Abstract
Proper 3′ end formation is critical for the production of functional mRNAs. Termination by RNA polymerase II is linked to mRNA cleavage and polyadenylation, but it is less clear whether earlier stages of mRNA production also contribute to transcription termination. We performed a genetic screen to identify mutations that decreased transcriptional readthrough of a defective GAL10 poly(A) terminator. A partial deletion of the GAL10 downstream region leads to transcription through the downstream GAL7 promoter, resulting in the inability of cells to grow on galactose. Mutations in elongation factors Spt4 and Spt6 suppress the readthrough phenotype, presumably by decreasing the amount of polymerase transcribing through the downstream GAL7 promoter. Interestingly, mutations in the mRNA-binding protein Npl3 improve transcription termination. Both in vivo and in vitro experiments suggest that Npl3 can antagonize 3′ end formation by competing for RNA binding with polyadenylation/termination factors. These results suggest that elongation rate and mRNA packaging can influence polyadenylation and termination.
Keywords: hnRNP, polyadenylation, termination, transcription
Introduction
In eukaryotes, transcription by RNA polymerase II (RNApII) and mRNA processing are highly concerted events. Nascent mRNAs are cotranscriptionally capped, spliced, and polyadenylated. Different phosphorylated states of the RNApII C-terminal domain (CTD) mediate coupling by acting as binding sites for the different mRNA processing factors (Cho et al, 1997; McCracken et al, 1997; Hirose and Manley, 1998; Ahn et al, 2004).
mRNA capping is linked to phosphorylation of CTD serine 5, a reaction catalyzed by the kinase subunit of the basal transcription factor TFIIH (Jove and Manley, 1984; Rasmussen and Lis, 1993; Cho et al, 1997; Yue et al, 1997; Cho et al, 1998; Ho and Shuman, 1999; Komarnitsky et al, 2000; Rodriguez et al, 2000; Schroeder et al, 2000). There is crosstalk between capping and the transition into transcription elongation (Cho et al, 1997; Ping and Rana, 2001; Kim et al, 2004a; Mandal et al, 2004). Several biochemical and genetic interactions between capping enzymes and the Spt4/5 complex (also known as DRB sensitivity-inducing factor or DSIF) have been documented (Wen and Shatkin, 1999; Pei and Shuman, 2002; Lindstrom et al, 2003).
The CTD of RNApII also couples events at the 3′ ends of genes (McCracken et al, 1997; Ahn et al, 2004; Kim et al, 2004b). Recruitment of polyadenylation factors to transcription complexes is mediated by a combination of two signals. One is the appearance of specific polyadenylation/termination sequences that are presumably recognized as RNA. In yeast, these sequences appear to be fairly degenerate. The second signal is phosphorylation of RNApII CTD on serine 2, a reaction dependent upon the Ctk1 kinase in yeast (Lee and Greenleaf, 1991; Cho et al, 2001; Skaar and Greenleaf, 2002; Ahn et al, 2004) or the analogous Cdk9 kinase in higher eukaryotes (Price, 2000; Shim et al, 2002; Ni et al, 2004). The phosphorylated CTD enhances transcript cleavage in vitro (Hirose and Manley, 1998). A cap-binding complex (CBC, composed of Cbp20 and Cbp80) binds to the cap structure, stabilizing the mRNA (Beelman and Parker, 1995) and promoting efficient 3′ end cleavage (Flaherty et al, 1997). This linkage, as well as dedicated RNA surveillance and degradation mechanisms, ensures that mRNAs have intact 5′ and 3′ ends.
In Saccharomyces cerevisiae, the galactose-responsive genes GAL1, GAL7, and GAL10 are strongly induced upon growth in media containing galactose as the sole carbon source (Johnston, 1987; Lohr et al, 1995). All three genes are tightly regulated and required for galactose utilization. An interesting feature of these genes is their proximity to one another in the yeast genome (Figure 1A). The GAL1 and GAL10 genes are divergently transcribed and share promoter elements within their intergenic region. GAL10 is upstream of GAL7 and both are transcribed in the same direction. Mutations in the intergenic region between GAL10 and GAL7 reduce proper termination of the GAL10 transcript, causing readthrough transcription that interferes with the downstream GAL7 promoter (St John and Davis, 1981; Greger and Proudfoot, 1998). Therefore, these mutants are unable to grow in media containing galactose as the sole carbon source.
Figure 1.

Characterization of the gal10Δ56 locus. (A) Schematic representation of the GAL1/GAL10/GAL7 locus depicting the increase of RNApII readthrough upon removal of the GAL10 poly(A) site (Δ56). Readthrough precludes binding of transcription initiation factors (black circle) and expression of downstream GAL7. The locations of primers used for ChIP analysis are shown underneath. A Northern blot showing the reduction in the level of GAL7 expression is also shown. RNA was prepared from wild-type (FY268) and GAL10 poly(A)-deleted strain (CKY185) induced with 2% galactose overnight. A 20 μg portion of RNA was loaded for each strain. The blot was probed for the indicated transcripts, including the stable SNR190 RNA (as a loading control). The readthrough transcript that hybridizes with both GAL10 and GAL7 sequences is marked (RT). (B) ChIP with anti-TBP antibody was performed in wild-type (FY268) and gal10Δ56 (YSB1768) strains that were induced with galactose for 30 min. PCR reactions of the GAL locus are shown and are represented graphically (quantitation is described in Materials and methods) in the diagram (black bars for the wild type and white bars for gal10Δ56). The upper band in each lane is the primer pair as numbered in panel A. The lower band is a PCR product for a nontranscribed region that serves as a negative control. The values shown represent averages of three independent ChIP experiments and error bars show standard deviation.
We carried out a genetic selection for mutations that could suppress a GAL10 poly(A) site deletion. Suppression can be mediated by an event that interferes with readthrough of the mutated terminator, thereby restoring adequate expression of GAL7, but must also maintain expression of GAL10. We expected that extragenic mutations isolated in our selection would either reduce elongation or improve termination of the GAL10 gene (Figure 1A). Four complementation groups of suppressors were identified. Two correspond to previously identified genes involved in elongation, SPT6 and SPT4 (Winston et al, 1984; Hartzog et al, 1998; Rondon et al, 2003; Endoh et al, 2004; Kaplan et al, 2004). We also identified suppressor mutations in the NPL3 gene. Npl3 was originally identified as an effector of rRNA biogenesis (Russell and Tollervey, 1992). Later, it was shown that Npl3 is an RNA-binding protein recruited cotranscriptionally to RNApII-transcribed genes, contributing to mRNA transport out of the nucleus (Lei et al, 2001). More recently, it was reported that Npl3 can act as a negative regulator of translation (Windgassen et al, 2004). Here, we provide genetic and biochemical evidence that Npl3 antagonizes termination of transcription.
