Abstract
We have previously reported a set of Moloney murine leukemia virus derived envelopes retargeted to the Pit-2 phosphate transporter molecule, by insertion of the Pit-2 binding domain (BD) at the N terminus of the ecotropic retroviral envelope glycoproteins (S. Valsesia-Wittmann et al., J. Virol. 70:2059-2064, 1996). The resulting chimeric envelopes share two BDs: an additional N-terminal BD (Pit-2 BD) and the BD of the ecotropic envelope (mCAT-1 BD). By inserting a variety of different amino acid spacers between the two binding domains, we showed that retroviruses can potentially use the targeted cell surface receptor Pit-2, the ecotropic retroviral receptor mCAT-1, or both receptors cooperatively for entry into target cell (S. Valsesia-Wittmann et al., EMBO J 6:1214–1223, 1997). An extreme example of receptor cooperativity was encountered when envelopes with specific proline-rich interdomain spacers (PRO spacers) were tested: both receptors had to be coexpressed at the surface of the targeted cells to cooperatively allow infection. Here, we characterized the role of PRO spacer in the cooperation of receptors. We have shown that the particular organization of the PRO spacer—a β-turn polyproline—was responsible for the cooperative effect. In the native configuration of the viruses, the structure masked the regions located downstream of the PRO spacer, thus the mCAT-1 BD. After interaction with the targeted Pit-2 receptor, the BD of the backbone envelope became accessible, and we demonstrated that interaction between the mCAT-1 BD and the mCAT-1 receptor is absolutely necessary. This interaction leads to natural fusion triggering and entry of viruses into targeted cells.
Most type C mammalian retrovirus envelopes share proline-rich regions located in the middle of their surface proteins (SUs). These regions provide a hinge separating two functional domains of the SU (10): the N-terminal receptor binding domain (BD) (3) and the C-terminal domain involved in post-binding entry events (16, 18). Several reports suggest that the proline-rich domain is not just a flexible linker but rather a functional domain; it was found (i), in some cases, to influence receptor recognition (3, 17); (ii) to be important for stabilization of SU-transmembrane protein (TM) interaction (10), and (iii) to affect virus fusiogenicity possibly by altering glycosylation (1). A direct role of the natural proline-rich region of the SU in stabilization of the conformation and fusiogenicity of the murine leukemia virus (MLV) envelope (13, 23) was recently demonstrated. Predictive structure analysis indicated that the proline-rich peptides present in these contructs may be organized as a β-turn polyproline. This particular structure has uncommon properties, such as the ability to oligomerize in quaternary structure and also to develop elastomeric forces (20).
When such a proline-rich domain was inserted between two BDs of a chimeric MLV envelope glycoprotein, generated by fusion of a Pit-2 BD at the N terminus of Moloney retrovirus envelope protein mCAT-1 BD, we showed that the entry of retroviruses into targeted cells required the presence of both receptors Pit-2 and mCAT-1 at the surface of the cells (21, 22). Binding assays indicated that the mCAT-1 BD of the virion-associated chimeric envelopes is partially or completely hidden (22). Moreover, substitution of proline-rich intergenic spacer (PRO spacer) by a characterized β-turn polyproline fragment from bovine elastin gave the same efficiency of cooperation of the receptors. Finally, we demonstrated that, after unmasking, secondary interaction between mCAT-1 and the backbone envelope is a crucial prerequisite for induction of the fusion process involving the TM subunit. These data are consistent with a model for receptor cooperativity in which binding to the targeted (Pit-2) receptor triggers conformational rearrangements of the envelope that lead to unmasking of the hidden mCAT-1 BD, thereby facilitating its interaction with the viral (mCAT-1) receptor, which leads to fusion.
MATERIALS AND METHODS
Cell lines.
The human fibroblastic TE671 cell line (ATCC CRL8805) was grown in Dulbecco modified Eagle medium (DMEM; Life Technologies) supplemented with 10% fetal bovine serum (Gibco-BRL). These cells express Pit-2 receptor and are susceptible to amphotropic MLV.
The TELCeB6 cell line was derived from the TE671 cell line after transfection and clonal selection of cells containing a plasmid expressing Moloney MLV (MMLV) gag and pol proteins. TELCeB6 cells produce noninfectious viral core particles, carrying an nlsLacZ reporter retroviral vector.
Normal Chinese hamster ovary (CHO) fibroblasts are resistant to infection by both amphotropic and ecotropic MLVs. CeRD9 was derived from the CHO cell line after transfection and clonal selection of cells containing a plasmid expressing the mCAT-1 receptor (12). The CeAR13 cell line was derived from transfection of CeRD9 clone after clonal selection of cells containing a plasmid expressing Pit-2 receptor and thus expressed both receptors (12). We generated CHO–Pit-2 cells by transfection and clonal selection of cells expressing Pit-2 receptor. All cells were grown in DMEM (Life Technologies) supplemented with 10% fetal bovine serum and proline (Life Technologies). The murine fibroblastic NIH 3T3 cell line expressing both mCAT-1 and Pit-2 receptor were grown in DMEM (Life Technologies) supplemented with 10% new born bovine serum (Life Technologies).
Plasmids, transfection, and virus production.
Plasmids encoding the ecotropic (FBMOSALF), amphotropic (FBASALF), and AMO chimeric (FBAMOSALF) envelopes were described elsewhere (6). Expression plasmids for AMOFx, AMO1, AMO1Fx, AMOG1Fx, AMOG2, AMOG2Fx, AMOG3Fx, AMOΔPRO2, AMOΔPRO3, AMOΔPRO4, and AMOPRO chimeric envelopes were as described elsewhere (21, 22).
