Abstract
The incidence of Mycobacterium marinum infection is on the rise; however, the existing drug treatment cycle is lengthy and often requires multi-drug combination. Therefore, there is a need to develop new and effective anti-M. marinum drugs. Cochliomycin A, a 14-membered resorcylic acid lactone with an acetonide group at C-5′ and C-6′, exhibits a wide range of antimicrobial, antimalarial, and antifouling activities. To further explore the effect of this structural change at C-5′ and C-6′ on this compound’s activity, we synthesized a series of compounds with a structure similar to that of cochliomycin A, bearing ketal groups at C-5′ and C-6′. The R/S configuration of the diastereoisomer at C-13′ was further determined through an NOE correlation analysis of CH3 or CH2 at the derivative C-13′ position and the H-5′ and H-6′ by means of a 1D NOE experiment. Further comparative 1H NMR analysis of diastereoisomers showed the difference in the chemical shift (δ) value of the diastereoisomers. The synthetic compounds were screened for their anti-microbial activities in vitro. Compounds 15–24 and 28–35 demonstrated promising activity against M. marinum, with MIC90 values ranging from 70 to 90 μM, closely approaching the MIC90 of isoniazid. The preliminary structure–activity relationships showed that the ketal groups with aromatic rings at C-5′ and C-6′ could enhance the inhibition of M. marinum. Further study demonstrated that compounds 23, 24, 29, and 30 had significant inhibitory effects on M. marinum and addictive effects with isoniazid and rifampicin. Its effective properties make it an important clue for future drug development toward combatting M. marinum resistance.
Keywords: 14-membered resorcylic acid lactones, Mycobacterium marinum, anti-Mycobacterium marinum, ketal groups, diastereoisomers
1. Introduction
Mycobacterium marinum (M. marinum), a nontuberculous mycobacterium (NTM), is one of the major contributors to extrapulmonary mycobacterial infections [1,2]. M. marinum infects human skin and soft tissues, leading to the development of a single papulonodular, verrucose, or ulcerated granulomatous lesion at the infected site [3,4,5,6,7]. Notably, such a local infection may spread to the tendon sheaths or joints [8]. In recent years, the incidence of M. marinum infection has shown an increasing trend [9]. In the United States, the incidence of this disease can reach 0.27 infections per 100,000 people, with the highest incidence observed among individuals who come into contact with fish and fish containers [2]. For M. marinum infection, drugs such as rifambutin, ethambutol, clarithromycin, and others are often used in combinations of two or three, and the duration of medication is generally 2 weeks to 18 months [10,11,12]. However, the existing drug treatments are less than satisfactory. The treatment period for M. marinum is lengthy, and multi-drug combinations are required. In addition, there is no standard for the treatment of M. marinum infection in terms of drug selection, dose, and concentration [13]. These problems have led to an urgent need to optimize treatments and develop novel and effective anti-M. marinum drugs.
Natural products are of outstanding significance in drug development, and numerous drugs or lead compounds are inspired by them [14,15]. In the decades from 1981 to 2019, more than 50% of small-molecule drugs developed globally were influenced by natural products [16,17]. Microorganisms are capable of producing secondary metabolites with novel structures and significant biological activity, and these metabolites have served as a source of inspiration for the development of many new drugs [18,19,20,21,22]. The natural products derived from marine microorganisms possess many unique properties and important value, distinct from terrestrial natural products available on land. Marine natural products exhibit rich diversity, extensive medicinal value, and biological activities [23,24].
Marine fungi have been shown to have complex and varied structures and can produce secondary metabolites with various biological activities that have considerable synthetic value and great significance. They hold significant promise in the research and development of new pharmaceuticals [25,26]. Our laboratory has also focused further attention and research on marine-derived fungi, with the expectation of finding and developing more natural products and derivatives with multifarious activities [27,28].
The 14-membered resorcylic acid lactones (RALs) are polyketides characterized by a 14-membered macrocyclic ring fused to a resorcylic acid residue, and they are classified as natural products [29,30]. In our previous research, we isolated a series of 14-membered RALs, including cochliomycins A–G, 5-Bromozeaenol, and 3, 5-Dibromozeaenol, from the marine-derived fungus Cochliobolus lunatus [29,31,32,33]. The 14-membered RALs have various biological activities such as antibacterial, antifouling, antimalarial, and antiviral activity [20,31,34,35,36,37].
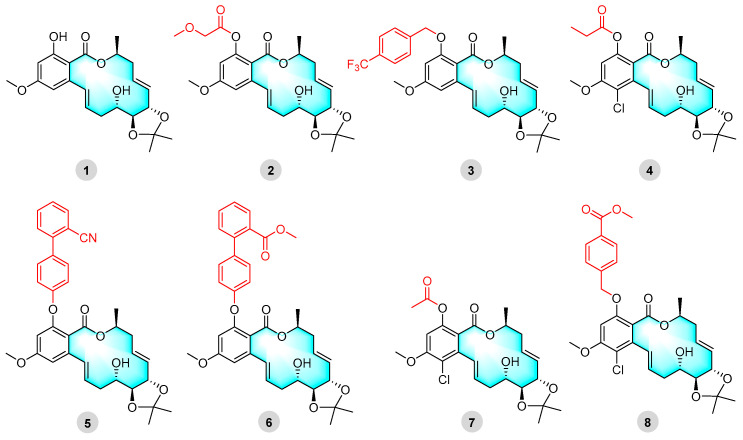
Cochliomycin A, a natural product of 14-membered RALs with an acetonylic group, can significantly reduce barnacle settlement at a concentration of 1.2 μg/mL [35]. In addition, it has the advantage of exhibiting low toxicity and is an environmentally effective antifouling compound [35]. In our previous study, the cochliomycin A derivative 2 showed strong selective algal inhibitory activity [20]. In particular, the selective inhibition of the diatoms of Navicula laevissima and Navicula exigua was close to that of SeaNine 211 [20]. Derivatives 5 and 6 showed strong antimalarial activity and non-toxicity when used against Plasmodium falciparum [20]. Furthermore, cochliomycin A derivatives also exhibited anti-M. marinum activity (Figure 1) [33,36].
Figure 1.
Cochliomycin A (1) and its derivatives (2–8) with strong antialgal and antiplasmodial activities or antibacterial activity.
Cochliomycin A and its derivatives with acetonide or deuterium acetonyl groups at C-5′ and C-6′ have potent antimalarial and antifouling activity [20]. It is thought that increasing the number of ketal groups at C-5′ and C-6′ would enhance the activity of cochliomycin A derivatives. In this study, we synthesized new compounds, 10–37, which are a series of marine-derived 14-membered RALs with ketal groups. The aim was to obtain 14-membered RAL derivatives with novel structures and evaluate their structure–activity relationships. The synthetic derivatives were screened for in vitro activity against M. marinum and five other bacteria and fungi. The activity-screening results showed that derivatives 15–24 and 28–35 exhibited selective inhibition against M. marinum.
2. Results and Discussion
2.1. Chemistry
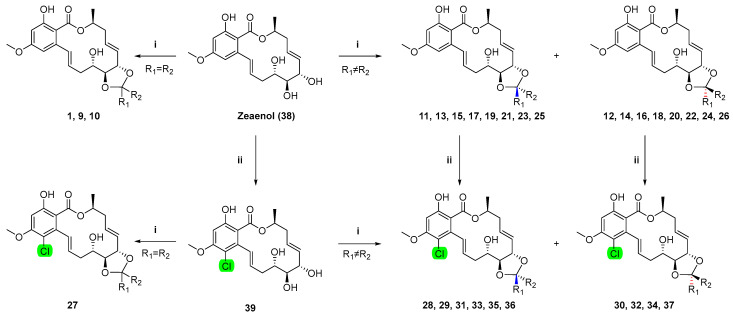
The crude extracts obtained from Cochliobolus lunatus (CHNSCLM-0009) were separated via silica gel column chromatography. After further recrystallization, about 5.4 g of zeaenol (38) was obtained [20]. Using zeaenol (38) as the initial raw material, derivatives 10–37 with novel ketal groups were semi-synthesized through either a one-step reaction or a two-step chemical reaction (Scheme 1).
Scheme 1.
The synthetic route. i p-TsOH, K2CO3, R1COR2, 25 °C, 2–4 h; ii SO2Cl2, 0 °C.
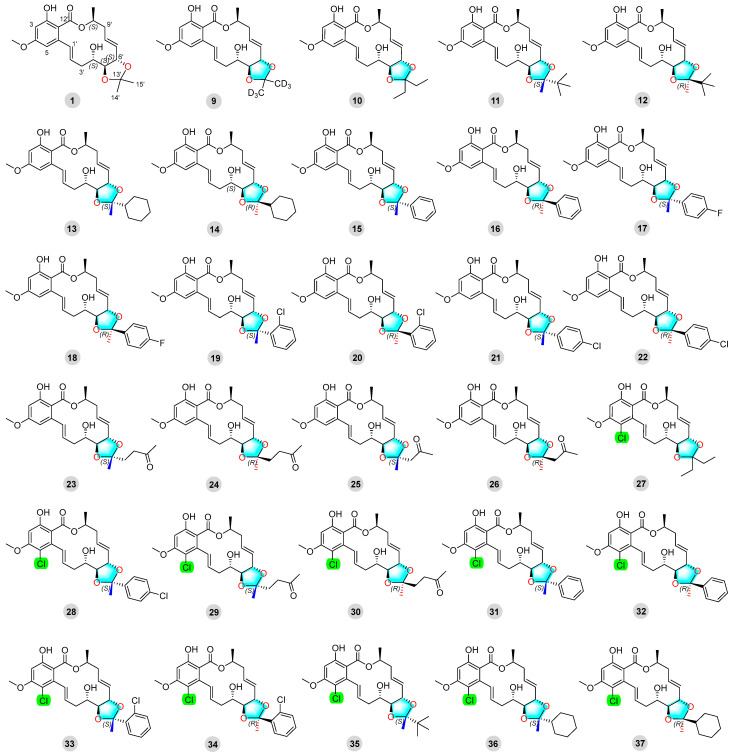
Twenty-eight newly synthesized compounds, 10–37, with ketal groups are shown in Figure 2.
Figure 2.
Cochliomycin A (1) and its derivatives (9–37).