Results
Suppressors of a transcription terminator deletion
A deletion of 55 base pairs (bp) within the intergenic region between the GAL10 and GAL7 genes causes readthrough transcription from GAL10 into the GAL7 promoter (Greger and Proudfoot, 1998; Kaplan et al, 2004). Reduction in GAL7 expression results in a gal− phenotype. Chromatin immunoprecipitation (ChIP) for TATA-binding protein (TBP) confirmed that binding of basal transcription factors to the downstream GAL7 promoter was compromised in the gal10Δ56 strain (Figure 1B). Control primers for the entire GAL cluster showed that TBP binding to the GAL1 and GAL10 promoters was not affected (Figure 1B). Using 3′-RACE, we detected some polyadenylated transcripts terminated immediately downstream of the deleted poly(A) sequence (data not shown). We believe that this termination is likely not sufficient since GAL10/GAL7 readthrough transcript is increased and GAL7 mRNA is reduced (Figure 1A).
Beginning with this gal10Δ56 strain (kindly provided by C Kaplan), mutations were selected that could suppress the lack of growth in galactose. In this scheme, suppression requires both re-establishment of expression of the downstream GAL7 gene and also maintenance of GAL10 expression. Three categories of suppressors were predicted: (1) intragenic mutations that either improve GAL10 termination or increase GAL7 promoter activity, (2) extragenic mutations that reduce readthrough due to a transcription elongation defect, or (3) extragenic mutations that reduce readthrough through enhanced termination. Our prediction of class 2 suppressors was supported by the ability of the gal10Δ56 strain to grow slowly on galactose plates when the elongation-inhibiting drugs mycophenolic acid (MPA) or 6-azauracil (6-AU) (Exinger and Lacroute, 1992; Shaw and Reines, 2000) are added (Figure 2).
Figure 2.

6-AU and MPA partially suppress the gal10Δ56 galactose growth defect. Serial 10-fold dilutions of a gal10Δ56 strain (YSB1768) were spotted on 2% galactose plates supplemented with 6-AU (10, 15, or 25 μg/ml) or MPA (15 μg/ml) as indicated. Plates were incubated for 4 days at 30°C. gal10Δ56 strains grown on 2% galactose or 2% glucose plates are shown as controls.
Cells carrying the gal10Δ56 allele were mutagenized and mutants were isolated that were able to grow on galactose media. Four independent complementation groups of suppressors of the galactose− phenotype (referred to as sog1–4) were identified (Table I). Attempts to clone the sog4 gene have so far been unsuccessful. The suppressor genes sog2 (two alleles) and sog3 (two alleles) were identified through candidate plasmid complementation as SPT4 and SPT6, respectively. We note that SPT4 and SPT6 mutants have previously been shown to suppress the readthrough of the gal10Δ56 allele (Kaplan et al, 2004). Plasmids complementing sog1 were isolated from a genomic library and were found to contain the NPL3 gene. Linkage and plasmid complementation confirmed that NPL3 is allelic with sog1.
Table 1.
Phenotypes of suppressor alleles
| Complementation groups | Ts | Cs | Caffeine (15 mM) | Spt phenotype | Dominance | Genes |
|---|---|---|---|---|---|---|
| sog1 | − | +/− | − | + | Recessive | NPL3 |
| sog2 | + | + | + | − | Recessive | SPT4 |
| sog3 | − | + | + | − | Recessive | SPT6 |
| sog4 | + | + | + | − | Recessive | NA |
| +, wild-type phenotype; −, mutant phenotype. | ||||||
Suppression by spt4 is caused by defective elongation
The identification of suppressors sog2 and sog3 as SPT4 and SPT6 confirmed our genetic predictions, since both Spt4 and Spt6 modulate transcription elongation (Swanson and Winston, 1992; Hartzog et al, 1998; Wada et al, 1998). Our suppressing allele of SPT4 was designated spt4-301 (Figure 3C). Previously characterized spt4 alleles were tested to see if they also suppress the gal10Δ56 galactose growth phenotype. Four previously isolated alleles of spt4, including spt4Δ∷URA3, were crossed into the gal10Δ56 background. Genetic analysis showed that spt4-289 strongly suppresses the gal10Δ56 gal− phenotype. spt4-6, spt4-3, or spt4Δ∷URA3(Winston et al, 1984) restores growth on galactose (data not shown), but growth is slow (it should be noted that these spt4 mutations cause slow growth on their own). Next, we analyzed the transcription patterns of the suppressing spt4 alleles. RNA was isolated from cells (wild type, gal10Δ56, spt4-289, spt4-289 in the gal10Δ56 background, and our spt4-301 in gal10Δ56) that had been grown in galactose. Northern blotting shows that the level of readthrough transcription is decreased in both spt4-289 and spt4-301 as compared to gal10Δ56 (Figure 3A and data not shown). Accordingly, GAL7 transcription is restored. ChIP of TBP on the GAL locus at 30 min (Figure 3B) and 4 h (not shown) postinduction showed that TBP occupancy at the GAL7 promoter was restored in the spt4-301 suppressor strain, although not to wild-type levels. This lower occupancy of TBP in the spt4-301 strain correlates with slower growth on galactose. Therefore, suppression is partial but sufficient for the selection.
Figure 3.
