also formed with near-nanomolar affinities. The situation in which the reactants correspond with the open state of the monomer is shown in Fig. 6B. Because, in this case, the estimated cadherin concentrations (in the micromolar range; see above) would be orders of magnitude greater than their dimerization affinities, essentially every cadherin in an interface (i.e., 1,000 in our estimate) would form a dimer. The consequence of having a large number of dimers, each with binding energies on the order of -10 kcal/mol, would be that essentially any two cells expressing cadherins would be irreversibly stuck together. Binding specificity would be lost. Thus, it is essential for cadherin binding affinities to be very low and for concentrations to be kept in a range in which only a small number of dimers are formed (Fig. 6). Only under these conditions does it appear to be possible to achieve high kinetic specificity, even among closely related family members.
Because the number of dimers increases quadratically with cadherin surface density, it is easy to imagine cases in which the adhesive forces between cells expressing cadherins at very different concentrations is quite large, which would have important consequences for cell-adhesive specificity. If we assume a density of 300 cadherins per μm2 (100,000 cadherins per cell), the model predicts ≈160 homophilic dimers per cell pair and ≈30 dimers for heterophilic adhesion. The latter number predicts approximately -90 kcal/mol for heterophilic cell–cell adhesion energies so that we might expect cells expressing different types of cadherins to become kinetically trapped. In contrast, cells containing only 10,000 cadherins are predicted to form approximately one or two dimers for the case of homophilic adhesion and zero dimers for heterophilic adhesion. The sensitivity of cell–cell adhesion strength to cadherin densities in this range of expression levels may help reconcile the conflicting results reported in the literature (see above). Specifically, it is clear that the extent to which sorting is observed in cell-aggregation experiments will strongly depend on concentrations (7, 26) and mixing conditions (5). The sensitivity of adhesion to expression levels highlights the difficulty in matching in vivo conditions in in vitro experiments.
Other Mechanisms for Achieving High Specificity and Low Affinity
Based on this discussion, we might expect domain swapping to be characteristic of other proteins involved in cell–cell adhesion. Bjorkman and coworkers have suggested that domain swapping may occur in the hemolin (27) family of insect adhesion proteins and in the L1 family of mammalian adhesion proteins (28). The mechanism suggested by these researchers involves entire domains rather than individual strands, but we suggest that the underlying rationale of enabling the formation of highly specific and low-affinity molecular complexes remains the same. As is the case for cadherins, the term high specificity is used here only with respect to nonfamily members. Specificity within a family requires a mechanism that exploits multiple protein interactions, as described in the previous section.
Other mechanisms to achieve specificity and low affinity have been discussed. In some cases, binding specificity is coupled to a chemical reaction such as peptide synthesis (29) or cleavage by sequence-specific ribozymes (30, 31). In the case of ribozyme cleavage, recognition sequences that are too long will reduce sequence discrimination because both correct and mismatched sequences will tend to bind (30). The crucial parameter in both cases relates to the relative sizes of the off constants and the catalytic rate constants. In essence, rate constants must be “tuned” so that the correct sequence will react and the wrong sequence will dissociate.
Tuning is achieved in the cadherin system by using domain swapping to reduce affinities so that dissociation constants are larger than cadherin concentrations. More generally, any mechanism in which favorable free-energy contributions to monomer stability must be overcome in order for a complex to be formed can be used to lower affinities even for well formed, seemingly high-affinity interfaces. Of course, there will always be some strain energy associated with binding, but in some cases, this strain may be associated with a specific mechanism such as domain swapping, or the loss of entropy when an unstructured peptide binds to its cognate recognition domain in a signaling pathway. One mechanism that has attracted considerable attention is when regions of proteins are required to fold to bind. This general phenomenon is often seen in proteins involved in regulatory functions such as transcriptional control (32). We note that the “cost” of protein folding is likely to be entropic, whereas the “cost” of domain swapping is likely to be enthalpic. However, the underlying logic is in setting up a competitive situation in which forces that favor a monomer, whether their origin is enthalpic or entropic, can be overcome only by a highly specific oligomeric interface.
