Abstract
Here we show that the PIASy protein is specifically required for mitotic modification of Topoisomerase-II by SUMO-2 conjugation in Xenopus egg extracts. PIASy was unique among the PIAS family members in its capacity to bind mitotic chromosomes and recruit Ubc9 onto chromatin. These properties were essential, since PIASy mutants that did not bind chromatin or failed to recruit Ubc9 were functionally inactive. We observed that PIASy depletion eliminated essentially all chromosomal accumulation of EGFP–SUMO-2-conjugated species, suggesting that it is the primary E3-like factor for mitotic chromosomal substrates of SUMO-2. PIASy-dependent SUMO-2-conjugated species were highly concentrated on the inner centromere, and inhibition of PIASy blocked anaphase sister chromatid segregation in egg extracts. Taken together, our observations suggest that PIASy is a critical regulator of mitotic SUMO-2 conjugation for Topoisomerase-II and other chromosomal substrates, and that its activity may have particular relevance for centromeric functions required for proper chromosome segregation.
Keywords: chromatin, mitosis, PIASy, SUMO, Topoisomerase-II
Introduction
SUMO proteins are a conserved family of ubiquitin-related peptides that become covalently conjugated to target proteins in a manner similar to ubiquitin (Johnson, 2004). Budding yeast possesses a single SUMO protein, Smt3p. There are three human SUMO paralogues: SUMO-1 is about 45% identical to SUMO-2 and SUMO-3, which are 96% identical to each other. Newly translated SUMO proteins are initially processed to yield a C-terminal di-glycine motif. The processed forms are activated in an ATP-dependent manner, forming a thioester linkage with their activating (E1) enzyme, Uba2/Aos1. They are subsequently transferred to an intermediate thioester-linked complex with their conjugating (E2) enzyme, Ubc9. Finally, an isopeptide bond is formed between SUMO proteins and substrates through the cooperative action of Ubc9 and SUMO ligases (E3 enzymes). Since SUMO proteins can be deconjugated from their substrates by highly active isopeptidases (Melchior et al, 2003), it is likely that SUMO modification is dynamic in vivo.
PIAS/Siz family proteins are conserved SUMO ligases found in both yeast and vertebrate cells that mediate the conjugation of a wide variety of substrates (reviewed in Melchior et al, 2003; Johnson, 2004). There are three PIAS family proteins in budding yeast (Siz1p, Siz2p and Mms21p), and five PIAS family members in vertebrates (PIAS1, PIAS3, PIASxα, PIASxβ and PIASy) (Melchior et al, 2003; Johnson, 2004; Zhao and Blobel, 2005). PIAS proteins have a number of motifs that are conserved among family members, including an N-terminal SAP domain, a RING-like motif (SP-ring domain) and a C-terminal domain that is rich in serine and acidic amino acids (S/DE domain). The SAP domain binds AT-rich DNA sequences, and promotes localization to the nuclear matrix (Aravind and Koonin, 2000; Okubo et al, 2004). The SP-ring domain resembles ring-finger motif found in many ubiquitin E3 ligases, and has been shown to associate with Ubc9 (Hochstrasser, 2001). The function of the S/DE domain is not well defined, although it may confer interaction with SUMO proteins (Minty et al, 2000).
The conjugation of Smt3 to yeast Topoisomerase-II (Top2p) is likely to play an important role in chromosome segregation: Bachant et al (2002) reported that metaphase is prolonged in mutants that lack an Smt3p isopeptidase (smt4-Δ), and that such mutants also show defects in centromeric cohesion. Top2p was implicated in this phenotype, because top2 mutants lacking Smt3p modification sites significantly suppressed smt4-Δ centromeric cohesion defects. We have previously shown that Xenopus Topoisomerase-II is modified exclusively by SUMO-2 during mitosis under normal circumstances (Azuma et al, 2003). This modification is maximal in metaphase, followed by rapid deconjugation during anaphase. The differential extraction properties of modified and unmodified Topoisomerase-II suggest that SUMO-2 conjugation may mobilize a subpopulation of Topoisomerase-II from mitotic chromatin in a manner that is important for chromosome segregation in vertebrate cells. Inhibition of de novo SUMO conjugation using a dominant-negative form of Ubc9 caused the failure of anaphase sister chromatid segregation in Xenopus egg extracts, consistent with the notion that SUMO-2 conjugation of Topoisomerase-II or other substrates is important for remodeling of mitotic chromosomes at the metaphase–anaphase transition.
Here we show that the PIASy protein is essential for SUMO-2 conjugation to Topoisomerase-II in Xenopus egg extracts. Immunodepletion of PIASy from egg extracts abolished this modification. PIASy was uniquely able to rescue Topoisomerase-II conjugation in depleted extracts. PIASy was also essential for the recruitment of Ubc9 to the mitotic chromosomes. PIASy mutants that failed to recruit Ubc9 to mitotic chromatin similarly failed to restore Topoisomerase-II conjugation to PIASy-depleted egg extracts, suggesting that Ubc9 binding and chromatin recruitment may be important aspects of PIASy's mechanism. We examined the overall pattern of SUMO-2 association with mitotic chromosomes through incorporation of a fluorescently tagged SUMO-2 protein (EGFP–SUMO-2). EGFP–SUMO-2 prominently accumulated on the inner centromeres (ICs) of mitotic chromosomes in a PIASy-dependent manner, suggesting that PIASy is the primary E3-like factor for mitotic chromosomal substrates of SUMO-2. Notably, inhibition of PIASy blocked sister chromatid segregation during anaphase. Together, our observations suggest that PIASy is a critical regulator of mitotic SUMO-2 conjugation for Topoisomerase-II and perhaps other chromosomal substrates, and that its activity may have particular relevance for centromeric functions required for proper chromosome segregation.
Results
Chromatin-associated factors support SUMO-2 modification of Topoisomerase-II
Topoisomerase-II becomes conjugated to SUMO-2 in mitotic Xenopus egg extracts in a chromatin-dependent manner (Azuma et al, 2003). We examined the dynamics of this modification in M-phase-arrested Xenopus egg extracts (CSF extracts; Kornbluth et al, 2001) (Supplementary Figure 1). Inhibition of SUMO conjugation through the addition of a dominant-negative mutant of Ubc9 (dnUbc9; Azuma et al, 2003) resulted in the rapid loss of SUMO-2-conjugated Topoisomerase-II (SUMO-2·Topoisomerase-II), suggesting that isopeptidases are highly active during mitosis and that ongoing conjugation is required to maintain a steady-state level of SUMO-2·Topoisomerase-II.
In order to examine which components of the conjugation machinery are associated with mitotic chromatin, we utilized CSF extracts containing dnUbc9 as a mitotic source of unconjugated, chromatin-bound Topoisomerase-II. The chromatin was isolated by centrifugation through a sucrose cushion, sheared by sonication, and incubated with SUMO conjugation pathway enzymes plus a recombinant fusion of the green fluorescent protein to SUMO-2 (EGFP–SUMO-2). When mitotic chromatin was incubated with purified E1 (20 nM/reaction) and E2 (300 nM/reaction), EGFP–SUMO-2 was conjugated to form high-molecular-weight species, including EGFP–SUMO-2·Topoisomerase-II (Figure 1A, brackets). If either E1 or E2 enzymes were lacking from the reaction, no conjugation of EGFP–SUMO-2 occurred and the free pool of EGFP–SUMO-2 remained unchanged throughout the reaction (Figure 1A, lower panel, arrow). These findings show that although the chromatin did not contain sufficient E1 or E2 activity to support Topoisomerase-II conjugation, all required E3-like factors were present within the chromatin fraction and sufficient to fully support conjugation.
Figure 1.