To generate expression plasmids for AMOEL3, AMOEL3V, AMOEL3VA, AMOEL3I, and AMOEL3AI, we reconstituted, for each spacer, a double-stranded DNA by hybridization of sense and antisense oligonucleotides corresponding to the entire sequence of the spacer elastin since no templates were available. We mixed equimolar amounts of both oligonucleotides, heated at 95°C in a thermoblock for a few seconds, and then allowed them to cool slowly to room temperature in order to allow hybridization. Double-stranded products of 63 bp each were then digested with EaeI and cloned into NotI-linearized FBAMOSALF plasmid (6). The correct sequence of final clones was systematically confirmed by sequencing.
To generate the others constructs, a general method of PCR amplification, using a high-fidelity polymerase (Vent polymerase; Biolabs) was performed as previously described (21). The correct sequence of PCR products, as well as the final constructs, were systematically confirmed by sequence analysis.
To generate AMOGS3 plasmid, an upper nucleotide (805Fc, 5′-TCCAATTCCTTCCAAGGGGC) located just upstream of the XhoI site of the 4070A env gene (nucleotide 594) was used in combination with an oligonucleotide providing a BamHI site (LowGS3, 5′-TATAGGATCCCCCACCTCCAGATCCGCCACCTCCCACATTAAGGACCTGCCGGGTCAGGA) to generate by PCR amplification a 159-bp fragment encompassing the GS3 interdomain spacer on the FBAMOSALF env gene as template. In parallel, a second PCR amplification was performed with an oligonucleotide (LMOAΔPRO3, 5′-TATGTGCGGCCGCGTCTGGCAGAACGGGGTTTGG) located downstream of the BspEI site of the FBMOSALF env gene in combination with an oligonucleotide providing a BamHI site (UpGS3, 5′-CATCGGATCCGGTGGTGGCGGATCGGCCGCACCTCATCAAGTCTAT) to generate a 623-bp fragment also encompassing the GS3 spacer domain on the FBAMOSALF env gene template. These two PCR-amplified fragments were digested, respectively, with XhoI-BamHI and BamHI-BspEI and cloned into XhoI-BspEI-digested FBAMOSALF plasmid.
MOD84K plasmid was generated by two PCR amplifications. (i) The first used oligonucleotide Xba-ATG (5′-ACCATCCTCTGGACGACATG) located on the XbaI site and an ATG initiation codon of FBMOSALF env gene in combination with an oligonucleotide providing the substitution of D to K and creating an XhoI site (Low XhoD84K, 5′-ATTTTCTCGAGCAGCCTGGGCTGCTGCCCC). The PCR amplification product of 360 bp was then digested by XbaI and XhoI. (ii) The second used oligonucleotide LMOAΔPRO3 in combination with an oligonucleotide providing the substitution of D to K and creating an XhoI (Up XhoD84K, 5′-AGGCTGCTCGAGAAAATGCGAAGAACCTTTAACCTCCC). The 470-bp PCR product was then digested with XhoI-BamHI. The two digested PCR fragments were cloned into the XbaI-BamHI FBMOSALF plasmid expressing the MO envelope.
To generate the AMOD84K mutants, the same strategy was used with oligonucleotide 805FC instead of Xba-ATG in combination with the oligonucleotide Low XhoD84K to generate a 430-bp fragment and also the same 470-bp PCR fragment resulting from LMOAΔPRO3-Up XhoD84K amplification. Both fragments were digested by XhoI and XhoI-BamHI, respectively, and cloned into an XhoI-BamHI FBAMOSALF plasmid.
To generate the AMOΔ3PROD84K and AMOPROD84K constructs, the NotI-BamHI fragment from AMOD84K plasmid sharing the D-to-K mutation was cloned into the NotI-BamHI AMOΔ3PRO and AMOPRO plasmids.
To generate AMOGIXD84K, the 200-bp XhoI-BstEII fragment from the AMOG1X plasmid, corresponding to the G1X spacer, was cloned into the XhoI-BstEII AMOD84K plasmid.
To generate expression plasmids for MOAPRO, an upper oligonucleotide (UpMOAPRO, 5′-ACTGGGGCTTACGTTTGT-3-′) located just upstream of the BamHI site of the MMLV env gene was used in combination with a lower oligonucleotide providing a NotI site (LMOAPRO, 5′-ATCGAGGTCACCGCGGCCGCGGGACCCCGAGTCCCCATAGGGCCC-3′) to generate by PCR a 200-bp fragment encompassing the PRO interdomain spacer on the FBMOSALF env gene as a template. This PCR fragment was double digested with BamHI and NotI. In parallel, a 2-kbp NdeI-BamHI fragment from the FBMOSALF plasmid was purified, providing the 5′ end of the MO env gene. These two inserts were then cloned into the NdeI-NotI-digested FBEASALF plasmid (7).
The MOAΔPRO plasmid was generated by performing a PCR amplification with UpMOAPRO oligonucleotide in combination with the LMOAΔPRO3 lower oligonucleotide, providing a shorter PRO spacer. After BamHI-NotI digestion, this insert was cloned into the BamHI-NotI-linearized MOAPRO plasmid.
TELCeB6 cells were transfected with envelope expression plasmids by calcium phosphate precipitation as previously described (6). Transfected cells were selected with phleomycin (50 μg/ml), and phleomycin-resistant colonies were pooled.
Virus-containing supernatants were collected after an overnight production from freshly confluent env-transfected TELCeB6 cells in regular medium.
Immunoblots.