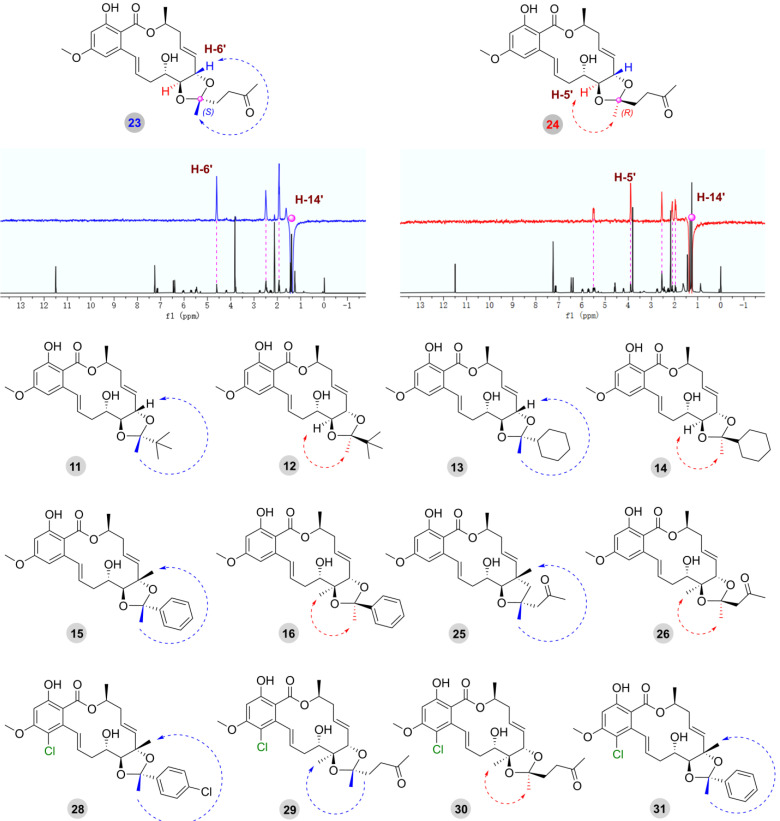
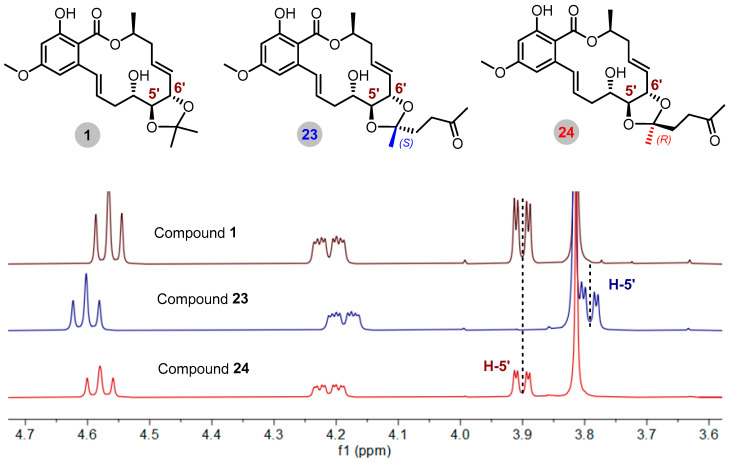
It is worth noting that the R/S configuration of the C-13′ position was the key point for structural identification. The final determination of the R/S configuration was achieved by analyzing the correlation between the CH3 or CH2 at the C-13′ position and the H-5′ and H-6′ of the adjacent chiral center (Figure 3). Compounds 23 and 24 were used to illustrate the detailed structural determination by irradiating H-14′ (δH 1.40) in compound 23, which resulted in an enhancement of the signal for H-6′ (δH 4.60), suggesting that H-14′ and H-6′ should be placed on the same side of the ring. Conversely, irradiating H-14′ (δH 1.32) in compound 24 enhanced the signal of H-5′ (δH 3.90), indicating that H-14′ and H-5′ should be placed on the same side of the ring. Therefore, the absolute configuration at the C-13′ position of compound 23 was determined to be 13′S, and the absolute configuration at C-13′ position of compound 24 was determined to be 13′R. The absolute configuration of the C-13′ position of the other derivatives (11−16, 25, 26, and 28−31) was confirmed through analysis of the same experiments (Figure 3, Supplementary Figures S1–S98).
Figure 3.
NOE analysis of compounds 23 and 24 and key NOE correlations of other compounds containing ketal groups.
Meanwhile, the absolute configuration at the C-13′ position was also distinguished by changes in the corresponding chemical shift (δ) value of H-5′. In the 1H NMR spectrum, the chemical shifts of H-5′ signals were more affected by the shielding effect compared with the data on compound 1 when the absolute configuration of C-13′ position is S. The chemical shift of H-5′ in the S configuration for derivative 23 shifted from 3.90 to 3.79 ppm. On the contrary, there was no difference regarding the chemical shift of H-5′ in the R configuration for derivative 24 compared with the data of compound 1 (δH 3.90 in 24, δH 3.90 in 1) (Figure 4). The key 1H NMR data (δ) for diastereoisomers are presented in Table 1, and key 13C NMR data (δ) for diastereoisomers are presented in Table S2.
Figure 4.
Stacking of partial 1H NMR spectra of diastereomeric derivatives (with 23 and 24 as examples).
Table 1.
Key 1H NMR data (δ) for diastereoisomers a.
| Compound | 4′ | 5′ | 6′ |
|---|---|---|---|
| 11 | 4.19, m | 3.68, dd (8.5, 2.5) | 4.62, t (8.5) |
| 12 | 4.32, m | 3.93, dd (8.5, 2.3) | 4.49, t (8.5) |
| 13 | 4.20, m | 3.74, dd (8.4, 2.4) | 4.58, t (8.4) |
| 14 | 4.27, m | 3.91, dd 8.4, 2.3) | 4.49, t (8.4) |
| 15 | 4.32, m | 3.73, dd (7.3, 2.2) | 4.70, dd (9.5, 7.3) |
| 16 | 4.22, m | 4.03, dd (8.3, 2.2) | 4.59, t (8.3) |
| 17 | 4.31, m | 3.71, dd (7.3, 2.2) | 4.69, dd (9.5, 7.3) |
| 18 | 4.22, m | 4.02, dd (8.4, 2.2) | 4.58, t (8.4) |
| 19 | 4.34, m | 3.73, dd (7.3, 2.2) | 4.72, dd (9.4, 7.3) |
| 20 | 4.26, m | 4.02, dd (8.4, 2.3) | 4.55, t (8.4) |
| 21 | 4.30, m | 3.69, dd (7.3, 2.2) | 4.69, dd (9.4, 7.3) |
| 22 | 4.22, m | 4.02, dd (8.3, 2.2) | 4.56, t (8.3) |
| 23 | 4.19, m | 3.79, dd (8.5, 2.4) | 4.60, t (8.5) |
| 24 | 4.21, m | 3.90, dd (8.3, 2.1) | 4.58, t (8.3) |
| 25 | 4.23, m | 3.89, dd (8.4, 2.3) | 4.62, t (8.4) |
| 26 | 4.26, m | 3.96, dd (7.6, 2.1) | 4.62, t (7.6) |
| 29 | 4.17, m | 3.71, dd (8.7, 2.5) | 4.59, t (8.7) |
| 30 | 4.20, m | 3.81, dd (8.6, 2.2) | 4.56, t (8.6) |
| 31 | 4.30, m | 3.65, dd (9.0, 2.4) | 4.69, t (9.0) |
| 32 | 4.20, m | 3.94, dd (8.6, 2.2) | 4.53, t (8.6) |
| 33 | 4.32, m | 3.64, dd (8.1, 2.3) | 4.70, t (8.1) |
| 34 | 4.24, m | 3.95, dd (8.7, 2.4) | 4.51, t (8.7) |
| 36 | 4.17, m | 3.67, dd (8.7, 2.5) | 4.57, t (8.7) |
| 37 | 4.25, m | 3.82, dd (8.7, 2.4) | 4.45, t (8.7) |
a Solvent: CDCl3.
2.2. Evaluation of Biological Activity
2.2.1. Anti-M. marinum and Other-Antimicrobial Activity
The treatment period for M. marinum is long and requires multi-drug combination therapy. However, there is a lack of criteria for the optimal antimicrobial regimen and duration of treatment after M. marinum infection [13,38]. In this study, we assessed the antibacterial and antifungal activities of 30 14-membered RAL derivatives. We found that most of the compounds demonstrated significant antibacterial activity, as shown in Table 2. The remaining derivatives (MIC90 > 200 µM) were categorized as less effective, as shown in Table 2. Derivatives 15–24 and 28–35 exhibited promising activity against M. marinum compared to the positive isoniazid with an MIC90 of 40 μM. It is worth noting that most of the compounds displayed a degree of antibacterial selectivity.
Table 2.
Antimicrobial activity of representative compounds of the RALs with ketal groups 1.
| Compound | MIC90 (µM) | |||||
|---|---|---|---|---|---|---|
| M. marinum | S. aureus | E. coli | P. aeruginosa | C. albicans | V. vulnificus | |
| 11 | >200 | >100 | >100 | >100 | >100 | >100 |
| 15 | 80 | 25 | >100 | >100 | >100 | >100 |
| 16 | 80 | >100 | >100 | >100 | >100 | >100 |
| 17 | 80 | >100 | >100 | >100 | >100 | >100 |
| 18 | 70 | >100 | >100 | >100 | >100 | >100 |
| 19 | 70 | >100 | >100 | >100 | >100 | >100 |
| 20 | 70 | >100 | >100 | >100 | >100 | >100 |
| 21 | 80 | 25 | >100 | >100 | >100 | >100 |
| 22 | 80 | >100 | nt | >100 | >100 | nt |
| 23 | 80 | >100 | >100 | >100 | >100 | >100 |
| 24 | 70 | >100 | >100 | >100 | >100 | 25 |
| 28 | 70 | >100 | >100 | >100 | >100 | >100 |
| 29 | 70 | 50 | >100 | >100 | >100 | >100 |
| 30 | 80 | >100 | 25 | >100 | >100 | >100 |
| 31 | 80 | >100 | >100 | >100 | >100 | >100 |
| 32 | 80 | >100 | nt | >100 | >100 | nt |
| 33 | 80 | >100 | nt | >100 | >100 | nt |
| 34 | 90 | >100 | nt | >100 | >100 | nt |
| 35 | 80 | >100 | nt | >100 | >100 | nt |
| Isoniazid | 40 | nt | nt | nt | nt | nt |
| Rifampicin | 10 | nt | nt | nt | nt | nt |
| Ciprofloxacin | nt | 3.13 | 0.10 | 1.56 | nt | nt |
| Amphotericin B | nt | nt | nt | nt | 0.84 | nt |
| Chloramphenicol | nt | nt | nt | nt | nt | 9.75 |
1 Results are the average of three independent experiments, each performed in duplicate. Standard deviations were less than ±10%. nt = not tested.
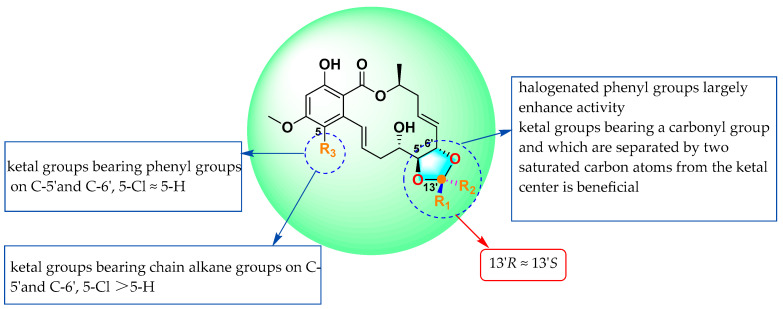
By summarizing the MIC90 data and structures of the above 30 compounds, we obtained preliminary insights into structure–activity relationships (SARs) (Figure 5): (1) through a comparison of compounds 11 and 35, it was found that the 14-membered RALs had no obvious activity after the introduction of ketal groups bearing chain alkane groups, but the activity levels increased significantly after the further introduction of chlorine atoms at C-5; (2) by comparing the MICs of 23, 24, 25, and 26, it was observed that the introduction of ketal groups that bear a carbonyl group and are separated by two saturated carbon atoms from the ketal center at C-5′ and C-6′ significantly enhanced anti-M. marinum activity; (3) by comparing the MICs of compounds 15–22, it was observed that the introduction of ketal groups bearing phenyl groups or phenyl groups with halogen atoms (F/Cl) at C-5′ and C-6′ largely enhanced activity; and (4) after the introduction of ketal groups bearing phenyl groups at C-5′ and C-6′ and the further introduction of chlorine atoms at C-5, the anti-M. marinum activity of the products changed little compared with that before introduction (15–22, 28, and 31–34).
Figure 5.
Summary of the structure–activity relationships.