Last, we note that tuning dissociation constants to be larger than the concentrations of interacting species facilitates the evolution of closely related family members that do not interfere with each other's function. The relative effects of small differences in binding affinity, generated for example by changes in only a single hydrogen bond, will be greatest when the fraction of free monomers is much larger than the fraction of bound species. In the case of cadherins, intrafamily specificity at the cellular level is then achieved through the exploitation of multiple weak interactions, a phenomenon that appears to be quite general in its importance.
Acknowledgments
We thank Melanie Bennett, Nir Ben Tal, Jane Dyson, David Eisenberg, Wayne Hubbell, Tom Jessell, Diana Murray, and Peter Wright for insightful discussions and helpful comments on the manuscript. This work was supported in part by National Institutes of Health Grants GM-30518 (to B.H.H.) and GM-062270 (to L.S.), Israel Science Foundation Grant 227/02 (to A.B.-S.), and the U.S.–Israel Binational Science Foundation Grant 2002-75 (to A.B.-S.). The Fritz Haber Center is supported by the Minerva Foundation (Munich).
Author contributions: C.P.C., L.S., and B.H.H. designed research; C.P.C., S.P., and A.B.-S. performed research; C.P.C., L.S., and B.H.H. analyzed data; A.B.-S. contributed new reagents/analytic tools; and C.P.C. and B.H.H. wrote the paper.
References
- 1.Vleminckx, K. & Kemler, R. (1999) BioEssays 21, 211-220. [DOI] [PubMed] [Google Scholar]
- 2.Takeichi, M. (1995) Curr. Opin. Cell Biol. 7, 619-627. [DOI] [PubMed] [Google Scholar]
- 3.Pla, P., Moore, R., Morali, O. G., Grille, S., Martinozzi, S., Delmas, V. & Larue, L. (2001) J. Cell. Physiol. 189, 121-132. [DOI] [PubMed] [Google Scholar]
- 4.Niessen, C. M. & Gumbiner, B. M. (2002) J. Cell Biol. 156, 389-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duguay, D., Foty, R. A. & Steinberg, M. S. (2003) Dev. Biol. 253, 309-323. [DOI] [PubMed] [Google Scholar]
- 6.Patel, S. D., Chen, C. P., Bahna, F., Honig, B. & Shapiro, L. (2003) Curr. Opin. Struct. Biol. 13, 690-698. [DOI] [PubMed] [Google Scholar]
- 7.Steinberg, M. S. & Takeichi, M. (1994) Proc. Natl. Acad. Sci. USA 91, 206-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boggon, T. J., Murray, J., Chappuis-Flament, S., Wong, E., Gumbiner, B. M. & Shapiro, L. (2002) Science 296, 1308-1313. [DOI] [PubMed] [Google Scholar]
- 9.Haussinger, D., Ahrens, T., Aberle, T., Engel, J., Stetefeld, J. & Grzesiek, S. (2004) EMBO J. 23, 1699-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapiro, L., Fannon, A. M., Kwong, P. D., Thompson, A., Lehmann, M. S., Grubel, G., Legrand, J. F., Als-Nielsen, J., Colman, D. R. & Hendrickson, W. A. (1995) Nature 374, 327-337. [DOI] [PubMed] [Google Scholar]
- 11.Malicki, J., Jo, H. & Pujic, Z. (2003) Dev. Biol. 259, 95-108. [DOI] [PubMed] [Google Scholar]
- 12.Tamura, K., Shan, W. S., Hendrickson, W. A., Colman, D. R. & Shapiro, L. (1998) Neuron 20, 1153-1163. [DOI] [PubMed] [Google Scholar]
- 13.Ozawa, M. (2002) J. Biol. Chem. 277, 19600-19608. [DOI] [PubMed] [Google Scholar]
