Abstract
RNA editing increases during development in more than 20 transcripts encoding proteins involved in rapid synaptic neurotransmission in Drosophila central nervous system and muscle. Adar (adenosine deaminase acting on RNA) mutant flies expressing only genome-encoded, unedited isoforms of ion-channel subunits are viable but show severe locomotion defects. The Adar transcript itself is edited in adult wild-type flies to generate an isoform with a serine to glycine substitution close to the ADAR active site. We show that editing restricts ADAR function since the edited isoform of ADAR is less active in vitro and in vivo than the genome-encoded, unedited isoform. Ubiquitous expression in embryos and larvae of an Adar transcript that is resistant to editing is lethal. Expression of this transcript in embryonic muscle is also lethal, with above-normal, adult-like levels of editing at sites in a transcript encoding a muscle voltage-gated calcium channel.
Keywords: ADAR, Drosophila, ion channel, RNA editing
Introduction
The ADAR (adenosine deaminase acting on RNA) enzymes deaminate specific adenosines in transcripts to inosines. Inosine is the base-pairing equivalent of guanosine during translation or cDNA synthesis (Basilio et al, 1962). When the conversion of adenosine to inosine changes codon meaning, it can have dramatic effects on the properties of the encoded protein. Editing of the Q/R site in vertebrate transcripts encoding the glutamate-gated receptor B subunit (GluR-B) affects calcium permeability of AMPA receptors (Sommer et al, 1991), and may slow receptor assembly (Greger et al, 2003). Similarly, RNA editing in transcripts encoding the G-protein-coupled serotonin (5-HT2C) receptor (Burns et al, 1997) produces isoforms with reduced coupling to the target G protein (Price et al, 2001).
ADARs recognise short stretches of imperfectly paired RNA duplex formed by pairing of an exon with an editing site complementary sequence (ECS) that is usually located in an adjacent intron (for review, see Keegan et al, 2001). ADAR proteins contain either two or three dsRNA binding domains at the amino terminus and the catalytic deaminase domain is present in the carboxy terminus of the protein (for review, see Keegan et al, 2004). The deaminase domain contains three zinc-chelating motifs and motif I includes an essential glutamate residue. ADAR protein with this glutamate mutated is inactive but is able to compete with active proteins for editing sites in vitro and in vivo (Gallo et al, 2003). The Drosophila ADAR protein forms dimers on RNA substrates through the amino terminus and the first dsRNA binding domain (Gallo et al, 2003). Dimerisation is essential for catalysis, and different isoforms of Drosophila ADAR form heterodimers. The variety of isoforms and their possible interactions suggest complex regulation of RNA editing.
In Drosophila melanogaster, there is a single Adar gene, mutations in which produce flies that survive to adulthood but display major defects in walking and mating (Palladino et al, 2000b). Locomotion defects in Adar mutant flies are present from eclosion and are succeeded by age-dependent neurodegeneration in the brain (Palladino et al, 2000a). In embryos, Adar is expressed strongly throughout the central nervous system (CNS), but it is also expressed more weakly outside the nervous system in mesoderm and endoderm (Palladino et al, 2000a). Like many Drosophila genes, Adar is expressed from an embryonic promoter and later from an adult promoter activated at metamorphosis. Adar is expressed in the brain of adult flies (Ma et al, 2001), but the complete adult expression pattern has not been described. All of the transcripts that are known to be edited in Drosophila are expressed in neurons but some also show muscle expression. The known editing targets in Drosophila include the cacophony (cac) (Smith et al, 1996) and paralytic (para) (Hanrahan et al, 2000) transcripts encoding the large α1 subunits of the main voltage-gated calcium channel and voltage-gated sodium channel in the CNS, respectively. Editing at all sites in these target transcripts is eliminated in Adar mutant flies (Palladino et al, 2000b). Recently, a further 16 edited transcripts have been described (Hoopengardner et al, 2003), and the list of edited transcripts is likely to be still incomplete (Stapleton et al, 2002). Adar mutants have also been isolated under the name hypnos-2 (hypoxia, anoxia sensitive), in a screen for mutants that recover from anoxic stupor more slowly than wild type (Ma et al, 2001).
To investigate how the expression of different ADAR isoforms at different developmental stages and in different tissues contributes to the regulation of RNA editing in Drosophila, we characterised the editing activities of ADAR isoforms in vitro and used the GAL4-UAS system (Brand and Perrimon, 1993) to express cDNA rescue constructs encoding different ADAR isoforms in Adar mutant flies. We find that ADAR isoforms predominant in adult flies are more active than those from embryos and larvae and that editing occurs in muscle as well as within the CNS. We extensively characterise the effect on protein function of one editing event in which ADAR edits its own transcript to produce an ADAR isoform that is less active. At the physiological level, Adar transcript editing appears to be a form of negative autoregulation. An Adar cDNA engineered to resist RNA editing causes lethality when ubiquitously expressed. We present evidence that this lethality is mediated through excessive editing of target transcripts in embryos and larvae.
We show that editing levels at sites in the cacophony transcript are lowest in embryos, rise during larval development and are highest in adult flies, but editing is never complete. A similar increase in editing during development occurs at editing sites in the Ca alpha 1D transcript (Zheng et al, 1995; Eberl et al, 1998; Hoopengardner et al, 2003), which encodes an L-type voltage-gated calcium channel expressed in both neurons and muscle. In this transcript, editing at some sites is essentially 100% efficient in adult flies, in muscle as well as in neurons.
Results
ADAR edits the cacophony transcript and the Adar transcript in vitro
RNA editing itself contributes to the generation of ADAR variants in Drosophila. The Adar transcript is edited so that a serine (S) residue close to zinc-chelating motif II in the active site in the deaminase domain is replaced with glycine (G), hence the name Adar S/G site (Palladino et al, 2000a) (Figure 1A). Editing at this site increases during development, being almost completely absent in embryos and rising to 40% in adults. Oddly, RNA editing occurs in the transcript encoding the ADAR 3/4 splice form that predominates after induction of the adult promoter at metamorphosis but not in the 3a splice form that is expressed at all stages (Palladino et al, 2000a). To identify an ECS for the editing site in Adar exon 7, the flanking introns were sequenced but no ECS was found. However, Adar exon 7 itself can fold into a highly duplex structure (Figure 1B) that is conserved even at the third base position in codons in Drosophila pseudoobscura (RA Reenan, unpublished data).
Figure 1.
