Abstract
Histone acetyltransferases have been shown to participate in many essential cellular processes, particularly those associated with activation of transcription. SAGA (Spt-Ada-Gcn5 acetyltransferase) and SLIK (SAGA-like) are two highly homologous multisubunit histone acetyltransferase complexes that were originally identified in the yeast Saccharomyces cerevisiae. Here, we identify the protein Sgf73/Sca7 as a component of SAGA and SLIK, and a homologue of the human SCA7-encoded protein ataxin-7, which, in its polyglutamine expanded pathological form, is responsible for the neurodegenerative disease spinocerebellar ataxia 7 (SCA7). Our findings indicate that yeast Sca7 is necessary for the integrity and function of both SAGA and SLIK, and that the human ataxin-7 is able to compliment the loss of Sca7 in yeast. A polyglutamine-expanded version of ataxin-7 assembles a SAGA complex that is depleted of critical proteins that regulate the ability of SAGA to acetylate nucleosomes. These observations have significant implications for the function of the human Sca7 protein in disease pathogenesis.
Keywords: chromatin, transcription, SCA7
The close interaction between DNA and histone proteins in the context of chromatin presents a fundamental problem for processes such as transcription and repair in which direct access to the DNA template is required. Chromatin-modifying complexes regulate chromatin function by transferring different moieties to histones, most frequently to their tail regions. Patterns of histone acetylation are associated with the transcriptional capacity of chromatin (1). SAGA and SLIK/SALSA are highly conserved transcriptional coactivator complexes that preferentially acetylate histones H3 and H2B (2, 3) and are involved in the regulation of DNA polymerase II-transcribed genes in yeast (4). These two Gcn5-dependent histone acetyltransferase (HAT) complexes have also been shown to contain many transcription-related proteins (1, 5). Homologous STAGA, PCAF, and TFTC complexes have been isolated from mammalian cells, indicating that SAGA complexes play a pivotal role in transcription in eukaryotes (6–8).
Here, we identify the yeast homologue of the human SCA7 gene product, ataxin-7, as a component of the SAGA and SLIK complexes. SCA7 is one of eight known autosomal dominant neurodegenerative disorders, including Huntington's disease and several other spinocerebellar ataxias, caused by CAG (encoding glutamine, Q) nucleotide repeat expansions (9, 10). In SCA7, cerebellar ataxia is associated with macular dystrophy, retinal degeneration, progressive loss of photoreceptor and bipolar cells, and eventual blindness (11, 12). The glutamine-repeat number expands from <35 (nonpathogenic) to an extreme of 300 (pathogenic) copies, although 75% of normal alleles encode proteins with only 10 repeats (13). However, the physiological role of the normal ataxin-7 protein is unknown. The distribution of ataxin-7 mRNA and protein do not correlate with the selective neuronal cell loss observed in patients; therefore, the selective pattern of degeneration contrasts with ubiquitous ataxin-7 expression (14). Thus, understanding the normal function of the Sca7 protein is likely to be critical for understanding how disease pathogenesis arises.
Materials and Methods
Yeast Strains and Plasmids. The WT, sca7Δ, Δ+ 10Q, and Δ+ 60Q strains used for large-scale SAGA and SLIK preparations were derivatives of BY4741 and BY4742 (Research Genetics, Huntsville, AL). TAP tagging was performed in the yeast strain BY4742, as described in ref. 15. Gal4-VP16 toxicity assays were performed by using the VP16 expression plasmid pSB201 (CEN, Leu) (16). Human SCA7–10Q and SCA7–75Q were obtained in the mammalian expression vector pCDNA3.1/HisA (10). They were cloned into the yeast expression vector pRS416 (CEN, Ura). CAG repeat lengths were confirmed by sequencing, and Sca7–75Q contracted to 60Q during the cloning process.
