Abstract
Chronic hepatitis C virus (HCV) infections can be cured only in a fraction of patients treated with alpha interferon (IFN-α) and ribavirin combination therapy. The mechanism of the IFN-α response against HCV is not understood, but evidence for a role for viral nonstructural protein 5A (NS5A) in IFN resistance has been provided. To elucidate the mechanism by which NS5A and possibly other viral proteins inhibit the cellular antiviral program, we have constructed a subgenomic replicon from a known infectious HCV clone and demonstrated that it has an approximately 1,000-fold-higher transduction efficiency than previously used subgenomes. We found that IFN-α reduced replication of HCV subgenomic replicons approximately 10-fold. The estimated half-life of viral RNA in the presence of the cytokine was about 12 h. HCV replication was sensitive to IFN-α independently of whether the replicon expressed an NS5A protein associated with sensitivity or resistance to the cytokine. Furthermore, our results indicated that HCV replicons can persist in Huh7 cells in the presence of high concentrations of IFN-α. Finally, under our conditions, selection for IFN-α-resistant variants did not occur.
Hepatitis C virus (HCV) causes persistent infection in approximately 80% of infected adults and variable and severe liver disease in an estimated 70% of those who cannot clear the virus (1). HCV is an enveloped, positive-stranded RNA virus encoding a polyprotein that is proteolytically processed into 10 polypeptides. Four of them are enzymes: cysteine and serine proteases, an ATP-dependent helicase, and an RNA-directed RNA polymerase (17). While these enzymes are used as potential targets for virus-specific antiviral therapies, their genes exhibit high variability among the different HCV genotypes, and, most likely, drug-resistant variants will evolve during antiviral therapy. Currently available combination therapy with alpha interferon (IFN-α) and ribavirin is effective in less than 50% of treated patients. Although the mechanism controlling the IFN response in patients is likely to be complex, there is evidence that nonstructural (NS) protein 5A (NS5A) evolves to confer resistance against IFN-α during antiviral therapy (5, 6). This resistance is believed to be a consequence of a specific interaction between NS5A and protein kinase R (PKR), an important mediator of the antiviral program induced by IFN-α.
Unfortunately, efforts to investigate the molecular mechanisms responsible for IFN resistance were hampered by the lack of tissue culture systems permissive for the replication and production of infectious HCV from available cDNA clones. Recently, Lohmann and colleagues (13) reported that a subgenomic replicon containing a neomycin phosphotransferase gene (neoR) in lieu of the viral structural genes replicated in Huh7 cells (see Fig. 1A). However, from the low frequency with which Huh7 cells supported replication, it is possible that the selected isolate is defective and requires genetic changes for efficient genome synthesis. This possibility has now been confirmed in this and other reports demonstrating that a selection for replicons occurs in transfected cells with mutations at different positions in the NS region (3, 12, 13). Furthermore, investigations about the role of NS5A or other viral proteins in IFN resistance depend on the possibility of testing HCV isolates obtained from IFN responders as well as from nonresponders.
FIG. 1.
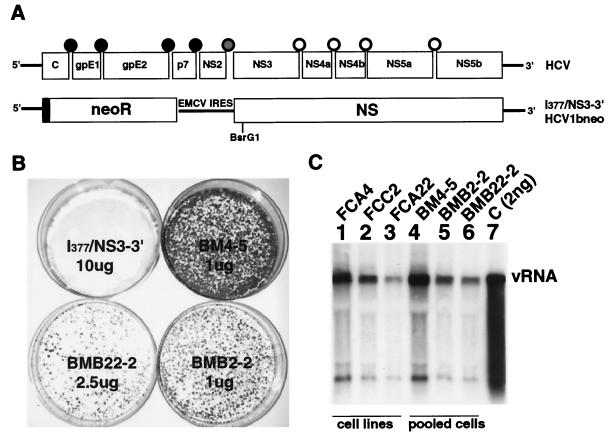
Replication of subgenomic replicons in Huh7 cells. (A) Physical map of subgenomic HCV replicons. Top, physical map of HCV with the processed polypeptides and the flanking untranslated regions. Black, gray, and white circles, cleavage sites for the host signalase, the NS2/3 protease, and the NS3 protease on the polyprotein, respectively. Bottom, structure of subgenomic replicon I377/NS3-3′ as described by Lohmann et al. (13). Black rectangle, 12 amino acids derived from the HCV core protein. Restriction site BsrG1, used for the construction of HCV1bneo, is indicated. (B) Replication of adapted mutants. Huh7 cells were electroporated with the indicated amounts of in vitro-transcribed RNA. The mutations present in subgenomic RNAs BM4-5, BMB22-2, and BMB2-2 are listed in Table 1. G418-resistant colonies were stained with crystal violet 16 days after electroporation. (C) Northern blot analysis of total RNA extracted from cell lines FCA4, FCC2, and FCA22 and from pooled G418-resistant cells transfected with the indicated RNAs. Total RNA (5 μg) obtained from each plate was loaded onto an agarose-formaldehyde gel. In vitro-transcribed subgenomic RNA served as a control (C) for the hybridization reaction. Genomic HCV RNA (vRNA) was detected with a riboprobe spanning the neor gene.