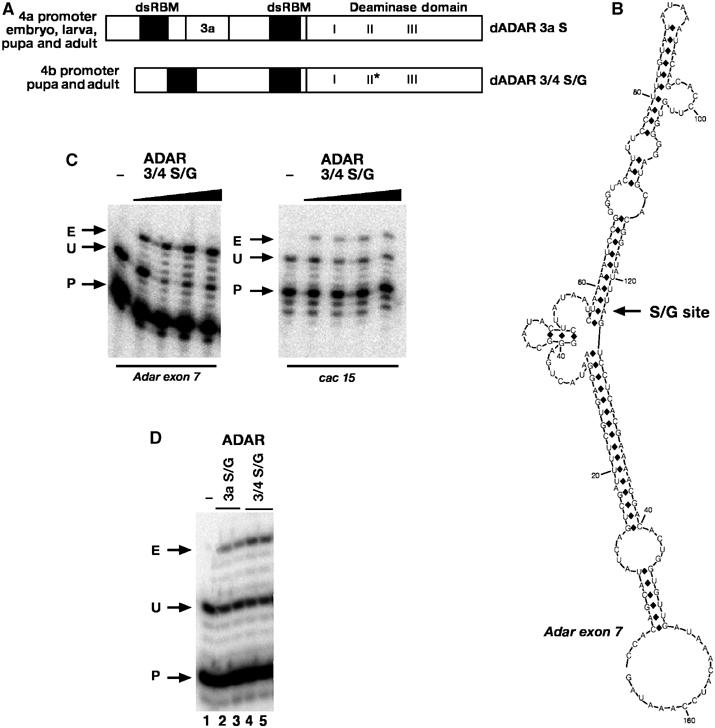
Adar protein isoforms expressed, predicted dsRNA structure of Adar exon 7 and in vitro editing of sites in Adar and cacophony transcripts. (A) Structures of ADAR protein isoforms and timings of expression. The Adar 4a promoter is expressed at all stages, whereas the Adar 4b promoter is strongly induced at metamorphosis and is pupal/adult specific. 4a transcripts are spliced predominantly to produce the 3a splice forms, but 4b transcripts produce only the 3/4 splice form that predominates in adult flies. Exon notations are as described by Palladino et al (2000b). Inclusion of exon 3a arises from splicing to an alternative 5′ splice donor and results in incorporation of 37 extra amino acids between the two double-stranded RNA binding motifs (dsRBMs). Motifs I, II and III in the deaminase domain contain conserved zinc-chelating cysteine and histidine residues. Motif I contains the sequence CHAE and the glutamate residue is essential for catalysis. The S/G RNA editing event in Adar exon 7, indicated by an asterisk, changes a conserved serine codon seven codons after motif II to a glycine codon. (B) RNA structure prediction for all 166 bases of exon 7 of the Adar transcript using the mfold program (Zuker et al, 1999). The edited adenosine 123 is indicated by an arrow labelled S/G site. Editing occurs in transcripts spliced to produce the 3/4 isoform. (C) Poisoned primer extension comparing editing in vitro of Adar exon 7 and cac 15 substrates by the same increasing concentrations of the ADAR 3/4 S/G protein mix. The left lane of each panel has no ADAR protein and 1, 3, 6.25 and 12.5 μl of purified protein was used in the ADAR protein lanes. Arrows indicate radiolabelled primer (P) and the unedited (U) and edited (E) primer extension products. (D) Poisoned primer extension assay on Adar exon 7 comparing purified ADAR 3a S/G or ADAR 3/4 S/G proteins. Lane 1 has no protein, lanes 2 and 3 are duplicate lanes containing ADAR 3a S/G that edits Adar exon 7 transcript to 15% whereas lanes 4 and 5 are duplicate lanes containing ADAR 3/4 S/G with the same concentration of protein as in lanes 2 and 3 that edits the substrate to 33%. Arrows indicate radiolabelled primer (P) and the unedited (U) and edited (E) primer extension products.
To determine whether Drosophila ADAR edits the Adar transcript in vitro, ADAR 3/4 protein was expressed in the yeast Pichia pastoris and purified. RNA substrates corresponding to cacophony exon 15+intron 15 (cac15) or Adar exon 7 (Figure 1B) were generated by in vitro transcription, purified and incubated with ADAR proteins. RNA editing was measured using a poisoned primer extension assay (Figure 1C). The RNA editing site in cac 15 was chosen for in vitro editing assays because the edited A is not embedded in a run of A's and the site is efficiently edited.
A purified recombinant Drosophila ADAR 3/4 S/G isoform mix (see below) edits the cac 15 site to approximately 20% at saturation, whereas the same protein concentrations edit the Adar exon 7 S/G site with 70% efficiency (Figure 1C). Editing of the Adar exon 7 S/G site to 70% is higher than at any other site that has been measured in vitro and is even higher than the 40% editing that is found in vivo at this position. In addition, the ADAR 3/4 isoform (lanes 4 and 5) edits the Adar exon 7 S/G site more efficiently than the ADAR 3a isoform does in vitro (Figure 1D, lanes 2 and 3).
Adar transcript editing generates an ADAR isoform with reduced activity
Editing of Adar transcript occurs primarily in adult flies when RNA editing in general is highest. The edited isoform could be contributing to either the increase in overall ADAR activity in adult flies or to an autoregulation acting to restrain editing. Producing a pure preparation of unedited ADAR S protein is complicated by the fact that Adar exon 7 is sufficient as a substrate for RNA editing. Editing of the Adar transcript occurs even in mRNA produced from Adar cDNA constructs expressed in Pichia (Figure 2A, lower chromatogram) that produce an ADAR 3/4 S/G protein mixture. Pichia and other yeasts lack endogenous ADARs or A to I editing activity and an Adar cDNA expressing just the deaminase domain of Drosophila ADAR is not edited (Figure 2A, upper chromatogram). Producing pure unedited ADAR S protein requires an editing-resistant Adar S mRNA. To produce such an ineditable Adar S mRNA, the edited serine codon was changed to another serine codon that does not have an A at the edited first position (Figure 2C). Generating the edited isoform ADAR G using the edited cDNA is trivial.
Figure 2.
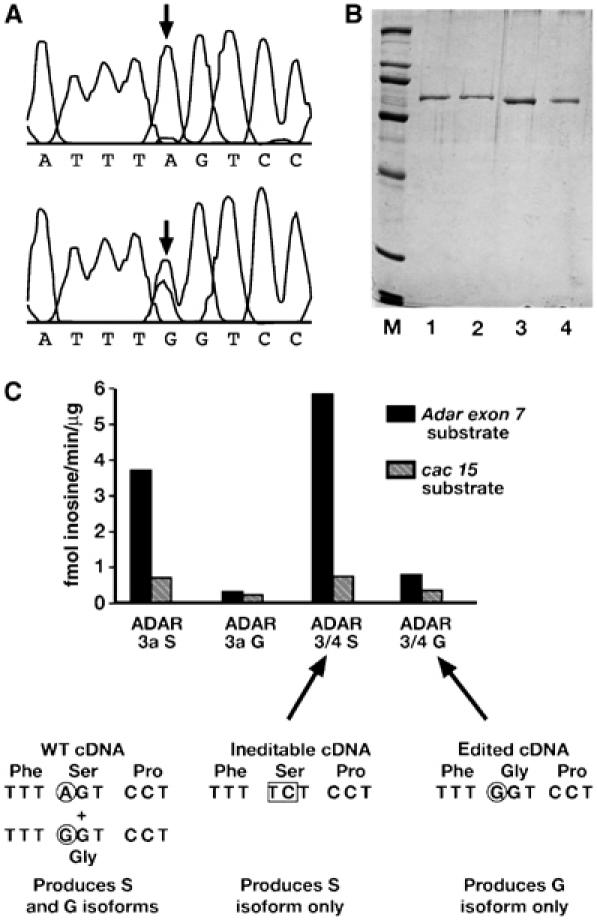
ADAR edits the Adar transcript to generate an edited ADAR isoform with reduced editing activity. (A) Editing of the Adar transcript expressed in Pichia. Sequences of Adar exon 7 RT–PCR product pools from Pichia expressing an active ADAR 3/4 protein (lower chromatogram) or an inactive ADAR protein encoding only the ADAR deaminase domain (upper chromatogram). An arrow marks the edited position. The resulting purified ADAR S/G protein will be a mixture of edited and unedited isoforms as shown in Figure 1C. (B) An SDS polyacrylamide gel of the purified homogeneous unedited S and edited G isoforms of ADAR used in the in vitro assays stained with GelCode Blue Stain Reagent (Pierce). BioRad High Molecular weight markers are on the left; lane 1 is ADAR 3a S, lane 2 is ADAR 3a G, lane 3 is ADAR 3/4 S and lane 4 is ADAR 3/4 G. (C) Strategy for circumventing editing of the Adar transcript during protein expression in P. pastoris. To produce a pure preparation of the genome-encoded, unedited ADAR S isoform, the edited serine codon is mutated to a different serine codon that does not have A at the edited position. The mRNA expressed from this construct is ineditable and the encoded protein is the S isoform. Specific editing activities of purified unedited S or edited G isoforms of ADAR on cac 15 and Adar exon 7 transcripts are shown. The specific activity is the amount of inosine generated per minute per microgram ADAR protein on the specific substrates, measured using poisoned primer extension assays.