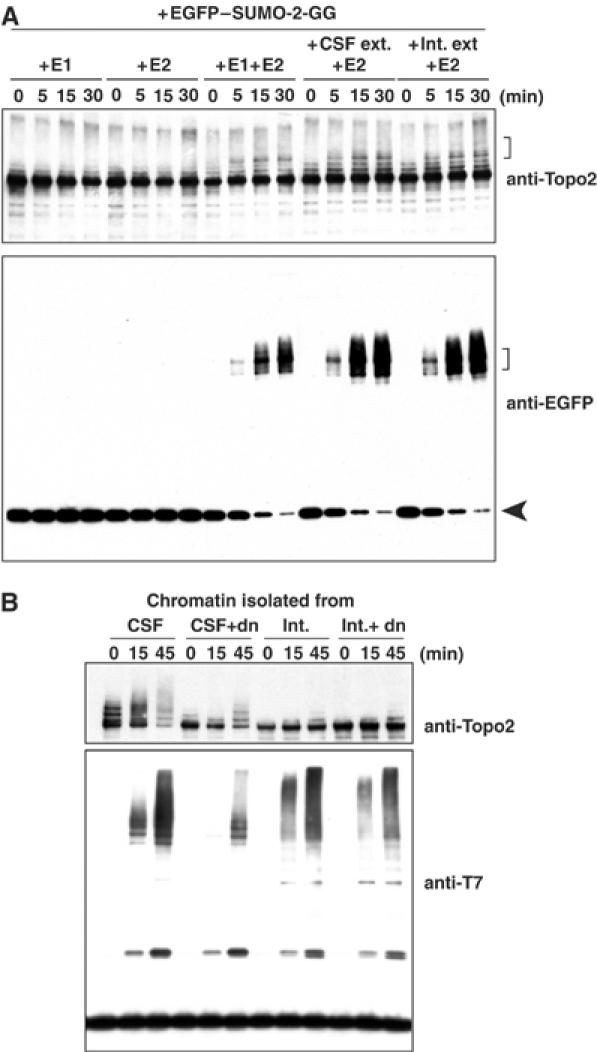
Mitotic chromatin is competent to promote SUMO-2 modification of Topoisomerase-II in vitro. (A) Mitotic chromatin promotes Topoisomerase-II conjugation. Mitotic chromatin was assembled in CSF extract containing dnUbc9, and isolated as described in Materials and methods. The chromatin was incubated with purified Uba2/Aos1 (20 nM/reaction; ‘+E1'), purified Ubc9 (300 nM/reaction; ‘+E2') or both (‘+E1 +E2'), as indicated. Alternatively, the chromatin was incubated with M-phase egg extract (1/100 volume of reaction mixture; ‘+CSF ext. +E2') or interphase egg extract (1/100 volume of reaction mixture; ‘+Int. ext. +E2') plus purified Ubc9. Aliquots were taken from the reaction mixtures at the indicated times (in minutes), and conjugation of EGFP–SUMO-2 to Topoisomerase-II was analyzed by Western blotting with anti-EGFP and anti-TopoII antibodies. Brackets indicate EGFP–SUMO-2-conjugated Topoisomerase-II, and arrowhead indicates unconjugated EGFP–SUMO-2. (B) Interphase chromatin does not support Topoisomerase-II conjugation. Chromatin was prepared from CSF or interphase extracts in the presence or absence of dnUbc9. After isolation, the chromatin was incubated in a reaction mixture containing purified Uba2/Aos1, purified Ubc9 and T7–SUMO-2-GG. At the indicated times (min), aliquots were taken and incorporation of T7–SUMO-2 to Topoisomerase-II was analyzed by Western blotting with anti-Topo2 and anti-T7 antibodies.
In the absence of purified E1, both CSF and interphase extracts promoted SUMO-2 modification very efficiently (Figure 1A, right lanes), indicating that extracts from either cell cycle phase served equally well as sources of E1 activity. This observation argues that mitotic regulation is not exerted through Uba2/Aos1. Similarly, we observed that either form of extract worked equally well as a source of E2 activity (data not shown), indicating that Ubc9 is also not a major point of regulation. Notably, when the reactions were performed using chromatin assembled in interphase extracts, Topoisomerase-II was not strongly conjugated with SUMO-2, although there was robust conjugation of other high-molecular-weight targets (Figure 1B). These observations indicate that mitotic Topoisomerase-II conjugation to SUMO-2 requires a chromatin-associated E3-like factor.
PIASy is required for mitotic SUMO-2 conjugation to Topoisomerase-II
We took a candidate-based approach to the identification of chromatin-associated proteins that promote Topoisomerase-II conjugation, focusing on PIAS family proteins. We assumed that the machinery for modification of a conserved substrate would be similarly conserved, and only the PIAS/Siz class of SUMO ligases has been documented to function in both vertebrates and yeast (Melchior et al, 2003; Johnson, 2004). In particular, we examined PIASy because it has been reported to preferentially promote SUMO-2 conjugation (Sachdev et al, 2001; Chun et al, 2003).
We cloned the Xenopus homologue of PIASy (Supplementary Figure 2), which is 86% identical to its human counterpart. We produced affinity-purified polyclonal rabbit antibodies that specifically recognized human PIASy (anti-PIASyhs) (Supplementary Figure 3), as well as either the N-terminus (anti-PIASyxl-N) or C-terminus (anti-PIASyxl-C) of the Xenopus PIASy protein. We used these antibodies for immunodepletion of CSF extracts, and confirmed by Western blotting that over 99% of PIASy was removed from extracts through this procedure (data not shown). Sperm chromatin was allowed to assemble into mitotic chromosomes within the depleted extracts. Chromosomes assembled in the PIASy-depleted CSF extracts were morphologically indistinguishable from chromosomes assembled in control extracts (data not shown). The chromosomes were purified and analyzed by Western blotting (Figure 2A). Comparable total levels of Topoisomerase-II were associated with chromosomes from the depleted and control reactions, but there was no detectable SUMO-2·Topoisomerase-II in the absence of PIASy (second and third panels). Similarly, chromatin assembled in PIASy-depleted extracts did not support conjugation of EGFP–SUMO-2 with Topoisomerase-II in the semi-purified assay system described in Figure 1 (Figure 2B).
Figure 2.