As shown in this report, we constructed a subgenomic replicon composed of sequences derived from an infectious HCV clone that exhibits a transduction efficiency approximately 3 orders of magnitude higher than those for initially described isolates (14). Furthermore, we investigated the response of HCV replicons to the antiviral program induced by IFN-α in Huh7 cells and asked whether IFN can cure cells from infection and whether IFN-sensitive variants can be selected in tissue culture cells. We found that HCV replication is sensitive to IFN-α independently of whether the replicon is derived from a (putatively) IFN-sensitive or -resistant cDNA clone and observed that even long-term treatment of cell cultures with the cytokine cannot clear the virus from the cells and does not seem to yield IFN-resistant variants.
MATERIALS AND METHODS
Cell culture.
Huh7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Gibco-BRL) supplemented with 10% fetal bovine serum, l-glutamine, nonessential amino acids, penicillin, and streptomycin.
Construction of recombinant plasmids.
HCV genotype 1b replicon I377/NS3-3′ (GenBank accession no. AJ242652) was assembled and cloned from chemically synthesized DNA oligomers (3). HCV1bneo was engineered by replacing a BsrGI-ScaI fragment in I377/NS3-3′ with a BsrGI-XbaI fragment from plasmid HCV-N (2). BM4-5 was constructed by replacing an EcoRI (position 5083)-XhoI (position 5570) fragment of I377/NS3-3′ with the corresponding fragment, which was cloned by reverse transcriptase PCR (RT-PCR) with RNA isolated from cell line FCA4. BMB2-2 and BM22-2 were made by replacing an SspI (position 4045)-EcoRI (position 5083) fragment in I377/NS3-3′ with the corresponding fragment, which was cloned by RT-PCR with RNA isolated from cell lines FCC2 and FCA22, respectively. Plasmid 1bneo/delS was derived from HCV1bneo by deletion of nucleotides 6955 to 6957 (HCV-N; accession no. AF139594) using oligomer-directed site-specific mutagenesis. I377/NS3-3′X was constructed by filling in the XhoI site in I377/NS3-3′ (position 5570) with Klenow DNA polymerase. The nucleotide sequences of the substituted fragments were determined with an ABI sequencer.
In vitro transcription and purification of RNA.
Plasmids were linearized with ScaI (I377/NS3-3′ and its derivatives), XbaI (HCV1bneo), or MluI (D2Rneo) and transcribed in vitro with the MEGAscript kit (Ambion, Austin, Tex.) in accordance with the manufacturer's protocol. For a 20-μl reaction mixture, transcription was stopped by the addition of 4 U of DNase I and the mixture was incubated for 45 min at 37°C. RNA was extracted with 1 ml of TRIzol reagent (Gibco-BRL) and precipitated with 500 μl of isopropanol. RNA pellets were washed once with 75% ethanol and dissolved in RNase-free water. To remove the residual amount of template DNA, RNA preparations were extracted once with acid phenol, precipitated with ethanol, and resuspended in RNase-free water (9).
RNA transfection.
Subconfluent Huh7 cells were trypsinized and washed once with complete DMEM and once with serum-free DMEM-F12 medium. Cell pellets were resuspended in serum-free DMEM-F12 medium at a density of 107 cells/ml. To 200 μl of the cell suspensions in an electroporation cuvette (0.2-cm gap; BTX, San Diego, Calif.) 1 to 10 μg of in vitro-transcribed RNA was added. The cells were immediately electroporated with an ECM 630 apparatus (BTX) set to 200 V and 1,000 μF. After electroporation the cell suspension was kept for 5 min at room temperature and then diluted into DMEM supplemented with 10% fetal bovine serum and nonessential amino acids and seeded into a 10-cm-diameter petri dish. After 24 h, G418 was added to obtain a final concentration of 1 mg/ml, and medium was changed every other day. G418-resistant colonies became visible after 2 to 3 weeks.