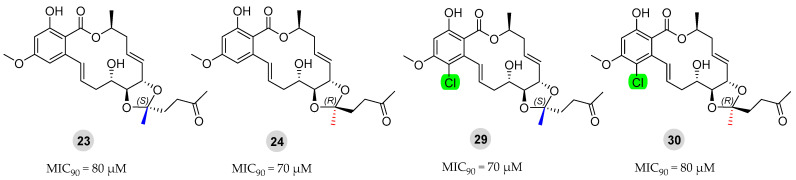
2.2.2. Anti-M. marinum Effects of Compounds 23, 24, 29, and 30 in Combination with Positive Drugs
Currently, the clinical management of M. marinum infections typically involves combination therapy, wherein two or three drugs are administered concurrently [33,39]. Compounds 23, 24, 29, and 30 had significant anti-M. marinum activity, close to that of the positive drug isoniazid, as shown in Figure 6. Compounds 23, 24, 29, and 30 were evaluated in conjunction with two standard antibiotics (isoniazid and rifampicin) using the checkerboard method for drug sensitivity testing [33]. The MIC90 values obtained from these combinations are presented in Table 3 and Table 4.
Figure 6.
Chemical structures of derivatives 23, 24, 29, and 30 and their anti-M. marinum activity.
Table 3.
Anti-M. marinum effects of compounds 23, 24, 29, and 30 in combination with the positive drug isoniazid.
| Isoniazid MIC90 (µM) | Compounds MIC90 (µM) | FICI 1 | Mode of Action | |||
|---|---|---|---|---|---|---|
| Alone | Combined | Alone | Combined | |||
| 23 | 40 | 20 | 80 | 40 | 1 | additive |
| 24 | 40 | 20 | 70 | 8.75 | 0.625 | additive |
| 29 | 40 | 20 | 70 | 8.75 | 0.625 | additive |
| 30 | 40 | 20 | 80 | 40 | 1 | additive |
1 The mode of action was determined via fractional inhibitory concentration index (FICI): (1) FICI ≤ 0.5, synergistic effect; (2) 0.5 < FICI ≤ 1, additive effect; (3) 1 < FICI ≤ 2, irrelevant; (4) FICI > 2, antagonistic effect.
Table 4.
Anti-M. marinum effects of compounds 23, 24, 29, and 30 in combination with the positive drug rifampicin.
| Rifampicin MIC90 (µM) | Compounds MIC90 (µM) | FICI 1 | Mode of Action | |||
|---|---|---|---|---|---|---|
| Alone | Combined | Alone | Combined | |||
| 23 | 10 | 5 | 80 | 20 | 0.75 | additive |
| 24 | 10 | 5 | 70 | 8.75 | 0.625 | additive |
| 29 | 10 | 5 | 70 | 8.75 | 0.625 | additive |
| 30 | 10 | 5 | 80 | 20 | 0.75 | additive |
1 The mode of action was determined via fractional inhibitory concentration index (FICI): (1) FICI ≤ 0.5, synergistic effect; (2) 0.5 < FICI ≤ 1, additive effect; (3) 1 < FICI ≤ 2, irrelevant; (4) FICI > 2, antagonistic effect.
The findings indicate that all the compounds effectively reduced the required dosages of the standard antibiotics while exhibiting antibacterial activity. Notably, at a concentration of 8.75 μM for compounds 24 and 29, M. marinum’s sensitivity to both rifampicin and isoniazid increased twofold, demonstrating a significant addictive effect. It is important to highlight that following combination therapy, the dosages of both compounds and standard antibiotics decreased; this reduction contributed to mitigating resistance in M. marinum to some extent.
3. Materials and Methods
3.1. General Experimental Procedures
Column chromatography (CC) was performed using silica gel (Qingdao Haiyang Chemical Group Co., Qingdao, China; 200–300 mesh) and Sephadex LH-20 (Amersham Biosciences, Amersham, UK). TLC silicone plate (Yantai Zifu Chemical Group Co., Yantai, China; G60, F-254) was used to meet the needs of thin-layer chromatography analysis. Semi-preparative HPLC was performed using a Waters 1525 system with a semi-preparative C18 column (Amsterdam, The Netherlands; Kromasil, 5 µm, 10 × 250 mm) and a Waters 2996 photodiode array detector with a flow rate of 2.0 mL/min. NMR spectra were recorded using a Bruker Advance NEO 400. Chemical shifts δ were measured in ppm, using TMS as the internal standard, and coupling constants (J) were measured in Hz. A Micromass Q-TOF mass spectrometer was used to detect HRESIMS spectra.
3.2. Fungal Material
The fungal strain Cochliobolus lunatus (CHNSCLM-0009) was isolated from a piece of tissue from the inner part of the gorgonian coral Dichotella gemmacea (GX-WZ-20080034) collected from the Weizhou coral reef in the South China Sea in September 2008. Through 16S rRNA gene analysis, the fungus was identified as Clostridium selenospora with the access code ZJ2008002. The fungal strain is currently in storage at the Key Laboratory of Marine Drugs, the Ministry of Education of China, School of Medicine and Pharmacy, Ocean University of China, Qingdao, China.
3.3. Fermentation, Extraction, and Isolation
Cochliobolus lunatus (CHNSCLM-0009) was cultivated under liquid fermentation conditions.
Fermentation was carried out using 500 mL flasks, each filled with 200 mL liquid medium (soluble starch (2 g), NaNO3 (1 g), NaOAc (0.2 g), 1% salinity). The flasks were cultivated at 28 °C for 10 days on a rotary shaker at 120 rpm [20,40]. The fermentation liquid in each flask was extracted three or four times with the same volume of EtOAc. The combined EtOAc solution was evaporated until dryness under a vacuum to obtain 19 g crude extract. Ethyl acetate and petroleum ether were selected as eluents, and the crude extracts were separated via silica gel column chromatography (CC). Zeaenol (38) was obtained at a 4:1 (v/v) ratio of petroleum ether/ethyl acetate eluent. After recrystallization with ethyl acetate/petroleum ether/methanol reagent, 5.4 g of zeaenol (38) was obtained.
3.4. General Synthetic Methods for Compounds 10–37
Here, we describe in detail the steps of synthesizing the new compounds 10–37. Compounds 10–37 were identified using NMR and HRESIMS data, and more details are provided in the Supplementary Data.
3.4.1. General Procedure for the Synthesis of 10–26
Zeaenol (38, 30 mg, 82.33 µmol), ketone reagent (2 mL, Supplementary Table S1), and p-TsOH (trace) in dry CH2Cl2 (2 mL) were stirred at 25 °C for 2–4 h. During the reaction, TLC was used to monitor the reaction progress. The reaction solution was extracted with H2O and CH2Cl2, and the organic layer was evaporated until dry to obtain the crude product. Compounds 10–14 and 17–22 were obtained by separating the crude products on silica gel CC (200–300 mesh) using petroleum ether and ethyl acetate as eluents. After this separation process, we used semipreparative HPLC (65%–85% CH3CN-H2O) to obtain pure compounds 15, 16, and 23–26.
3.4.2. General Procedure for the Synthesis of 28, 31, 33, and 34
Compound 39 (30 mg, 73.36 µmol), ketone reagent (2 mL, Supplementary Table S1), and p-TsOH (trace) in dry CH2Cl2 (2 mL) were stirred at 25 °C for 2–4 h. During the reaction, TLC was used to monitor the reaction progress. The reaction solution was extracted with H2O and CH2Cl2, and the organic layer was evaporated until dry to obtain the crude product. Compounds 28, 31, 33, and 34 were obtained by separating the crude products on silica gel CC (200–300 mesh) using petroleum ether and ethyl acetate as eluents.
3.4.3. General Procedure for the Synthesis of 27, 29, 30, 32, 35, and 37
Compound 10 (30 mg, 69.41 µmol) and SO2Cl2 (10 μL) were dissolved in CH2Cl2 (2 mL). The SO2Cl2 mixture was slowly dripped into the reaction system under ice-bath conditions and stirred for 1–3 h. The reaction was detected via TLC and stopped with ice water. The reaction solution was extracted with H2O and CH2Cl2, and the organic layer was evaporated until dry to obtain the crude product. Compound 27 was obtained by separating the crude products on silica gel CC (200–300 mesh) using petroleum ether and ethyl acetate as eluents. The synthesis and separation of compounds 29, 30, 32, 35, and 37 were the same as those noted above. Notably, in obtaining compounds 29 and 30, we further used semipreparative HPLC (65–85% CH3CN-H2O).
3.4.4. Characterization Data of Compounds 10–37
The planar structures of the new compounds 10–37 were determined using NMR data and HRESIMS spectrum, and the data are as follows. The 1D-NOE was used to determine its spatial structure, and other details were included in the Supplementary Data.
Compound 10: white, solid; yield, 93.0%; 1H NMR (400 MHz, CDCl3) δ 11.50 (1H, s), 7.16 (1H, dd, J = 15.4, 2.4 Hz), 6.47 (1H, d, J = 2.6 Hz), 6.40 (1H, d, J = 2.6 Hz), 6.01 (1H, m), 5.72 (1H, ddd, J = 15.4, 10.5, 3.1 Hz), 5.54–5.43 (2H, overlapped), 4.56 (t, J = 8.4 Hz, 1H), 4.25 (m, 1H), 3.85 (dd, J = 8.4, 2.4 Hz, 1H), 3.82 (s, 3H), 2.76 (m, 1H), 2.58–2.38 (overlapped, 3H), 2.30 (m, 1H), 1.69 (qd, J = 7.5, 1.9 Hz, 2H), 1.61 (d, J = 7.5 Hz, 2H), 1.44 (3H, d, J = 6.4 Hz), 0.94 (3H, t, J = 7.5 Hz), 0.87 (3H, t, J = 7.5 Hz). 13C NMR (100 MHz, CDCl3) δ 170.9 (C), 164.9 (C), 164.1 (C), 142.3 (C), 134.2 (CH), 132. (CH), 129.7 (CH), 126.5 (CH), 112.3 (C), 107.3 (CH), 104.6 (C), 100.2 (CH), 81.6 (CH), 75.5 (CH), 70.7 (CH), 69.2 (CH), 55.6 (OCH3), 38.1 (CH2), 36.1 (CH2), 30.5 (CH2), 30.0 (CH2), 19.3 (CH3), 8.4 (CH3), 8.2 (CH3). HRESIMS m/z 433.2221 [M + H]+ (calcd for C24H33O7+, 433.2221).
Compound 11: white, solid; yield, 79.4%; 1H NMR (400 MHz, CDCl3) δ 11.48 (1H, s), 7.14 (1H, dd, J = 15.4, 2.4 Hz), 6.45 (1H, d, J = 2.6 Hz), 6.39 (1H, d, J = 2.6 Hz), 6.05 (1H, m), 5.68 (1H, ddd, J = 15.4, 10.3, 3.0 Hz), 5.53–5.38 (2H, overlapped), 4.62 (1H, t, J = 8.5 Hz), 4.19 (1H, m), 3.81 (3H, s), 3.68 (1H, dd, J = 8.5, 2.5 Hz), 2.75 (1H, m), 2.58–2.39 (3H, overlapped), 2.32 (1H, m), 1.44 (3H, d, J = 6.4 Hz), 1.35 (3H, s), 0.93 (9H, s). 13C NMR (100 MHz, CDCl3) δ 170.9 (C), 164.8 (C), 164.1 (C), 142.5 (C), 134.2 (CH), 131.2 (CH), 130.0 (CH), 126.5 (CH), 113.9 (C), 107.4 (CH), 104.6 (C), 100.2 (CH), 82.9 (CH), 75.1 (CH), 70.7 (CH), 68.7 (CH), 55.6 (OCH3), 39.3 (CH2), 38.2 (CH2), 35.7 (C), 25.4 (CH3 × 3), 21.1 (CH3), 19.4 (CH3). HRESIMS m/z 447.2367 [M + H]+ (calcd for C25H35O7+, 447.2377).