Virus producer cells were lysed in a 20 mM Tris-HC1 buffer (pH 7.5) containing 1% Triton X-100, 0.05% sodium dodecyl sulfate (SDS), 5 mg of sodium deoxycholate per ml, 150 mM NaCl, and 1 mM phenylmethylsulfonyl fluoride. Lysates were incubated for 10 min at 4°C and then centrifuged for 10 min at 10,000 × g to pellet the nuclei. Supernatants were then frozen at −70°C until further analysis. Virus samples were obtained by ultracentrifugation of viral supernatants (10 ml) in a SW41 Beckman rotor (30,000 rpm, 1 h, 4°C). Pellets were suspended in 100 μl of phosphate-buffered saline (PBS) and frozen at −70°C. Samples (30 μg for cell lysates or 10 μl for purified viruses) were mixed 5:1 (vol/vol) in a 375 mM Tris-HCl (pH 6.8) buffer containing 6% SDS, 30% β-mercaptoethanol, 10% glycerol, and 0.06% bromophenol blue; boiled for 3 min, and then run on SDS–10% acrylamide gels. After protein transfer onto nitrocellulose filters, immunostaining was performed in Tris base saline (pH 7.4) (TBS) with 5% milk powder and 0.1% Tween 20. The Antibodies (Quality Biotech, Inc.) were goat antisera raised against either Rausher leukemia virus (RLV) gp70-SU protein or RLV p30-CA protein and were diluted 1/1,000 and 1/10,000, respectively. Blots were developed using horseradish peroxidase-conjugated rabbit anti-goat immunoglobulin antibodies (Dako) and an enhanced chemiluminescence kit (Amersham Life Science).
Binding assays.
Target cells were washed in PBS and detached by a 10-min incubation at 37°C with 0.02% EDTA in PBS. Cells were washed in PBA (PBS with 2% fetal calf serum and 0.1% sodium azide). Then, 5 × 105 cells were incubated in PBA with viruses for 45 min at 37°C. Cells were then washed with PBA and incubated for 45 min at 4°C with monoclonal antibody 83A25, which recognizes a common epitope present on the ecotropic and amphotropic MLV surface unit (SU) and allows the detection of SU proteins that are either soluble or associated with the virus (8), or 9E8, which is specific of the transmembrane unit of the MLV envelope (TM), and allows the detection of MLV envelope only when associated with the virus (virus binding). Cells were washed twice with PBA and incubated with anti-rat immunoglobulin fluorescein isothiocyanate-conjugated antibodies (Dako). At 5 min before the two final washes in PBA, cells were counterstained with 20 μg of propidium iodide per ml. The specific fluorescence of the cells was analyzed with a flow cytometer (FACSCalibur; Becton Dickinson).
Infection assays.
Target cells were seeded in 24-well plates at a density of 5 × 104 cells per well. Viral supernatant dilutions containing 5 μg of Polybrene per ml were added, and cells were incubated for 3 to 5 h at 37°C. Viral supernatant was then removed, and cells were incubated in regular medium for 48 h. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining and viral titer determination as lacZ infectious units (iu)/milliliter were performed as previously described (6).
RESULTS
We have previously reported a set of chimeric MMLV-derived envelopes designed to retarget the Pit-2 phosphate transporter molecule by insertion into MMLV envelope of the first 205 amino acids of the amphotropic MLV 4070A SU corresponding to the BD (4070A BD) (Fig. 1) (22). We have shown that the infection of target cells by virions bearing these envelopes is strictly dependent on the nature of the interdomain spacer separating the 4070A BD from the MMLV backbone envelope. When a proline-rich motif of 60 amino acids was inserted as a spacer, generating the AMOPRO envelope, infection was dependent on the presence of both receptors (Pit-2 and mCAT-1) at the surface of the target cells, suggesting that these receptors cooperate during virus entry (Fig. 2). In contrast, when truncated forms of the proline-rich motif (AMOΔPRO2, AMOΔPRO3, or AMOΔPRO4 chimeric envelopes) were used, infection occurred only when the targeting receptor was expressed at the surface of cells but not when receptor corresponding to the backbone envelope was expressed alone. These data are consistent with a dynamic role of the proline-rich motif inserted into chimeric MLV-derived env in receptor cooperation during retroviral entry.
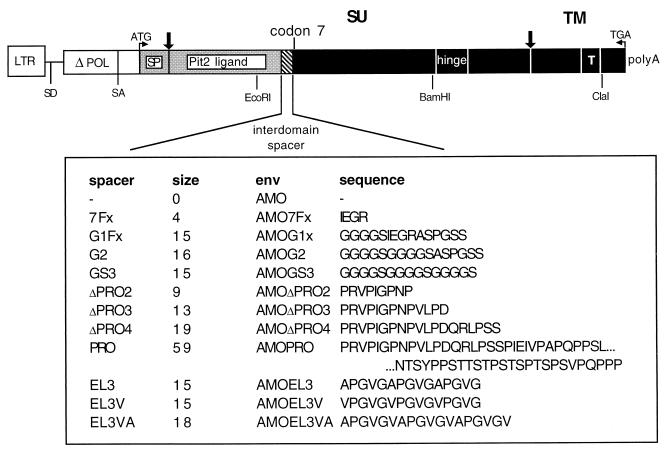
FIG. 1.
Schematic representation of envelope chimeras. The plasmid construct coding for chimeric envelope glycoproteins in which the Pit-2 BD was fused to MMLV SU is displayed. The general format is shown diagrammatically, and the amino acid sequence of the interdomain spacer is shown in detail. Vertical arrows, protein cleavage sites; SU, surface protein; TM, transmembrane protein; SP, envelope signal peptide; T, transmembrane domain; Pit-2 ligand, MLV-A receptor BD; black boxes, MMLV-derived envelope sequences; gray boxes, MLV-A-derived envelope sequences; white boxes, other MLV-derived envelope sequence. All env genes were expressed using the same promoter (LTR) and polyadenylation sites (polyA) from a subgenomic mRNA using retroviral splice donor (SD) and acceptor (SA) sites.