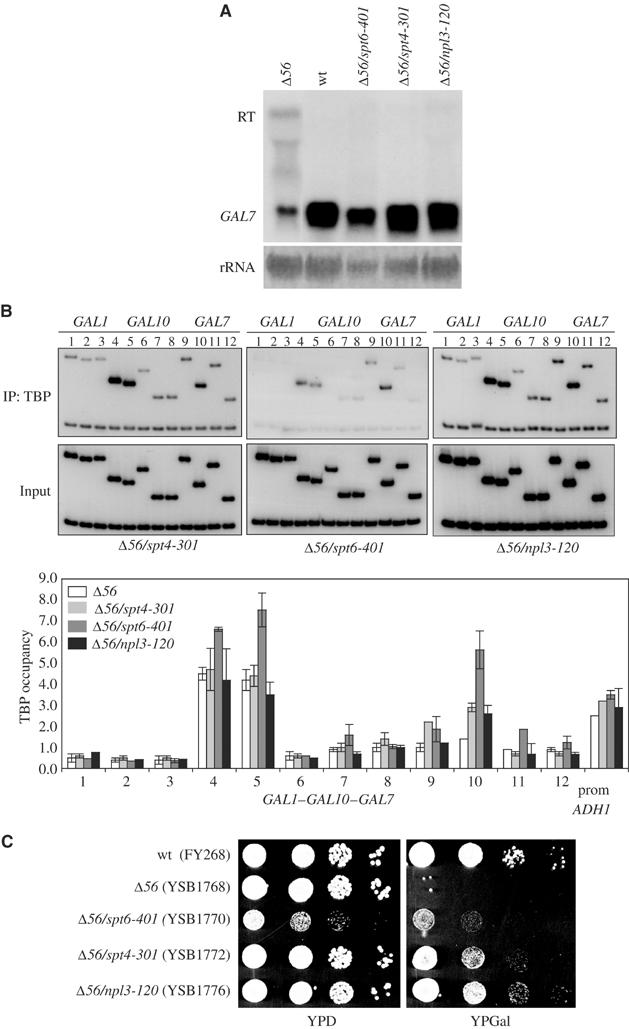
Suppression of readthrough at gal10Δ56. (A) RNA samples from gal10Δ56 strains with no suppressor (YSB1768), spt4-301 (YSB1772), spt6-401 (YSB1770), or npl3-120 (YSB1776) suppressing alleles were analyzed for GAL7 expression by Northern analysis. RT designates the readthrough transcript. In the bottom strip, the blot was stained for rRNA as a loading control. (B) ChIPs for TBP were performed for gal10Δ56 strains as described in panel A. Strains were induced with galactose for 30 min and primers are as in Figure 1. Representative PCR reactions are shown and results (average of two independent ChIP experiments) are represented graphically underneath. The ADH1 promoter was used as an unaffected control and shown at the right of the graph. (C) Serial dilutions of wild-type (FY268), gal10Δ56 (YSB1768), and gal10Δ56 strains containing the suppressing alleles spt6-401 (YSB1770), spt4-301 (YSB1772), and npl3-120 (YSB1776) were spotted onto YPD or YPGal plates and grown for 2 days at 30°C.
To probe the mechanism of the spt4-301 suppression, in vitro transcription extracts were prepared and analyzed for both elongation and termination defects. We used the previously described templates pG-Leu-CYCds (here designated pG−) and pG-Leu-CYCpAmax (designated pG−+pA) to measure elongation and termination (Steinmetz and Brow, 2003). These templates contain two G-less cassettes separated by the CYC1 downstream region either without (pG−) or with (pG−+pA) the poly(A) and termination sequences included. Although poly(A) site-dependent termination is not completely efficient in the yeast extract, a similar mammalian system produced quantitatively similar results (Tran et al, 2001). Using extracts from spt4-301 and the parent strain, the ratios of the G-less cassettes were compared (Table II). With a template lacking a poly(A) site, the ratio of the downstream to upstream G-less cassette is decreased for spt4-301 (38% readthrough to the second cassette) compared to wild type (68% readthrough). This result indicates that defects in Spt4 result in less efficient elongation, a result also seen in mammalian transcription systems (Wada et al, 1998; Kim et al, 2003) and in a yeast system (Rondon et al, 2003). The magnitude of the decrease was not changed by the presence of the poly(A) site (26% in the mutant versus 56% in the wild type), indicating that the spt4-301 mutation does not have an additional major effect on termination or polyadenylation. Based on this result, we believe that less efficient elongation in spt4-301 cells suppresses the termination defect in gal10Δ56 by reducing readthrough into the GAL7 promoter.
Table 2.
Transcription readthrough caused by different mutations
| Strain | pG− (no terminator) | pG−+pA | ||
|---|---|---|---|---|
| GAL10 background | ||||
| NPL3 (YSB1799) | 0.58 | ±0.03 | 0.33 | ±0.07 |
| npl3-120 (YSB1800) | 0.39 | ±0.06 | 0.13 | ±0.03 |
| gal10Δ56 background | ||||
| NPL3/SPT4/SPT6 (YSB1768) | 0.68 | ±0.02 | 0.56 | ±0.01 |
| npl3-210 (YSB1771) | 0.63 | ±0.07 | 0.28 | ±0.03 |
| spt4-301 (YSB1772) | 0.38 | ±0.02 | 0.26 | ±0.05 |
| spt6-401 (YSB1770) | 0.11 | ±0.07 | 0.05 | ±0.03 |
| Values shown are average of three independent experiments±standard deviation. | ||||
A severe elongation defect in spt6 mutant cells suppresses gal10Δ56
Two alleles of spt6 were isolated in the genetic selection (Figure 3C and data not shown), one of which (spt6-401) was selected for further analysis. Similar to the previously characterized allele spt6-50, spt6-401 is sensitive to 6-AU (Hartzog et al, 1998). As assayed by ChIP, TBP occupancy at the GAL7 promoter is restored near wild-type levels in spt6-401 cells (Figure 3B). Similar results were obtained when chromatin was immunoprecipitated with antibodies against the TFIIH subunit Tfb3 (not shown). TBP occupancy at GAL-induced promoters (GAL1, GAL2, GAL10, and GAL7), and also the constitutive ADH1, PYK1, and GAL3, was similar in both spt6-401 and wild-type cells (not shown), indicating that the effect at gal10Δ56 is not due to general changes in TBP occupancy.
Extracts from spt6-401 cells were assayed for elongation and termination defects in vitro as described above for spt4-301. Compared to wild type, extracts from spt6-401 cells display a reduction in the ratio of the second to first G-less cassette (11% readthrough in the mutant versus 68% in the wild-type extract) using the template lacking a poly(A) site, indicating less efficient elongation (Table II). An even greater reduction in readthrough is seen when the poly(A) and termination sequences are present (5% in the mutant versus 56% in the wild-type extract), possibly suggesting increased termination in the mutant extract. Therefore, suppression of gal10Δ56 in spt6-401 cells is likely to be mediated through a defect in elongation that also enhances termination by RNApII. Similar conclusions have been made by Kaplan et al (2004).
Increased termination efficiency in npl3 suppressors
The Npl3 protein contains two RNA recognition motifs (RRM1 and RRM2) and a glycine/arginine-rich repeat (GAR) (Birney et al, 1993). Mutations found in three npl3 suppressor alleles map near the RRM2 motif (L225S (isolated twice) and E244K). All alleles of npl3 identified in our selection are sensitive to high temperature, caffeine, and 6-AU (Figure 4 and Table I). These phenotypes, as well as the reported association of Npl3 with transcription elongation complexes (Lei et al, 2001; Lei and Silver, 2002), led us to suspect that the mutations in npl3 that suppress gal10Δ56 were doing so through effects on transcription. As with the other suppressors, the GAL locus was analyzed for TBP occupancy by ChIP using a representative allele of npl3. The distribution of TBP in npl3 suppressor alleles along the locus is similar to that of spt4 suppressor cells. The level of TBP occupancy in npl3-120 is increased at the GAL7 promoter when compared to gal10Δ56 (Figure 3B), suggesting an effect on transcription. Therefore, the suppressing effects of the npl3 mutants are unlikely to be mediated at later steps involving Npl3, such as RNA export or stability.