Purification of SAGA and SLIK Complexes. Whole-cell extracts (WCE) were prepared by glass bead-disruption from 4 liters each of WT, sca7Δ, Δ + 10Q, and Δ + 60Q yeast strains grown to mid-log phase in yeast extract/peptone/dextrose medium as described in ref. 15. WCE were bound to a Mono Q HR 16/10 column (GE Health Care). SAGA and SLIK were eluted with a 500-ml linear gradient from 100 to 500 mM NaCl, and peak chromatographic fractions were determined by HAT assays and Western blotting. WCE from 1 liter each of WT and SCA7-TAP strains were prepared in the same manner, bound to 200 μl of calmodulin resin (Stratagene), and eluted as described in ref. 15. For complex isolation used for silver staining in Fig. 5D, SAGA from WT and Δ + 60Q was purified from WCE from 1 liter of yeast over successive Ni-NTA agarose, Mono Q HR 16/10, Superose 6 HR 10/30, and Mini Q 3–2/3 columns (GE Health Care) as described in ref. 3. Silver staining was also done as described in ref. 17.
Fig. 5.
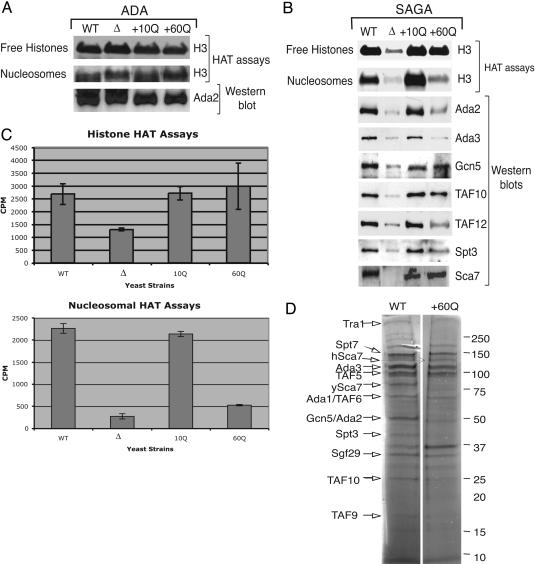
The Δ + 60Q strain has a defect in nucleosomal acetylation and altered levels of some SAGA proteins. (A) Fluorogram of HAT assays with histone or nucleosome substrates, and anti-Ada2 Western blots, using peak ADA complex fractions from WT, sca7Δ, Δ + 10Q, and Δ + 60Q strains. Consistent with ADA not containing Sca7, it is unaffected by addition of either SCA7 allele. (B) Fluorogram of HAT assays using SAGA from WT, sca7Δ, Δ + 10Q, and Δ + 60Q strains, performed with free histone or nucleosome substrates. Western blots with the indicated antisera show corresponding SAGA protein levels. Note that yeast Sca7 runs around 75 kDa, and human Sca7–10Q and hSca7–60Q run around 130 kDa. (C) Histograms displaying cpm of 3H incorporation onto free histones or nucleosomes after liquid HAT assays (15). Bars indicate standard errors. (D) Silver stain gel of partially purified SAGA from WT and Δ + 60Q strains. Molecular masses are indicated on the right, and predicted SAGA components by size are indicated on the left.
Western Blot Analysis and HAT Assays. Proteins from Mono Q columns were immunoblotted with rabbit Ada2, Ada3, TAF12, Spt3, TAF10, Sca7, and Gcn5 or protein A (to detect Sca7-TAP) antisera. Fluorography and liquid HAT assays were performed with HeLa free histones and nucleosomes as described in ref. 15.
β-Galactosidase Assays. β-Galactosidase assays were performed in WT, sca7Δ, and ada2Δ yeast strains as described in ref. 16. Yeast strains were transformed with a vector containing a GAL1 promoter element fused to LacZ and a second low-copy expression vector containing Gal4-VP16. If SAGA is present, Gal4-VP16 bound to the GAL1 promoter drives LacZ expression.
Results
Here, we report the identification of a 73-kDa protein as a component of SLIK and SAGA. The yeast protein Sgf73/Sca7 (18) was identified by mass spectrometry (data not shown) as a component of both purified SAGA and SLIK (3). Note that Sgf73/Sca7 migrates with Spt20, and that Spt7 is truncated in SLIK (3). This protein contains a highly conserved domain that is homologous between proteins from several organisms (Fig. 1A) including human ataxin-7. The yeast and human proteins share 21% identity over the entire length of the proteins and 50% identity over 50 aa (Fig. 1 A) (19).