Proteins were purified following overexpression in P. pastoris (Figure 2B), and their specific activities measured by poisoned-primer extension assays on cac 15 and Adar exon 7 transcripts (Figure 2C). The edited isoform is less active in RNA editing on both substrates. An eight-fold decrease in editing the Adar transcript was observed, whereas the decrease in specific activity in editing the cac transcript was approximately three-fold. The effect of editing is seen in the context of either the ADAR 3a or ADAR 3/4 splice forms; the difference between splice forms is less than the effect of editing but the ADAR 3/4 splice form is more active. These assays were performed with two different purified preparations of each ADAR protein, and the specific activities measured over a range of protein concentrations were the same in both preparations. Therefore, the ADAR exon 7 site is a high-affinity site for the ADAR enzyme and by editing it, subsequent translation produces an enzyme that is less active in editing. One effect of own-transcript editing is to limit RNA editing activity. The embryonic isoform ADAR 3a can also edit its own transcript in vitro (Figures 1D and 2C); however, this editing is not observed in wild-type flies. Editing at the Adar (S/G) site is also observed in the UAS-Adar 3a transgenic fly lines (data not shown).
Developmental regulation of site-specific RNA editing activity
Given these differences in editing efficiency in vitro between different ADAR isoforms, is there a correlation between ADAR isoform expression and editing of other transcripts in vivo? We chose to study editing of the cacophony (cac) transcript. This was the first transcript reported to be edited in Drosophila and 10 different sites are edited to give codon changes (Smith et al, 1996). We wished to determine if there is developmental regulation of cacophony transcript editing in vivo that reflects the expression of the more active ADAR isoforms in adults. The cac gene encodes the pore-forming α1 subunit of a voltage-gated calcium channel expressed in the CNS (Smith et al, 1996; Peixoto et al, 1997). The protein is 1851, amino acids long containing four internal repeats (I–IV) each with six proposed membrane-spanning segments (S1–S6). Figure 3A shows the locations within the predicted protein of amino-acid residues altered by RNA editing. Each editing site is named by the exon in which it is located.
Figure 3.
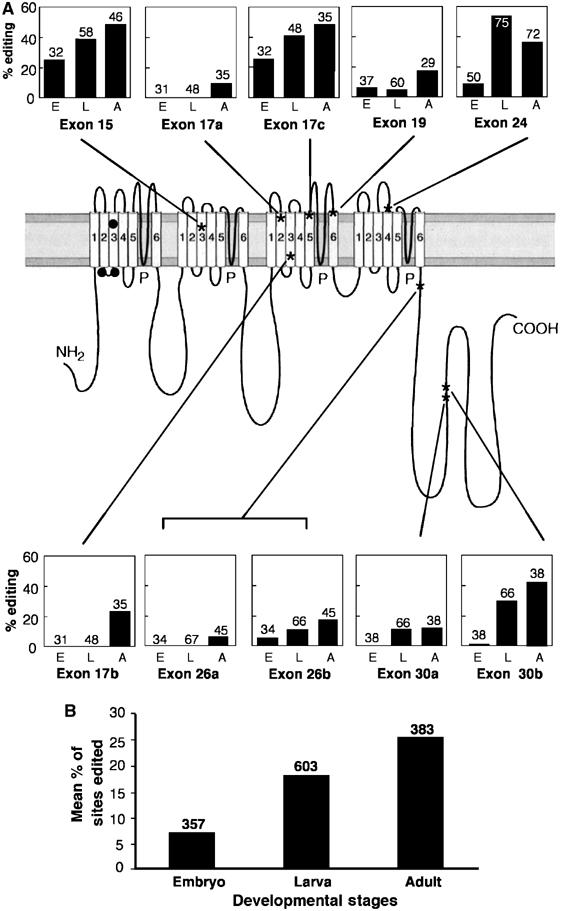
Developmental regulation of site-specific RNA editing in the cacophony transcript. (A) Percentage of individual sequenced cDNA clones edited at 10 sites in the cacophony transcript in embryos (E), larvae (L) and adult flies (A). The number of individual clones sequenced in each case is indicated above the error bars, which show the standard error of the percentage. Each editing site is named by the exon in which it is located. The locations within the protein of amino-acid residues changed by RNA editing are indicated on a conventional structure for this class of channel (Catterall, 2000). The cac 15 site is within the paddle structure involved in voltage gating. Black filled circles indicate three residue changes resulting from five RNA editing events in the Ca-alpha 1D transcript encoding a muscle voltage-gated calcium channel (see Figure 6). (B) Average editing level at all tested sites in the cacophony transcript at each developmental stage. The number of individual clones sequenced is indicated above the bars.
The developmental profile of RNA editing in the cac transcript is shown as the percentage of edited clones among individual sequenced cDNA clones from total RNA of embryos (E), larvae (L) and adult flies (A) (Figure 3A and B). All sites show an increase in RNA editing through development, consistent with increasing levels of ADAR expression through development and with the Adar mutant phenotype, which is most evident in adult flies. One editing site in the amino terminus was not studied as it is translationally silent and a second site in this region has been recently identified, raising the number of editing sites to 12 (Smith et al, 1996; Kawasaki et al, 2002). A similar developmental increase in editing has been reported for the para transcript (Hanrahan et al, 2000). In addition, we have also analysed approximately 10 other transcripts and this developmental increase in editing is observed in most cases (data not shown).
In transgenic flies, the ADAR 3/4 isoform efficiently rescues both Adar locomotion defects and editing of cac transcripts
Are the large differences in editing activity between the different ADAR isoforms in vitro reflected in their ability to rescue the Adar mutant phenotype? To test rescue of the Adar mutant phenotype by ADAR isoforms in vivo, cDNAs encoding ADAR 3a or ADAR 3/4 were fused to a truncated hsp70 promoter with five GAL4 binding sites upstream, in the vector pUAST (Phelps and Brand, 1998), and transgenic Drosophila lines were generated. The ADAR isoforms were expressed by crossing three independent transgenic lines for each UAS-Adar construct to another line that is heterozygous for the Adar 1F4 deletion and also has an actin-5C-GAL4 driver construct (Ito et al, 1997) that expresses GAL4 in all cells. We chose this GAL4 driver that directs ubiquitous expression of ADAR because an initial screen of drivers showed that this driver gave the most efficient rescue.