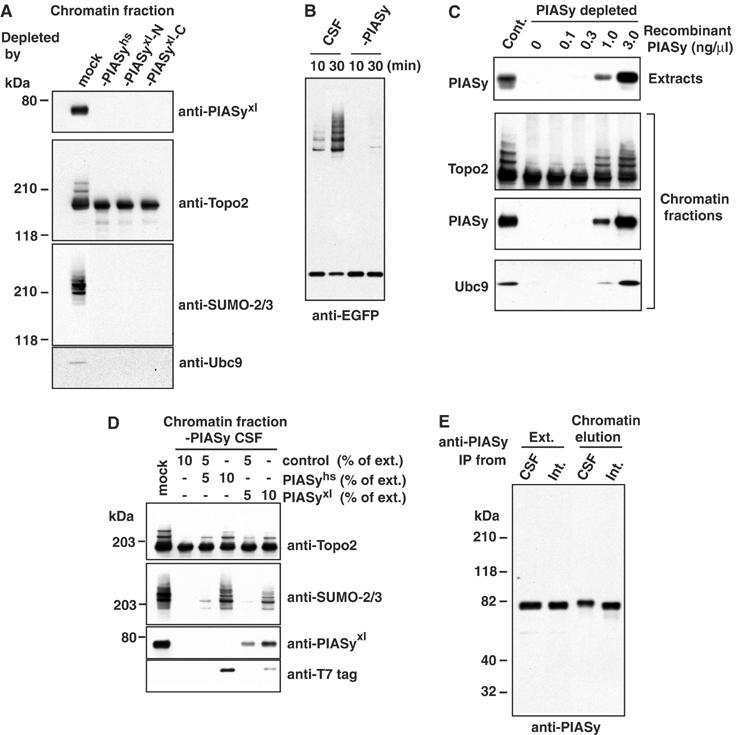
PIASy protein is essential for mitotic SUMO-2 conjugation to Topoisomerase-II. (A) PIASy is essential for SUMO-2 conjugation of Topoisomerase-II and for Ubc9 recruitment to chromatin. CSF extracts were immunodepleted, using antibodies against human PIASy (-PIASyhs), an N-terminal fragment of Xenopus PIASy (-PIASyxl-N) or a C-terminal fragment of Xenopus PIASy (-PIASyxl-C). Depleted extracts were incubated with 5000 sperm nuclei/μl. After 45 min, chromatin from each reaction was isolated and analyzed by Western blotting with the antibodies indicated to the right of each panel. (B) Mitotic chromatin lacking PIASy does not support Topoisomerase-II conjugation in vitro. CSF egg extracts were depleted of PIASy using affinity-purified antibodies, or mock-treated with nonspecific IgG. Sperm chromatin was added to each of the extracts, and allowed to assemble in the presence of dnUbc9. The assembled chromatin was isolated and incubated in reaction mixtures containing E1, E2 and EGFP–SUMO-2, as in Figure 1. Incorporation of EGFP–SUMO-2 was analyzed by Western blotting with anti-EGFP antibody. (C) Recombinant Xenopus PIASy protein restores Topoisomerase-II modification in PIASy-depleted extracts. His6-tagged Xenopus PIASy protein was expressed in bacteria and purified affinity chromatography and gel filtration. The indicated concentrations of purified protein were added to CSF extract that had been previously depleted using anti-PIASyxl-N antibodies. Samples from each reaction were analyzed by Western blotting with anti-PIASy antibody to compare the amount of endogenous protein and recombinant protein (top panel). After 45 min, chromatin from each reaction was isolated and analyzed by Western blotting with the antibodies indicated to the left of each panel (lower three panels). (D) Rescue of Topoisomerase-II modification with human and Xenopus PIASy proteins. Human (PIASyhs) and Xenopus (PIASyxl) PIASy proteins were translated in reticulocyte lysates. The translation reactions were added to CSF extract that had been previously depleted using anti-PIASyxl-N antibodies. The amount of reticulocyte lysate added was adjusted to 10% of the final reaction volume by mixing PIASy-expressed reticulocyte lysate with unprogrammed reticulocyte lysate (control), as indicated. After 45 min, chromatin from each reaction was isolated and analyzed by Western blotting with the antibodies indicated to the right of each panel. Note that both the human and Xenopus proteins were T7-tagged on their N-termini. (E) Reduced electrophoretic mobility of PIASy associated with mitotic chromatin. PIASy was immunoprecipitated from complete extracts (Ext.) or from fractions obtained by salt elution from chromatin (chromatin elution). The precipitated proteins were analyzed by Western blotting with anti-PIASy antibody. Precipitates from both M-phase (CSF) and interphase (Int.) extracts are shown.
To confirm that the absence of SUMO-2·Topoisomerase-II directly reflected the depletion of PIASy, we added recombinant Xenopus PIASy (Figure 2C) or PIASy that had been translated in reticulocyte lysates (Figure 2D) to depleted extracts containing sperm chromatin. When added at levels comparable to the endogenous PIASy protein, recombinant PIASy fully restored SUMO-2·Topoisomerase-II levels (Figure 2C). Similarly, reticulocyte lysates expressing human or Xenopus PIASy rescued SUMO conjugation of Topoisomerase-II, while unprogrammed reticulocyte lysates did not (Figure 2D). These observations confirm that PIASy is essential for mitotic conjugation of Topoisomerase-II to SUMO-2. Interestingly, PIASy associated with mitotic chromatin showed a small but reproducible shift in its electrophoretic mobility in comparison to either free PIASy from mitotic extracts or to chromatin-bound PIASy from interphase egg extracts (Figure 2E). This shift indicates that PIASy is subject to a chromatin-dependent post-translational modification in mitotic Xenopus egg extracts. It is attractive to speculate that such mechanisms may help promote PIASy activity on mitotic chromatin.
SUMO-2 conjugation of Topoisomerase-II is specific for PIASy
To determine whether other PIAS family members could support conjugation of Topoisomerase-II to SUMO-2, we added reticulocyte lysates expressing human PIAS3, PIASxα PIASxβ, PIASy or mouse PIAS1 to PIASy-depleted CSF extracts containing sperm chromatin (Figure 3A). As before, human PIASy promoted robust SUMO-2 conjugation to Topoisomerase-II. PIASxα promoted Topoisomerase-II modification, but far less efficiently than PIASy. PIAS1, PIAS3 and PIASxβ did not rescue Topoisomerase-II conjugation. These results indicate that PIASy is uniquely required for SUMO-2 conjugation to Topoisomerase-II in mitotic extracts.
Figure 3.
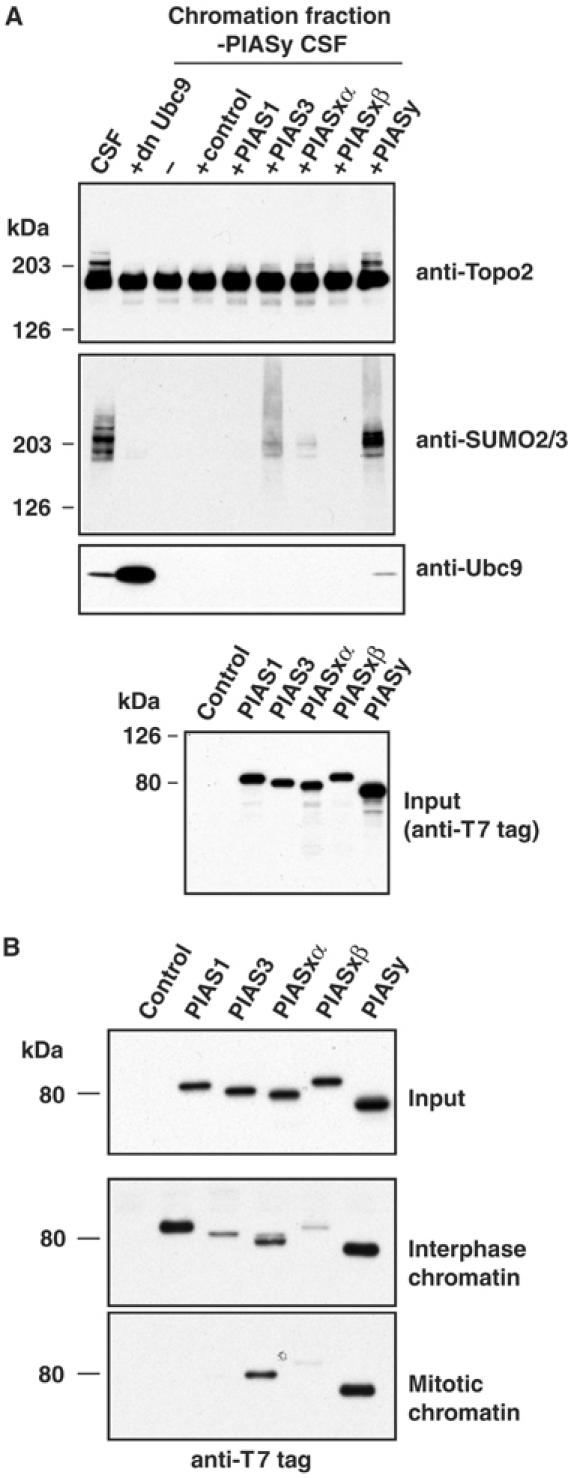
Topoisomerase-II conjugation specifically requires PIASy. (A) Other PIAS proteins do not efficiently promote Topoisomerase-II modification. Human, PIAS3, PIASxα, PIASxβ PIASy and mouse PIAS1 were translated in reticulocyte lysates. The translation reactions or unprogrammed reticulocyte lysates (control) were added to CSF extracts with 5000 sperm nuclei/μl (10% final reaction volume). All reactions were incubated for 45 min to allow chromatin remodeling and chromosome condensation. Chromatin from the reactions was isolated and analyzed by Western blotting with the antibodies indicated to the right of the upper three panels. The bottom panel shows a Western blot of the reticulocyte translation assays. Since all of the human PIAS proteins had T7 tags on their N-terminus, this provides quantitation of the relative amounts of each protein added to the CSF extracts. (B) Regulated association of PIAS proteins with mitotic and interphase chromatin. T7-tagged mammalian PIAS proteins were expressed in reticulocyte lysates and mixed with either interphase or CSF extracts in the presence of 5000 sperm nuclei/μl at room temperature. After 45 min, chromatin fractions were isolated, and the association of PIAS proteins was analyzed by Western blotting with anti-T7 antibodies.