Figure 4.
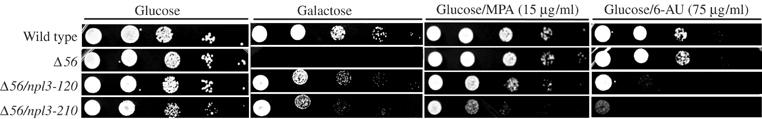
Suppressing alleles of NPL3 are sensitive to 6-AU and MPA. Serial dilutions of cells were spotted on YP plates containing 2% glucose, 2% galactose alone, or 2% galactose supplemented with 6-AU (75 μg/ml) or MPA (15 μg/ml). Strains assayed were wild type (FY268), gal10Δ56 with no suppression (YSB1768), npl3-120 (YSB1776), or npl3-210 (YSB1771).
To explore whether the npl3 suppression was due to elongation or termination effects, the in vitro transcription assay described above was used (Figure 5A). On a template without a poly(A) site between cassettes, npl3-120 extract had a slightly reduced ratio of readthrough to the second cassette (39% mutant versus 58% in wild type, less than a two-fold difference). When a poly(A) site is included between cassettes, the difference is more pronounced (13% readthrough in the mutant versus 33% in wild type). Therefore, this allele may affect both elongation and termination. In a second set of transcription experiments, the npl3-210 allele (assayed in the original gal10Δ56 background) only showed an effect on the template containing the terminator insert (28 versus 56% readthrough) (Table II). In both experiments, the Npl3 mutants actually had more efficient termination, suggesting that Npl3 may antagonize a step necessary for termination.
Figure 5.

Npl3 antagonizes termination in vitro. (A) Mutations in NPL3 enhance transcription termination in vitro. At top is a schematic diagram of transcription templates pG− and pG−+pA (Steinmetz and Brow, 2003). Two G-less cassettes are separated by a spacer with no insert (pG−) or containing the CYC1 termination sequence (pG−+pA). Whole-cell extracts (WCEs) from NPL3 (YSB1799, lanes 1 and 2) and npl3-120 (YSB1800, lanes 3 and 4) strains were used in transcription reactions with either pG− (lanes 1 and 3) or pG−+pA (lanes 2 and 4). The percent of polymerases that read through (i.e. the ratio of downstream G-less cassette 2 to upstream G-less cassette 1 normalized for labeled U content) is shown below each lane. (B) Recombinant wild-type Npl3 reverses the termination-enhancing effect of npl3-210. Npl3 was purified from E. coli and added to transcription extracts from npl3-210 (YSB1771). Templates were either pG− (lanes 1–4) or pG−+pA (lanes 5–8). Heat-inactivated Npl3 (HI, incubated at 95°C for 5 min) was added to WCEs as negative control and is shown in lanes 4 and 8. The percentage of polymerases that read through is shown under each lane.
To test this idea directly, recombinant Npl3 was added back to mutant transcription extracts to see whether it would promote transcription readthrough. Figure 5B shows a representative experiment using npl3-210 extract supplemented with increasing concentrations of native or heat-inactivated Npl3 added. Using the template without a poly(A) site, additional Npl3 does not change the readthrough (always 50–60%) compared to the negative control (100HI). However, using the template with a poly(A) site, additional Npl3 increases the ratio of the second G-less cassette to the first. Reactions containing 100 ng of heat-denatured Npl3 show about 23% readthrough, while 10–100 ng of native Npl3 increase readthrough to greater than 40%. Therefore, Npl3 antagonizes termination in vitro.
To test whether the npl3 suppressors also exhibit termination defects in vivo, a series of termination reporter plasmids (as described in Materials and methods) were used (Hyman et al, 1991). All contained a LacZ reporter gene downstream of the rp51 intron. Plasmids with the ADH2 terminator (pL101) or CYC1 terminator (pL201) cloned into the intron show much less expression of the downstream β-galactosidase gene compared to the construct with no terminator insert (pHZ18Δ2Sma). These plasmids were transformed into NPL3 or npl3-120 strains and then spotted into SC-gal plates and allowed to grow overnight. Strains were then replica plated onto filters for the β-galactosidase assay (Figure 6). For further quantitation, the β-galactosidase assays were also carried out with protein extracts derived from liquid cultures (Figure 6, bottom panel). Expression levels for the construct with no terminator were slightly lower in the npl3-120 strain (1.192 versus 2.236 β-gal units/μg protein, less than a two-fold effect). In contrast, the constructs containing the terminators had much lower levels of β-galactosidase in the npl3-120 strain. The ADH2-3′end showed a 23-fold decrease in expression (0.003 U/μg in the mutant versus 0.079 wild type) and the CYC1-3′end had a 7.5-fold decrease (0.034 versus 0.254) in the mutants relative to wild-type cells (Figure 6). Since these strong effects are only seen when a terminator sequence is present, it appears that termination is more efficient in the npl3 suppressor strain. Considering these results and the in vitro experiments, we suggest that the gal10Δ56 allele is suppressed by partial loss of an Npl3 function that antagonizes termination.
Figure 6.