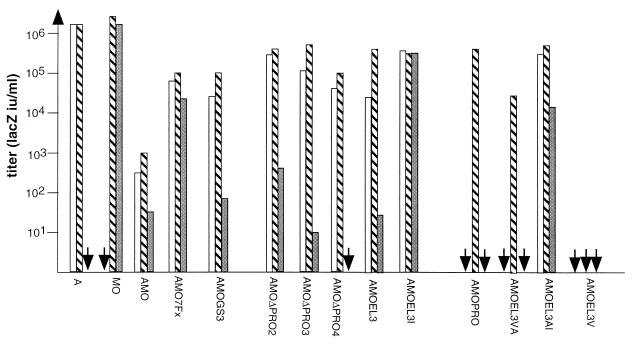
FIG. 2.
Infection by virions expressing targeting envelopes. Virions were used to infect CHO cells expressing Pit-2 receptor (CHO–Pit-2 cells; white bars), mCAT-1 receptor (CeRD9; gray bars), or both receptors (CeAR13; striped bars). Data correspond to the average of 20 repeats (each duplicate), and no significant difference from one experiment to another was observed (standard deviation of 0.25 log, 99% confidence interval of the mean).
β-Turn helix plays a critical role in receptor cooperation.
The proline-rich region of 4070A MLV is predicted by the Chou-Fasman analysis (5) to form a regular arrangement of 11 β-turns induced by the majority of the prolines in this region (13, 22). This highly ordered structure, consisting of multiple reverse turns, is consistent with the polyproline β-turn helix form of a secondary structure (15).
As shown for the dynamic β-spirals of bovine elastin, such a repetitive β-turn motif in the MLV env proline-rich region may display unusual physical properties, such as self-assembly into oligomeric quaternary structures, increasing structural order with increasing temperature, and the development of elastomeric forces coincident with molecular ordering (20).
In order to see if the properties of the polyproline β-turn helices can account for the behavior of retroviruses coated with AMOPRO and AMOΔPRO envelopes, we tried to replace PRO spacers by a nonviral β-spiral peptide from bovine elastin (4, 20). Three synthetic peptides were chosen for this approach: (i) an 18-amino-acid spacer corresponding to three repeats of the hexapeptide APGVGV, named EL3VA (APGVGVAPGVGVAPGVGV), or (ii) a 15-amino-acid peptide corresponding to three repeats of a pentapeptide VPGVG, named EL3V (VPGVGVPGVGVPGVG), which were both shown to correspond to the main repeating sequences identified in the tropoelastin, the precursor of fibrous elastin, or (iii) three repeats of the pentapeptide APGVG motif, called EL3 (APGVGAPGVGAPGVG).
These peptides were predicted to have propensities to form a β-spiral and can be classified from weakest to strongest in β-turn helix propensity as EL3, EL3VA, and EL3V.
In order to check if the size of the spacer might influence receptor cooperation efficiency, a length control unfolded spacer of 15 amino acids was designed: GS3, which corresponds to three repeats of four glycine residues and one serine residue. This spacer was cloned into the AMO env to generate the AMOGS3 envelope.
The resulting envelopes, AMOEL3VA, AMOEL3V, AMOEL3, and AMOGS3, were transfected into TelCeB6 cells that provide MLV core and an nls-LacZ vector (6).
By Western blot analysis of cell lysates of transfected cells or supernatants of these cells, we demonstrated that chimeric EL3 envelopes were processed and incorporated into MLV particles (Fig. 3A and B). We also showed that they were all able to bind to the Pit-2 receptor (Fig. 4). To address the question of the infectivity of retroviruses generated, cells expressing either Pit-2 or mCAT-1 receptor alone (CHO–Pit-2 and CerD9, respectively) or Pit-2 and mCAT-1 (CeAR13) were infected. When AMOGS3 envelopes were used as a control (Fig. 2), minimum or no significant cooperation of receptors was seen.
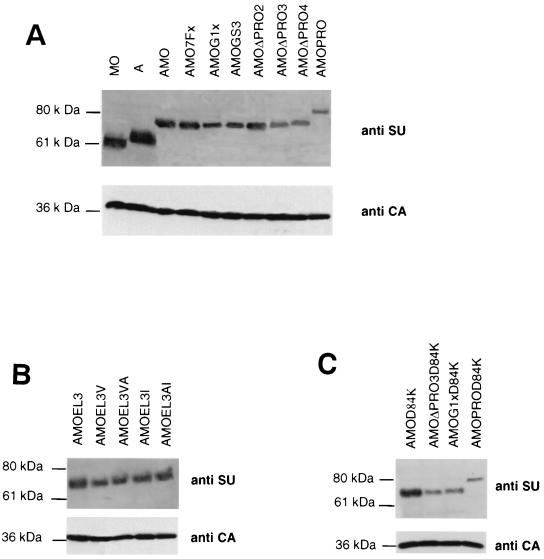
FIG. 3.
Viral incorporation of chimeric envelope glycoproteins. Immunoblots of pellets of viral particles from TELCeB6 cells transfected with the envelopes. The blot was stained with an SU antiserum and cut at 46 kDa in order to stain the lower part with a p30 antiserum to detect the CA protein as a viral production control.
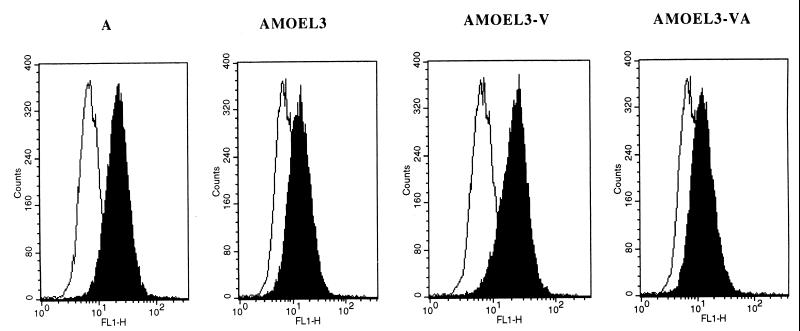
FIG. 4.