To determine why only PIASy promoted robust mitotic SUMO-2 modification of Topoisomerase-II, we compared the chromatin association of T7-tagged versions of each of the PIAS proteins that were produced in reticulocyte lysates. After a 45-min incubation in interphase or mitotic extracts with sperm chromatin, we confirmed the formation of interphase nuclei and condensed mitotic chromosomes, respectively. Chromatin was isolated from each of the reactions, and analyzed by Western blotting using antibodies directed against the T7 tag (Figure 3B, lower two panels). All of the PIAS proteins bound to interphase chromatin with varying efficiencies, but only PIASy and PIASxα bound to mitotic chromatin. This observation shows that the chromatin binding of individual PIAS proteins is differentially regulated during the cell cycle. The absence of chromatin binding may be sufficient to explain the inability of PIAS1, PIAS3 and PIASxβ to restore SUMO-2 conjugation in PIASy-depleted egg extracts. By contrast, the weak capacity of PIASxα to modify Topoisomerase-II was not simply explained by inability to bind mitotic chromatin.
PIASy recruits Ubc9 to mitotic chromosomes
We noted that a pool of Ubc9 is recruited to the mitotic chromatin fraction (Figures 2A, C and 3A). Although Ubc9 was not co-depleted from egg extracts with PIASy, it was not recruited onto chromatin in the absence of PIASy (Figure 2A). However, recruitment could be restored by recombinant PIASy (Figure 2C) or reticulocyte lysates expressing human PIASy (Figure 3A). These observations suggest that Ubc9 associates to chromatin in a PIASy-dependent manner. We performed two sets of experiments to determine whether this association is important for the modification of Topoisomerase-II: First, we examined the capacity of other PIAS proteins to recruit Ubc9 to the chromatin. As might be expected from their failure to bind mitotic chromatin, PIAS1, PIAS3 and PIASxβ did not restore Ubc9 recruitment in PIASy-depleted extracts (Figure 3A, third panel). More interestingly, although PIASxα bound to chromatin, it did not recruit Ubc9 onto the chromatin at levels comparable to PIASy. This finding indicates that the weak activity shown by PIASxα in promoting the mitotic conjugation of Topoisomerase-II may be due to its limited capacity to promote Ubc9 association with mitotic chromosomes.
Second, we mutated PIASy to test whether its activity in promoting Topoisomerase-II conjugation could be distinguished from its capacity to mediate Ubc9 recruitment: These mutations included deletion of the SAP domain, deletion of a C-terminal fragment that included S/DE domain and a point mutation in the SP-ring domain (Figure 4A). When the mutants were translated in vitro and added to PIASy-depleted extracts, none of them restored the SUMO-2 conjugation to Topoisomerase-II (Figure 4B, upper two panels), showing each of the conserved domains was necessary for PIASy function. We assayed chromosomes isolated from the same reactions by Western blotting, to determine whether the mutants associated with chromatin and whether they were able to recruit Ubc9. Blotting with antibodies against the T7 tag in each of the translated proteins (Figure 4B, second panel from bottom) showed that truncation of the SAP domain abolished chromatin binding of PIASy, consistent with the previous observation that the SAP domain binds AT-rich DNA sequences (Aravind and Koonin, 2000; Okubo et al, 2004). Probably as a direct consequence of its inability to bind to chromatin, this mutant did not recruit Ubc9 to mitotic chromosomes. By contrast, both the S/DE domain truncation and the SP-ring point mutant bound chromatin efficiently (Figure 4B), demonstrating that chromatin binding is necessary but not sufficient for promotion of Topoisomerase-II conjugation. Remarkably, both of these mutants failed to restore the recruitment of Ubc9 into the chromosomal fraction, suggesting that Ubc9 recruitment is closely linked to the PIASy's role in Topoisomerase-II conjugation.
Figure 4.
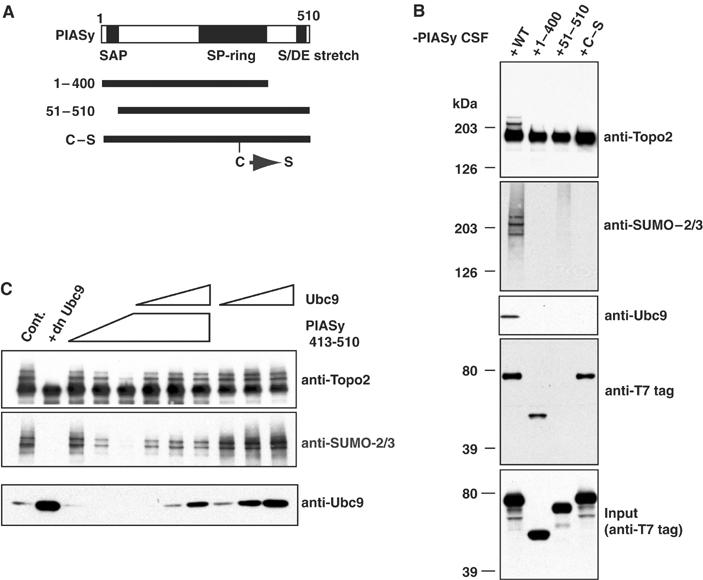
Three conserved domains of PIASy are required for SUMO-2 modification of Topoisomerase-II. (A) Representation of conserved domains within PIASy and mutants analyzed. (B) PIASy domains required for chromatin binding and Ubc9 recruitment. Reticulocyte lysates expressing wild-type or mutant human PIASy proteins were added into PIASy-depleted extracts and chromatin fractions were analyzed by Western blotting with the indicated antibodies. (C) Exogenous C-terminal fragment of PIASy blocks Ubc9 recruitment to chromatin. Increasing amounts of human PIASy C-terminal fragment (PIASyhs413–510) and/or Ubc9 were added to CSF extracts containing 5000 sperm nuclei/μl (PIASyhs413–510=0.5, 1, 2 μM; Ubc9=0.2, 0.6, 2 μM). After isolation of the chromatin fraction from these reactions, SUMO-2 conjugation to Topoisomerase-II and Ubc9 recruitment to the chromatin fraction were analyzed by Western blotting with the indicated antibodies.