Fig. 1.
Confirmation of Sca7 association with SAGA and SLIK and identification of its homologues. (A) Protein sequence alignment (boxshade, www.ch.embnet.org/software/BOX_form.html) showing the most highly conserved region of ySca7 and its homologues. Black boxes represent identity and gray boxes represent similarity between amino acids at given positions. (B) WCE from untagged WT and SCA7-TAP strains were bound to and eluted from calmodulin beads, and the elution fractions were analyzed by Western blotting. Shown are Western blots using protein A, TAF12, and Ada2 antibodies. I, input; FT, flow-through; W, wash; E, elution.
Yeast Sca7/Sgf73, hereafter referred to as Sca7, is a previously uncharacterized component of SAGA and SLIK. To confirm the association of Sca7 with other components of SAGA and SLIK, a C-terminal TAP-tagged SCA7 strain was created (15). WCE were prepared from WT and SCA7-TAP strains, bound to calmodulin, and eluted with EGTA (3). The elution fractions were analyzed by Western blotting with an anti-protein A antibody to detect the Sca7 fusion protein and antibodies to detect the SAGA and SLIK components Ada2 and TAF12 (17). Fig. 1B shows the coprecipitation of Sca7 with Ada2 and TAF12 in the SCA7-TAP strain but not in the WT untagged strain, suggesting that Sca7 is a stable component of SLIK and SAGA.
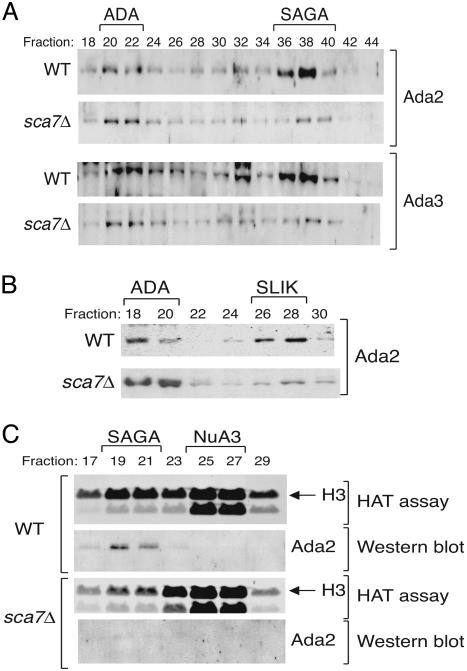
To determine what effect the loss of Sca7 had on SAGA and SLIK, WCE were prepared from WT and sca7Δ strains and bound to Ni2+-NTA agarose. The eluates were fractionated over a Mono Q anion exchange column to isolate SAGA, or flow-through extracts were fractionated to isolate SLIK (3). To assess the integrity of these complexes, the fractions were analyzed by Western blotting. Fig. 2 A and B shows that the normal fractionation of SAGA and SLIK is significantly compromised in the sca7Δ strain. However, the ADA complex, which contains a subset of Ada proteins found in SAGA and SLIK, including Ada2 and Ada3, as well as the enzymatic subunit Gcn5 (20), remains intact (Fig. 2 A and B). Consistent with a loss of typical SAGA fractionation in the sca7Δ strain, there was also a corresponding loss of SAGA-associated HAT activity upon further fractionation over a Superose 6 size exclusion column (Fig. 2C). The unrelated Sas3-dependent NuA3 HAT complex (21) remains unaffected by the deletion of Sca7. This finding indicates that Sca7 is specifically important for the HAT activity and normal fractionation of the SAGA and SLIK complexes.
Fig. 2.