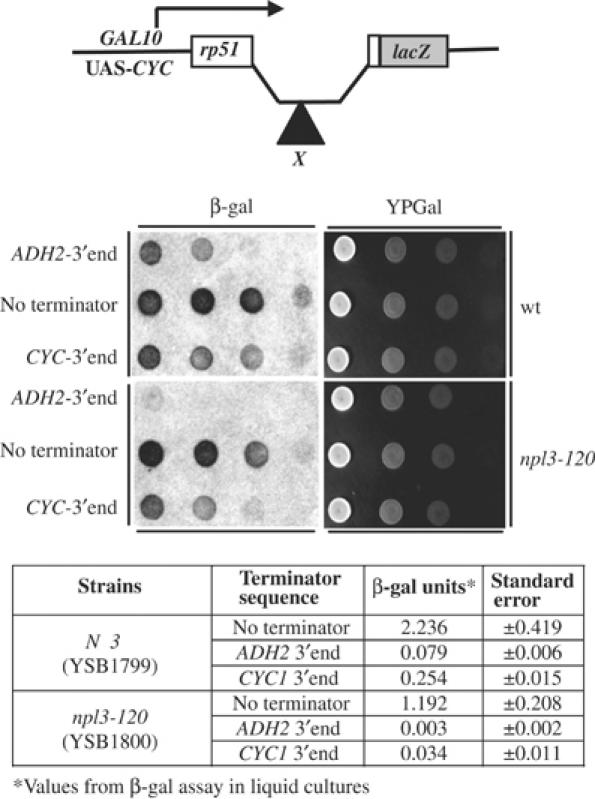
An npl3 mutation enhances transcription termination in vivo. The top panel shows a schematic diagram of reporter constructs. The parent construct (pHZ18Δ2Sma) contains an intron upstream of a lacZ reporter gene. Inserted within the intron (designated by X) is either the ADH2-3′end (pL101) or the CYC1-3′end (pL201). NPL3 (YSB1799) and npl3-120 (YSB1800) strains were transformed with the indicated constructs and spotted in serial dilutions on YP plates containing galactose (right panels). These were replica plated onto filters and assayed for β-galactosidase activity (β-gal, left panels). The reduced intensity of the color indicates less readthrough of the terminator. The same strains were assayed quantitatively from liquid cultures and the results are presented as units per microgram protein in the table.
To test whether the npl3-120 suppressor affected the recruitment of the polyadenylation/termination machinery, ChIP experiments were carried out on the gal10Δ56 locus (Figure 7A). Crosslinking of RNApII (assayed with the antibody 8WG16) did not reveal any gaps between GAL10 and GAL7 that might signify termination, presumably because the two genes are too close together. Npl3 levels were slightly reduced in the suppressor strain. In the wild-type parent strain, the polyadenylation/termination factor Rna15 crosslinked near the GAL10 3′ end. This crosslinking was greatly reduced in the gal10Δ56 strain, as expected from the increased readthrough at this locus. Strikingly, in the npl3-120 suppressor strain, Rna15 crosslinking was restored to levels at least as high as in the wild-type strain. To determine whether this effect was specific to the GAL10/7 locus or was a general effect, Rna15 levels at the ADH1, PYK1, and PMA1 genes were assayed (Figure 7B). Although levels of RNApII were similar in the wild-type and npl3-120 strains (Figure 7B and data not shown), crosslinking of Rna15 was markedly increased in the suppressor strain. Although the PMA1 terminator is efficient, we noted that RNApII crosslinking was reduced in the npl3-120 strain downstream of the cleavage site (primers 6 and 7), consistent with improved termination. These results strongly support a model in which Npl3 normally acts to antagonize cotranscriptional recruitment of polyadenylation/termination factors to the mRNA.
Figure 7.

Increased recruitment of polyadenylation/termination factor Rna15 at the 3′ end of genes in the npl3-120 strain. (A) ChIP was performed in wild-type (FY268), gal10Δ56 (YSB1768), and gal10Δ56, npl3-120 (YSB1777) strains. The same batch of chromatin from each strain was immunoprecipitated with antibodies against RNApII (8WG16), Npl3, or Rna15. PCR products are shown of the immunoprecipitates (IP) and input chromatin (INP) using primers as shown at top. Quantitations of results are shown to the right. (B) Results from ChIP of Rna15 at the ADH1, PYK1, and PMA1 genes are shown in graphs. Positions of PCR primers are shown schematically above each graph; polyadenylation sites are indicated by arrows. ChIP of RNApII for PMA1 is shown below the corresponding Rna15 ChIP. Note that for the RNApII quantitation, values were normalized to primer set 2 in the wild-type strain, which was arbitrarily set at 100.
Discussion
Using a genetic selection based on a deletion in the gal10 poly(A) site, we isolated mutations in three genes that decrease readthrough into the downstream GAL7 promoter. This genetic approach has some similarity to two previously described screens used to identify transcription factors: the Ty/solo δ insertion screen (Winston et al, 1984) and the cyc1-512 screen (Zaret and Sherman, 1982; Winston et al, 1984). In the Ty screen, a transcript from an inserted transposon element disrupts transcription from a nearby cellular gene. Suppression typically reduces the transcription from the Ty promoter or instead favors the downstream promoter, allowing reactivation of the selectable downstream gene. In Sherman's cyc1-512 screen, a deletion of 38 bp upstream of the normal poly(A) site results in unstable readthrough transcripts and downregulation of CYC1. Using conditions that require strong CYC1 expression, mutations that restore adequate expression of CYC1 were isolated in this screen (e.g. CBC1 and UPF1).
Our approach differs from both screens in that both the upstream and downstream promoters (GAL10 and GAL7) must remain transcriptionally active, since both are required for growth on galactose. This approach presumably reduces the isolation of suppressors affecting transcription initiation and should instead produce mutations that affect the readthrough transcript. As predicted, the selection resulted in the isolation of mutations that either enhance termination or impede elongation.
The isolation of elongation factor mutants such as spt4 and spt6 in the selection was expected, and agrees with a similar finding from Kaplan et al (2004). Mutations in these factors probably reduce the number of transcripts reading through from GAL10. By reducing the rate of elongation in the gal10Δ56 background, there is likely to be an increased opportunity for basal factors to bind to and initiate transcription from the downstream GAL7 promoter. This effect can be observed in the increased crosslinking of TBP to the GAL7 promoter in the suppressor mutants. Note that the spt6 mutant actually shows better TBP binding at the GAL7 promoter than spt4, but weaker suppression (Figure 3). This is presumably due to the additional severe elongation defect when Spt6 is defective. Since Spt5 has been shown to enhance capping (Wen and Shatkin, 1999; Mandal et al, 2004), and Spt4 and Spt5 are interacting partners of DSIF, we cannot formally rule out an additional effect of capping upon termination in the spt4-mediated suppression gal10Δ56.
More surprising was the isolation of Npl3 mutants as suppressors of defective termination. Previous indirect evidence links Npl3 to mRNA processing events. Npl3 is an RNA-binding protein (Russell and Tollervey, 1995) and may also bind directly to CBC components (Shen et al, 2000). A functional interaction between Npl3 and 3′ end processing proteins (Hrp1 and Rna15) has been suggested by genetic interactions. Mutations in HRP1 and RNA15 suppress the Ts− phenotype of npl3-1 (Henry et al, 1996). Hrp1 and Rna15 are required for cleavage and polyadenylation of mRNAs (Chen and Moore, 1992; Kessler et al, 1996). Kessler et al (1997) reported that recombinant Npl3 did not affect cleavage or polyadenylation in a purified in vitro system. However, both our in vivo and in vitro results indicate that Npl3 antagonizes transcription termination, which is linked to cleavage and polyadenylation. Our results are not contradictory, since in the Kessler experiments cleavage/polyadenylation was not coupled to transcription.