Binding assays on Pit-2 receptor. Human TE671 cells expressing Pit-2 receptors were used as a target. The background of fluorescence was provided by incubating the cells with DMEM only (white histograms). Binding assays (black histograms) were performed with some of the envelopes shown in Fig. 1. The envelope glycoprotein content of the different samples was normalized by immunoblot on a viral supernatant.
For the EL3-based envelope, the results revealed a different cooperation efficiency depending on the nature of the β-spiral peptide inserted. We first showed that substitution of the PRO spacer by the less-stable β-spiral peptide (EL3) leads to a cooperation of receptors similar to the one obtained for AMOΔPRO3 envelope: chimeric virions can infect cells only if the target Pit-2 receptor was expressed, with very low efficiency when mCAT-1 receptor was present and with increased efficiency when both receptors are expressed. Once more, when the intermediate-strength β-spiral peptide (EL3VA) was used as a spacer, the infection of target cells was strictly dependent on the presence of the two receptors, a result surprisingly similar to that seen with the 60-amino-acid PRO spacer. These results demonstrated the critical role of the nature of the β-turn helix in the cooperation of receptors.
However, with the EL3V spacer, which had the strongest propensity to form a β-spiral (whereas the chimeric protein is normally and correctly processed), no infection was observed, even in the presence of both receptors, thus demonstrating that insertion of very stable β-spiral spacers blocks the transmission of conformational changes of the protein after binding to the Pit-2 receptor.
These data suggested that the substitution of PRO spacers by β-turn polyproline helix from bovine elastin leads to a cooperation of receptors and that the efficiency of cooperation depends on the stability of the helix.
Deletion of proline residue in the β-turn helix abolishes cooperation.
In order to confirm the dynamic role of the env structure in receptor cooperation, we replaced the repeated proline residues with isoleucine, which prevents β-turn helix formation. By PCR mutation we replaced the three proline residues present in the EL3 spacer of AMOEL3 and AMOEL3-VA and derived envelopes named AMOEL3-I and AMOEL3-AI, respectively. As expected, these mutations had no effect on the processing and virion incorporation of the corresponding envelopes (Fig. 3B).
To assess the effect of such mutations on infectivity, infection assays was performed on target cells expressing either one or two receptors. The results obtained are reported in Fig. 2. We demonstrated that a proline-to-isoleucine substitution (AMOEL3AI versus AMOEL3VA) abolished receptor cooperation. Unlike AMOEL3VA, the AMOEL3AI virions were able to infect cells expressing either mCAT-1 or Pit-2 with an efficiency at least of 104 iu/ml. Viral titers were 1 log greater on cells expressing both receptor CeAR13s than the AMOEL3VA ones.
For the AMOEL3 envelope, the mutation of proline to isoleucine enhanced the infection efficiency by at least 1 log on cells expressing Pit-2 receptors alone and by 4 logs on cells expressing mCAT-1. These data suggest that the structure of the spacers as a β-turn polyproline helix is crucial for receptor cooperation efficiency.
Interaction with both receptors is required for fusion transduction.
Our results are consistent with a model of two-step interaction for the entry of the targeted virions into cells. To address the requirement, in the second step, of MMLV backbone envelope interaction with the mCAT-1 receptor, we generated a set of mCAT-1 binding-defective mutants. It has been previously demonstrated (14) that an Asp residue in position 84 in the MMLV wild-type env plays a critical role in binding to receptor. When Asp-84 is replaced with a Lys residue, the infection is blocked. By PCR amplification, we created the Asp-to-Lys mutation at position 84 of the mCAT-1 BD of AMO, AMOΔPRO3, AMOPRO, and AMOG1X, resulting in AMOD84K, AMOΔPRO3D84K, AMOPROD84K, and AMOG1XD84K, respectively. The resulting envelopes were tranfected into TELCeB6 cells, and we showed by Western blot analysis that the production, maturation, and incorporation into viral particles of each env glycoprotein were not significally affected by the mutation (Fig. 3C).
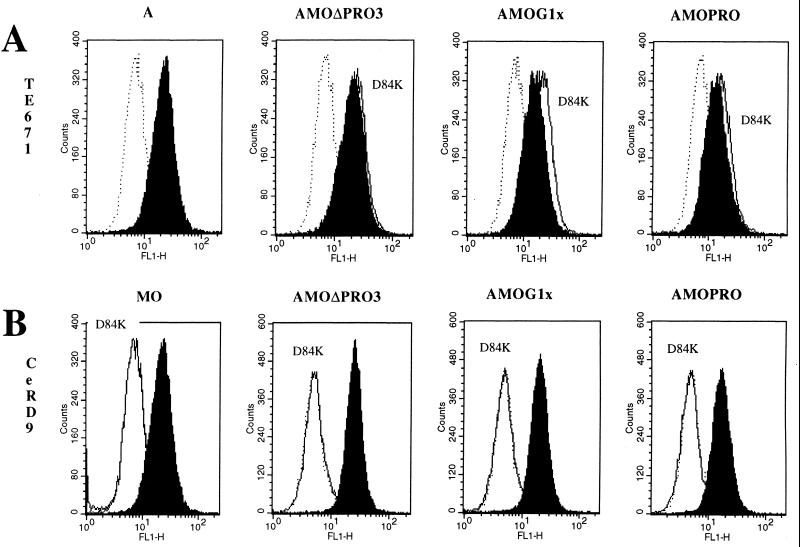
By flow cytometric analysis, we also confirmed that, whereas all of these mutants were able to bind to cells expressing Pit-2 receptors in a manner similar to that of env (Fig. 5A), none were able to bind to cells expressing mCAT-1 receptors alone (Fig. 5B).
FIG. 5.