Sca7Δ yeast display defects in SAGA and SLIK fractionation and function. (A) WCE from WT and sca7Δ strains were fractionated over a Mono Q anion exchange column (after Ni-NTA binding and elution) to separate SAGA and SLIK. Western blot analysis with Ada2 and Ada3 antibodies confirms loss of normal fractionation of the SAGA complex in the sca7Δ strain. (B) Nickel flow-through extracts were also fractionated over a Mono Q anion exchange column to isolate SLIK. Western blot analysis with an Ada2 antibody demonstrates loss of normal SLIK fractionation in the sca7Δ strain as well. (C) Fluorograms of HAT assays performed with free histone substrates and peak SAGA Mono Q fractions further purified over a Superose 6 column. Western blotting of the same fractions was performed with an Ada2 antibody.
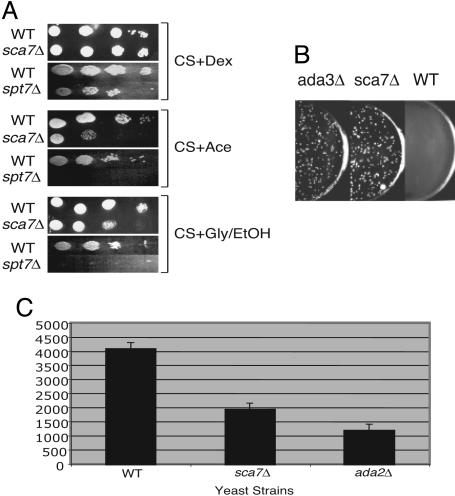
Slight growth defects were observed in the sca7Δ strain on media containing acetate as the carbon source, media containing glycerol and ethanol as the carbon sources (Fig. 3A), and synthetic complete media lacking inositol (data not shown). Inositol auxotrophy is typical of mutations in coactivators (indicative of a defect in general transcription), and the inability to use acetate as a carbon source indicates possible defects in the tricarboxylic acid or glyoxylate cycles, consistent with loss of SAGA and/or SLIK function (3, 22, 23).
Fig. 3.
Transcriptional and growth defects of sca7Δ yeast. (A) Ten-fold serial dilutions of WT, sca7Δ, and spt7Δ [as a control (3)] yeast strains were assayed for their ability to grow on synthetic complete medium containing dextrose, acetate, or glycerol plus ethanol carbon sources. (B) Yeast transformed with a high-copy Gal4-VP16 plasmid were plated on selective medium. Gal4-VP16 toxicity assays were performed in WT, ada3Δ, and sca7Δ strains. (C) Histogram depicting β-galactosidase assays using low-copy Gal4-VP16-dependent transcription of a Gal1-LacZ reporter construct transformed into the indicated yeast strains. Bars indicate standard errors.
The transcriptional coactivation capacity of the sca7 deletion strain was analyzed using an assay to screen for ADA gene function. All ADA gene products isolated to date are known to incorporate into the SAGA and SLIK complexes. This assay tests the cells' ability to survive overexpression of Gal4-VP16, which is toxic to WT cells. Overexpression of VP16 has been suggested to cause misdirection of SAGA to inappropriately activate a number of cellular genes, and to sequester general transcription factors away from productive transcription complexes (23). Mutations in SAGA that alter functional interaction with VP16 allow the cells to overcome the toxic growth defect and constitute an ADA phenotype. WT and sca7Δ yeast strains, along with an ada3Δ strain as a control, were transformed with a high-copy plasmid containing Gal4-VP16 (23). Fig. 3B shows that the sca7Δ strain behaves in the same manner as the ada3Δ strain in this assay, suppressing VP16 toxicity. This finding indicates that SCA7 is an ADA family member, consistent with its function as part of SAGA and SLIK. The sca7Δ yeast strain was also found to be deficient in low-copy Gal4-VP16-dependent expression of a LacZ reporter gene (Fig. 3C), similar to other ADA family members (23).