Compound 12: white, solid; yield, 24.1%; 1H NMR (400 MHz, CDCl3) δ 11.49 (1H, s), 7.16 (1H, dd, J = 15.4, 2.4 Hz), 6.48 (1H, d, J = 2.6 Hz), 6.40 (1H, d, J = 2.6 Hz), 5.98 (1H, m), 5.73 (1H, ddd, J = 15.4, 10.4, 3.0 Hz), 5.55–5.42 (2H, overlapped), 4.49 (1H, t, J = 8.5 Hz), 4.32 (1H, m), 3.93 (1H, dd, J = 8.5, 2.3 Hz), 3.82 (3H, s), 2.75 (1H, m), 2.57–2.38 (3H, overlapped), 2.30 (1H, m), 1.44 (3H, d, J = 6.4 Hz), 1.28 (3H, s), 1.00 (9H, s). 13C NMR (100 MHz, CDCl3) δ 170.9 (C), 164.9 (C), 164.1 (C), 142.2 (C), 134.2 (CH), 133.7 (CH), 129.3 (CH), 126.4 (CH), 113.5 (C), 107.3 (CH), 104.6 (C), 100.2 (CH), 80.8 (CH), 75.9 (CH), 70.6 (CH), 69.7 (CH), 55.6 (OCH3), 38.8 (CH2), 38.1 (CH2), 36.3 (C), 25.6 (CH3 × 3), 20.6 (CH3), 19.3 (CH3). HRESIMS m/z 447.2372 [M + H]+ (calcd for C25H35O7+, 447.2377).
Compound 13: white, solid; yield, 69.7%; 1H NMR (400 MHz, CDCl3) δ 11.50 (1H, s), 7.15 (1H, dd, J = 15.4, 2.4 Hz), 6.46 (1H, d, J = 2.6 Hz), 6.40 (1H, d, J = 2.6 Hz), 6.02 (1H, m), 5.70 (1H, ddd, J = 15.4, 10.4, 3.1 Hz), 5.53–5.39 (2H, overlapped), 4.58 (1H, t, J = 8.4 Hz), 4.20 (1H, m), 3.81 (3H, s), 3.74 (1H, dd, J = 8.4, 2.4 Hz), 2.75 (1H, m), 2.64–2.38 (3H, overlapped), 2.30 (1H, m), 1.83–1.60 (6H, overlapped), 1.44 (3H, d, J = 6.4 Hz), 1.32 (3H, s), 1.22–0.96 (5H, overlapped). 13C NMR (100 MHz, CDCl3) δ 170.9 (C), 164.9 (C), 164.1 (C), 142.4 (C), 134.2 (CH), 132.0 (CH), 129.8 (CH), 126.6 (CH), 112.0 (C), 107.3 (CH), 104.6 (C), 100.2 (CH), 82.1 (CH), 75.0 (CH), 70.7 (CH), 68.8 (CH), 55.6 (OCH3), 47.3 (CH2), 38.1 (CH2), 35.9 (C), 27.7 (CH), 27.5 (CH), 26.4 (CH), 26.4 (CH), 26.3 (CH), 22.6 (CH3), 19.4 (CH3). HRESIMS m/z 473.2522 [M + H]+ (calcd for C27H37O7+, 473.2534).
Compound 14: white, solid; yield, 33.2%; 1H NMR (400 MHz, CDCl3) δ 11.50 (1H, s), 7.16 (1H, dd, J = 15.4, 2.3 Hz), 6.47 (1H, d, J = 2.6 Hz), 6.40 (1H, d, J = 2.6 Hz), 5.98 (1H, m), 5.73 (1H, ddd, J = 15.4, 10.5, 3.1 Hz), 5.56–5.40 (2H, overlapped), 4.49 (1H, t, J = 8.4 Hz), 4.27 (1H, m), 3.91 (1H, dd, J = 8.4, 2.3 Hz), 3.81 (3H, s), 2.75 (1H, m), 2.56–2.37 (3H, overlapped), 2.29 (1H, m), 1.89–1.65 (6H, overlapped), 1.44 (3H, d, J = 6.4 Hz), 1.25 (3H, s), 1.22–1.01 (5H, overlapped). 13C NMR (101 MHz, CDCl3) δ 170.9 (C), 164.9 (C), 164.1 (C), 142.2 (C), 134.2 (CH), 133.4 (CH), 129.5 (CH), 126.5 (CH), 111.5 (C), 107.3 (CH), 104.6 (C), 100.2 (CH), 80.9 (CH), 75.5 (CH), 70.7 (CH), 69.4 (CH), 55.6 (OCH3), 47.3 (CH2), 38.0 (CH2), 36.2 (CH), 27.7 (CH2), 27.6 (CH2), 26.4 (CH2×2), 26.3 (CH2), 22.6 (CH3), 19.3 (CH3). HRESIMS m/z 473.2523 [M + H]+ (calcd for C27H37O7+, 473.2534).
Compound 15: white, solid; yield, 63.7%; 1H NMR (400 MHz, CDCl3) δ 11.49 (1H, s), 7.45–7.42 (2H, overlapped), 7.23–7.27 (3H, overlapped), 7.02 (1H, dd, J = 15.4, 2.4 Hz), 6.43 (1H, d, J = 2.6 Hz), 6.35 (1H, d, J = 2.6 Hz), 5.94 (1H, m), 5.68 (1H, ddd, J = 15.4, 10.5, 3.0 Hz), 5.42 (1H, m), 5.11 (1H, m), 4.70 (1H, dd, J = 9.5, 7.3 Hz), 4.32 (1H, m), 3.79 (3H, s), 3.73 (1H, dd, J = 7.3, 2.2 Hz), 2.79 (1H, m), 2.68 (1H, s), 2.39–2.19 (3H, overlapped), 1.65 (3H, s), 1.41 (3H, d, J = 6.5 Hz). 13C NMR (125 MHz, CDCl3) δ 170.6 (C), 164.9 (C), 164.0 (C), 144.4 (C), 141.9 (C), 134.0 (CH), 133.5 (CH), 129.0 (CH), 128.4 (CH × 2), 128.1 (CH), 125.7 (CH), 125.4 (CH × 2), 109.2 (C), 107.1 (CH), 104.4 (C), 100.2 (CH), 81.6 (CH), 76.5 (CH), 70.3 (CH), 69.1 (CH), 55.6 (OCH3), 37.8 (CH2), 36.3 (CH2), 29.1 (CH3), 19.0 (CH3). HRESIMS m/z 467.2081 [M + H]+ (calcd for C27H29O7+, 467.2064).
Compound 16: white, solid; yield, 43.1%; 1H NMR (400 MHz, CDCl3) δ 11.50 (1H, s), 7.51–7.48 (2H, overlapped), 7.40–7.36 (2H, overlapped), 7.30 (1H, m), 7.15 (1H, dd, J = 15.4, 2.3 Hz), 6.46 (1H, d, J = 2.6 Hz), 6.40 (1H, d, J = 2.6 Hz), 6.08 (1H, m), 5.71 (1H, ddd, J = 15.4, 10.6, 3.1 Hz), 5.62 (1H, m), 5.46 (1H, m), 4.59 (1H, t, J = 8.3 Hz), 4.22 (1H, m), 4.03 (1H, dd, J = 8.3, 2.2 Hz), 3.82 (3H, s), 2.65 (1H, m), 2.61–2.42 (2H, overlapped), 2.19 (1H, m), 1.90 (1H, m), 1.65 (3H, s), 1.45 (3H, d, J = 6.4 Hz). 13C NMR (125 MHz, CDCl3) δ 170.9 (C), 164.9 (C), 164.1 (C), 144.6 (C), 142.3 (C), 134.1 (CH), 131.8 (CH), 130.5 (CH), 128.9 (CH × 2), 128.3 (CH), 126.8 (CH), 124.6 (CH × 2), 109.1 (C), 107.4 (CH), 104.5 (C), 100.2 (CH), 82.9 (CH), 75.7 (CH), 70.7 (CH), 69.4 (CH), 55.6 (OCH3), 38.1 (CH2), 35.9 (CH2), 29.2 (CH3), 19.4 (CH3). HRESIMS m/z 467. 2057 [M + H]+ (calcd for C27H31O7+, 467.2064).
Compound 17: white, solid; yield, 64.8%; 1H NMR (400 MHz, CDCl3) δ 11.49 (1H, s), 7.43–7.39 (2H, overlapped), 7.03 (1H, dd, J = 15.4, 2.4 Hz), 7.01– 6.95 (2H, overlapped), 6.44 (1H, d, J = 2.6 Hz), 6.35 (1H, d, J = 2.6 Hz), 5.95 (1H, m), 5.68 (1H, ddd, J = 15.4, 10.5, 3.0 Hz), 5.43 (1H, m), 5.10 (1H, m), 4.69 (1H, dd, J = 9.5, 7.3 Hz), 4.31 (1H, m), 3.79 (3H, s), 3.71 (1H, dd, J = 7.3, 2.2 Hz), 2.79 (1H, m), 2.67 (1H, s), 2.38–2.35 (2H, overlapped), 2.25 (1H, m), 1.63 (3H, s), 1.41 (3H, d, J = 6.5 Hz). 13C NMR (100 MHz, CDCl3) δ 170.6 (C), 165.0 (C), 164.0 (C), 162.6 (C, d, J = 244.1 Hz), 141.8 (C), 140.3 (C, d, J = 3.2 Hz), 134.0 (CH), 133.3 (CH), 129.2 (CH), 127.3 (CH × 2, d, J = 8.1 Hz), 125.6 (CH), 115.2 (CH × 2, d, J = 21.5 Hz), 108.8 (C), 107.2 (CH), 104.4 (C), 100.2 (CH), 81.7 (CH), 76.5 (CH), 70.2 (CH), 69.0 (CH), 55.5 (OCH3), 37.8 (CH2), 36.3 (CH2), 29.2 (CH3), 19.0 (CH3). HRESIMS m/z 485.1974 [M + H]+ (calcd for C27H31O7F+, 485.1970).
Compound 18: white, solid; yield, 41.7%; 1H NMR (400 MHz, CDCl3) δ 11.49 (1H, s), 7.43–7.39 (2H, overlapped), 7.16 (1H, dd, J = 15.4, 2.3 Hz), 7.01–6.95 (2H, overlapped), 6.46 (1H, d, J = 2.6 Hz), 6.40 (1H, d, J = 2.6 Hz), 6.07 (1H, m), 5.71 (1H, ddd, J = 15.4, 10.5, 3.1 Hz), 5.60 (1H, m), 5.47 (1H, m), 4.58 (1H, t, J = 8.4 Hz), 4.22 (1H, m), 4.02 (1H, dd, J = 8.4, 2.2 Hz), 3.82 (3H, s), 2.67 (1H, m), 2.61–2.44 (2H, overlapped), 2.20 (1H, m), 1.90 (1H, s), 1.62 (3H, s), 1.45 (3H, d, J = 6.4 Hz). 13C NMR (100 MHz, CDCl3) δ 170.9 (C), 164.9 (C), 164.1 (C), 162.6 (C, d, J = 244.9 Hz), 142.3 (C), 140.4 (C, d, J = 3.5 Hz), 134.2 (CH), 131.6 (CH), 130.6 (CH), 126.7 (CH), 126.5 (CH × 2, d, J = 8.4 Hz), 115.7 (CH × 2, d, J = 21.5 Hz), 108.7 (C), 107.4 (CH), 104.5 (C), 100.2 (CH), 82.9 (CH), 75.8 (CH), 70.7 (CH), 69.2 (CH), 55.6 (OCH3), 38.0 (CH2), 35.9 (CH2), 29.2 (CH3), 19.4 (CH3). HRESIMS m/z 485.1973 [M + H]+ (calcd for C17H30O7F+, 485.1970).