Envelope binding assays. The background fluorescence was provided by incubating the cells with DMEM only (dotted line histograms). (A) Pit-2 binding control of the D84K mutant. Human TE671 cells expressing Pit-2 receptors were used as a target. Binding assays of D84K mutant envelopes (white histograms) were compared to chimeric envelope unmutated on residue 84 (black histograms). (B) mCAT-1 binding test of D84K mutant. CeRD9 cells expressing mCAT-1 receptor alone were used as a target. Binding assays of D84K mutant envelope binding (white histograms) were compared to unmutated recombinant envelope (black histograms). The envelope glycoprotein content of the different samples was normalized by immunoblot on a viral supernatant.
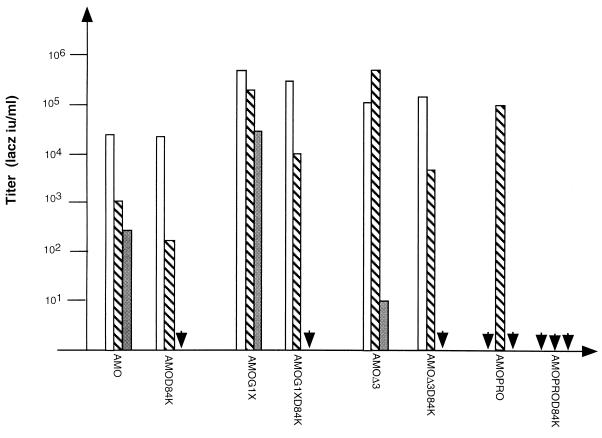
To check the infectious properties of the resulting mutants, an infection assay was performed on cells expressing mCAT-1, Pit-2, or both receptors as previously described. The results are reported in Fig. 6. As expected, AMO and AMOG1X viruses, which were able to infect cells that only express mCAT-1, failed to be infectious in the same cells when the D84K mutation was inserted, thus confirming the critical role of the Asp-84 residue. Like the controls AMOΔPRO3, and AMOPRO, AMOΔPRO3D84K and AMOPROD84K env-bearing viruses remained unable to infect cells which only expressed mCAT-1. The D84K mutation did not affect the entry of viruses through Pit-2, since AMOD84K, AMOΔPRO3D84K, and AMOG1XD84K were able to infect cells expressing Pit-2 receptors. AMOPROD84K, like the control AMOPRO env-bearing viruses, remained unable to infect cells when only Pit-2 is expressed.
FIG. 6.
Infection by virions expressing mCAT-1-deficient binding envelopes. Virions were used to infect CHO cells expressing Pit-2 receptor (CHO–Pit-2 cells; white bars), mCAT-1 receptor (CeRD9; gray bars), or both receptors (CeAR13; striped bars). Data correspond to the average of 20 repeats, and no significant difference from experiment to another was observed (standard deviation of 0.2 log, 99% confidence interval of the mean).
In CeAR13 cells which express both receptors, we observed for AMOD84K, AMOΔPRO3D84K, and AMOG1XD84K a 1-log reduction of viral titers compared to the corresponding wild-type chimeric envelopes.
These results are consistent with an infection occurring, in the normal configuration, in a ratio of 1 log through mCAT-1 receptors and a range of 103 to 104 through Pit-2, depending on the chimeric envelope. This suggests that the mechanisms of viral entry through these two receptors are different and that mCAT-1 is less efficient at allowing fusion when the mCAT-1 BD is not separated from the N-terminal domain of env by PRO spacers.
However, for AMOPRO viruses, for which infection was dependent on the presence of both receptors, inhibition of the interaction between mCAT-1 BD and its receptor completely prevented infection, thus demonstrating the crucial role of this second-step interaction for the entry of chimeric viruses into cells.
Reciprocal receptor cooperation.
To test if there is a preferential order of interaction with receptors, we used as the backbone envelope the 4070A amphotropic envelope. The ecotropic mCAT-1 BD was fused at the N terminus of the 4070A Env protein. These two BD were spaced either with ΔPRO3 or PRO spacers. The resulting plasmids, named MOAΔPRO3 and MOAPRO, were transfected into TELCeB6 cells as previously described.
These chimeric amphotropic backbone envelopes were normally processed and incorporated into viral particles (data not shown).
As with their counterpart, AMOΔPRO, the MOAΔPRO virions could only infect cells that express the targeted receptor alone (in that case CeRD9 cells expressing mCAT-1) and the CeAR13 cells expressing both receptors (Table 1). However, although AMOΔPRO virions were unable to infect cells that only expressed the receptor corresponding to the backbone envelope (CeRD9), MOAΔPRO virions remained able to infect, even with low efficiency, cells through Pit-2 receptors (CHO–Pit-2).
TABLE 1.
Titration of mCAT-1 targeting envelopes on CHO cells expressing receptors Pit-2 and/or mCAT-1
| Envelopea | Titer (LacZ iu/ml)b
|
||
|---|---|---|---|
| CeAR13 | CHO–Pit-2 | CeRD9 | |
| MO | 107 | <1 | 107 |
| A | 107 | 107 | <1 |
| MOAΔPRO | 2.5 × 105 | 1.3 × 102 | 9.5 × 104 |
| MOAPRO | 1.1 × 104 | <101 | 5.2 × 101 |
Envelopes are as depicted in Fig. 1 and expressed on LacZ virions.
Data correspond to an average of 10 repeats, each done in triplicate (standard deviation of 0.3 log, 99% confidence interval of the mean).
Similar to AMOPRO, MOAPRO failed to infect cells expressing the backbone receptor alone, whereas it was able to infect CeAR13 cells expressing both receptors. These data seem to indicate that the Pit-2 BD isn't functional and that a receptor cooperation is required for infection.
However, we observed that MOAPRO virions remained able to infect cells expressing only the targeted receptor (CeRD9), although with a very poor efficiency. These data suggest that even if receptor cooperation was efficient, it appeared to be less stringent than what we observed for AMOPRO virions.