All of these results are consistent with a model in which cotranscriptional binding of Npl3 to the nascent mRNA transcript antagonizes the recognition of RNA sequences by the polyadenylation/termination machinery. This may help suppress recognition of cryptic polyadenylation sequences, a function likely to be necessary given the weak consensus sequences for 3′ end formation in yeast. Since Npl3 appears antagonistic to termination, mutations in polyadenylation factors such as RNA15 or HRP1 could restore the balance between these opposing activities, explaining the observed suppression patterns (Henry et al, 1996).
Proper cotranscriptional packaging of mRNAs appears to be essential for proper recognition of polyadenylation/termination sequences. The RNA-binding protein Hrp1 is recruited cotranscriptionally (Komarnitsky et al, 2000) and promotes proper choice of polyadenylation sites (Kessler et al, 1997; Minvielle-Sebastia et al, 1998; Gross and Moore, 2001). Another hnRNP, the polypyrimidine tract-binding protein (PTB), was recently reported to play a role in RNA metabolism by decreasing formation of mRNA 3′ ends in mammalian cells (Castelo-Branco et al, 2004). Therefore, the interplay between different RNA-binding proteins appears to be critical for proper mRNA 3′ end recognition formation.
Materials and methods
Strains
Strains used in this report are listed in Supplementary Table 1.
Mutant isolation
Overnight cultures of MATa (CKY185) and MATα (YSB1768) cells, containing the Δ56 deletion downstream of GAL10, were grown in YPD (yeast extract/peptone/dextrose) and 500 μl was inoculated into 2 ml of YPD and allowed to grow for 2 h (early log phase). Cells were then pelleted, washed, and plated (5 × 106 cells/plate) onto YPGal (2% galactose)+antimycin A (1 μg/ml) plates. Antimycin A is included to inhibit the respiration necessary for nonfermentative growth. Plates were mutagenized with ultraviolet light (50 J/m2) and Gal+ colonies were allowed to grow at 30°C. Colonies from mating types a and α were isolated from the irradiated plates. In addition, several colonies that grew spontaneously from a nonirradiated plate were isolated. Suppressor candidate strains were tested for dominance/recessive tests by mating to parent strains of the opposite mating type followed by replica plating onto YPGal+antimycin A plates. A total of 19 dominant suppressors, likely cis-acting, were identified and were not characterized further.
Recessive mutants were assayed for additional phenotypes, including heat sensitivity (37°C), cold sensitivity (16°C), and caffeine sensitivity (YPD plus 15 mM caffeine). In addition, recessive mutants were screened for the Spt− phenotype, which can be caused by mutations in certain elongation factors, by plating on lys− and his− plates (Winston and Carlson, 1992). A total of 58 recessive mutants were assigned to four complementation groups designated sog1–4. Members of each group were transformed with plasmids containing SPT4, SPT5, or SPT6 (kindly provided by F Winston, Harvard University). Suppressors sog2 and sog3 were complemented with plasmids containing SPT4 and SPT6, respectively. Identities of the mutant genes were confirmed by segregation of crosses to an spt4Δ∷URA3 (YSB1775) strain and an spt6 (CKY122) strain.
The sog1 gene was cloned by complementation using a genomic DNA library (Rose et al, 1987). Approximately 3000 transformants of a sog1 mutant strain were selected using the library's URA3 marker. These were screened by replica plating for complementation of the gal− and heat-sensitive (37°C) phenotypes of the sog1 mutant. A total of 14 plasmids were recovered and rescreened for complementation. After restriction analysis, four plasmids were selected for sequencing. The sequences from four of the plasmids mapped to chromosome IV with an overlapping region 3463 bp upstream and 1654 bp downstream of the NPL3 gene. The sog1 suppressor was confirmed to be the NPL3 gene through a linkage test using an npl3Δ∷KANMX strain (Research Genetics) and by complementation with an NPL3-containing plasmid (kindly provided by P Silver, Harvard University; Henry et al, 1996). Three suppressor alleles of sog1/npl3 were sequenced and were found to encode either amino-acid substitution L225S or E244K.
RNA preparation and analyses
Cells were induced with 2% galactose overnight. Total RNA was prepared as described previously (Ausubel et al, 1988). Northern blot transfer and hybridization were performed as described previously (Swanson et al, 1991). 32P-labeled probes were generated by random hexamer labeling (Ausubel et al, 1988).
Reporter assay for termination readthrough
Wild type (YSB1799) and npl3-120 (YSB1800) were transformed with plasmids pL101, pL201, or pHZ18Δ2Sma (kindly provided by C Moore, Tufts University; Hyman et al, 1991). These plasmids contain the GAL10 UAS-CYC promoter driving a transcript containing the rp51 intron upstream of lacZ. Either the ADH2-3′end or the CYC1-3′end sequences are inserted into the rp51 intron in pL101 or pL201, respectively (Hyman et al, 1991). Transformants were processed as described previously (Runner and Brewster, 2003) with the following modifications. After overnight growth in synthetic complete media with galactose but lacking uracil (SC-gal-URA−) at 30°C, cells were pelleted and resuspended in 200 μl of SC-gal-URA− media. Four serial dilutions were spotted onto SC-gal-URA− plates. These were replica plated onto Whatman 1A filter paper on YPD or SC-gal-URA− plates and allowed to grow overnight at 30°C. Filters were assayed for lacZ activity as described previously (Burns et al, 1994).
For liquid cultures, wild-type (YSB1799) and npl3-120 (YSB1800) strains transformed with plasmids pL101, pL201, or pHZ18Δ2Sma were grown in SC-raf-URA− overnight. Cells were supplied with new SC-raf-URA− media and induced with 2% galactose for 6 h. Cells (2 × 108) were pelleted and lysed and the resulting extracts were used to measure β-galactosidase activity as described previously (Ausubel et al, 1988). For quantitation, β-galactosidase units were normalized to protein concentrations.