Compound 19: white, solid; yield, 65.4%; 1H NMR (400 MHz, CDCl3) δ 11.48 (1H, s), 7.60 (1H, m), 7.34 (1H, m), 7.22–7.18 (2H, overlapped), 7.04 (1H, dd, J = 15.4, 2.4 Hz), 6.45 (1H, d, J = 2.6 Hz), 6.35 (1H, d, J = 2.6 Hz), 5.95 (1H, m), 5.71 (1H, ddd, J = 15.4, 10.5, 3.0 Hz), 5.42 (1H, m), 5.10 (1H, m), 4.72 (1H, dd, J = 9.4, 7.3 Hz), 4.34 (1H, m), 3.79 (3H, s), 3.73 (1H, dd, J = 7.3, 2.2 Hz), 2.80 (1H, m), 2.68 (1H, s), 2.40–2.33 (2H, overlapped), 2.26 (1H, m), 1.78 (3H, s), 1.41 (3H, d, J = 6.5 Hz). 13C NMR (100 MHz, CDCl3) δ 170.7 (C), 164.9 (C), 164.0 (C), 141.9 (C), 140.6 (C), 134.0 (C), 132.9 (CH), 132.0 (CH), 131.6 (CH), 129.6 (CH), 129.3 (CH), 127.9 (CH), 126.8 (CH), 125.9 (CH), 108.7 (CH), 107.2 (CH), 104.4 (C), 100.2 (CH), 81.6 (CH), 76.3 (CH), 70.3 (CH), 69.0 (CH), 55.6 (OCH3), 37.8 (CH2), 36.4 (CH2), 26.5 (CH3), 19.0 (CH3). HRESIMS m/z 501.1692 [M + H]+ (calcd for C27H30O7Cl+, 501.1675).
Compound 20: white, solid; yield, 33.8%; 1H NMR (400 MHz, CDCl3) δ 11.49 (1H, s), 7.65 (1H, dd, J = 7.5, 2.0 Hz), 7.40 (1H, dd, J = 7.5, 1.7 Hz), 7.29 (1H, dd, J = 7.4, 1.7 Hz), 7.24 (1H, dd, J = 7.4, 2.1 Hz), 7.14 (1H, dd, J = 15.3, 2.3 Hz), 6.45 (1H, d, J = 2.6 Hz), 6.40 (1H, d, J = 2.6 Hz), 6.10 (1H, m), 5.75–5.56 (2H, overlapped), 5.48 (1H, m), 4.55 (1H, t, J = 8.4 Hz), 4.26 (1H, m), 4.02 (1H, dd, J = 8.4, 2.3 Hz), 3.82 (3H, s), 2.65 (1H, m), 2.60–2.45 (2H, overlapped), 2.19 (1H, m), 1.93 (1H, s), 1.77 (3H, s), 1.45 (3H, d, J = 6.4 Hz). 13C NMR (100 MHz, CDCl3) δ 170.9 (C), 164.9 (C), 164.1 (C), 142.3 (C), 140.9 (C), 134.2 (C), 131.8 (CH), 131.4 (CH), 131.3 (CH), 130.9 (CH), 129.7 (CH), 127.4 (CH), 127.0 (CH), 126.5 (CH), 108.8 (C), 107.4 (CH), 104.5 (C), 100.2 (CH), 82.9 (CH), 75.5 (CH), 70.6 (CH), 69.2 (CH), 55.6 (OCH3), 38.1 (CH2), 35.8 (CH2), 27.0 (CH3), 19.4 (CH3). HRESIMS m/z 501.1672 [M + H]+ (calcd for C27H30OCl+, 501.1675).
Compound 21: white, solid; yield, 56.6%; 1H NMR (400 MHz, CDCl3) δ 11.49 (1H, s), 7.39–7.36 (2H, overlapped), 7.29–7.26 (2H, overlapped), 7.03 (1H, dd, J = 15.4, 2.4 Hz), 6.43 (1H, d, J = 2.6 Hz), 6.35 (1H, d, J = 2.6 Hz),5.95 (1H, m), 5.68 (1H, ddd, J = 15.4, 10.5, 3.0 Hz), 5.44 (1H, m), 5.09 (1H, dd, J = 15.2, 9.4, 1.3 Hz), 4.69 (1H, dd, J = 9.4, 7.3 Hz), 4.30 (1H, m), 3.79 (3H, s), 3.69 (1H, dd, J = 7.3, 2.2 Hz), 2.79 (1H, m), 2.63 (1H, s), 2.38–2.19 (3H, overlapped), 1.62 (3H, s), 1.41 (3H, d, J = 6.5 Hz). 13C NMR (100 MHz, CDCl3) δ 170.6 (C), 165.0 (C), 164.0 (C), 143.0 (C), 141.8 (C), 134.1 (C), 134.0 (CH), 133.3 (CH), 129.3 (CH), 128.6 (CH × 2), 127.0 (CH × 2), 125.6 (CH), 108.7 (C), 107.2 (CH), 104.4 (C), 100.2 (CH), 81.7 (CH), 76.5 (CH), 70.2 (CH), 69.0 (CH), 55.6 (CH), 37.8 (OCH3), 36.3 (CH2), 29.0 (CH3), 19.0 (CH3). HRESIMS m/z 501.1685 [M + H]+ (calcd for C27H30O7Cl+, 501.1675).
Compound 22: white, solid; yield, 32.3%; 1H NMR (400 MHz, CDCl3) δ 11.49 (1H, s), 7.45–7.42 (2H, overlapped), 7.36–7.33 (2H, overlapped), 7.16 (1H, dd, J = 15.4, 2.3 Hz), 6.46 (1H, d, J = 2.6 Hz), 6.40 (1H, d, J = 2.5 Hz), 6.07 (1H, m), 5.71 (1H, ddd, J = 15.4, 10.5, 3.1 Hz), 5.60 (1H, m), 5.46 (1H, m), 4.56 (1H, t, J = 8.3 Hz), 4.22 (1H, m), 4.02 (1H, dd, J = 8.3, 2.2 Hz), 3.82 (3H, s), 2.66 (1H, m), 2.59–2.44 (2H, overlapped), 2.20 (1H, m), 1.88 (1H, d, J = 1.5 Hz), 1.61 (3H, s), 1.44 (3H, d, J = 6.3 Hz). 13C NMR (101 MHz, CDCl3) δ 170.9 (C), 164.9 (C), 164.1 (C), 143.0 (C), 142.3 (C), 134.2 (C), 134.1 (CH), 131.5 (CH), 130.7 (CH), 129.0 (CH × 2), 126.7 (CH), 126.2 (CH × 2), 108.6 (C), 107.4 (CH), 104.5 (C), 100.2 (CH), 82.9 (CH), 75.9 (CH), 70.7 (CH), 69.2 (CH), 55.6 (OCH3), 38.1 (CH2), 35.9 (CH2), 29.1 (CH3 ), 19.5 (CH3 ). HRESIMS m/z 501.1684 [M + H]+ (calcd for C27H30O7Cl+, 501.1675).
Compound 23: white, solid; yield, 52.8%; 1H NMR (400 MHz, CDCl3) δ 11.50 (1H, s), 7.15 (1H, dd, J = 15.4, 2.3 Hz), 6.46 (1H, d, J = 2.6 Hz), 6.40 (1H, d, J = 2.6 Hz), 6.03 (1H, m), 5.70 (1H, ddd, J = 15.4, 10.4, 3.0 Hz), 5.52–5.42 (2H, overlapped), 4.60 (1H, t, J = 8.5 Hz), 4.19 (1H, m), 3.82 (3H, s), 3.79 (1H, dd, J = 8.5, 2.4 Hz), 2.76 (1H, m), 2.58–2.38 (5H, overlapped), 2.29 (1H, m), 2.13 (3H, s), 1.93 (2H, t, J = 6.4 Hz), 1.44 (3H, d, J = 6.4 Hz), 1.40 (3H, s). 13C NMR (100 MHz, CDCl3) δ 208.2 (C), 170.9 (C), 164.9 (C), 164.1 (C), 142.2 (C), 134.3 (CH), 131.8 (CH), 130.3 (CH), 126.3 (CH), 109.4 (C), 107.4 (CH), 104.5 (C), 100.2 (CH), 82.3 (CH), 75.4 (CH), 70.6 (CH), 68.8 (CH), 55.6 (OCH3), 38.4 (CH2), 38.0 (CH2), 36.0 (CH2), 33.8 (CH3), 30.1 (CH2), 25.5 (CH3), 19.3 (CH3). HRESIMS m/z 461.2157 [M + H]+ (calcd for C25H33O8+, 461.2170).
Compound 24: white, solid; yield, 35.2%; 1H NMR (400 MHz, CDCl3) δ 11.50 (1H, s), 7.15 (1H, dd, J = 15.4, 2.3 Hz), 6.47 (1H, d, J = 2.6 Hz), 6.39 (1H, d, J = 2.6 Hz), 5.99 (1H, m), 5.73 (1H, ddd, J = 15.4, 10.6, 3.1 Hz), 5.57–5.41 (2H, overlapped), 4.58 (1H, t, J = 8.3 Hz), 4.21 (1H, m), 3.90 (1H, dd, J = 8.3, 2.1 Hz), 3.81 (3H, s), 2.75 (1H, m), 2.65–2.35 (5H, overlapped), 2.27 (1H, m), 2.17 (3H, s), 2.11 (1H, m), 1.96 (1H, m), 1.44 (3H, d, J = 6.4 Hz), 1.32 (3H, s). 13C NMR (100 MHz, CDCl3) δ 210.2 (C), 170.9 (C), 164.9 (C), 164.1 (C), 142.2 (C), 134.0 (CH), 133.1 (CH), 129.6 (CH), 126.8 (CH), 109.3 (C), 107.3 (CH), 104.5 (C), 100.2 (CH), 81.9 (CH), 77.4 (CH), 70.7 (CH), 69.0 (CH), 55.6 (OCH3), 38.5 (CH2), 38.0 (CH2), 36.6 (CH2), 33.7 (CH3), 29.8 (CH2), 25.8 (CH3), 19.3 (CH3). HRESIMS m/z 461.2155 [M + H]+ (calcd for C2H33O8+, 461.2170).
Compound 25: white, solid; yield, 54.4%; 1H NMR (400 MHz, CDCl3) δ 11.48 (1H, s), 7.15 (1H, dd, J = 15.4, 2.3 Hz), 6.46 (1H, d, J = 2.6 Hz), 6.40 (1H, d, J = 2.6 Hz), 6.03 (1H, m), 5.70 (1H, ddd, J = 15.4, 10.5, 3.0 Hz), 5.55–5.39 (2H, overlapped), 4.62 (1H, t, J = 8.4 Hz), 4.23 (1H, m), 3.89 (1H, dd, J = 8.4, 2.3 Hz), 3.81 (3H, s), 2.75 (3H, m), 2.57–2.38 (3H, overlapped), 2.29 (1H, m), 2.17 (3H, s), 1.48 (3H, s), 1.44 (3H, d, J = 6.4 Hz). 13C NMR (100 MHz, CDCl3) δ 205.6 (C), 170.9 (C), 164.9 (C), 164.1 (C), 142.2 (C), 134.3 (CH), 131.7 (CH), 130.4 (CH), 126.4 (CH), 107.8 (C), 107.4 (CH), 104.5 (C), 100.2 (CH), 82.1 (CH), 75.5 (CH), 70.6 (CH), 68.6 (CH), 55.6 (OCH3), 53.4 (CH2), 38.0 (CH2), 36.0 (CH2), 32.0 (CH3), 25.8 (CH3), 19.3 (CH3). HRESIMS m/z 447.2026 [M + H]+ (calcd for C24H31O8+, 447.2013).