Northern blot hybridization.
Total cellular RNA was extracted with TRIzol reagent (Gibco-BRL). Five to 10 μg of total RNA was electrophoresed through a 1.0% agarose gel containing 2.2 M formaldehyde and transferred to a nylon membrane and immobilized by UV cross-linking (Stratagene). Hybridization was carried out with [32P]UTP-labeled in vitro-transcribed RNA in a solution containing 50% deionized formamide, 5× SSC (750 mM sodium chloride, 750 mM sodium citrate), Denhardt's solution, 0.02 M sodium phosphate (pH 6.8), 0.2% sodium dodecyl sulfate (SDS), 100 μg of sheared denatured salmon sperm DNA/ml, and 100 μg of yeast RNA/ml for 16 h at 58°C. The membranes were washed once in 2× SSC–0.1% SDS for 30 min at room temperature and twice in 0.1× SSC–0.1% SDS for 30 min at 68°C. Membranes were exposed to X-ray film or to a phosphorimaging screen for quantitative analyses with the Fuji BAS 1000 system.
RT-PCR and DNA sequencing.
HCV replicons were isolated and cloned from established cell lines by PCR amplification of three fragments spanning the entire NS region including sequences from position 1387 to 7794. DNA synthesis was carried out with SuperScript II RT provided in a cDNA synthesis kit (Gibco-BRL). The DNA oligomers used as primers for the RT reactions mapped to positions 3876 to 3857, 5640 to 5618, and 7794 to 7760, respectively. The reaction mixtures were incubated for 1 h at 45°C. PCR was performed with an Advantage PCR kit (Clontech, Palo Alto, Calif.). One microliter of the cDNA reaction mixture was used for PCR with primers spanning positions 1387 to 3876, 3400 to 5610, and 5538 to 7794. The PCR products were purified by agarose gel electrophoresis and ligated with pGEMT Easy (Promega, Madison, Wis.). Three to six clones of each fragment were sequenced with an ABI automatic DNA sequencer, and a consensus sequence was established with the help of a sequence assembly program (Genetics Computer Group, Madison, Wis.).
In vitro translation
In vitro translation reactions were carried out with micrococcal-nuclease-treated rabbit reticulocyte lysates (Promega) in the presence of [35S]methionine in accordance with the manufacturer's protocol. Translation products were electrophoresed through SDS–10% polyacrylamide gels. The gel was fixed with 30% methanol and 10% glacial acetic acid, treated with NAMP100 Amplify solution (Amersham), dried, and exposed to X-ray film.
RESULTS
Adaptive mutations in cell lines expressing HCV subgenomes.
To conduct our studies, we used a chemically synthesized replica of the subgenome, I377/NS3-3′, described by Lohmann et al. (13) (Fig. 1A). This clone expresses an NS5A protein that is associated with an IFN-α-resistant phenotype (3). Electroporation of Huh7 cells in the presence of RNA transcribed from linearized I377/NS3-3′ plasmids resulted in the selection of G418-resistant colonies essentially as reported previously (13). From four independent experiments we estimated that approximately 1 to 5 out of 106 cells transfected with 10 μg of RNA survived the selection process. We expanded 20 clones into cell lines for the analysis of viral RNA. All 20 cell lines contained subgenomic viral RNA of positive as well as negative polarity (results not shown). RNA expression levels ranged from approximately 200 to 2,000 copies of plus strand RNA per cell, depending on the cell line examined (Fig. 1C, lanes 1 to 3). Moreover, we found that the accumulation of viral RNA was strongly influenced by cell density and, by inference, by the cell cycle. The levels of viral RNA in subconfluent cells were approximately 10-fold higher than those in confluent cells (results not shown).
In agreement with previous reports (3, 12), we identified consensus mutations in cDNAs obtained from replicons present in the four cell lines examined (Table 1). Two cell lines, FCA22 and FCC2, contained an identical mutation changing lysine 821 in NS4B to threonine. Line FCA4 contained only one consensus mutation, a deletion of serine 1176 in NS5A, and FCA1 contained several consensus mutations in NS3 as well as in NS5A. To examine whether the consensus mutations conferred an adapted phenotype, we constructed variants BM4-5, BMB22-2, and BMB2-2, carrying the consensus mutations observed in cell lines FCA4, FCA22, and FCC2, respectively (Table 1). Transfection of Huh7 cells with these mutants resulted in the appearance of many colonies, ranging from approximately 250 (BMB22-2) to 3,500 (BM4-5) colonies per μg of RNA (Fig. 1B, Table 2). This represented an increase of the transduction efficiency of 3 to 4 orders of magnitude compared to that for the parent I377/NS3-3′ construct (Table 2). Viral RNA levels in pools of cells transfected with the adapted mutants were similar to those in the parental cell lines, indicating that the nature of the mutations could influence the level of viral replication.