- 14.Pertz, O., Bozic, D., Koch, A. W., Fauser, C., Brancaccio, A. & Engel, J. (1999) EMBO J. 18, 1738-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett, M. J., Choe, S. & Eisenberg, D. (1994) Proc. Natl. Acad. Sci. USA 91, 3127-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, Y. & Eisenberg, D. (2002) Protein Sci. 11, 1285-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo Conte, L., Chothia, C. & Janin, J. (1999) J. Mol. Biol. 285, 2177-2198. [DOI] [PubMed] [Google Scholar]
- 18.Nooren, I. M. & Thornton, J. M. (2003) J. Mol. Biol. 325, 991-1018. [DOI] [PubMed] [Google Scholar]
- 19.Nicholls, A., Sharp, K. A. & Honig, B. (1991) Proteins 11, 281-296. [DOI] [PubMed] [Google Scholar]
- 20.Perret, E., Benoliel, A. M., Nassoy, P., Pierres, A., Delmas, V., Thiery, J. P., Bongrand, P. & Feracci, H. (2002) EMBO J. 21, 2537-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett, M. J., Schlunegger, M. P. & Eisenberg, D. (1995) Protein Sci. 4, 2455-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haussinger, D., Ahrens, T., Sass, H. J., Pertz, O., Engel, J. & Grzesiek, S. (2002) J. Mol. Biol. 324, 823-839. [DOI] [PubMed] [Google Scholar]
- 23.Nagar, B., Overduin, M., Ikura, M. & Rini, J. M. (1996) Nature 380, 360-364. [DOI] [PubMed] [Google Scholar]
- 24.Foty, R. A. & Steinberg, M. S. (2005) Dev. Biol. 278, 255-263. [DOI] [PubMed] [Google Scholar]
- 25.Miyaguchi, K. (2000) J. Struct. Biol. 132, 169-178. [DOI] [PubMed] [Google Scholar]
- 26.Friedlander, D. R., Mege, R. M., Cunningham, B. A. & Edelman, G. M. (1989) Proc. Natl. Acad. Sci. USA 86, 7043-7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su, X. D., Gastinel, L. N., Vaughn, D. E., Faye, I., Poon, P. & Bjorkman, P. J. (1998) Science 281, 991-995. [DOI] [PubMed] [Google Scholar]
- 28.Schurmann, G., Haspel, J., Grumet, M. & Erickson, H. P. (2001) Mol. Biol. Cell 12, 1765-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ninio, J. (1986) FEBS Lett. 196, 1-4. [DOI] [PubMed] [Google Scholar]
- 30.Herschlag, D. (1991) Proc. Natl. Acad. Sci. USA 88, 6921-6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin, P. Z. & Pyle, A. M. (1999) J. Mol. Biol. 291, 15-27. [DOI] [PubMed] [Google Scholar]
- 32.Dyson, H. J. & Wright, P. E. (2004) Chem. Rev. 104, 3607-3622. [DOI] [PubMed] [Google Scholar]
- 33.Barrientos, L. G., Louis, J. M., Botos, I., Mori, T., Han, Z., O'Keefe, B. R., Boyd, M. R., Wlodawer, A. & Gronenborn, A. M. (2002) Structure (Cambridge, U.K.) 10, 673-686. [DOI] [PubMed] [Google Scholar]
- 34.Hakansson, M., Svensson, A., Fast, J. & Linse, S. (2001) Protein Sci. 10, 927-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayes, M. V., Sessions, R. B., Brady, R. L. & Clarke, A. R. (1999) J. Mol. Biol. 285, 1857-1867. [DOI] [PubMed] [Google Scholar]
- 36.LeFevre, K. R. & Cordes, M. H. (2003) Proc. Natl. Acad. Sci. USA 100, 2345-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis, R. J., Scott, D. J., Brannigan, J. A., Ladds, J. C., Cervin, M. A., Spiegelman, G. B., Hoggett, J. G., Barak, I. & Wilkinson, A. J. (2002) J. Mol. Biol. 316, 235-245. [DOI] [PubMed] [Google Scholar]
- 38.Rousseau, F., Schymkowitz, J. W., Wilkinson, H. R. & Itzhaki, L. S. (2001) Proc. Natl. Acad. Sci. USA 98, 5596-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]