Chromatin immunoprecipitations
ChIPs were performed essentially as described previously (Komarnitsky et al, 2000). Anti-Npl3 serum was a gift from D Tollervey and anti-Rna15 serum was a gift from Claire Moore. Oligonucleotides used for PCR are listed in Supplementary Table 2. For α-TBP immunoprecipitations, antibody was preincubated for 60 min at room temperature with protein A-agarose or protein G-Sepharose CL-4B (Amersham) beads as indicated and beads were washed once with TE pH 8.0. Chromatin solution was then added and reactions incubated overnight at 4°C. Immunoprecipitates were washed, protease treated, and decrosslinked. Conditions for PCR were as described previously (Komarnitsky et al, 2000). PCR products were quantified by PhosphoImager as described previously (Kim et al, 2004b). Briefly, the efficiency of amplification for each region relative to a chromosome V nontranscribed control region was calculated from the input sample. This value was then used to normalize the specific signal obtained from each immunoprecipitation. Division of this normalized value by the background amplification of the nontranscribed control gives a relative value (x-fold compared to the nontranscribed internal control) that allows trend comparison across samples to be performed. Note that a value of 1.0 means that there is no crosslinking above background.
In vitro transcription assays
Cell pellets were crushed in liquid nitrogen as described previously (Schultz et al, 1991) and then resuspended in 1 ml of transcription buffer A (200 mM Tris pH 7.9, 390 mM NH4SO4, 10 mM MgSO4, 1 mM EDTA, 20% glycerol, 2 mM DTT, plus protein inhibitors (1 μg/ml pepstatin A, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml antipain, and 1 μg/ml benzamidine) per gram wet weight. These suspensions were then further subjected to glass bead disruption. Lysates were collected and treated as described previously (Keogh et al, 2002). Aliquots of whole-cell extract (13–45 mg/ml) were frozen in liquid nitrogen and stored at −80°C.
RNApII transcription reactions were carried out as described by Keogh et al (2002). Templates used were the plasmids pCYCds or pCYCpAmax (kindly provided by E Steinmetz and D Brow, University of Wisconsin Medical School; Steinmetz and Brow, 2003). RNA was resolved on a denaturing urea–acrylamide gel and quantified by PhosphoImager as described previously (Steinmetz and Brow, 2003). For each strain, the ratio of total counts of the downstream G-less cassette to total counts of upstream G-less cassette was normalized for the ratio of labeled U residues contained in each cassette (G-less1=102, G-less2=40 U residues) as described previously (Steinmetz and Brow, 2003).
Recombinant Npl3 protein
Escherichia coli strain BL21(DE3) was transformed with pSBEThis7-Npl3 (details available upon request). A 1 l culture (LB+kanamycin 25 μg/ml) grown at 25°C to OD600≈0.5 was induced by the addition of 1 mM IPTG. All steps from this point on were performed at 4°C and all buffers contained protease inhibitors (1 μg/ml pepstatin A, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml antipain, and 1 μg/ml benzamidine). Cells were harvested and lysates were prepared by sonication in binding buffer (10 mM Tris–HCl pH 7.9, 500 mM NaCl, 20 mM imidazole, 1 mM PMSF). The soluble extract was incubated at 4°C with gentle rolling for 2 h with 2 ml of Ni-NTA resin (Qiagen). The resin was spun down gently, washed twice with 20 ml of binding buffer plus 10 mM imidazole plus 1 mM PMSF, twice with 20 ml of binding buffer plus 20 mM imidazole plus 1 mM PMSF, and resuspended in 5 ml of binding buffer plus 20 mM imidazole plus 1 mM PMSF. Resin was applied to a column and eluted in ten 500 μl fractions with binding buffer plus 200 mM imidazole. Recombinant Npl3 was assayed by Coomassie staining and by Western blotting with anti-Npl3 (kindly provided by D Tollervey, University of Edinburg; Venema and Tollervey, 1995).
Supplementary Material
Supplementary Table 1
Supplementary Table 2
Acknowledgments
We are grateful to Fred Winston and C Kaplan for the construction of gal10Δ56 and other strains used in this study. We are also grateful to E Larschan, M Kim, C Sawa, M Keogh, and L Vasilieva for technical advice and helpful discussions. We thank E Steinmetz and D Brow for plasmids pCYCds and pCYCpAmax, L Hyman and C Moore for anti-Rna15 antibody and plasmids pL101, pL201, and pHZ18Δ2Sma, P Silver for plasmid pMHY3, and D Tollervey for anti-Npl3 antibody. M Bucheli was supported by fellowships from UNCF/Pfizer and the American Cancer Society (PF-03-224-01-GMC). This work is supported by grants GM46498 and GM56663 from the National Institutes of Health to SB.
References
- Ahn SH, Kim M, Buratowski S (2004) Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 13: 67–76 [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1988) Current Protocols in Molecular Biology. New York, NY: Green Publishing Associates/Wiley Interscience [Google Scholar]
- Beelman CA, Parker R (1995) Degradation of mRNA in eukaryotes. Cell 81: 179–183 [DOI] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR (1993) Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res 21: 5803–5816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns N, Grimwade B, Ross-Macdonald PB, Choi EY, Finberg K, Roeder GS, Snyder M (1994) Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev 8: 1087–1105 [DOI] [PubMed] [Google Scholar]
- Castelo-Branco P, Furger A, Wollerton M, Smith C, Moreira A, Proudfoot N (2004) Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol Cell Biol 24: 4174–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Moore C (1992) Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol Cell Biol 12: 3470–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S (2001) Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev 15: 3319–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Rodriguez CR, Takagi T, Buratowski S (1998) Allosteric interactions between capping enzyme subunits and the RNA polymerase II carboxy-terminal domain. Genes Dev 12: 3482–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Takagi T, Moore CR, Buratowski S (1997) mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev 11: 3319–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh M, Zhu W, Hasegawa J, Watanabe H, Kim DK, Aida M, Inukai N, Narita T, Yamada T, Furuya A, Sato H, Yamaguchi Y, Mandal SS, Reinberg D, Wada T, Handa H (2004) Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol Cell Biol 24: 3324–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exinger F, Lacroute F (1992) 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr Genet 22: 9–11 [DOI] [PubMed] [Google Scholar]
- Flaherty SM, Fortes P, Izaurralde E, Mattaj IW, Gilmartin GM (1997) Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc Natl Acad Sci USA 94: 11893–11898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger IH, Proudfoot NJ (1998) Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J 17: 4771–4779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S, Moore CL (2001) Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol Cell Biol 21: 8045–8055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA, Wada T, Handa H, Winston F (1998) Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev 12: 357–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry M, Borland CZ, Bossie M, Silver PA (1996) Potential RNA binding proteins in Saccharomyces cerevisiae identified as suppressors of temperature-sensitive mutations in NPL3. Genetics 142: 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Manley JL (1998) RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395: 93–96 [DOI] [PubMed] [Google Scholar]
- Ho CK, Shuman S (1999) Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell 3: 405–411 [DOI] [PubMed] [Google Scholar]