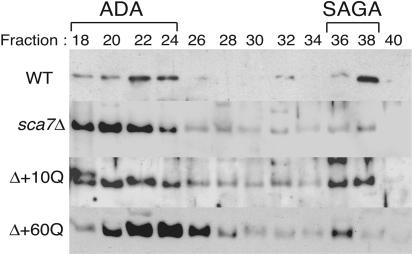
We next wanted to determine whether the human SCA7 gene was able to complement the loss of the yeast SCA7 gene. Therefore, we generated yeast vectors expressing human SCA7-encoded ataxin-7 protein containing either 10 (nonpathogenic) or 60 (pathogenic) glutamines (Q) (10). These plasmids were transformed independently into the sca7Δ yeast strain. SAGA complex was purified from these strains (termed Δ + 10Q, or simply 10Q, and Δ + 60Q, or 60Q) as described above. The fractions were analyzed by Western blotting to assess the restoration of normal fractionation of SAGA. Fig. 4 indicates that the SAGA fractionation lost in the sca7Δ yeast strain, as compared with WT, is restored by human ataxin-7 with both 10Q and 60Q.
Fig. 4.
Defect in normal SAGA fractionation in the sca7Δ strain is restored by the addition of human SCA7 with 10Q or 60Q. The human SCA7 gene encoding either 10 (10Q) or 60 (60Q) glutamine repeats was introduced into the sca7Δ yeast strain on a pR412 (CEN) expression vector. Extracts from the Δ+ 10Q and Δ + 60Q strains were fractionated over a Mono Q column in the same manner as the WT and deletion strain. Western blot analysis of column fractions was performed with an Ada2 antibody.
To determine whether aspects of SAGA function that were lost in the deletion strain were restored by the addition of the human genes, we tested what effect human ataxin-7 10Q and 60Q had on SAGA HAT activity in comparison with the ADA complex when expressed in the sca7Δ yeast strain. HAT assays were performed by using partially purified ADA or SAGA complex from the WT, sca7Δ, Δ + 10Q, and Δ + 60Q strains. The fractionation pattern (Fig. 4) and histone and nucleosomal HAT activity of partially purified ADA complex from peak chromatographic fractions from these strains remains unaltered by any of the sca7 mutations (Fig. 5A). However, when free histones are used as a substrate, the SAGA HAT activity that is diminished in the sca7Δ strain is restored to near WT levels in both the Δ + 10Q and Δ + 60Q strains (Fig. 5B). In contrast, the Δ + 60Q strain specifically is defective in acetylation when nucleosomes are used as a substrate (Fig. 5B). To quantify the amount of HAT activity, we performed liquid HAT assays using equivalent amounts of SAGA complex from the same strains (15). Liquid HAT assays allow the quantification of incorporation of radioactive acetate onto a substrate by scintillation counting. Fig. 5C confirms that SAGA from the 10Q and 60Q strains displays HAT activity at levels very similar to that of WT SAGA when free histones are used as a substrate, whereas when nucleosomes are used as a substrate SAGA from the 60Q strain can acetylate only about 25% as well as WT SAGA.
We postulated that a possible explanation for this defect was an altered association or function of the catalytic components of the complex. Therefore, Western blots were also performed on peak SAGA fractions to help determine whether complex integrity is compromised by Poly Q expansion. The amounts of the ADA complex present from each fractionation (Fig. 5A) served as a reference to normalize the amount of corresponding SAGA to use from each strain. Interestingly, levels of the HAT enzyme Gcn5 were roughly equivalent in the 10Q and 60Q SAGA complexes. However, the levels of key protein components essential to the HAT function of SAGA and SLIK, namely Ada2, Ada3, and TAF12, are diminished in the 60Q-containing complex (Fig. 5B). Although recombinant Gcn5 is unable to acetylate nucleosomal substrates, its association with Ada3 via the bridging factor Ada2 enables Gcn5 to modify nucleosomal histones (24). Furthermore, we have previously demonstrated that TAF12 (TAF68) is essential for SAGA to function as a nucleosomal HAT and for Spt3 to associate (17).
To further analyze the composition of SAGA from the WT and Δ + 60Q strains, partially purified SAGA was analyzed by silver stain (Fig. 5D). A largely unaltered composition of the complex was observed, with the reduced association of a small number of subunits in 60Q, including those predicted to represent Ada2 and Spt3. Therefore, an alteration of complex components including the depletion of critical proteins that regulate Gcn5-dependent nucleosome recognition and modification within SAGA gives significant insight into the possible mechanism of dysfunction for the poly(Q)-expanded ataxin-7 protein during SCA7 disease pathogenesis.