The S/DE domain of PIASy is required for binding to Ubc9
Deletion of the S/DE domain compromised the capacity of PIASy to mediate Ubc9 recruitment to chromosomes (Figure 4B). To investigate this point further, we monitored reactions containing increasing amounts of a recombinant, purified C-terminal PIASy fragment (a.a. 413–510; Figure 4C). This fragment effectively blocked both the conjugation of SUMO-2 to Topoisomerase-II (top, middle panels) and the recruitment of Ubc9 to the chromatin (bottom panel) in a dose-dependent fashion. This finding suggested that the C-terminal domain of PIASy plays a positive role in Ubc9 recruitment to chromatin, and that the C-terminal PIASy fragment may block recruitment by sequestering the free pool of Ubc9 protein. A strong prediction from this hypothesis is that addition of exogenous Ubc9 should overcome capacity of the fragment to inhibit Topoisomerase-II conjugation. Consistent with this idea, we found that exogenous Ubc9 protein was able to restore Topoisomerase-II conjugation and Ubc9 accumulation on chromosomes in the presence of the recombinant C-terminal fragment containing the S/DE domain.
SUMO-2 accumulation on the IC requires PIASy
In addition to its function as a regulator of Topoisomerase-II conjugation, we examined the global role of PIASy in regulating mitotic conjugation of SUMO-2. We determined the distribution of EGFP–SUMO-2 on spindles in mitotic Xenopus egg extracts (Figure 5A): EGFP–SUMO-2 concentrated over the chromosomes, with bright foci on the metaphase plate resembling centromeres. The foci were entirely eliminated in the presence of dnUbc9 (Figure 5A, right panels), showing that they resulted from the accumulation EGFP–SUMO-2-conjugated species. To establish the nature of these foci, we assembled chromosomes as in Figure 5A, but added nocodazole coincident with mitotic induction to prevent spindle assembly and allow isolation of individual chromosomes. The isolated chromosomes were stained using antibodies against the inner centromeric kinase Aurora B and the centromeric histone variant CENP-A (Figure 5B). Aurora B localization overlapped extensively with EGFP–SUMO-2, while CENP-A bracketed foci of EGFP–SUMO-2. These patterns indicate that SUMO-2-conjugated species localize to the IC (Cleveland et al, 2003). As with intact spindles, the inner centromeric accumulation of EGFP–SUMO-2 was entirely absent in the presence of dnUbc9 (Figure 5B, lower panels). It has previously been shown that GFP–SUMO-2 is distributed throughout mitotic HeLa cells (Ayaydin and Dasso, 2004). It is therefore possible that the accumulation of SUMO-2 on ICs may differ between species or between embryonic and somatic cells. Alternatively, it is possible that a limited population of inner centromeric GFP–SUMO-2 could not be clearly distinguished in those live imaging experiments due to a relatively high background of free GFP–SUMO-2 and diffusible conjugated species.
Figure 5.
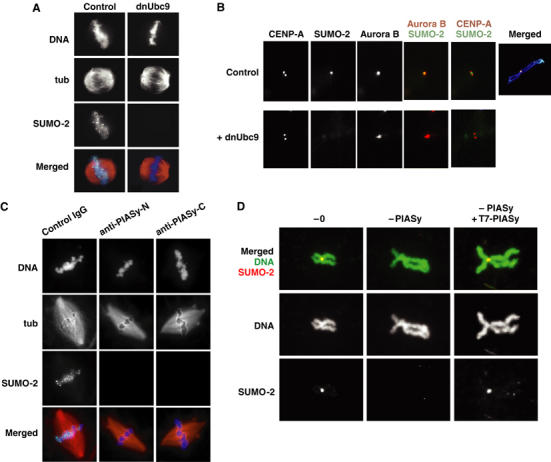
PIASy mediates SUMO-2 incorporation on the IC of mitotic chromosomes. (A) EGFP–SUMO-2 foci are conjugation-dependent. CSF extracts containing rhodamine-labeled tubulin (red) and EGFP–SUMO-2 (green) were released into interphase by CaCl2 addition. In all, 200 sperm/μl were added 5 min after CaCl2 and the extract was incubated at 23°C for 55 min. Re-entry into mitosis was induced by addition of fresh CSF extract (50% of reaction volume) with or without dnUbc9 (final concentration at 150 ng/μl). After 30 min, samples were removed and analyzed for microtubule and EGFP–SUMO-2 distribution, as well as by staining with Hoechst 33342 DNA dye (blue). (B) EGFP–SUMO-2 foci coincide with ICs. Reactions containing EGFP–SUMO-2 (green) were treated as in (A), except that nocodazole was added coincident with induction of M-phase re-entry with fresh CSF extracts. Samples from each reaction were spun onto coverslips and analyzed by immunofluorescence with antibodies against CENP-A and Aurora B (shown red in merged panels). For comparison, EGFP–SUMO-2 signal from the control extract is also shown in a merged image with Hoechst 33342 staining (far right panel). (C) EGFP–SUMO-2 foci require mitotic PIASy activity. Mitotic chromatin and spindles were assembled and analyzed as in (A). Anti-PIASy antibodies were added coincident with induction of M-phase re-entry with fresh CSF extract. (D) IC accumulation of EGFP–SUMO-2 is PIASy-dependent. EGFP–SUMO-2 (shown in red) was added to untreated CSF extracts, PIASy-depleted extracts or PIASy-depleted extracts supplemented with reticulocyte-translated human PIASy. The extracts were released into interphase by CaCl2 addition. Sperm nuclei addition, re-entry into mitosis and preparation of samples were as described in (B). Samples from each reaction were simultaneously stained with Hoechst 33342 (shown in green).
We examined whether PIASy was required for inner centromeric accumulation of EGFP–SUMO-2 (Figure 5C and D). We first assayed the assembly centromeric EGFP–SUMO-2 foci in egg extracts depleted of PIASy. We observed that these foci were entirely absent from spindles in depleted extracts (data not shown), suggesting that PIASy was essential for their formation. It was formally possible that the absence of EGFP–SUMO-2 foci might reflect a deficiency in PIASy function during the preceding interphase when the chromosomes were replicated, rather than insufficient mitotic SUMO-2 conjugation. To determine whether de novo PIASy-mediated conjugation is required during mitosis, we selectively inhibited PIASy upon the induction of mitosis, using antibodies against the N- and C-terminus of Xenopus PIASy (Figure 5C). Both antibodies blocked conjugation of Topoisomerase-II (Figure 6A), suggesting that they efficiently prevented PIASy-mediated mitotic SUMO-2 conjugation. Remarkably, both antibodies inhibited the formation of centromeric EGFP–SUMO-2 foci (Figure 5C), confirming that mitotic PIASy activity was required. Finally, we analyzed the assembly of individual chromosomes after PIASy depletion in nocodazole-treated extracts (Figure 5D). EGFP–SUMO-2 accretion on the IC was abolished in PIASy-depleted extracts, showing that PIASy was absolutely required to promote SUMO-2 conjugation of inner centromeric substrates. Addition of reticulocyte-translated PIASy restored the accumulation of EGFP–SUMO-2 on the IC, further supporting this conclusion (Figure 5D, right panels).
Figure 6.