- Hyman LE, Seiler SH, Whoriskey J, Moore CL (1991) Point mutations upstream of the yeast ADH2 poly(A) site significantly reduce the efficiency of 3′-end formation. Mol Cell Biol 11: 2004–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M (1987) A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev 51: 458–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jove R, Manley JL (1984) In vitro transcription from the adenovirus 2 major late promoter utilizing templates truncated at promoter-proximal sites. J Biol Chem 259: 8513–8521 [PubMed] [Google Scholar]
- Kaplan CD, Holland MJ, Winston F (2004) Interaction between transcription elongation factors and mRNA 3′-end formation at the Saccharomyces cerevisiae GAL10–GAL7 locus. J Biol Chem 280: 913–922 [DOI] [PubMed] [Google Scholar]
- Keogh MC, Cho EJ, Podolny V, Buratowski S (2002) Kin28 is found within TFIIH and a Kin28–Ccl1–Tfb3 trimer complex with differential sensitivities to T-loop phosphorylation. Mol Cell Biol 22: 1288–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler MM, Henry MF, Shen E, Zhao J, Gross S, Silver PA, Moore CL (1997) Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev 11: 2545–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler MM, Zhao J, Moore CL (1996) Purification of the Saccharomyces cerevisiae cleavage/polyadenylation factor I. Separation into two components that are required for both cleavage and polyadenylation of mRNA 3′ ends. J Biol Chem 271: 27167–27175 [DOI] [PubMed] [Google Scholar]
- Kim DK, Inukai N, Yamada T, Furuya A, Sato H, Yamaguchi Y, Wada T, Handa H (2003) Structure–function analysis of human Spt4: evidence that hSpt4 and hSpt5 exert their roles in transcriptional elongation as parts of the DSIF complex. Genes Cells 8: 371–378 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Jeong SH, Heo JH, Jeong SJ, Kim ST, Youn HD, Han JW, Lee HW, Cho EJ (2004a) mRNA capping enzyme activity is coupled to an early transcription elongation. Mol Cell Biol 24: 6184–6193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S (2004b) Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J 23: 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Greenleaf AL (1991) CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr 1: 149–167 [PMC free article] [PubMed] [Google Scholar]
- Lei EP, Krebber H, Silver PA (2001) Messenger RNAs are recruited for nuclear export during transcription. Genes Dev 15: 1771–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei EP, Silver PA (2002) Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes Dev 16: 2761–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, McCleery JA, Yates JR III, Hartzog GA (2003) Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol Cell Biol 23: 1368–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D, Venkov P, Zlatanova J (1995) Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J 9: 777–787 [DOI] [PubMed] [Google Scholar]
- Mandal SS, Chu C, Wada T, Handa H, Shatkin AJ, Reinberg D (2004) Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc Natl Acad Sci USA 101: 7572–7577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL (1997) The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361 [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L, Beyer K, Krecic AM, Hector RE, Swanson MS, Keller W (1998) Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J 17: 7454–7468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT (2004) Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol Cell 13: 55–65 [DOI] [PubMed] [Google Scholar]
- Pei Y, Shuman S (2002) Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J Biol Chem 277: 19639–19648 [DOI] [PubMed] [Google Scholar]
- Ping YH, Rana TM (2001) DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J Biol Chem 276: 12951–12958 [DOI] [PubMed] [Google Scholar]
- Price DH (2000) P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol 20: 2629–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT (1993) In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci USA 90: 7923–7927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez CR, Cho EJ, Keogh MC, Moore CL, Greenleaf AL, Buratowski S (2000) Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol Cell Biol 20: 104–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon AG, Garcia-Rubio M, Gonzalez-Barrera S, Aguilera A (2003) Molecular evidence for a positive role of Spt4 in transcription elongation. EMBO J 22: 612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Novick P, Thomas JH, Botstein D, Fink GR (1987) A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 60: 237–243 [DOI] [PubMed] [Google Scholar]
- Runner VM, Brewster JL (2003) A genetic screen for yeast genes induced by sustained osmotic stress. Yeast 20: 913–920 [DOI] [PubMed] [Google Scholar]
- Russell I, Tollervey D (1995) Yeast Nop3p has structural and functional similarities to mammalian pre-mRNA binding proteins. Eur J Cell Biol 66: 293–301 [PubMed] [Google Scholar]
- Russell ID, Tollervey D (1992) NOP3 is an essential yeast protein which is required for pre-rRNA processing. J Cell Biol 119: 737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder SC, Schwer B, Shuman S, Bentley D (2000) Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev 14: 2435–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MC, Choe SY, Reeder RH (1991) Specific initiation by RNA polymerase I in a whole-cell extract from yeast. Proc Natl Acad Sci USA 88: 1004–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Reines D (2000) Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion. Mol Cell Biol 20: 7427–7437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen EC, Stage-Zimmermann T, Chui P, Silver PA (2000) The yeast mRNA-binding protein Npl3p interacts with the cap-binding complex. J Biol Chem 275: 23718–23724 [DOI] [PubMed] [Google Scholar]
- Shim EY, Walker AK, Shi Y, Blackwell TK (2002) CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev 16: 2135–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar DA, Greenleaf AL (2002) The RNA polymerase II CTD kinase CTDK-I affects pre-mRNA 3′ cleavage/polyadenylation through the processing component Pti1p. Mol Cell 10: 1429–1439 [DOI] [PubMed] [Google Scholar]
- St John TP, Davis RW (1981) The organization and transcription of the galactose gene cluster of Saccharomyces. J Mol Biol 152: 285–315 [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Brow DA (2003) Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol Cell Biol 23: 6339–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MS, Malone EA, Winston F (1991) SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol 11: 4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MS, Winston F (1992) SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics 132: 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran DP, Kim SJ, Park NJ, Jew TM, Martinson HG (2001) Mechanism of poly(A) signal transduction to RNA polymerase II in vitro. Mol Cell Biol 21: 7495–7508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, Tollervey D (1995) Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast 11: 1629–1650 [DOI] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, Buratowski S, Handa H (1998) DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev 12: 343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Shatkin AJ (1999) Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev 13: 1774–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windgassen M, Sturm D, Cajigas IJ, Gonzalez CI, Seedorf M, Bastians H, Krebber H (2004) Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol Cell Biol 24: 10479–10491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Carlson M (1992) Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet 8: 387–391 [DOI] [PubMed] [Google Scholar]
- Winston F, Chaleff DT, Valent B, Fink GR (1984) Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics 107: 179–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin AJ (1997) Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci USA 94: 12898–12903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Sherman F (1982) DNA sequence required for efficient transcription termination in yeast. Cell 28: 563–573 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1
Supplementary Table 2