Compound 26: white, solid; yield, 36.3%; 1H NMR (400 MHz, CDCl3) δ 11.48 (1H, s), 7.14 (1H, dd, J = 15.4, 2.4 Hz), 6.48 (1H, d, J = 2.6 Hz), 6.39 (1H, d, J = 2.6 Hz), 6.01 (1H, m), 5.76 (1H, ddd, J = 15.4, 10.6, 3.1 Hz), 5.54–5.41 (2H, overlapped), 4.62 (1H, t, J = 7.6 Hz), 4.26 (1H, m), 3.96 (1H, dd, J = 7.6, 2.1 Hz), 3.81 (3H, s), 2.97 (1H, d, J = 14.1 Hz), 2.82–2.71 (2H, overlapped), 2.57–2.37 (2H, overlapped), 2.28–2.18 (5H, overlapped), 1.44 (3H, d, J = 6.4 Hz), 1.40 (3H, s). 13C NMR (100 MHz, CDCl3) δ 207.2 (C), 170.9 (C), 164.9 (C), 164.1 (C), 142.2 (C), 133.9 (CH), 133.0 (CH), 130.0 (CH), 126.8 (CH), 107.7 (C), 107.2 (CH), 104.5 (C), 100.2 (CH), 82.1 (CH), 76.0 (CH), 70.5 (CH), 69.1 (CH), 55.6 (OCH3), 51.2 (CH2), 37.9 (CH2), 36.6 (CH2), 32.9 (CH3), 27.3 (CH3), 19.2 (CH3). HRESIMS m/z 447.1998 [M + H]+ (calcd for C24H31O8+, 447.2013).
Compound 27: white, solid; yield, 58.8%; 1H NMR (400 MHz, CDCl3) δ 11.39 (1H, s), 6.63 (1H, dd, J = 16.2, 1.3 Hz), 6.47 (1H, s), 6.00 (1H, m), 5.54–5.33 (3H, overlapped), 4.54 (1H, t, J = 8.6 Hz), 4.23 (1H, m), 3.91 (3H, s), 3.77 (1H, dd, J = 8.6, 2.5 Hz), 2.86 (1H, m), 2.62–2.31 (4H, overlapped), 1.75–1.60 (4H, overlapped), 1.39 (3H, d, J = 6.4 Hz), 0.93 (3H, t, J = 7.5 Hz), 0.88 (3H, t, J = 7.5 Hz). 13C NMR (100 MHz, CDCl3) δ 170.7 (C), 162.5 (C), 160.0 (C), 140.0 (C), 131.1 (CH), 130.7 (CH), 130.5 (CH), 129.0 (CH), 114.1 (C), 112.5 (CH), 106.4 (C), 99.5 (CH), 81.2 (CH), 75.7 (CH), 71.5 (CH), 68.6 (CH), 56.6 (OCH3), 37.2 (CH2), 34.8 (CH2), 30.5 (CH2), 30.3 (CH2), 19.4 (CH3), 8.3 (CH3), 8.3 (CH3). HRESIMS m/z 467.1817 [M + H]+ (calcd for C24H32O7Cl+, 467.1831).
Compound 28: white, solid; yield, 54.5%; 1H NMR (400 MHz, CDCl3) δ 11.45 (1H, s), 7.40–7.37 (2H, overlapped), 7.29–7.27 (2H, overlapped), 6.56 (1H, d, J = 14.8 Hz), 6.42 (1H, s), 5.96 (1H, m), 5.43 (1H, ddd, J = 14.8, 8.5, 3.8 Hz), 5.35 (1H, m), 5.17 (1H, m), 4.68 (1H, t, J = 8.1 Hz), 4.29 (1H, m), 3.88 (3H, s), 3.62 (1H, dd, J = 8.1, 2.3 Hz), 2.90 (1H, m), 2.63 (1H, m), 2.51 (1H, m), 2.42–2.25 (2H, overlapped), 1.62 (3H, s), 1.37 (3H, d, J = 6.5 Hz). 13C NMR (100 MHz, CDCl3) δ 170.5 (C), 162.6 (C), 160.0 (C), 143.0 (C), 139.7 (C), 134.0 (C), 131.7 (CH), 130.2 (CH), 130.1 (CH), 129.1 (CH), 128.6 (CH × 2), 126.8 (CH × 2), 114.0 (C), 108.7 (CH), 106.2 (C), 99.6 (CH), 81.1 (CH), 76.7 (CH), 71.3 (CH), 68.4 (CH), 56.6 (OCH3), 37.1 (CH2), 34.8 (CH2), 28.9 (CH3), 19.1 (CH3). HRESIMS m/z 535.1283 [M + H]+ (calcd for C27H29O7Cl2+, 535.1285).
Compound 29: white, solid; yield, 53.9%; 1H NMR (400 MHz, CDCl3) δ 11.41 (1H, s), 6.62 (1H, d, J = 16.1 Hz), 6.47 (1H, s), 6.02 (1H, m), 5.54–5.34 (3H, overlapped), 4.59 (1H, t, J = 8.7 Hz), 4.17 (1H, m), 3.91 (3H, s), 3.71 (1H, dd, J = 8.7, 2.5 Hz), 2.86 (1H, m), 2.62–2.47 (4H, overlapped), 2.47–2.31 (2H, overlapped), 2.13 (3H, s), 1.95 (2H, t, J = 7.4 Hz), 1.40–1.38 (6H, overlapped). 13C NMR (125 MHz, CDCl3) δ 208.3 (C), 170.6 (C), 162.5 (C), 160.0 (C), 139.9 (C), 131.1 (CH), 130.4 (CH), 130.2 (CH), 129.1 (CH), 114.1 (C), 109.5 (CH), 106.4 (C), 99.6 (CH), 81.8 (CH), 75.6 (CH), 71.4 (CH), 68.2 (CH), 56.6 (OCH3), 38.4 (CH2), 37.2 (CH2), 34.7 (CH2), 33.9 (CH3), 30.1 (CH2), 25.6 (CH3), 19.4 (CH3). HRESIMS m/z 495.1765 [M + H]+ (calcd for C25H32O8Cl+, 495.1780).
Compound 30: white, solid; yield, 32.3%; 1H NMR (400 MHz, CDCl3) δ 11.37 (1H, s), 6.63 (1H, dd, J = 16.1, 1.6 Hz), 6.46 (1H, s), 5.98 (1H, m), 5.52 (1H, ddd, J = 16.1, 9.1, 3.5 Hz), 5.47–5.34 (2H, overlapped), 4.56 (1H, t, J = 8.6 Hz), 4.20 (1H, m), 3.91 (3H, s), 3.81 (1H, dd, J = 8.6, 2.2 Hz), 2.87 (1H, m), 2.61–2.51 (4H, overlapped), 2.46–2.28 (2H, overlapped), 2.17 (3H, s), 2.10 (1H, m), 1.96 (1H, m), 1.96 (3H, d, J = 6.3 Hz), 1.33 (3H, s). 13C NMR (100 MHz, CDCl3) δ 209.8 (C), 170.7 (C), 162.4 (C), 160.0 (C), 140.0 (C), 131.4 (CH), 131.1 (CH), 130.4 (CH), 128.9 (CH), 114.1 (C), 109.4 (CH), 106.5 (C), 99.5 (CH), 81.4 (CH), 75.9 (CH), 71.4 (CH), 68.5 (CH), 56.6 (OCH3), 38.5 (CH2), 37.2 (CH2), 35.4 (CH2), 33.7 (CH3), 29.9 (CH2), 25.7 (CH3), 19.5 (CH3). HRESIMS m/z 495.1761 [M + H]+ (calcd for C25H32O8Cl+, 495.1780).
Compound 31: white, solid; yield, 58.5%; 1H NMR (400 MHz, CDCl3) δ 11.44 (1H, s), 7.49–7.41 (2H, overlapped), 7.35–7.27 (3H, overlapped), 6.55 (1H, d, J = 17.4 Hz), 6.42 (1H, s), 5.95 (1H, m), 5.43 (1H, ddd, J = 16.1, 8.5, 3.8 Hz), 5.33 (1H, m), 5.19 (1H, m), 4.69 (1H, t, J = 9.0 Hz), 4.30 (1H, m), 3.88 (3H, s), 3.65 (1H, dd, J = 9.0, 2.4 Hz), 2.90 (1H, m), 2.67 (1H, s), 2.50 (1H, m), 2.40–2.27 (2H, overlapped), 1.65 (3H, s), 1.37 (3H, d, J = 6.5 Hz). 13C NMR (100 MHz, CDCl3) δ 170.5 (C), 162.6 (C), 160.0 (C), 144.3 (C), 139.8 (C), 131.9 (CH), 130.3 (CH), 130.0 (CH), 129.0 (CH), 128.4 (CH × 2), 128.1 (CH), 125.3 (CH × 2), 114.0 (C), 109.1 (CH), 106.2 (C), 99.5 (CH), 81.0 (CH), 76.6 (CH), 71.3 (CH), 68.4 (CH), 56.5 (OCH3), 37.1 (CH2), 34.8 (CH2), 29.0 (CH3), 19.1 (CH3). HRESIMS m/z 501.1664 [M + H]+ (calcd for C27H30O7Cl+, 501.1675).
Compound 32: white, solid; yield, 37.6%; 1H NMR (400 MHz, CDCl3) δ 11.35 (1H, s), 7.49 (2H, d, J = 7.5 Hz), 7.38 (2H, t, J = 7.5 Hz), 7.31 (1H, d, J = 7.2 Hz), 6.62 (1H, dd, J = 16.1, 2.4 Hz), 6.47 (1H, s), 6.04 (1H, m), 5.58–5.41 (3H, overlapped), 4.53 (1H, t, J = 8.6 Hz), 4.20 (1H, m), 3.94 (1H, dd, J = 8.6, 2.2 Hz), 3.91 (3H, s), 2.76 (1H, m), 2.65–2.41 (3H, overlapped), 2.23 (1H, m), 1.65 (3H, s), 1.39 (3H, d, J = 6.3 Hz). 13C NMR (100 MHz, CDCl3) δ 170.7 (C), 162.4 (C), 160.0 (C), 144.6 (C), 140.0 (C), 131.3 (CH), 130.9 (CH), 130.0 (CH), 128.9 (CH × 3), 128.2 (CH), 124.5 (CH × 2), 114.1 (C), 109.1 (CH), 106.5 (C), 99.5 (CH), 82.4 (CH), 75.9 (CH), 71.3 (CH), 68.8 (CH), 56.6 (OCH3), 37.2 (CH2), 34.7 (CH2), 29.3 (CH3), 19.6 (CH3). HRESIMS m/z 501.1662 [M + H]+ (calcd for C27H30O7Cl+, 501.1675).
Compound 33: white, solid; yield, 55.7%; 1H NMR (400 MHz, CDCl3) δ 11.41 (1H, s), 7.62 (1H, dd, J = 6.0, 3.6 Hz), 7.35 (1H, dd, J = 6.0, 3.6 Hz), 7.24–7.20 (2H, overlapped), 6.56 (1H, dd, J = 16.1, 1.4 Hz), 6.42 (1H, s), 5.95 (1H, m), 5.45 (1H, ddd, J = 16.2, 8.7, 3.8 Hz), 5.32 (1H, m), 5.18 (1H, m), 4.70 (1H, t, J = 8.1 Hz), 4.32 (1H, m), 3.88 (3H, s), 3.64 (1H, dd, J = 8.1, 2.3 Hz), 2.90 (1H, m), 2.68 (1H, s), 2.51 (1H, m), 2.42–2.25 (2H, overlapped), 1.79 (3H, s), 1.37 (3H, d, J = 6.4 Hz). 13C NMR (100 MHz, CDCl3) δ 170.5 (C), 162.5 (C), 160.0 (C), 140.6 (C), 139.8 (C), 132.0 (C), 131.6 (CH), 131.4 (CH), 130.4 (CH), 130.2 (CH), 129.6 (CH), 129.0 (CH), 127.7 (CH), 126.7 (CH), 114.0 (C), 108.7 (CH), 106.2 (C), 99.5 (CH), 81.0 (CH), 76.4 (CH), 71.4 (CH), 68.3 (CH), 56.6 (OCH3), 37.1 (CH2), 34.9 (CH2), 26.5 (CH3), 19.1 (CH3). HRESIMS m/z 535.1282 [M + H]+ (calcd for C27H29O7Cl2+, 535.1285).