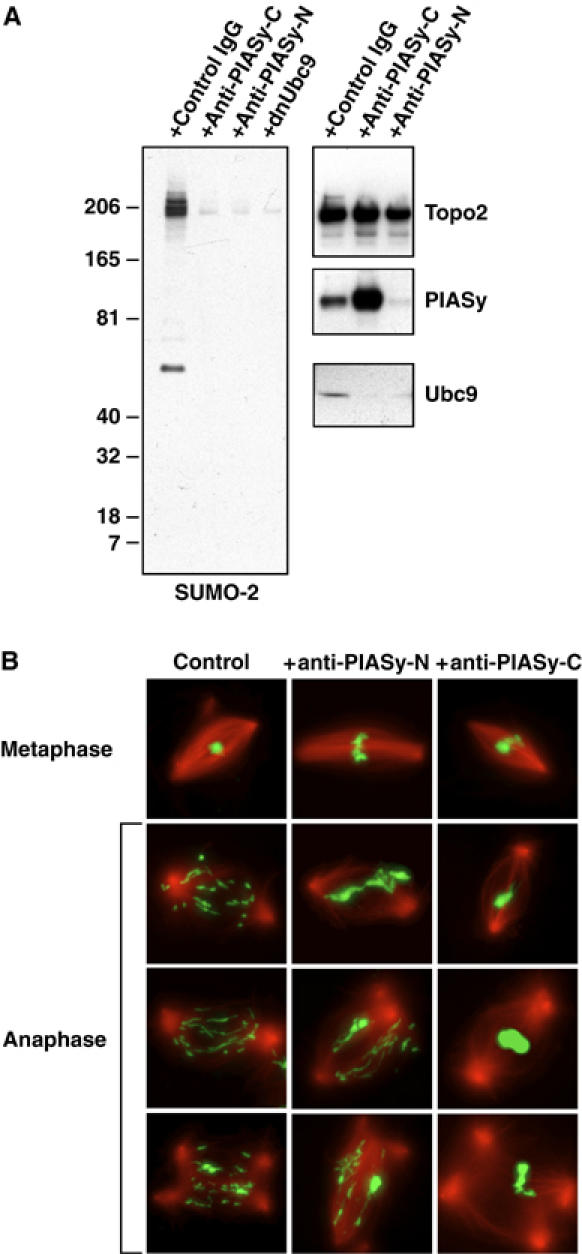
Perturbation of PIASy function in mitosis by anti-PIASy antibodies causes chromosome segregation defects. (A) Anti-PIASyxl-N and anti-PIASyxl-C block SUMO-2 conjugation on mitotic chromatin. CSF extract was released into interphase by CaCl2 addition. In all, 2000 sperm/μl were added 5 min after CaCl2, and the extract was incubated at 23°C for 55 min. Re-entry into mitosis was induced with fresh CSF extract (50% of reaction volume) containing control IgG, antibodies against the C-terminus of Xenopus PIASy (anti-PIASyxl-C), antibodies against the N-terminus of Xenopus PIASy (anti-PIASyxl-N) (final antibody concentration in each case=0.3 μg/μl), or dnUbc9 (final concentration at 150 ng/μl). Mitotic chromatin was isolated from the control reaction and each of the antibody-containing reactions, as indicated. The chromatin was subjected to Western blotting with antibodies indicated to the right of each panel. (B) Anti-PIASyxl-N and anti-PIASyxl-C block sister chromatid segregation in anaphase. Reactions as in (A) were supplemented with rhodamine-labeled tubulin. 30 min after the induction of metaphase by CSF extracts, anaphase was induced with an addition of CaCl2. Aliquots were removed prior to CaCl2 addition (upper row) or 13 min after CaCl2 addition (bottom three rows). The samples were analyzed for DNA morphology using Hoechst 33342 DNA dye (green) and for microtubule structures (red) after fixation.
Disruption of PIASy blocks chromosome segregation
Since inhibition of all SUMO conjugation during mitosis by dnUbc9 disrupts chromosome segregation in Xenopus egg extracts (Azuma et al, 2003), we wished to examine whether the subset of SUMO-2 substrates modified by PIASy played a role in chromosome segregation (Figure 6). When added at the onset of mitosis, antibodies against both the N- and C-terminus of PIASy were as effective as dnUbc9 in blocking the SUMO-2 conjugation of Topoisomerase-II (left panel). Notably, the antibodies blocked PIASy function through different mechanisms. The N-terminus of PIASy is required for its association to chromatin (Figure 4). Consistent with this finding, anti-PIASyxl-N blocked the recruitment of PIASy into the chromatin fraction, as well as recruitment of Ubc9. By contrast, the C-terminus of PIASy is required for its interaction with Ubc9 (Figure 4); anti-PIASyxl-C allowed abundant recruitment of PIASy onto the chromosomes, but blocked the recruitment of Ubc9.
As expected, spindles assembled normally in antibody-treated egg extracts (Figure 6B, upper panels) (Azuma et al, 2003). We examined chromosome segregation 13 min after anaphase was induced in each of the reactions by release of CSF arrest with CaCl2. Control reactions showed separated sister chromatids migrating poleward (left panels), while segregation was inhibited in dnUbc9-treated reactions (data not shown; Azuma et al, 2003). Significantly, antibodies against both the N- and C-terminus of PIASy caused substantial defects in chromosome segregation. Chromosomes remained within a single, condensed chromatin mass in reactions treated with anti-PIASyxl-C (right panels), suggesting a dramatic failure of sister chromatid segregation. In the anti-PIASyxl-N-treated reactions, we typically observed that large portions of the chromatin remained near the center of the anaphase spindles, with strands stretching toward each of the poles. Together, the capacity of both antibodies to block chromosome through interference with different domains of PIASy strongly indicates that the modification of PIASy targets is essential for chromosome segregation.
Discussion
We sought to find factors responsible for mitotic chromatin-dependent SUMO-2 conjugation of Topoisomerase-II (Azuma et al, 2003). Our initial studies suggested that mitotic accumulation of SUMO-2·Topoisomerase-II did not result from alterations of SUMO E1 or E2 activities, but rather from the activation of another chromatin-associated factor. PIASy fulfills the criteria for such a factor, since it binds to mitotic chromosomes and is absolutely required for Topoisomerase-II conjugation. PIASy was unique among the PIAS family members in its capacity to both bind mitotic chromosomes and recruit Ubc9 into the chromatin-associated fraction. These properties were essential, since PIASy mutants that did not associate with chromatin or that failed to recruit Ubc9 were unable to restore Topoisomerase-II conjugation in PIASy-depleted mitotic extracts. PIASy depletion eliminated the accumulation of SUMO-2-conjugated species on mitotic chromosomes, suggesting that it is the primary E3-like factor required for chromosomal substrates during mitosis. Notably, PIASy-dependent SUMO-2-conjugated species were not broadly distributed throughout the chromosomes, but rather concentrated on the IC. Moreover, inhibition of PIASy leads to a failure of sister chromatid separation, suggesting that modification of inner centromeric components is crucial for chromosome segregation in Xenopus egg extracts.
The accumulation of SUMO-2·Topoisomerase-II in Xenopus egg extracts requires chromatin (Azuma et al, 2003). Our findings suggest a simple mechanism through which PIASy may promote the appropriate, chromatin-dependent modification of Topoisomerase-II: Unlike many ubiquitin E2 enzymes, Ubc9 interacts with the SUMO modification consensus sites in target proteins directly, albeit at a relatively low affinity, such that SUMO conjugation can occur without E3 enzymes in the presence of high concentrations of Ubc9 (Bernier-Villamor et al, 2002). PIASy binds chromatin through its SAP domain, and recruits Ubc9 through its SP-ring and C-terminal domains (Figure 4). Through these interactions, PIASy may facilitate modification of Topoisomerase-II on chromatin by simple elevation of local Ubc9 concentrations.
ICs play highly specialized roles in mitotic vertebrate cells, acting as critical sites of chromatid cohesion throughout metaphase (Hauf et al, 2001; Cleveland et al, 2003). We had previously observed that blocking SUMO conjugation using dnUbc9 disrupts chromosome segregation in egg extracts (Azuma et al, 2003). Our observations indicate that PIASy-mediated conjugation particularly controls inner centromeric events that are required for successful sister chromatid segregation: PIASy is essential for the accumulation of SUMO-2 conjugated proteins on the IC (Figure 5), and inhibition of PIASy with antibodies directed against two separate regions of PIASy prevented sister chromatid segregation in egg extracts during anaphase (Figure 6). The chromosome segregation defects elicited by the antibodies showed subtle differences. Anti-PIASyxl-C caused a more profound defect, with essentially all chromatin remaining in a single mass during anaphase, while anti-PIASyxl-N-treated extracts showed chromatin masses that were stretched but not separated. Although the capacity of anti-PIASyxl-C to cause increased accumulation of PIASy on chromosomes raised the possibility that anti-PIASyxl-C might block segregation through its own association to chromatin, similar issues do not arise for reactions containing anti-PIASyxl-N. Anti-PIASyxl-N antibodies interfere with PIASy in a manner that entirely prevents its recruitment to chromosomes. In any case, independence of the mechanisms through which these antibodies inhibit PIASy (Figure 6A) strongly supports the overall conclusion that PIASy is indispensable for accurate chromosome segregation.