Rescue of the Adar mutant phenotype in male flies was quantitated using open field locomotion across lines in a gridded plate to measure restoration of walking ability. This is the simplest test that can be applied to the Adar mutant flies (three 2 min measurements on each of 10 or more flies for each transgenic UAS-Adar line). The data are presented as the average number of lines crossed in the assay period. More specific behavioural tests are unsuitable because they require normal locomotion.
Figure 4A shows rescue of the X-chromosome Adar 1F4 locomotion defect in male progeny by either the UAS-Adar 3a or UAS-Adar 3/4 constructs with the actin-5C-GAL4 driver. Adar mutant flies are very defective in the locomotion test (Figure 4A) (Palladino et al, 2000b). Clearly, the ADAR 3/4 isoform gives a more effective rescue than the ADAR 3a isoform and this correlates with their in vitro editing activities (Figure 1D). Locomotion rescue correlates very well with in vitro editing by ADAR splice forms because a very large number of locomotion measurements were carried out (90 or more measurements of locomotion for each UAS-Adar construct). The Adar wild-type strain used for comparison in these experiments contains the actin-5C-GAL4 driver construct, which reduces locomotion compared to Canton S without affecting editing of cac 15 or cac 17c sites significantly.
Figure 4.
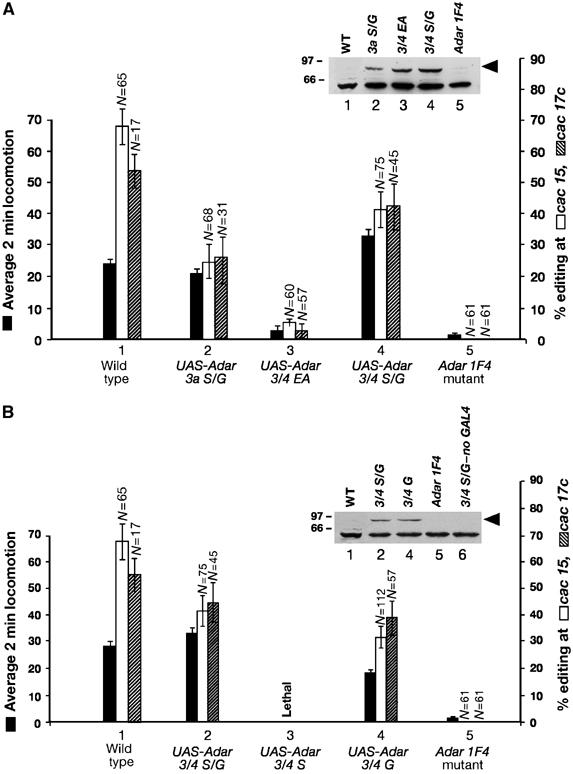
Phenotypic rescue and lethality associated with expression of ADAR isoforms in Drosophila. (A) The ADAR 3/4 isoform that is enriched in adult flies rescues Adar IF4 locomotion defects and RNA editing more efficiently than the ADAR 3a isoform. Open field locomotion measurements on male Adar wild-type; actin-5C-GAL4 flies (1) or mutant male Adar 1F4 flies or rescued male flies of the general genotype y1, Adar 1F4, w; act-5C-GAL4; UAS-Adar isoform. Average 2 min locomotion measurements are three measurements on each of 10 flies from three independent transgenic lines for each UAS-Adar construct. Standard errors are indicated. Percentage editing at the cac 15 site and at the cac 17c site, plotted on the right Y-axis, is presented beside the locomotion rescue data for each isoform. The percentage editing at the cac editing sites was determined by sequencing individual cDNAs from wild-type, Adar 1F4 or rescued flies and the number of individual cDNA clones sequenced is above the error bar. Standard error of the percentage is indicated. The inset shows a Western blot with an anti-FLAG antibody of normalised fly extracts of wild-type, Adar 1F4 and rescue genotypes. The lanes in the inset are numbered to correspond to the rescue lines in the main figure. The arrow indicates ADAR and the band underneath is nonspecific. (B) Expression of a Drosophila Adar transcript resistant to editing is lethal. Free range locomotion assays on Adar IF4 mutant flies expressing UAS-Adar constructs under the control of an actin-5C-GAL4 driver and rescue of editing at the cac15 and cac 17 sites measured as in panel A. The inset shows a Western blot with an anti-FLAG antibody of normalised fly extracts of wild-type, Adar 1F4 and the viable rescue genotype. The lanes in the inset are numbered to correspond to the rescue lines in the main figure; however, there is no lane 3 as it is lethal and extract from male y1, Adar 1F4, w; Cy; UAS-Adar 3/4 flies is included as it shows that very little protein is expressed in the absence of the GAL4 driver.
Rescue of RNA editing at the cac 15 site and the cac 17c site was measured by sequencing individual cDNA clones from two or more independent RT–PCR reactions on RNA from rescue flies of one transgenic line for each construct (Figure 4A). The number of individual clones sequenced is presented above the error bars. We chose to measure in vivo editing in the cac transcript as it was the first transcript that was found to be site-specifically edited in Drosophila; however, we do not believe that the unedited cac transcript is the cause of the Adar mutant phenotype. Rescue of editing is substantial but it does not reach or exceed wild-type levels. In addition, sites in a number of other edited transcripts are also edited at levels approaching but not exceeding wild-type editing levels in the UAS-Adar 3/4 rescue line (data not shown). The expressed ADAR proteins bear FLAG and 6xHis epitope tags and Western blots with anti-FLAG antibody have been performed in the rescue lines to confirm that expression levels of constructs are comparable (Figure 4A, inset), given the variations between lines that arise due to position effects on transgene insertions. The variation between lines will not affect locomotion rescue data, which are based on multiple lines, but needs to be considered for in vivo editing, which is based on single lines. Nevertheless, the differences in locomotion rescue and the level of editing observed in vivo between Adar 3a S/G and Adar 3/4 S/G are similar to that observed in vitro (Figure 1D). The difference in locomotion rescue between the ADAR splice forms is greater when a less effective rescue driver is substituted (data not shown).
ADAR must act prior to splicing at many sites. Editing sites are frequently found close to splice sites within predicted RNA duplexes that include a splice junction. Loss of vertebrate ADAR2 causes accumulation of an incompletely spliced GluR-B (Higuchi et al, 2000). It is possible that RNA editing could assist splicing by simply binding the duplex at splice sites and recruiting helicases to disrupt the duplex. Therefore, we wished to confirm that the Adar mutant phenotype in Drosophila is due to loss of adenosine deaminase activity. A UAS-ADAR 3/4 EA construct encoding a protein with a mutation in the active site glutamate (Lai et al, 1995) gave minimal rescue of locomotion, and very low levels of editing were found at the cac 15 site and the cac 17c site in RNA from these flies even though this protein is efficiently expressed (Figure 4A). Therefore, deamination is required for rescue of the Adar mutant phenotype; the deaminated base itself could also facilitate splicing in cases where it destabilises a dsRNA editing structure that occludes a splice junction.