Discussion
The SAGA family of HAT complexes has been exquisitely conserved between yeast and humans (17), and Sca7 protein homologues have been identified in numerous species (Fig. 1 A). In strong support of our observations, human ataxin-7 is also a component of the mammalian HAT complexes STAGA and TFTC, and similar observations have been made in the presence of an expanded version in mammalian cells [see companion article (25) and ref. 26]. Studies using a mouse model indicate that STAGA complexes are recruited to retinal cone-rod homeobox (CRX) protein target promoters but that reduced levels of acetylated H3 on promoter/enhancer regions of photoreceptor genes occurs concomitant with onset of retinal degeneration in SCA7 polyglutamine expanded mice (25).
Numerous studies have reported that expanded versions of human ataxin-7 precipitate in nuclear inclusions (NIs) (10). In fact, a common occurrence in SCA7 patients is the formation of NIs that contain aggregates of an N-terminal truncated mutant form of ataxin-7 in the brain and retina (9, 27). NIs are hypothesized to play a role in sequestration of some toxic soluble components or to be toxic themselves. However, although NIs are frequently associated with disease pathogenesis, aggregates have also been observed in the cytoplasm (28) and in neuron populations resistant to SCA7-related degeneration (27). More recently, NIs in a neuronal Huntington model have been described as a “coping” response and thought to reduce the level of neuronal cell death (29). The selective pattern of neurodegeneration in these disorders may therefore be caused by several factors. For example, dysfunction in transcriptional activation and coactivation is also speculated to play a role in poly(Q) disease pathogenesis because of the observation that CBP (CREB binding protein), a mammalian HAT, has been localized to aggregates containing proteins with expanded poly(Q) tracts (9, 30). This finding suggests a possible connection between disruption of HAT complex function and poly(Q) toxicity. Importantly, both forms of the human SCA7 gene, normal and expanded, allow reassembly of the yeast SAGA complex (Fig. 4). Therefore, the pathology of SCA7 disease may not simply be the consequence of precipitation of expanded Sca7 protein but also of putative dysfunction of soluble mutant protein.
It has been hypothesized that because of its association with a HAT complex, the normal function of Sca7 is in transcriptional regulation (27). Our studies suggest that a substantial amount of ataxin-7–60Q remains soluble and associates with SAGA. Therefore, our data implicates that the expanded glutamine tract Sca7 protein assembles a dysfunctional and putatively dominant-negative SAGA complex, which may be recruited to specific promoters, has equivalent amounts of Sca7 associated but lacks critical levels of the Ada2, Ada3, and TAF12 proteins that regulate nucleosome acetylation by Gcn5. This result is important because of the previously mentioned observation that SCA7 disease pathogenesis has been associated with the formation of NIs containing aggregates of the expanded protein. A recent study addressing the involvement of the ubiquitin proteosome system (UPS) in SCA7 disease pathogenesis determined that in a mouse model expressing a large number of CAG repeats (associated with infantile onset in humans), no impairment of the UPS could be detected despite establishment of disease. Furthermore, these studies indicated a protective role against neuronal dysfunction for poly(Q) nuclear inclusions (31). It is important to note that SAGA also has been suggested to be a target of an expanded huntingtin protein fragment expressed in yeast (32). Expanded huntingtin causes a down-regulation of certain SAGA-dependent genes, consistent with loss of SAGA function. It will be interesting to determine whether a number of poly(Q) disorders are the result, in part, of SAGA dysfunction.
SCA7 disease may therefore be a result of a combination of observations: formation of nuclear inclusions as well as perturbed chromatin modification and transcriptional regulation. SAGA and SLIK, together with TFIID, regulate most DNA polymerase II-transcribed genes in yeast. However, Gcn5 as a part of SAGA, and SLIK is essential for the expression of only a subset of these but is dominant in the regulation of stress responsive genes (33). Loss of expression of certain SAGA-dependent neuronal genes may therefore contribute to the manifestation of this disorder. Importantly, histone deacetylase inhibition may provide a mechanism for reversing loss of SAGA acetylation patterns in diseased cells and could provide a possible therapy for these disorders.