It is not yet clear whether PIASy plays a critical role during mitosis in other contexts. Surprisingly, PIASy knockout mice are physiologically normal and fertile (Wong et al, 2004), and they show levels of SUMO-1 and SUMO-3 conjugation indistinguishable from control mice. It is possible that other PIAS family members can compensate for the absence of PIASy in mice to a greater extent that we observe in mitotic Xenopus extracts (Figure 3A). Alternatively, it is possible that mechanisms of cohesion release exist in mice that allow bypass of the requirement for SUMO conjugation. Similar to mouse PIASy knockouts, mutants lacking the Schizosaccharomyces pombe PIAS protein Pli1p are viable and able to progress through mitosis. However, pli1 mutants show pronounced defects in silencing of centromeric heterochromatin, as well as greatly increased rates of minichromosome loss and sensitivity to the microtubule poison thiabendazole (Xhemalce et al, 2004). Collectively, these phenotypes suggest that SUMO modification has an important role in centromeric function in S. pombe, probably through maintenance of heterochromatic structures, but that cells compensate for the loss of Pli1p through alternative mechanisms. Most strikingly, the closest homologue of PIASy in Drosophila, Su(var)2–10, encodes an essential protein (Hari et al, 2001). Su(var)2–10 was initially identified as a Suppressor of Position-Effect Variegation (Wustmann et al, 1989). Mutations in this gene cause both minichromosome and endogenous chromosome inheritance defects (Hari et al, 2001): Su(var)2–10 mutants show aberrant condensation of mitotic chromosomes, as well as faulty chromosome organization within interphase nuclei, leading to the proposal that Su(var)2–10 controls multiple aspects of chromosome structure and function throughout the cell cycle.
In summary, PIASy is a critical regulator of mitotic SUMO-2 conjugation of Topoisomerase-II and perhaps other chromosomal proteins. PIASy-mediated conjugation appears to have particular relevance for IC functions in Xenopus extracts, in a manner that is indispensable for sister chromatid segregation.
Materials and methods
Recombinant protein expression, cloning and antibodies
cDNAs encoding human PIASxα, PIASxβ, PIASy and mouse PIAS1 were kindly provided by Ke Shuai (UCLA). Human PIAS3 cDNA and Xenopus PIASy were, respectively, cloned from human testis QUICK-Clone cDNA (BD Biosciences) and Xenopus tadpole cDNA (kindly provided by T Amano, NICHD) using PCR amplification, and were verified by DNA sequencing. All full-length cDNAs were subcloned into pET28 using EcoRI and XhoI restriction sites. Full-length PIAS proteins were expressed in reticulocyte lysates (TNT quick T7, Promega).
cDNA fragments encoding human PIASy (amino acids 413–510, PIASyhs413–510), Xenopus PIASy (amino acids 63–143; PIASyxl63–143 and 410–458; PIASyxl410–458) were subcloned into pGEX4T-1 (Amersham-Pharmacia Biotech) and pET28a (Novagen) using EcoRI and XhoI restriction sites. The recombinant proteins encoded by these cDNAs were expressed in Escherichia coli and purified by tag-based purification according to the manufacturer's protocol, followed by conventional protein chromatography.
Polyclonal antibodies against SUMO-2/3, human PIASy and Xenopus PIASy were generated in rabbits by injection with a recombinant SUMO-2, GST-PIASyhs413–510, His6-T7-PIASyxl63–143 and His6-T7-PIASyxl410–458, respectively. The rabbit antisera were subjected to affinity purification on NHS Sepharose columns with their covalently bound antigens. Affinity-purified chicken anti-Xenopus CENP-A antibodies were as described (kind gift of PT Stukenberg, University of Virginia Medical School; McCleland et al, 2003). Rabbit anti-Topoisomerase-II was the kind gift of PL Jones (University of Illinois at Urbana-Champaign, Urbana, IL; Luke and Bogenhagen, 1989). Goat anti-rabbit and goat anti-chicken antibodies coupled to Alexa Fluor 594 and 488 were from Molecular Probes.
Unless otherwise specified, other reagents were obtained from Sigma-Aldrich Chemical Company.
Xenopus egg extracts
Sperm chromatin preparations and low-speed extracts of Xenopus eggs arrested in metaphase by CSF were prepared as described (Kornbluth et al, 2001). Where required, CSF extracts were driven into interphase by addition of 0.6 mM CaCl2. For mitotic chromosome assembly or interphase nuclear assembly, demembranated sperm chromatin were added to CSF or interphase extracts, and incubated for 40 min at room temperature. Chromatin isolation was performed as previously described (Azuma et al, 2003). Unless otherwise specified, all reactions contained 5000 sperm nuclei/μl.
Protein purification
The E1 complex (Aos1/Uba2 heterodimer) was purified from Xenopus egg extract using the same methods previously described for E1 purification from HeLa cells (Azuma et al, 2001). Preparation of recombinant SUMO-2, wild type and dnUbc9 were as previously described (Azuma et al, 2003). The full-length Xenopus PIASy protein, with His6 and T7 tags at the N-terminus, was expressed in BL21(DE3) at 18°C in the presence of 5% glycerol and 2.5% ethanol. The PIASy protein was purified from the soluble fraction of the bacterial lysate using Talon affinity resin (Clonetech), followed by Superdex 200 gel filtration and Mono-Q chromatography (Amersham-Pharmacia Biotech).
Immunodepletion, immunoprecipitation and immunofluorescence
For immunodepletion, affinity-purified antibodies were mixed with Dynabeads-ProteinA (Dynal) overnight at 4°C. Antibody-bound beads were blocked with CSF-XB buffer (10 mM Hepes-KOH (pH 7.7), 100 mM KCl, 2 mM MgCl2, 5 mM EGTA and 50 mM sucrose) plus 5% BSA for 30 min at room temperature, and washed with CSF-XB buffer twice. CSF extracts were mixed with antibody beads, and incubated for 10 min at room temperature, followed by 20 min on ice. After repeating this cycle one additional time, depleted extracts were kept on ice until the reaction was started by addition of sperm nuclei. For add-back experiments, reticulocyte lysates expressing the indicated proteins were mixed with depleted extracts. The amounts of reticulocyte lysate were adjusted to 10% of the final reaction volume by mixing with unprogrammed reticulocyte lysate.
For immunoprecipitation, 0.5 μg affinity-purified antibodies were covalently coupled to 25 μl of Protein A-Sepharose FF (Amersham-Pharmacia Biotech) by crosslinking with DMP (Pierce) according to the manufacturer's protocol. CSF (200 μl) or interphase extract was diluted five-fold with extract buffer (10 mM Hepes-KOH (pH 7.2), 16 mM β-glycerophosphate, 1 mM MgCl2, 1 mM DTT, 5% glycerol) containing 100 mM NaCl, 0.5% NP-40, 0.2% Tween 20, 1 μM okadaic acid and protease inhibitors (5 μg/ml of each leupeptin, pepstatin and chymostatin, and 4 mM of pAEBSF). Antibody beads were mixed with the samples and rotated at 4°C for 1 h. After incubation, the beads were then washed three times with extract buffer, and the proteins associated with the beads were eluted with 0.1 M glycine, pH 2.0. Eluted proteins were subjected to Western blotting analysis.