DISCUSSION
Proline-rich regions are known to play a critical role in several pro- and eucaryotic systems (9, 15, 20). Depending on their structure, they contribute directly or indirectly to the binding properties, the structure and stability, or the functionality of proteins that contain such proline-rich regions. Thus, the proline-rich region seems to be a functional domain rather than a spacing region. We showed that insertion of a 60-residue PRO peptide between two BDs of chimeric MMLV-derived retroviral glycoproteins limited their ability to infect cells: infection became strickly dependent on the presence of both receptors, corresponding to the two BDs of the chimeric envelope glycoproteins at the surface of the cells.
Predictive structure analysis indicates that this PRO spacer may be organized as a β-turn helix. We showed that this structure can account for the behavior of the chimeric envelope since the substitution of PRO spacers by nonviral peptides, shown to be organized as a β-turn helix, induces receptor cooperation. Further, we demonstrated that the efficiency of receptors cooperation is modulated (i) by the number of turns forming the β-helix and (ii) within one β-turn of the helix by the number and nature of the amino acid spacing the PRO residues.
We showed that the most efficient cooperation was observed with the chimeric envelope containing the whole PRO region and that, at least, three to four β-turns are required to mask the backbone ecotropic envelope. Even for an equal given number of β-turns, while three repeats of PGVGAP corresponding to EL3 spacer “cooperates” as well as three turns of the PRO spacer (ΔPRO3), three repeats of PGVGVP (corresponding to EL3V) completely blocked cooperation. We speculate that the greater stability of the β-turn arrangement is not compatible with the refolding of the spacer after interaction between the additional N-terminal binding domain and its receptor. These data seem to indicate that spacers need to share a stability not too strong in order to transmit “signal” after interaction trough changes of conformation.
We also showed that the 18-amino-acid EL3VA spacer (APGVGV motif) presents the same “cooperative” properties as the 60-residue PRO spacers, and we can assume that this spacer has an improved arrangement of the β-turns in terms of structure and stability.
Taken together, these data suggest that the composition, length, and strength that determines the stability of the β-turn helix is crucial for the efficiency of receptor cooperation. As in the naive viral SU, the BD located downstream of the PRO spacer is masked by the presence of PRO spacers (22); these data are consistent with two-step receptor cooperation as follows: (i) binding to a targeted surface molecule induces conformational changes in the chimeric glycoprotein envelope via the β-turn helix and (ii) this, in turn, allows interaction with the fusion-competent retroviral receptor.
It is interesting to draw a parallel between the masking of the receptor binding region located downstream of the PRO spacer in the chimeric envelope and the masking of the fusion peptide by the proline-rich region naturally present between the BD of the SU and TM subunits in wild-type MLV envelope glycoproteins. Viruses have been selected to develop efficient and specific systems to block the activity of their fusion peptide until they bind specifically to the right receptor, and we can argue that the natural proline-rich region of wild-type MLV glycoproteins share a similar function.
This two-step entry of a virus into a cell, in a natural, efficient way, is attractive for the design of a new generation of retroviral vectors suitable for gene therapy. Indeed, we previously demonstrated that “one-step targeting”, by insertion of new BDs at the N terminus of the MMLV glycoprotein (6, 21) is not efficient in terms of the transduction of human cells. So, depending on the cells to be targeted, the next strategy will be to design a chimeric targeting envelope based on the backbone envelope for which human cells express the most efficient receptors in terms of transduction of the fusion signal. This might be performed with (i) an amphotropic envelope for which the ubiquitous receptor Pit-2 is expressed at the surface of nearly all human cells, (ii) a gibbon ape leukemia virus envelope for which Pit-1 receptors are largely expressed at the surface of hematopoietic precursors, or (iii) a hemagglutinin envelope of influenza viruses which recognizes the ubiquitous sialic acid (10a).
We demonstrated that reciprocal Pit-2–mCAT-1 cooperation with MOAPRO virions is possible, although infection still occurs partially directly through the targeted receptor. These observations suggested that even if ecotropic and amphotropic envelopes share similar topologies and properties, their exploitation of receptor potential seem to be slightly different.
In the MOAPRO construct, the PRO spacer inserted between the two BDs was derived from ecotropic strain, whereas for the AMOPRO construct the PRO spacer was derived from amphotropic ones.
The efficiency of masking of the PRO spacer appears to be lower for the amphotropic backbone envelope compared to the ecotropic one. This observation can be correlated with the predicted structure of these two regions: the amphotropic helix shares more stable and more numerous β-turns than ecotropic ones. We speculate that amphotropic envelopes are more fusogenic than ecotropic ones and, as a prerequisite, the inhibition of the fusion domain is more effective in order to restrict the fusion process unless specific binding to the receptor occurs.
A new goal will be to generalize this two-step targeting phenomena to the design of vectors suitable for gene therapy purposes. First, the backbone envelope has to be improved depending on the human cells to be targeted. We have to choose the most suitable backbone envelope for which the target cells express the most efficient retroviral receptors. Second, the spacer has to be optimized in order to design the most efficient β-turn helix polyproline in terms of masking and also unmasking. Lastly, the most specific ligand able to target the virus to specific cell surface molecules will have to be developed.
The improvement of these three components will give rise to a new concept for gene therapy retroviral vector design.
ACKNOWLEDGMENTS
This work was supported by Agence Nationale pour la Recherche contre le SIDA (ANRS), the European Community, AFIRST, Association Française contre les Myopathies (AFM), Association pour la Recherche contre le Cancer (ARC), SIDACTION, and Institut National de la Santé et de la Recherche Médicale (INSERM).
I thank Michèle Ottmann-Terrangle, David Camerini, and Fabian Wild for critical reading of the manuscript.