Ubiquitous expression of the genome-encoded isoform of ADAR is lethal in Drosophila
If the function of self-editing in Adar is to limit overall RNA editing activity, then expressing pure unedited ADAR 3/4 S isoform in Drosophila would bypass this regulation and could be toxic. The ineditable Adar 3/4 S and the edited Adar 3/4 G cDNA constructs used for expression in Pichia were introduced into the Drosophila pUAST vector. Transgenic fly lines bearing UAS-Adar 3/4 S and UAS-Adar 3/4 G constructs were generated, and crossed to the actin 5C-GAL4 driver to compare their ability to rescue the Adar 1F4 locomotion defect (Figure 4B). Crossing flies bearing the ineditable UAS-Adar 3/4 S construct to flies bearing an actin-5C-GAL4 driver construct produced no progeny having both constructs.
The complete lethality obtained with the ineditable UAS-Adar 3/4 S construct is an exacerbation of a partial loss of viability seen with the UAS-Adar 3/4 S/G construct. Among male progeny of the rescue crosses that carry both the Adar 1F4 mutation and the relevant UAS-Adar construct, the number of actin 5C-GAL4 driver flies obtained, expressed as a percentage of the number of Cy balancer flies obtained that lack the GAL4 driver, was UAS-Adar 3/4 EA 72%, UAS-Adar 3/4 G 69%, UAS-Adar 3/4 S/G 20% and UAS-Adar 3/4 S 0%. Decreases in viability correlate with levels of ADAR 3/4 S isoform expressed. When editing of Adar exon 7 was examined in UAS-Adar 3a S/G and UAS-Adar 3/4 S/G rescue flies (Figure 4A), we found that the transcript expressed from the transgene was 70% edited, that is, only 30% of the transcripts still expressed the ADAR 3/4 S protein. The ineditable UAS-Adar 3/4 S construct is lethal because it produces more of the genome-encoded form.
Rescue of the Adar 1F4 locomotion defect was less efficient with the UAS-Adar 3/4 G construct than with the unedited UAS-Adar S/G construct that generates a mixture of edited and unedited isoforms. The weaker rescue is not due to weaker expression of ADAR from the UAS-Adar 3/4 G construct (Figure 4B, inset). Editing of the cac15 and cac 17c sites was efficient in these rescue flies. The UAS-Adar 3/4 S construct lines give viable progeny when crossed to other GAL4 driver lines such as Cha-GAL4 (see below; Figure 5). Western blots confirm that the ADAR 3/4 S protein is produced at levels equivalent to the other ADAR isoforms in lines that rescue lethality (Supplementary data). With Cha-GAL4 and other drivers where the comparison can be made, the UAS-Adar 3/4 G construct rescues less efficiently than UAS-Adar 3/4 S, as expected from the in vitro data.
Figure 5.
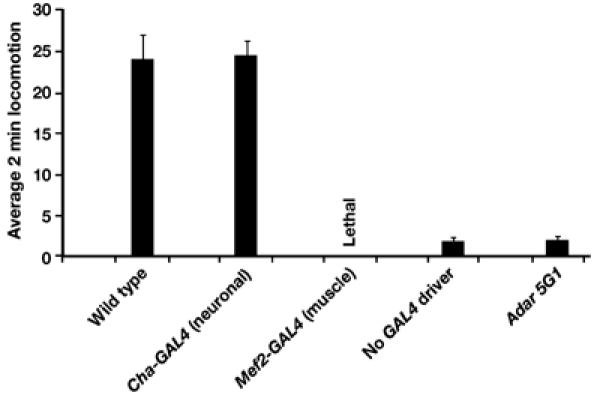
Locomotion rescue and lethality caused by expressing the UAS-Adar 3/4 S construct in the Adar 5G1 background under control of the CNS-specific Cha-GAL4 19B driver (Salvaterra and Kitamoto, 2001) or the muscle-specific Mef2-GAL4 driver (Ranganayakulu et al, 1996) or in the absence of any GAL4 driver. The Cha-GAL4 driver directs a very efficient rescue compared to male Adar wild-type; Cha-GAL4 flies whereas the Mef2-GAL4 driver causes lethality. In the absence of a GAL4 driver, there is no rescue compared to Adar 5G1 alone.
Tissue specificity of Adar rescue and UAS-Adar 3/4 S lethality
The GAL4-UAS system allows the tissue-specific requirements for Adar locomotion rescue and for the lethal effect of UAS-Adar 3/4 S expression to be defined. A complication is that low levels of ADAR expression are sufficient to partially rescue locomotion defects in Adar mutants because we see some GAL4-independent rescue in transgenic UAS-Adar 3/4 S/G lines. The UAS-Adar 3/4 S construct however shows no such GAL4-independent rescue. The UAS-Adar 3/4 S lines are therefore suitable for testing a range of different GAL4 drivers both for the tissue specificity of Adar locomotion rescue and of Adar S lethality.
An Adar 5G1/FM6 line that is also homozygous for UAS-Adar 3/4 S was available. Females of this line were crossed to various GAL4 driver lines that express GAL4 in neurons, muscles or glial cells to generate rescued male progeny. In the absence of a GAL4 driver construct, Adar 5G1; UAS-Adar 3/4 S is as locomotion-defective as Adar 5G1 alone (Figure 5A, rightmost columns). We tested the ability of GAL4 driver lines that express GAL4 throughout the CNS to direct efficient locomotion rescue with UAS-Adar 3/4 S. The best rescue was obtained with Cha-GAL4 driver that expresses strongly in cholinergic neurons (Salvaterra and Kitamoto, 2001), in the pattern of the neurotransmitter synthesis and transport protein choline acetyltransferase. Rescue of the Adar locomotion defect was efficient with the Cha-GAL4 driver when compared to an Adar wild-type; Cha-GAL4 control strain, which shows reduced locomotion compared to Canton S. Apparently, strong expression of the UAS-Adar 3/4 S construct in acetylcholine-synthesising neurons is sufficient to give considerable rescue of the Adar 5G1 locomotion defect. When crossed to UAS-GFP S65T to visualise the strength and localisation of GAL4 expression, the Cha-GAL4 driver gives stronger GAL4 expression than many other neuronal drivers and may rescue more efficiently than those drivers partly for that reason also.
One muscle-specific driver, dMef2-GAL4 (Ranganayakulu et al, 1996), that is expressed very strongly in muscles and also in the heart and in the mushroom bodies in the brain caused lethality with UAS-Adar 3/4 S (Figure 5A). Is this a result of ectopic ADAR expression outside the CNS and outside the normal expression range of ADAR or does editing also normally occur in muscle? To answer this question, we examined editing of the neuron- and muscle-expressed voltage-gated calcium channel Ca-alpha 1D transcript that is edited at five sites in exon 5, which encodes transmembrane segment S3 and the preceding intracellular loop in the first repeat domain of this channel (transmembrane segment I-S3, refer to Figure 3). This transcript shows high levels of editing in adult flies (Figure 5), with some sites virtually 100% edited. Editing is equally efficient in RNA extracted from dissected dorsal thorax, which consists almost entirely of flight muscle (Figure 6A). Editing is absent or much lower in embryos and larvae (Figure 6B).
Figure 6.