Acknowledgments
We thank Harry Orr and Shelley Berger for plasmids. P.A.G. was the recipient of a Burroughs Wellcome Fund Career Award in Biomedical Sciences. This work was supported by National Center for Research Resources Grant RR11823 (to J.R.Y.) and National Institute of Diabetes and Digestive and Kidney Diseases Grant DK58646 (to P.A.G.).
Author contributions: S.J.M., J.R.Y., and P.A.G. designed research; S.J.M., M.G.P.-G., and D.S. performed research; S.J.M., M.G.P.-G., D.S., J.R.Y., and P.A.G. analyzed data; and S.J.M. and P.A.G. wrote the paper.
Abbreviations: HAT, histone acetyltransferase; WCE, whole-cell extracts.
References
- 1.Torok, M. S. & Grant, P. A. (2004) Adv. Protein Chem. 67, 181-199. [DOI] [PubMed] [Google Scholar]
- 2.Sterner, D. E., Belotserkovskaya, R. & Berger, S. L. (2002) Proc. Natl. Acad. Sci. USA 99, 11622-11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pray-Grant, M. G., Schieltz, D., McMahon, S. J., Wood, J. M., Kennedy, E. L., Cook, R. G., Workman, J. L., Yates, J. R., III, & Grant, P. A. (2002) Mol. Cell. Biol. 22, 8774-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee, M. S., Son, M. Y., Park, J. I., Park, C., Lee, Y. C., Son, C. B., Kim, Y. S., Paik, S. G., Yoon, W. H., Park, S. K., et al. (2001) Cancer Lett. 172, 165-170. [DOI] [PubMed] [Google Scholar]
- 5.Pray-Grant, M. G., Daniel, J. A., Schiletz, D., Yates, J. R., III, & Grant, P. A. (2004) Nature 433, 434-438. [DOI] [PubMed] [Google Scholar]
- 6.Brand, M., Yamamoto, K., Staub, A. & Tora, L. (1999) J. Biol. Chem. 274, 18285-18289. [DOI] [PubMed] [Google Scholar]
- 7.Martinez, E., Palhan, V. B., Tjernberg, A., Lymar, E. S., Gamper, A. M., Kundu, T. K., Chait, B. T. & Roeder, R. G. (2001) Mol. Cell. Biol. 21, 6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogryzko, V. V., Schiltz, R. L., Russanova, V., Howard, B. H. & Nakatani, Y. (1996) Cell 87, 953-959. [DOI] [PubMed] [Google Scholar]
- 9.Yvert, G., Lindenberg, K. S., Devys, D., Helmlinger, D., Landwehrmeyer, G. B. & Mandel, J. L. (2001) Hum. Mol. Genet. 10, 1679-1692. [DOI] [PubMed] [Google Scholar]
- 10.Kaytor, M. D., Duvick, L. A., Skinner, P. J., Koob, M. D., Ranum, L. P. & Orr, H. T. (1999) Hum. Mol. Genet. 8, 1657-1664. [DOI] [PubMed] [Google Scholar]
- 11.Del-Favero, J., Krols, L., Michalik, A., Theuns, J., Lofgren, A., Goossens, D., Wehnert, A., Van den Bossche, D., Van Zand, K., Backhovens, H., et al. (1998) Hum. Mol. Genet. 7, 177-186. [DOI] [PubMed] [Google Scholar]
- 12.David, G., Durr, A., Stevanin, G., Cancel, G., Abbas, N., Benomar, A., Belal, S., Lebre, A. S., Abada-Bendib, M., Grid, D., et al. (1998) Hum. Mol. Genet. 7, 165-170. [DOI] [PubMed] [Google Scholar]
- 13.Stevanin, G., Giunti, P., Belal, G. D., Durr, A., Ruberg, M., Wood, N. & Brice, A. (1998) Hum. Mol. Genet. 7, 1809-1813. [DOI] [PubMed] [Google Scholar]
- 14.Lebre, A. S., Jamot, L., Takahashi, J., Spassky, N., Leprince, C., Ravise, N., Zander, C., Fujigasaki, H., Kussel-Andermann, P., Duyckaerts, C., et al. (2001) Hum. Mol. Genet. 10, 1201-1213. [DOI] [PubMed] [Google Scholar]