TABLE 1.
Summary of nucleotide sequence analysis of cDNAs derived from replicons in Huh7 cell lines
| Clone | Mutation ina:
|
Frequencyb | Adapted mutant | |
|---|---|---|---|---|
| cDNA (nt) | Encoded proteinc | |||
| FCA1 | G to A (1875) | None (25) NS3 | 6/6 | |
| A to G (2330) | E to G (177) NS3 | 4/6 | ||
| C to G (5487) | D to E (1229) NS5A | 3/3 | ||
| A to G (5695) | I to V (1299) NS5A | 3/3 | ||
| FCA4 | Del. TCA (5327–5329) | Del. S (1176) NS5A | 3/3 | BM4-5 |
| FCA22 | T to C (1777) | None (NAe) EMCV IRES | 3/3 | |
| A to C (4262) | K to T (821) NS4B | 3/3 | BMB22-2 | |
| C to T (6881) | A to V (1694) NS5B | 2/3 | ||
| FCC2 | A to T (4089) | A (763) NS4B | 3/3 | BMB2-2d |
| A to C (4262) | K to T (821) NS4B | 3/3 | ||
Nucleotide and amino acid positions are according to the sequence of the HCV subgenomic replicon I377/NS3-3′ (GenBank accession no. AJ242652). The numbering of the amino acids begins with the artificial methionine of the NS region. nt, nucleotide(s); del., deletion.
Fraction of PCR-amplified cDNA clones that carry the mutation.
Order of entries is as follows: mutation (amino acid position) protein designation.
Contains both mutations in FCC2.
NA, not applicable.
TABLE 2.
Summary of RNA transfection experiments
| Construct | Genotype | No. of colonies/μg of RNA |
|---|---|---|
| I377/NS3-3′ | 1b | 0.1–0.5 |
| BM4-5 | 1b | 3,455 |
| BMB2-2 | 1b | 644 |
| BM22-2 | 1b | 251 |
| HCV1bneo | 1b | 235–470 (352) |
| D2Rneo | 1a | NDa |
ND, not detected.
Replication of a subgenomic replicon derived from an infectious HCV clone.
This study and previous ones relied on an HCV clone that has not yet been tested for infectivity in a chimpanzee. Hence, it is possible that the low transduction efficiency observed with I377/NS3-3′ is due to a defect in the NS protein-coding region that must be corrected to achieve efficient RNA replication in Huh7 cells. In addition, it is not known whether HCV clones encoding NS5A associated with an IFN-α-sensitive phenotype can replicate in Huh7 cells. Therefore, we designed two additional constructs using known infectious HCV clones representing HCV subtypes 1a and 1b (HCV1a and -1b). In the HCV1a-based replicon, the 5′ and 3′ untranslated regions as well as the entire NS protein-coding region were derived from infectious HCV clone H77 (10). In the HCV1b-based replicon, HCV1bneo, the entire NS protein-coding region of I377/NS3-3′, beginning at a BsrG1 restriction site located 225 nucleotides downstream from the 5′ end of the NS3 coding sequence, was replaced with the corresponding region of infectious clone HCV-N (Fig. 1A) (2). Due to the design of the construct, the NS3 protein encoded by HCV1bneo differed from NS3 of HCV-N at two amino acids (amino acid [aa] 27, Arg in place of Lys; aa 73, Thr in place of Ala).