In the case of chromatin extraction, the antibody beads were prepared similarly. Chromatin fractions from 200 μl of either mitotic or interphase extracts were extracted for 15 min on ice, using extraction buffer containing 500 mM NaCl. The extracted fractions were centrifuged at 250 000 g for 60 min in 2°C (Beckman Optima TL centrifuge). Further incubation, washing and analysis steps were carried out as described above for samples precipitated from extracts.
For the immunofluorescence analysis in Figure 6, 100 μl of CSF extracts containing EGFP–SUMO-2 were released into interphase by 0.6 mM CaCl2 addition. In all, 200 sperms/μl were added 5 min after CaCl2, and the extracts were incubated at 23°C for 55 min to allow complete DNA replication. Re-entry into mitosis was induced by addition of 50 μl of fresh CSF extract. Nocodazole (10 μg/ml) and/or dnUbc9 (150 ng/μl final concentration) were also added at this point, where indicated. To assess spindle morphology (Figure 6A and B), the extract was also supplemented with rhodamine-labeled tubulin (25 mg/ml final concentration; Cytoskeleton, Inc.); after re-induction of mitosis, the extracts were incubated for 40 min, and treated as described (Desai et al, 1999).
To assess individual chromosomes (Figure 6C and D), reactions were allowed to proceed for 40 min after the addition of fresh CSF extract. After this incubation, 50 μl was removed from each reaction, diluted with 200 μl of buffer A (0.8 × CSF-XB buffer, containing 10 mM β-glycerolphosphate and 250 mM sucrose), and incubated for 10 min at 23°C. In all, 300 μl of buffer A containing 2% paraformaldehyde was added to this sample and incubated for a further 40 min. Samples were spun onto coverslips, postfixed by methanol and processed for immunostaining with anti-Aurora B, -CENP-A or -T7 antibody (Arnaoutov and Dasso, 2003).
In vitro SUMO conjugation assay
Chromatin was assembled in interphase or mitotic egg extracts, and isolated as described (Azuma et al, 2003). After isolation, it was re-suspended in a volume of 0.5 × CSF-XB buffer equivalent to one-fifth of the original assembly reaction, and sheared by brief sonication. SUMO modification reactions were performed with 20 μl of sheared chromatin in 80 μl of reaction mixture that also contained 1 mM ATP, 7.5 mM creatine phosphate and 0.5 mg/ml of creatine phosphokinase in 0.5 × CSF-XB buffer. E1 (20 nM) and E2 (300 nM) enzymes were added as indicated in Figure 1. The reactions also contained 300 nM of either EGFP–SUMO-2 or 500 nM of T7–SUMO-2 (both processed forms with di-glycine at C-terminus), as indicated. The reaction mixtures were incubated at room temperature. At the indicated times, aliquots were removed, mixed with SDS–PAGE sample buffer and boiled. SUMO modification was analyzed by Western blotting with anti-EGFP, anti-T7 and anti-Topo2 antibodies.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Acknowledgments
We thank Ke Shuai for mammalian PIAS clones, PL Jones and PT Stukenberg for anti-Topoisomerase and anti-CENP-A antibodies, respectively, and BB Quimby for critical reading of this manuscript. TA was supported during the conduct of these studies by a postdoctoral fellowship from the Japan Society for the Promotion of Science.
References
- Aravind L, Koonin EV (2000) SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci 25: 112–114 [DOI] [PubMed] [Google Scholar]
- Arnaoutov A, Dasso M (2003) The Ran GTPase regulates kinetochore function. Dev Cell 5: 99–111 [DOI] [PubMed] [Google Scholar]
- Ayaydin F, Dasso M (2004) Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol Biol Cell 15: 5208–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y, Arnaoutov A, Dasso M (2003) SUMO-2/3 regulates topoisomerase II in mitosis. J Cell Biol 163: 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y, Tan SH, Cavenagh MM, Ainsztein AM, Saitoh H, Dasso M (2001) Expression and regulation of the mammalian SUMO-1 E1 enzyme. FASEB J 15: 1825–1827 [DOI] [PubMed] [Google Scholar]
- Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ (2002) The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cells 9: 1169–1182 [DOI] [PubMed] [Google Scholar]
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD (2002) Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108: 345–356 [DOI] [PubMed] [Google Scholar]
- Chun TH, Itoh H, Subramanian L, Iniguez-Lluhi JA, Nakao K (2003) Modification of GATA-2 transcriptional activity in endothelial cells by the SUMO E3 ligase PIASy. Circ Res 92: 1201–1208 [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407–421 [DOI] [PubMed] [Google Scholar]
- Desai A, Murray A, Mitchison TJ, Walczak CE (1999) The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol 61: 385–412 [DOI] [PubMed] [Google Scholar]
- Hari KL, Cook KR, Karpen GH (2001) The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev 15: 1334–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Waizenegger IC, Peters JM (2001) Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science 293: 1320–1323 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M (2001) SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell 107: 5–8 [DOI] [PubMed] [Google Scholar]
- Johnson ES (2004) Protein modification by SUMO. Annu Rev Biochem 73: 355–382 [DOI] [PubMed] [Google Scholar]
- Kornbluth S, Yang J, Powers M (2001) Analysis of the cell cycle using Xenopus egg extracts. In Current Protocols in Cell Biology, Bonifacino JS, Dasso M, Lippincott-Schwartz J, Harford JB, Yamada KM (eds), pp 11.11.11–11.11.13. New York, NY: John Wiley & Sons, Inc. [DOI] [PubMed] [Google Scholar]
- Luke M, Bogenhagen DF (1989) Quantitation of type II topoisomerase in oocytes and eggs of Xenopus laevis. Dev Biol 136: 459–468 [DOI] [PubMed] [Google Scholar]
- McCleland ML, Gardner RD, Kallio MJ, Daum JR, Gorbsky GJ, Burke DJ, Stukenberg PT (2003) The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev 17: 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F, Schergaut M, Pichler A (2003) SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem Sci 28: 612–618 [DOI] [PubMed] [Google Scholar]
- Minty A, Dumont X, Kaghad M, Caput D (2000) Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J Biol Chem 275: 36316–36323 [DOI] [PubMed] [Google Scholar]
- Okubo S, Hara F, Tsuchida Y, Shimotakahara S, Suzuki S, Hatanaka H, Yokoyama S, Tanaka H, Yasuda H, Shindo H (2004) NMR structure of the N-terminal domain of SUMO ligase PIAS1 and its interaction with tumor suppressor p53 and A/T-rich DNA oligomers. J Biol Chem 279: 31455–31461 [DOI] [PubMed] [Google Scholar]
- Sachdev S, Bruhn L, Sieber H, Pichler A, Melchior F, Grosschedl R (2001) PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev 15: 3088–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KA, Kim R, Christofk H, Gao J, Lawson G, Wu H (2004) Protein inhibitor of activated STAT Y (PIASy) and a splice variant lacking exon 6 enhance sumoylation but are not essential for embryogenesis and adult life. Mol Cell Biol 24: 5577–5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wustmann G, Szidonya J, Taubert H, Reuter G (1989) The genetics of position-effect variegation modifying loci in Drosophila melanogaster. Mol Gen Genet 217: 520–527 [DOI] [PubMed] [Google Scholar]
- Xhemalce B, Seeler JS, Thon G, Dejean A, Arcangioli B (2004) Role of the fission yeast SUMO E3 ligase Pli1p in centromere and telomere maintenance. EMBO J 23: 3844–3853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Blobel G (2005) A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA 102: 4777–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3