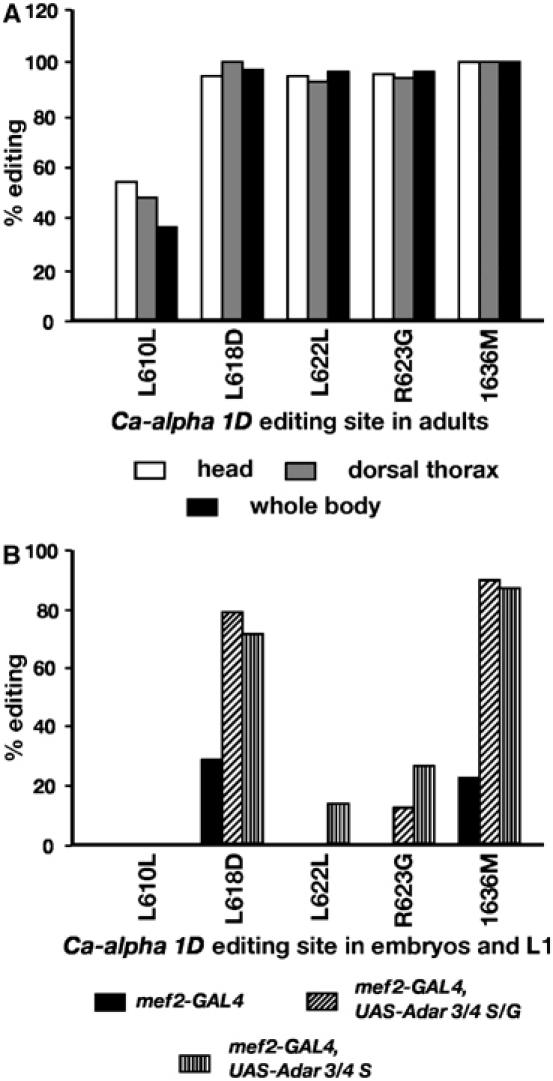
Editing normally occurs in muscle and toxic levels of ADAR activity cause hyperediting of specific editing sites in a muscle transcript. (A) Editing in the Ca-alpha 1D transcript encoding an L-type voltage-gated calcium channel normally occurs in muscle. Percentage editing at five sites in the Ca-alpha 1D transcript based on multiple sequence chromatograms of several RT–PCR product pools from RNA isolated from head (neuron), dorsal thorax (muscle) or whole body of adult Canton S flies is shown. The editing sites are named by the position of the altered base and the codon change introduced. Locations of residues changed by editing in this channel are indicated on the generic calcium channel structure in Figure 3. (B) Hyperediting of sites in the Ca-alpha 1D transcript in embryos and L1 larvae expressing either no additional ADAR or ADAR 3/4 S/G or ADAR 3/4 S isoforms under the control of the muscle-specific Mef2-GAL4 driver. Editing was analysed by bulk sequencing RT–PCR products derived from RNA isolated from 48 h collections of embryos and larvae.
One possibility is that lethality due to excess ADAR activity in muscles could arise from hyperediting of Ca-alpha 1D and other transcripts during the embryonic and larval stages of development. Obtaining a pure population of embryos and larvae dying as a result of UAS-Adar overexpression is possible using the Mef2-GAL4 driver, which is a homozygous viable GAL4 driver line. We crossed the homozygous Mef 2-GAL4 driver line to wild-type Canton S or to homozygous UAS-Adar lines and collected embryos and first instar larvae of the genotype Mef2-GAL4/+; UAS-Adar 3/4 isoform. These embryos and larvae have an intact chromosomal Adar gene. In the absence of any additional ADAR, the L618D and I636M sites in Ca-alpha 1D are edited at low levels and other sites are not edited, as seen in progeny of the cross to Canton S (labelled Mef2-GAL4 in Figure 6B). When extra ADAR is expressed from the UAS-Adar 3/4 S/G construct, an arrangement that gives viable adult flies, a considerable increase in editing, almost to levels typical of adult flies, is seen at the L618D and I636M sites in these embryos and larvae. When extra ADAR is expressed from the UAS-Adar 3/4 S construct, no adult flies are obtained; the same strong increase in editing is seen at the two strong sites and editing now also occurs at two weaker sites, L622L and R623G (Figure 6B). We conclude that hyperediting occurs as a result of increasing ADAR expression early in development and that this is the likely cause of death. Although for experimental reasons we can easily observe hyperediting in Ca-alpha 1D transcripts, we do not know whether hyperediting of this transcript or some other transcript is the cause of lethality.
Discussion
We report here that, in adult Drosophila, ADAR edits its mRNA with 40% efficiency to encode a less active isoform. We propose that this own-transcript editing is a form of negative autoregulation. When this editing is prevented in vivo so that only the genome-encoded ADAR is expressed, then levels of total ADAR protein otherwise sufficient for phenotypic rescue become lethal. This early lethality is associated with hyperediting of sites in at least one target transcript.
Previously, it has been shown that mammalian ADAR2, which is the orthologue of Drosophila ADAR (J Brindle and LP Keegan, unpublished data), edits a 3′ splice site in its own pre-mRNA so that an additional 47 nucleotides are included in the mRNA (Rueter et al, 1999). This editing is also considered to be a negative autoregulatory mechanism due to frameshifts and inefficient translation initiation from internal methionines that lead to a reduction in protein levels from the edited transcript. Therefore, both Drosophila ADAR and ADAR2 appear to have evolved completely different own-transcript RNA editing sites to control ADAR activity. In another example of convergent evolution of RNA editing sites, the same codon change is introduced by editing at the same position in Drosophila and vertebrate voltage-gated potassium channels in different channel families (Bhalla et al, 2004).
ADAR editing requires only a short RNA duplex to target it; nevertheless, the evolution of an RNA duplex within Drosophila Adar exon 7 is surprising and suggests that editing of the mature Adar mRNA may be required. Adar exon 7 probably folds into a very stable secondary structure, as editing of this minisubstrate in vitro to 70% is higher than for any other transcript so far tested in vitro. The Adar exon 7 structure that supports RNA editing is conserved in other Drosophila species but not in the mosquito (RA Reenan and MA O'Connell, unpublished data). If transient increases or decreases in ADAR activity occur in response to unknown regulatory signals in Drosophila, then Adar editing of mature Adar mRNA could respond immediately to restore the correct level of RNA editing. In vertebrates, modulation of ADAR2 activity in response to changes in neurotransmitter levels has been suggested (Gurevich et al, 2002a, 2002b).
The conversion of serine to glycine has an unexplained dramatic effect on RNA editing activity and this requires further study. This amino acid is conserved in all ADAR2-type enzymes, whereas ADAR1-type enzymes have a conserved aspartic acid at the same position (Keegan et al, 2004); one possibility is that the serine is phosphorylated but we are unable to detect this. When the serine is mutated to aspartic acid to introduce a negative charge as found in ADAR1, dADAR is completely inactive (MA O'Connell, unpublished results). Defining the function of this serine residue in the deaminase domain awaits the crystallisation of ADAR.
As alternative splicing is critical for generating proteomic diversity, it will be important in the future to determine the functional differences between protein isoforms in an animal model. Here, we demonstrate that Drosophila is an excellent animal model to study proteomic diversity. We observe an excellent correlation between the activities of the different ADAR isoforms that have been overexpressed in yeast and assayed in vitro and their in vivo activities as tested in flies. Even though ADAR 3/4 S is very active in vitro, it is not intuitive that this would be toxic in Drosophila when it is ubiquitously expressed, considering that ADAR 3/4 G, which differs by one amino acid, has no toxic effects. It is improbable that many isoforms of other proteins will have such dramatic effects on activity in vivo; however, some will. Drosophila may be the proteomics choice of model organism in future, considering the resource costs of generating equivalent numbers of transgenic mice. While the present work required a great deal of effort to overcome the variations due to position effects on transgene insertions, these limitations arising from the Drosophila transformation method may be relieved by using new genetic techniques that reduce position effects on transgene expression or that express variants from a common site in the chromosome (Gloor, 2004).