- 15.McMahon, S. J., Doyon, Y., Cote, J. & Grant, P. A. (2004) Methods Enzymol. 377, 154-167. [DOI] [PubMed] [Google Scholar]
- 16.Berger, S. L., Pina, B., Silverman, N., Marcus, G. A., Agapite, J., Regier, J. L., Triezenberg, S. J. & Guarente, L. (1992) Cell 70, 251-265. [DOI] [PubMed] [Google Scholar]
- 17.Grant, P. A., Schieltz, D., Pray-Grant, M. G., Steger, D. J., Reese, J. C., Yates, J. R., III, & Workman, J. L. (1998) Cell 94, 45-53. [DOI] [PubMed] [Google Scholar]
- 18.Sanders, S. L., Jennings, J., Canutescu, A., Link, A. J. & Weil, P. A. (2002) Mol. Cell. Biol. 22, 4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheel, H., Tomiuk, S. & Hofmann, K. (2003) Hum. Mol. Genet. 12, 2845-2852. [DOI] [PubMed] [Google Scholar]
- 20.Eberharter, A., Sterner, D. E., Schieltz, D., Hassan, A., Yates, J. R., III, Berger, S. L. & Workman, J. L. (1999) Mol. Cell. Biol. 19, 6621-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.John, S., Howe, L., Tafrov, S. T., Grant, P. A., Sternglanz, R. & Workman, J. L. (2000) Genes Dev. 14, 1196-1208. [PMC free article] [PubMed] [Google Scholar]
- 22.Larschan, E. & Winston, F. (2001) Genes Dev. 15, 1946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horiuchi, J., Silverman, N., Pina, B., Marcus, G. A. & Guarente, L. (1997) Mol. Cell. Biol. 17, 3220-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balasubramanian, R., Pray-Grant, M. G., Selleck, W., Grant, P. A. & Tan, S. (2002) J. Biol. Chem. 277, 7989-7995. [DOI] [PubMed] [Google Scholar]
- 25.Palhan, V. B., Chen, S., Peng, G.-H., Tjernberg, A., Gamper, A. M., Fan, Y., Chait, B. T., La Spada, A. R. & Roeder, R. G. (2005) Proc. Natl. Acad. Sci. USA 102, 8472-8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helmlinger, D., Hardy, S., Sasorith, S., Klein, F., Robert, F., Weber, C., Miguet, L., Potier, N., Van-Dorsselaer, A., Wurtz, J. M., et al. (2004) Hum. Mol. Genet. 13, 1257-1265. [DOI] [PubMed] [Google Scholar]
- 27.Yvert, G., Lindenberg, K. S., Picaud, S., Landwehrmeyer, G. B., Sahel, J. A. & Mandel, J. L. (2000) Hum. Mol. Genet. 9, 2491-2506. [DOI] [PubMed] [Google Scholar]
- 28.Lindenberg, K. S., Yvert, G., Muller, K. & Landwehrmeyer, G. B. (2000) Brain Pathol. 10, 385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arrasate, M., Mitra, S., Schweitzer, E. S., Segal, M. R. & Finkbeiner, S. (2004) Nature 431, 805-810. [DOI] [PubMed] [Google Scholar]
- 30.Hughes, R. E. (2002) Curr. Biol. 12, R141-R143. [DOI] [PubMed] [Google Scholar]
- 31.Bowman, A. B., Yoo, S. Y., Dantuma, N. P. & Zoghbi, H. Y. (2005) Hum. Mol. Genet. 14, 679-691. [DOI] [PubMed] [Google Scholar]
- 32.Hughes, R. E., Lo, R. S., Davis, C., Strand, A. D., Neal, C. L., Olson, J. M. & Fields, S. (2001) Proc. Natl. Acad. Sci. USA 98, 13201-13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huisinga, K. L. & Pugh, B. F. (2004) Mol. Cell 13, 573-585. [DOI] [PubMed] [Google Scholar]