Compound 34: white, solid; yield, 31.9%; 1H NMR (400 MHz, CDCl3) δ 11.36 (1H, s), 7.64 (1H, dd, J = 7.5, 2.0 Hz), 7.40 (1H, dd, J = 7.5, 1.7 Hz), 7.28 (1H, dd, J = 7.5, 1.7 Hz), 7.23 (1H, dd, J = 7.5, 2.0 Hz), 6.61 (1H, dd, J = 16.1, 1.5 Hz), 6.47 (1H, s), 6.06 (1H, m), 5.60–5.46 (2H, overlapped), 5.43 (1H, m), 4.51 (1H, t, J = 8.7 Hz), 4.24 (1H, m), 3.95 (1H, dd, J = 8.7, 2.4 Hz), 3.91 (3H, s), 2.76 (1H, m), 2.60 (1H, m), 2.47 (1H, m), 2.24 (1H, m), 2.01 (1H, s), 1.77 (3H, s), 1.39 (3H, d, J = 6.3 Hz). 13C NMR (100 MHz, CDCl3) δ 170.7 (C), 162.4 (C), 160.0 (C), 140.8 (C), 139.9 (C), 131.8 (CH), 131.7 (CH), 131.5 (CH), 130.7 (CH), 129.7 (CH × 2), 129.0 (CH), 127.4 (CH), 126.9 (CH), 114.1 (C), 108.9 (CH), 106.4 (C), 99.5 (CH), 82.4 (CH), 75.7 (CH), 71.3 (CH), 68.6 (CH), 56.6 (OCH3), 37.2 (CH2), 34.6 (CH2), 27.0 (CH3), 19.5 (CH3). HRESIMS m/z 535.1285 [M + H]+ (calcd for C27H29O7Cl2+, 535.1285).
Compound 35: white, solid; yield, 49.3%; 1H NMR (400 MHz, CDCl3) δ 11.42 (1H, s), 6.60 (1H, dd, J = 16.1, 1.2 Hz), 6.47 (1H, s), 6.05 (1H, m), 5.52–5.34 (3H, overlapped), 4.61 (1H, t, J = 8.7 Hz), 4.17 (1H, m), 3.91 (3H, s), 3.63 (1H, dd, J = 8.7, 2.6 Hz), 2.86 (1H, m), 2.64–2.52 (2H, overlapped), 2.49–2.30 (2H, overlapped), 1.39 (3H, d, J = 6.4 Hz), 1.35 (3H, s), 0.94 (9H, s). 13C NMR (100 MHz, CDCl3) δ 170.7 (C), 162.5 (C), 159.9 (C), 140.1 (C), 130.9 (CH), 130.5 (CH), 129.9 (CH), 129.0 (CH), 114.1 (C), 114.1 (CH)106.4 (C), 99.5 (CH), 82.2 (CH), 75.4 (CH), 71.4 (CH), 68.1 (CH), 56.6 (OCH3), 39.3 (CH2), 37.3 (CH2), 34.4 (C), 25.4 (CH3 × 3), 20.9 (CH3), 19.4 (CH3). HRESIMS m/z 461.2519 [M + H]+ (calcd for C26H37O7+, 461.2534).
Compound 36: white, solid; yield, 62.5%; 1H NMR (400 MHz, CDCl3) δ 11.42 (1H, s), 6.61 (1H, dd, J = 16.1, 1.4 Hz), 6.46 (1H, s), 6.02 (1H, m), 5.53–5.35 (3H, overlapped), 4.57 (1H, t, J = 8.7 Hz), 4.17 (1H, m), 3.91 (3H, s), 3.67 (1H, dd, J = 8.7, 2.5 Hz), 2.86 (1H, m), 2.64–2.51 (2H, overlapped), 2.48–2.30 (2H, overlapped), 1.84–1.61 (6H, overlapped), 1.39 (3H, d, J = 6.4 Hz), 1.32 (3H, s), 1.22–0.97 (5H, overlapped). 13C NMR (100 MHz, CDCl3) δ 170.7 (C), 162.5 (C), 160.0 (C), 140.0 (C), 130.7 (CH), 130.6 (CH), 130.4 (CH), 129.0 (CH), 114.1 (C), 112.0 (CH), 106.4 (C), 99.5 (CH), 81.6 (CH), 75.2 (CH), 71.5 (CH), 68.2 (CH), 56.6 (OCH3), 47.4 (CH2), 37.2 (CH2), 34.6 (C), 27.6 (CH), 27.4 (CH), 26.4 (CH), 26.4 (CH), 26.3 (CH), 22.7 (CH3), 19.4 (CH3). HRESIMS m/z 507.21 [M + H]+ (calcd for C27H36O7Cl+, 507.2144).
Compound 37: white, solid; yield, 41.7%; 1H NMR (400 MHz, CDCl3) δ 11.38 (1H, s), 6.63 (1H, dd, J = 16.1, 1.4 Hz), 6.46 (1H, s), 5.97 (1H, m), 5.51 (1H, ddd, J = 16.1, 8.9, 3.7 Hz), 5.47–5.36 (2H, overlapped), 4.45 (1H, t, J = 8.7 Hz), 4.25 (1H, m), 3.91 (3H, s), 3.82 (1H, dd, J = 8.7, 2.4 Hz), 2.86 (1H, m), 2.60–2.52 (2H, overlapped), 2.48–2.28 (2H, overlapped), 1.89–1.62 (6H, overlapped), 1.38 (3H, d, J = 6.4 Hz), 1.26 (3H, s), 1.10 (5H, overlapped). 13C NMR (100 MHz, CDCl3) δ 170.7 (C), 162.4 (C), 160.0 (C), 140.0 (C), 131.5 (CH), 130.7 (CH), 130.4 (CH), 129.1 (CH), 114.1 (C), 111.7 (CH), 106.5 (C), 99.5 (CH), 80.4 (CH), 75.8 (CH), 71.4 (CH), 68.8 (CH), 56.6 (OCH3), 47.4 (CH2), 37.2 (CH2), 34.9 (CH), 27.7 (CH2×2), 26.4 (CH2×2), 26.3 (CH2), 22.7 (CH3 ), 19.5 (CH3 ). HRESIMS m/z 507.2162 [M + H]+ (calcd for C27H36O7Cl+, 507.2144).
3.5. Antimicrobial Activity
The broth dilution method was used to screen antibacterial activity in vitro. Broth micro- or macro-dilution is the most basic method for antibiotic susceptibility testing [38,40]. In the screening of anti-M. marinum activity, the commonly used isoniazid and rifampicin were selected as positive controls. Ciprofloxacin and chloramphenicol were positive in antibacterial tests. Amphotericin B was used as a positive drug in the antifungal test. The strains were inoculated into the corresponding medium, cultured at 32 °C for 8 h, and diluted to 105 CFU/mL with the corresponding blank medium, and then a 198 μL bacterial solution and a 2 μL sample were added into the 96-well plates, using DMSO as a negative control. The treated 96-well plates were also cultured at 32 °C for 24 h or 48 h.
3.6. Statistical Analysis
GraphPad Prism 8 software was used for data analysis. Data are represented using the means ± SD. Data can only be considered statistically significant when p < 0.05. Data are presented as the means of three experiments.
4. Conclusions
In conclusion, 28 new derivatives with ketal groups were synthesized through a one-to-two-step chemical semi-synthetic reaction, enriching the structural diversity of the 14-membered RALs. In vitro activity evaluation of the synthesized derivatives showed that compounds 15–24 and 28–35 showed effective anti-M. marinum activity. Preliminary structure–activity relationship analysis of the compounds showed that the introduction of ketal groups with aromatic rings in C-5′ and C-6′ significantly enhanced the activity of the compounds. The 14-membered RALs showed no significant activity after the introduction of chain ketal groups at the C-5′ and C-6′ positions, but the activity increased significantly after the further introduction of chlorine atoms at C-5. The introduction of ketal groups to the hydroxyl groups at C-5′ and C-6′ and the ketone reaction, in which two carbonyl groups in the molecule are separated by two saturated carbon atoms, significantly increased anti-M. marinum activity. Further combined-administration experiments showed that compounds 23, 24, 29, and 30 enhanced the anti-M. marinum activity of the positive drug. Therefore, compounds 23, 24, 29, and 30 have effective anti-M. marinum activity in vitro, providing a new idea for the development of new anti-M. marinum drugs.
Acknowledgments
On the auspicious occasion of her 95th birthday, this paper is dedicated to Youyou Tu, the esteemed recipient of the 2015 Nobel Prize in Physiology or Medicine, in recognition of her groundbreaking discovery of Artemisinin, which has saved millions of lives worldwide. We thank Syngenta for the fellowships awarded to Yan-Wei Wu and Qun Zhang. We also thank Cong Wang at the School of Medicine and Pharmacy, Ocean University of China, for the NMR test.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/md22100431/s1. Table S1: Different ketone reagents used to generate compounds 10–37. Table S2: Key 13C NMR data (δ) for diastereoisomers. Figure S1–S98: 1H NMR, 13C NMR, 1D NOE, and HRESIMS results for compounds 10–37.