In Drosophila, the number of transcripts known to be edited to cause recoding events is much greater than in vertebrates; the list of vertebrate targets may now be nearly complete and includes a large amount of editing of Alu elements embedded in transcripts (Kim et al, 2004; Levanon et al, 2004). The number of recoding sites per target transcript is also higher in Drosophila, suggesting that flies make considerable use of RNA editing as an easy and efficient method of generating proteomic diversity. RNA editing contributes to an enormous potential diversity of transcripts from the cacophony gene encoding the large, pore-forming alpha subunit of the main, non-L-type, voltage-gated calcium channel in the CNS. This protein has a critical role at synapses on axon termini where it responds to depolarising signals by allowing calcium entry to induce vesicle fusion and neurotransmitter release. Pairwise choices due to alternative splicing events identified in this study (data not shown), and previously (Peixoto et al, 1997; Gallo et al, 2002), predict 25 splice forms. Pairwise choices of edited or unedited isoforms at each editing site where editing changes codon meaning could multiplex this with potentially 211 edited isoforms. Transcript diversity in cacophony and other RNA editing targets may not reach the full possible maximum level as is the case for the Adar transcript itself where the edited form of the Adar 3a transcript is not observed. It is not surprising that the misregulation of RNA editing that we have induced experimentally in this study, leading to this proteomic complexity being expressed at the wrong developmental stage, causes lethality.
The highest levels of editing are seen in adult flies, consistent with the Adar mutant phenotype that includes adult locomotion defects and age-dependent neurodegeneration (Palladino et al, 2000b; Ma et al, 2001). ADAR mutant phenotypes in mice and Caenorhabditis elegans also demonstrate that editing is essential for viability and for proper functioning of the nervous system (Higuchi et al, 2000; Palladino et al, 2000b; Tonkin et al, 2002; Hartner et al, 2004; Wang et al, 2004). We show here that editing also normally occurs outside the nervous system in Drosophila. Loss of RNA editing in muscles may contribute to the locomotion defects in Adar mutant flies. RNA editing outside the nervous system is also necessary in vertebrates; ADAR1 mutant mouse embryos undergo widespread apoptosis due to loss of editing in unknown target transcripts (Hartner et al, 2004; Wang et al, 2004). The primary requirement for RNA editing in Drosophila may be in the abundant cholinergic neurons in the brain, consistent with efficient rescue using a Cha-GAL4 driver, with patterns of neurodegeneration seen in Adar mutants (Palladino et al, 2000b; Ma et al, 2001), and with electrophysiological measurements linking slow recovery from anoxic stupor to defects in brain interneurons (Ma et al, 2001). Also, several transcripts encoding acetylcholine receptor alpha and beta subunits are edited in Drosophila (Grauso et al, 2002; Hoopengardner et al, 2003). Adar expression is much lower in muscle than in CNS in embryos and it is possible that toxicity arising from ADAR overexpression in embryos could be due to hyperediting of the Ca-alpha 1D transcript encoding an L-type voltage-gated calcium channel. In vertebrates, this channel class is found in muscles and has a critical role on the postsynaptic side of the neuro muscular junction where it responds to depolarisation by allowing calcium entry into muscle cells and subsequent release of calcium from intracellular stores for muscle contraction (Catterall, 2000). We do not, however, know what the effects of editing on most of the channels are nor do we know which channels are critical for either rescue or hyperediting toxicity.
In this study, we describe some of the RNA editing events that we have found to be almost 100% efficient in adult flies. In mice, the GluR-B Q/R site is edited with virtually 100% efficiency. An edited GluR-B construct is able to rescue all the defects in a mouse ADAR2 mutant (Higuchi et al, 2000), and no functional requirement has been found for the unedited GluR-B isoform, which is not found even early in development. For the sites that are 100% edited in adult Drosophila, the situation is somewhat different, that is, the unedited transcripts are expressed significantly earlier in development and our results suggest that these forms are required at this stage. Lethality due to increased ADAR activity is associated with increasing editing at some sites in an embryonic ion channel transcript close to levels seen in adults. In contrast to the case of the GluR-B Q/R site, we suggest that flies require unedited and edited isoforms at different stages. It may be possible to rescue the Adar mutant phenotype in flies by expressing cDNA constructs encoding edited versions of target ion channel transcripts; however, the corresponding unedited transcripts may also be required.
The fruit fly undergoes complete metamorphosis to build two very different-looking complex organisms using a gene set that is small and relatively nonredundant compared to that of vertebrates. Many fly genes show complex patterns of embryonic and adult expression, with different enhancers, promoters, RNA splicing patterns and, finally, different RNA editing patterns in adult transcripts. Customising fast neurotransmission to the requirements of the different life-cycle stages by RNA editing is the most baroque of the gene-sparing strategies so far described in Drosophila.
Materials and methods
ADAR protein expression in P. pastoris
For details, see Supplementary data.
In vitro RNA editing assays
For details, see Supplementary data.
UAS-Adar fly strains and rescue experiments
UAS-Adar lines were generated by injection of pUAST-Adar constructs using standard procedures. Most lines were generated by coinjection of the UAS-Adar construct and a helper plasmid that expresses P-element transposase transiently into w1118 eggs followed directly by balancing of stable transformant lines. For the UAS-Adar 3/4 unedited cDNA, the eggs used for injection were of the (del 2-3) 68C strain that expresses P-element transposase constitutively. Injected flies were crossed to w1118. Transformants initially had variegated eye colour as transposition continues after the initial integration. Different stable inserts separated from the transposase source by further backcrosses to w1118 were recognised by consistent rather than variegated eye colour and balanced. In the case of pUAST-ADAR 3a (S/G), only one transformant line with an insert on the X-chromosome was initially obtained and this was mobilised by crosses to the Jumpstarter stock to generate multiple lines.
The Adar deletion lines are previously described (Palladino et al, 2000b). The Adar mutations on the X-chromosome were crossed to an actin-5C-Gal4 25FO1 driver insert on chromosome II to generate GAL4 driver lines of the form y, Adar (1F4 or 5G1), w/w, FM6; (miniw+) actin-5C-Gal4/SM5 Cy. Rescue was quantified in 2- to 4-day-old male flies of the genotype y, Adar 1F4, w; (miniw+) actin-5C-Gal4; (miniw+)UAS-Adar and in sibling males of the genotype y, Adar 1F4, w; SM5 Cy; (miniw+)UAS-Adar to determine whether the rescue was GAL4 dependent. Locomotion measurements were performed on three independent transformant lines for each construct except UAS-Adar 3/4 for which there were two lines.
Drosophila GAL4 driver lines used in this study
The actin-5C-GAL4 25FO1 line derives from a flip-out of a yellow gene and is a very strong driver that has been used in other studies on the nervous system (Ito et al, 1997). The Cha-GAL4 19B, UAS-GFP.S65T T2 (Salvaterra and Kitamoto, 2001) and Mef2-GAL4 on chromosome III (Ranganayakulu et al, 1996) lines have been described previously .
Quantitating RNA editing activity in vivo
For details, see Supplementary data.