REFERENCES
- 1.Andersen K B. A domain of murine retrovirus surface protein gp70 mediates cell fusion, as shown by a novel SC-1 cell fusion system. J Virol. 1994;68:3175–3182. doi: 10.1128/jvi.68.5.3175-3182.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae Y, Kingsman S M, Kingsman A J. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J Virol. 1997;71:2092–2099. doi: 10.1128/jvi.71.3.2092-2099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battini J L, Danos O, Heard J M. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J Virol. 1995;69:713–719. doi: 10.1128/jvi.69.2.713-719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhandary K, Senadhi S E, Prasad K U, Prasad K, Urry D W, Vijay-Kumar S. Conformation of a cyclic decapeptide analog of a repeat pentapeptide sequence of elastin: cyclo-bis(valyl-prolyl-alanyl-valyl-glycyl) Int J Peptide Protein Res. 1990;36:122–127. doi: 10.1111/j.1399-3011.1990.tb00954.x. [DOI] [PubMed] [Google Scholar]
- 5.Chou P Y, Fasman G D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:45–148. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- 6.Cosset F-L, Morling F J, Takeuchi Y, Weiss R A, Collins M K L, Russell S J. Retroviral retargeting by envelopes expressing an N-terminal binding domain. J Virol. 1995;69:6314–6322. doi: 10.1128/jvi.69.10.6314-6322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosset F-L, Takeuchi Y, Battini J L, Weiss R A, Collins M K L. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans L H, Morrison R P, Malik F G, Portis J, Britt W. A neutralizable epitope common to the envelope glycoproteins of ecotropic, polytropic, xenotropic, and amphotropic murine leukemia viruses. J Virol. 1990;64:6176–6183. doi: 10.1128/jvi.64.12.6176-6183.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fontenot J D, Tjandra N, Ho C, Andrews P C, Montelaro R C. Structure and self-assembly of a retrovirus (FeLV) proline-rich neutralization domain. J Biomol Struct Dyn. 1994;11:821–837. doi: 10.1080/07391102.1994.10508035. [DOI] [PubMed] [Google Scholar]
- 10.Gray K D, Roth M J. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J Virol. 1993;67:3489–3496. doi: 10.1128/jvi.67.6.3489-3496.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Hatziioannou T, Delahaye E, Martin F, Russell S J, Cosset F L. Retroviral display of functional binding domains fused to the amino terminus of influenza hemagglutinin. Hum Gene Ther. 1999;10:1533–1544. doi: 10.1089/10430349950017860. [DOI] [PubMed] [Google Scholar]
- 11.Kabat D. Molecular biology of Friend viral erythroleukemia. Curr Top Microbiol Immunol. 1989;148:1–42. doi: 10.1007/978-3-642-74700-7_1. [DOI] [PubMed] [Google Scholar]
- 12.Kozak S L, Siess D C, Kavanaugh M P, Miller A D, Kabat D. The envelope glycoprotein of an amphotropic murine retrovirus binds specifically to the cellular receptor/phosphate transporter of susceptible species. J Virol. 1995;69:3433–3440. doi: 10.1128/jvi.69.6.3433-3440.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavillette D, Maurice M, Roche C, Russell S J, Sitbon M, Cosset F-L. A proline-rich motif downstream of the receptor binding domain modulates conformation and fusogenicity of murine retroviral envelopes. J Virol. 1998;72:9955–9965. doi: 10.1128/jvi.72.12.9955-9965.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacKrell A J, Soong N W, Curtis C M, Anderson W F. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J Virol. 1996;70:1768–1774. doi: 10.1128/jvi.70.3.1768-1774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsushima C N, Creutz E, Kretsinger R H. Polyproline, beta-turn helices. Novel secondary structures proposed for the tandem repeats within rhodopsin, synaptophysin, synexin, gliadin, RNA polymerase II, hordein, and gluten. Proteins. 1990;7:125–155. doi: 10.1002/prot.340070204. [DOI] [PubMed] [Google Scholar]
- 16.Nussbaum O, Roop A, Anderson W F. Sequences determining the pH dependence of viral entry are distinct from the host range-determining region of the murine ecotropic and amphotropic retrovirus envelope proteins. J Virol. 1993;67:7402–7405. doi: 10.1128/jvi.67.12.7402-7405.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ott D, Rein A. Basis for receptor specificity of nonecotropic murine leukemia virus surface glycoprotein gp70SU. J Virol. 1992;66:4632–4638. doi: 10.1128/jvi.66.8.4632-4638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinter A, Chen T-E, Lowy A, Cortez N G, Siligari S. Ecotropic murine leukemia virus-induced fusion of murine cells. J Virol. 1986;57:1048–1054. doi: 10.1128/jvi.57.3.1048-1054.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeuchi Y, Cosset F L, Lachmann P J, Okada H, Weiss R A, Collins M K L. Type C retrovirus inactivation by human complement is determined by both the viral genome and the producer cell. J Virol. 1994;68:8001–8007. doi: 10.1128/jvi.68.12.8001-8007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urry D W. Entropic elastic processes in protein mechanisms. II. Simple (passive) and coupled (active) development of elastic forces. J Protein Chem. 1988;7:1–34. doi: 10.1007/BF01025240. [DOI] [PubMed] [Google Scholar]
- 21.Valsesia-Wittmann S, Morling F J, Nilson B H K, Takeuchi Y, Russell S J, Cosset F-L. Improvement of retroviral retargeting by using amino acid spacers between an additional binding domain and the N terminus of Moloney murine leukemia virus SU. J Virol. 1996;70:2059–2064. doi: 10.1128/jvi.70.3.2059-2064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valsesia-Wittmann S, Morling F J, Nilson B H K, Takeuchi Y, Russell S J, Cosset F-L. Receptor cooperation in retrovirus entry: recruitment of an auxiliary entry mechanism after retargeted binding. EMBO J. 1997;6:1214–1423. doi: 10.1093/emboj/16.6.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zavorotinskaya T, Albritton L M. Suppression of a fusion defect by second site mutations in the ecotropic murine leukemia virus surface protein. J Virol. 1999;73:5034–5042. doi: 10.1128/jvi.73.6.5034-5042.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]