Author Contributions
J.-N.Y. contributed to preparation of all compounds and writing—original draft and writing—review and editing; C.-F.W. contributed to related work concerning writing—original draft and writing—review and editing; X.-L.Z. contributed to NOE detection; Y.-J.C. contributed to work related to bioactivity; Y.-W.W. contributed to the examination of the nuclear magnetic data; Q.Z. contributed to NOE data checking and the normalization of charts and tables; C.-L.S. contributed to the structural identification and synthesis of compounds; Y.-C.G. and M.-Y.W. were the project leaders, organizing and guiding the experiments and manuscript writing. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data are contained within the article or Supplementary Material.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This work was supported by the Special Funds of Shandong Province for Qingdao National Laboratory of Marine Science and Technology (No. 2022QNLM030003), Shandong Province Special Fund “Frontier Technology and Free Exploration” from Laoshan Laboratory (No. 8-01), the State Key Laboratory for Chemistry and Molecular Engineering of Medicinal Resources, Guangxi Normal University (No. CMEMR2023-B16), the National Key Research and Development Program of China (No. 2022YFC2601305), the Taishan Scholars Program of China, and the Innovation Center for Academicians of Hainan Province.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Holden I.K., Kehrer M., Andersen A.B., Wejse C., Svensson E., Johansen I.S. Mycobacterium marinum infections in Denmark from 2004 to 2017: A retrospective study of incidence, patient characteristics, treatment regimens and outcome. Sci. Rep. 2018;8:6738. doi: 10.1038/s41598-018-24702-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubry A., Mougari F., Reibel F., Cambau E. Mycobacterium marinum. Microbiol. Spectr. 2017;5:735–752. doi: 10.1128/microbiolspec.TNMI7-0038-2016. [DOI] [PubMed] [Google Scholar]
- 3.Petrini B. Mycobacterium marinum: Ubiquitous agent of waterborne granulomatous skin infections. Eur. J. Clin. Microbiol. Infect. Dis. 2006;25:609–613. doi: 10.1007/s10096-006-0201-4. [DOI] [PubMed] [Google Scholar]
- 4.Jernigan J.A., Farr B.M. Incubation period and sources of exposure for cutaneous Mycobacterium marinum infection: Case report and review of the literature. Clin. Infect. Dis. 2000;31:439–443. doi: 10.1086/313972. [DOI] [PubMed] [Google Scholar]
- 5.Steinbrink J., Alexis M., Angulo-Thompson D., Ramesh M., Alangaden G., Miceli M.H. Mycobacterium marinum remains an unrecognized cause of indolent skin infections. Cutis. 2017;100:331–336. [PubMed] [Google Scholar]
- 6.Johnson M.G., Stout J.E. Twenty-eight cases of Mycobacterium marinum infection: Retrospective case series and literature review. Infection. 2015;43:655–662. doi: 10.1007/s15010-015-0776-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gauthier D.T., Rhodes M.W. Mycobacteriosis in fishes: A review. Vet. J. 2009;180:33–47. doi: 10.1016/j.tvjl.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 8.Aubry A., Chosidow O., Caumes E., Robert J., Cambau E. Sixtythree cases of Mycobacterium marinum infection: Clinical features, treatment, and antibiotic susceptibility of causative isolates. Arch. Intern. Med. 2002;162:1746–1752. doi: 10.1001/archinte.162.15.1746. [DOI] [PubMed] [Google Scholar]
- 9.Haworth C.S., Banks J., Capstick T., Fisher A.J., Gorsuch T., Laurenson I.F., Leitch A., Loebinger M.R., Milburn H.J., Nightingale M., et al. British Thoracic Society guidelines for the management of non-tuberculous mycobacterial pulmonary disease (NTM-PD) Thorax. 2017;72:ii1–ii64. doi: 10.1136/thoraxjnl-2017-210927. [DOI] [PubMed] [Google Scholar]
- 10.Canetti D., Riccardi N., Antonello R.M., Nozza S., Sotgiu G. Mycobacterium marinum: A brief update for clinical purposes. Eur. J. Intern. Med. 2022;105:5–19. doi: 10.1016/j.ejim.2022.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Rallis E., Koumantaki-Mathioudaki E. Treeatment of Mycobacterium marinum cutaneous infections. Expert. Opin. Pharmacother. 2007;8:2965–2978. doi: 10.1517/14656566.8.17.2965. [DOI] [PubMed] [Google Scholar]
- 12.Gu A.k., Zhang Y. Research progress of Mycobacterium marinum. Chin. J. Infect. Control. 2022;21:1048–1052. [Google Scholar]
- 13.Alffenaar J.W., Märtson A.G., Heysell S.K., Cho J.G., Patanwala A., Burch G., Kim H.Y., Sturkenboom M.G.G., Byrne A., Marriott D., et al. Therapeutic Drug Monitoring in Non-Tuberculosis Mycobacteria Infections. Clin. Pharmacokinet. 2021;60:711–725. doi: 10.1007/s40262-021-01000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atanasov A.G., Zotchev S.B., Dirsch V.M., Supuran C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug. Discov. 2021;20:200–216. doi: 10.1038/s41573-020-00114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lachance H., Wetzel S., Kumar K., Waldmann H. Charting, navigating, and populating natural product chemical space for drug discovery. J. Med. Chem. 2012;55:5989–6001. doi: 10.1021/jm300288g. [DOI] [PubMed] [Google Scholar]
- 16.Zhao J.X., Yue J.M. Frontier studies on natural products: Moving toward paradigm shifts. Sci. China Chem. 2023;66:928–942. doi: 10.1007/s11426-022-1512-0. [DOI] [Google Scholar]
- 17.Newman D.J., Cragg G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020;83:770–803. doi: 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- 18.Hai Y., Cai Z.M., Li P.J., Wei M.Y., Wang C.Y., Gu Y.C., Shao C.L. Trends of antimalarial marine natural products: Progresses, challenges and opportunities. Nat. Prod. Rep. 2022;39:969–990. doi: 10.1039/D1NP00075F. [DOI] [PubMed] [Google Scholar]
- 19.Carroll A.R., Copp B.R., Davis R.A., Keyzers R.A., Prinsep M.R. Marine natural products. Nat. Prod. Rep. 2022;39:1122–1171. doi: 10.1039/D1NP00076D. [DOI] [PubMed] [Google Scholar]
- 20.Xu W.F., Wu N.N., Wu Y.W., Qi Y.X., Wei M.Y., Pineda L.M., Ng M.G., Spadafora C., Zheng J.Y., Lu L., et al. Structure modification, antialgal, antiplasmodial, and toxic evaluations of a series of new marine-derived 14-membered resorcylic acid lactone derivatives. Mar. Life Sci. Technol. 2022;4:88–97. doi: 10.1007/s42995-021-00103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou X.M., Wang C.Y., Gerwick W.H., Shao C.L. Marine natural products as potential anti-tubercular agents. Eur. J. Med. Chem. 2019;165:273–292. doi: 10.1016/j.ejmech.2019.01.026. [DOI] [PubMed] [Google Scholar]
- 22.Shao C.L., Wu H.X., Wang C.Y., Liu Q.A., Xu Y., Wei M.Y., Qian P.Y., Gu Y.C., Zheng C.J., She Z.G., et al. Potent antifouling resorcylic acid lactones from the gorgonian-derived fungus Cochliobolus lunatus. J. Nat. Prod. 2011;74:629–633. doi: 10.1021/np100641b. [DOI] [PubMed] [Google Scholar]
- 23.Li R., Zhou Y., Zhang X.X., Yang L.J., Liu J.Y., Wightman S.M., Lv L., Liu Z.Q., Wang C.Y., Zhao C.Y. Identification of marine natural product Pretrichodermamide B as a STAT3 inhibitor for efficient anticancer therapy. Mar. Life Sci. Technol. 2023;5:94–101. doi: 10.1007/s42995-022-00162-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y.H., Mándi A., Li H.L., Li X.M., Li X., Meng L.H., Yang S.Q., Shi X.S., Kurtán T., Wang B.G. Isolation and characterization of three pairs of verrucosidin epimers from the marine sediment-derived fungus Penicillium cyclopium and configuration revision of penicyrone A and related analogues. Mar. Life Sci. Technol. 2023;5:223–231. doi: 10.1007/s42995-023-00173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blunt J.W., Copp B.R., Keyzers R.A., Munro M.H., Prinsep M.R. Marine natural products. Nat. Prod. Rep. 2016;33:382–431. doi: 10.1039/C5NP00156K. [DOI] [PubMed] [Google Scholar]
- 26.Jia Y.L., Wei M.Y., Chen H.Y., Guan F.F., Wang C.Y., Shao C.L. (+)- and (−)-Pestaloxazine A, a Pair of Antiviral Enantiomeric Alkaloid Dimers with a Symmetric Spiro[oxazinane-piperazinedione] Skeleton from Pestalotiopsis sp. Org. Lett. 2015;17:4216–4219. doi: 10.1021/acs.orglett.5b01995. [DOI] [PubMed] [Google Scholar]
- 27.Han Y.Q., Zhang Q., Xu W.F., Hai Y., Chao R., Wang C.F., Hou X.M., Wei M.Y., Gu Y.C., Wang C.Y. Targeted isolation of antitubercular cycloheptapeptides and an unusual pyrroloindoline-containing new analog, asperpyrroindotide A, using LC-MS/MS-based molecular networking. Mar. Life Sci. Technol. 2023;5:85–93. doi: 10.1007/s42995-022-00157-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J., Xu L., Zhang X.Q., Liu X., Zhang Z.X., Zhu Q.M., Liu J.Y., Iqbal M.O., Ding N., Shao C.L. Discovery of a natural small-molecule AMP-activated kinase activator that alleviates nonalcoholic steatohepatitis. Mar. Life Sci. Technol. 2023;5:196–210. doi: 10.1007/s42995-023-00168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jana N., Nanda S. Resorcylic acid lactones (RALs) and their structural congeners: Recent advances in their biosynthesis, chemical synthesis and biology. New J. Chem. 2018;42:17803–17873. doi: 10.1039/C8NJ02534G. [DOI] [Google Scholar]
- 30.Liu Q.A., Shao C.L., Gu Y.C., Blum M., Gan L.S., Wang K.L., Chen M., Wang C.Y. Antifouling and Fungicidal Resorcylic Acid Lactones from the Sea Anemone-Derived Fungus Cochliobolus lunatus. J. Agric. Food. Chem. 2014;62:3183–3191. doi: 10.1021/jf500248z. [DOI] [PubMed] [Google Scholar]
- 31.Xu W.F., Xue X.J., Qi Y.X., Wu N.N., Wang C.Y., Shao C.L. Cochliomycin G, a 14-membered resorcylic acid lactone from a marine-derived fungus Cochliobolus lunatus. Nat. Prod. Res. 2021;35:490–493. doi: 10.1080/14786419.2019.1633646. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X.Q., Spadafora C., Pineda L.M., Ng M.G., Sun J.H., Wang W., Wang C.Y., Gu Y.C., Shao C.L. Discovery, Semisynthesis, Antiparasitic and Cytotoxic Evaluation of 14-Membered Resorcylic Acid Lactones and Their Derivatives. Sci. Rep. 2017;7:11822. doi: 10.1038/s41598-017-12336-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jing Q.Q., Yin J.N., Cheng Y.J., Zhang Q., Cao X.Z., Xu W.F., Shao C.L., Wei M.Y. Study on the Anti-Mycobacterium marinum Activity of a Series of Marine-Derived 14-Membered Resorcylic Acid Lactone Derivatives. Mar. Drugs. 2024;22:135. doi: 10.3390/md22030135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W., Shao C.L., Chen M., Liu Q.A., Wang C.Y. Brominated resorcylic acid lactones from the marine-derived fungus Cochliobolus lunatus induced by histone deacetylase inhibitors. Tetrahedron Lett. 2014;55:4888–4891. doi: 10.1016/j.tetlet.2014.06.096. [DOI] [Google Scholar]
- 35.Wang K.L., Zhang G., Sun J., Xu Y., Han Z., Liu L.L., Shao C.L., Liu Q.A., Wang C.Y., Qian P.Y. Cochliomycin A inhibits the larval settlement of Amphibalanus amphitrite by activating the NO/cGMP pathway. Biofouling. 2016;32:35–44. doi: 10.1080/08927014.2015.1121245. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Q., Lv L.X., Wang W.H., Wei M.Y., Gu Y.C., Shao C.L. Recent Advances of Bioactive Marine Natural Products in Drug Discovery. J. Ocean Univ. China. 2024;23:1297–1318. doi: 10.1007/s11802-024-5975-4. [DOI] [Google Scholar]
- 37.Koul A., Arnoult E., Lounis N., Guillemont J., Andries K. The challenge of new drug discovery for tuberculosis. Nature. 2011;469:483–490. doi: 10.1038/nature09657. [DOI] [PubMed] [Google Scholar]
- 38.Balouiri M., Sadiki M., Ibnsouda S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016;6:71–79. doi: 10.1016/j.jpha.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen W.J., Bao F.F., Pan Q., Liu T.T., Xue X.T., Liu H., Zhang F.R. A Series of 35 Cutaneous Infections Caused by Mycobacterium marinum in Han Chinese Population. J. Trop. Med. 2023;2023:5514275. doi: 10.1155/2023/5514275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Y., Wu P., Wu R., Li H.L., Duan Y., Cai C.N., Liu Z.X., She P.F., Zhang D. Simeprevir restores the anti-Staphylococcus activity of polymyxins. AMB Express. 2023;13:122. doi: 10.1186/s13568-023-01634-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are contained within the article or Supplementary Material.