Supplementary Material
Supplementary Data 1
Supplementary Data 2
Acknowledgments
We thank K Matthews and K Cook and the Bloomington Drosophila Stock Center for collecting GAL4 and UAS lines and others from whom lines were sourced directly, including N Bonini, A Brand, A Jarman, U Kloter, E Olson, P Salvaterra, M Samson and S Voss. We also thank J Caceres, H Cooke, J Long, N Hastie, J Rosenthal and D Scadden and anonymous reviewers for constructive comments on the manuscript and S Bruce for illustrations. This work was supported by the Medical Research Council.
References
- Basilio C, Wahba AJ, Lengyel P, Speyer JF, Ochoa S (1962) Synthetic polynucleotides and the amino acid code. Proc Natl Acad Sci USA 48: 613–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla T, Rosenthal JJ, Holmgren M, Reenan R (2004) Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat Struct Mol Biol 11: 950–956 [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB (1997) Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387: 303–308 [DOI] [PubMed] [Google Scholar]
- Catterall WA (2000) Structure and regulation of voltage-gated Ca2+ channels. Annu Rev Cell Dev Biol 16: 521–555 [DOI] [PubMed] [Google Scholar]
- Eberl DF, Ren D, Feng G, Lorenz LJ, Van Vactor D, Hall LM (1998) Genetic and developmental characterization of Dmca1D, a calcium channel alpha1 subunit gene in Drosophila melanogaster. Genetics 148: 1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo A, Keegan LP, Ring GM, O'Connell MA (2003) An ADAR that edits transcripts encoding ion channel subunits functions as a dimer. EMBO J 22: 3421–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo A, Thomson E, Brindle J, O'Connell MA, Keegan LP (2002) Micro-processing events in mRNAs identified by DHPLC analysis. Nucleic Acids Res 30: 3945–3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor GB (2004) Gene targeting in Drosophila. Methods Mol Biol 260: 97–114 [DOI] [PubMed] [Google Scholar]
- Grauso M, Reenan RA, Culetto E, Sattelle DB (2002) Novel putative nicotinic acetylcholine receptor subunit genes, Dalpha5, Dalpha6 and Dalpha7, in Drosophila melanogaster identify a new and highly conserved target of adenosine deaminase acting on RNA-mediated A-to-I pre-mRNA editing. Genetics 160: 1519–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger IH, Khatri L, Kong X, Ziff EB (2003) AMPA receptor tetramerization is mediated by q/r editing. Neuron 40: 763–774 [DOI] [PubMed] [Google Scholar]
- Gurevich I, Englander MT, Adlersberg M, Siegal NB, Schmauss C (2002a) Modulation of serotonin 2C receptor editing by sustained changes in serotonergic neurotransmission. J Neurosci 22: 10529–10532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, Schmauss C (2002b) Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron 34: 349–356 [DOI] [PubMed] [Google Scholar]
- Hanrahan CJ, Palladino MJ, Ganetzky B, Reenan RA (2000) RNA editing of the Drosophila para Na(+) channel transcript. Evolutionary conservation and developmental regulation. Genetics 155: 1149–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH (2004) Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem 279: 4894–4902 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH (2000) Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406: 78–81 [DOI] [PubMed] [Google Scholar]
- Hoopengardner B, Bhalla T, Staber C, Reenan R (2003) Nervous system targets of RNA editing identified by comparative genomics. Science 301: 832–836 [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D (1997) The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development 124: 761–771 [DOI] [PubMed] [Google Scholar]
- Kawasaki F, Collins SC, Ordway RW (2002) Synaptic calcium-channel function in Drosophila: analysis and transformation rescue of temperature-sensitive paralytic and lethal mutations of cacophony. J Neurosci 22: 5856–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan LP, Gallo A, O'Connell MA (2001) The many roles of an RNA editor. Nat Rev Genet 2: 869–878 [DOI] [PubMed] [Google Scholar]
- Keegan LP, Leroy A, Sproul D, O'Connell MA (2004) Adenosine deaminases acting on RNA (ADARs): RNA-editing enzymes. Genome Biol 5: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A (2004) Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res 14: 1719–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Drakas R, Nishikura K (1995) Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J Biol Chem 270: 17098–17105 [DOI] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, Olshansky M, Rechavi G, Jantsch MF (2004) Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol 22: 1001–1005 [DOI] [PubMed] [Google Scholar]
- Ma E, Gu XQ, Wu X, Xu T, Haddad GG (2001) Mutation in pre-mRNA adenosine deaminase markedly attenuates neuronal tolerance to O2 deprivation in Drosophila melanogaster. J Clin Invest 107: 685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino MJ, Keegan LP, O'Connell MA, Reenan RA (2000a) dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. RNA 6: 1004–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino MJ, Keegan LP, O'Connell MA, Reenan RA (2000b) A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell 102: 437–449 [DOI] [PubMed] [Google Scholar]
- Peixoto AA, Smith LA, Hall JC (1997) Genomic organization and evolution of alternative exons in a Drosophila calcium channel gene. Genetics 145: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps CB, Brand AH (1998) Ectopic gene expression in Drosophila using GAL4 system. Methods 14: 367–379 [DOI] [PubMed] [Google Scholar]
- Price RD, Weiner DM, Chang MS, Sanders-Bush E (2001) RNA editing of the human serotonin 5-HT2C receptor alters receptor-mediated activation of G13 protein. J Biol Chem 276: 44663–44668 [DOI] [PubMed] [Google Scholar]
- Ranganayakulu G, Schulz RA, Olson EN (1996) Wingless signaling induces nautilus expression in the ventral mesoderm of the Drosophila embryo. Dev Biol 176: 143–148 [DOI] [PubMed] [Google Scholar]
- Rueter SM, Dawson TR, Emeson RB (1999) Regulation of alternative splicing by RNA editing. Nature 399: 75–80 [DOI] [PubMed] [Google Scholar]
- Salvaterra PM, Kitamoto T (2001) Drosophila cholinergic neurons and processes visualized with Gal4/UAS-GFP. Gene Expr Patterns 1: 73–82 [DOI] [PubMed] [Google Scholar]
- Smith LA, Wang XJ, Peixoto AA, Neumann EK, Hall LM, Hall JC (1996) A Drosophila calcium channel α1 subunit gene maps to a genetic locus associated with behavioural and visual defects. J Neurosci 16: 7868–7879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Köhler M, Sprengel R, Seeburg PH (1991) RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67: 11–19 [DOI] [PubMed] [Google Scholar]
- Stapleton M, Carlson J, Brokstein P, Yu C, Champe M, George R, Guarin H, Kronmiller B, Pacleb J, Park S, Wan K, Rubin GM, Celniker SE (2002) A Drosophila full-length cDNA resource. Genome Biol 3, RESEARCH0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin LA, Saccomanno L, Morse DP, Brodigan T, Krause M, Bass BL (2002) RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J 21: 6025–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K (2004) Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem 279: 4952–4961 [DOI] [PubMed] [Google Scholar]
- Zheng W, Feng G, Ren D, Eberl DF, Hannan F, Dubald M, Hall LM (1995) Cloning and characterization of a calcium channel alpha 1 subunit from Drosophila melanogaster with similarity to the rat brain type D isoform. J Neurosci 15: 1132–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M, Mathews DH, Turner DH (eds) (1999) Algorithms and Thermodynamics for RNA Secondary Structure Prediction: A Practical Guide. Dordrecht: Kluwer Academic Publishers [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data 1
Supplementary Data 2