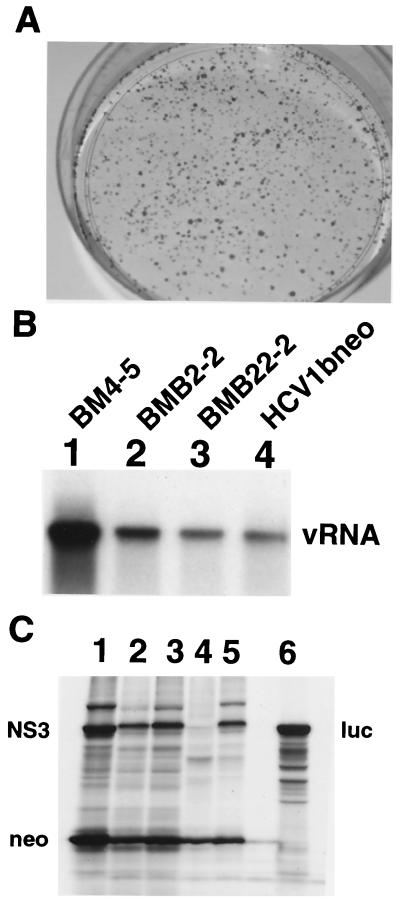
While transfection of Huh7 cells with HCV1a-based replicon D2Rneo did not yield any G418-resistant colonies, transfection with HCV1bneo resulted in the selection of approximately 350 colonies per μg of RNA (Fig. 2A; Table 2). Hence the transduction efficiency observed with HCV1bneo was similar to those of adapted variants BMB22-2 and BMB2-2 described above and about 700- to 3,500-fold higher than that observed with RNA derived from I377/NS3-3′. An analysis of HCV1bneo RNA present in G418-resistant cells derived from a pool of an estimated 2,000 colonies revealed about 200 copies of RNA per cell, similar to the levels observed in pools from cells transfected with BMB22-2 and BMB2-2 (Fig. 2B). Similar results were obtained with subgenomic replicon HCVNneo, where the entire NS protein-coding region was derived from HCV-N (results not shown).
FIG. 2.
Replication of a subgenomic replicon derived from an infectious HCV1b clone. (A) Huh7 cells were electroporated with RNA from HCV1bneo, and colonies were stained 16 days after the transfection. (B) Northern blot analysis of total RNA isolated from pooled cells transfected with the indicated RNAs. Genomic HCV RNA (vRNA) was detected as described for Fig. 1. (C) In vitro translation of viral RNA transcribed from plasmids I377/NS3-3′, HCV1bneo, HCVNneo, D2Rneo, and D2Ineo (lanes 1 to 5) . Luciferase was translated from pLuc as a positive control (lane 6). The positions of HCV NS3 and the neomycin phosphotransferase (neo) are indicated.
These results showed that a subgenomic replicon based on an infectious HCV1b clone can efficiently establish stable replicons in transfected Huh7 cells. On the other hand the results indicated that an HCV1a-derived clone was defective in this system. In an attempt to explain this difference between the two subtypes, we performed in vitro translation reactions with RNA derived from I377/NS3-3′, HCV1bneo, HCVNneo, and D2Rneo. Unexpectedly, we noticed a significant difference between the translation efficiencies of the NS polypeptides of the two subtypes (Fig. 2C). With the HCV1b-based constructs, translation levels of the NS proteins were similar to those of the neomycin gene, whereas with D2Rneo translation of the viral proteins was significantly reduced. Thus, our results suggested that sequences in the NS region may somehow influence the activity of the encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES). In apparent agreement with such a possibility, a secondary-structure analysis of sequences spanning the IRES sequence and the first 200 nucleotides of NS3 revealed that the HCV1a clone could potentially form a stem-loop structure with the EMCV IRES (results not shown). Indeed, substitution of the first 227 nucleotides of the NS region in D2Rneo with the corresponding fragment of HCV1b rescued the translation of the NS proteins (Fig. 2C, lane 5). However recombinant subgenomic replicon D2Ineo did not yield colonies after transfection of Huh7 cells, suggesting that either some other features of the HCV1a isolate interfered with the establishment of stable replicons or the chimeric NS3 protein was not supporting replication in transfected Huh7 cells.
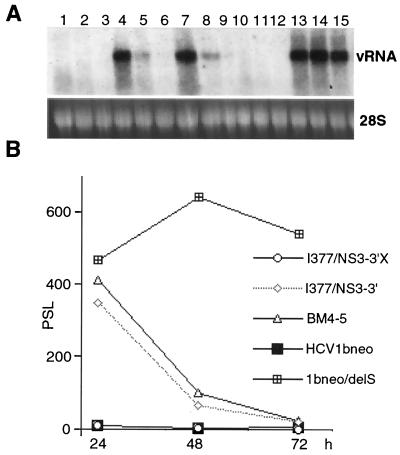
To determine whether adaptive mutations identified with I377/NS3-3′-based replicons could increase the replication levels of HCV-N-based subgenomes, we introduced the adaptive mutation of replicon BM4-5, leading to the deletion of serine 1176 in NS5A, into HCV1bneo. In vitro-transcribed RNA from the resulting replicon, 1bneo/delS, was transfected along with RNA from HCV1bneo, I377/NS3-3′, and BM4-5 into Huh7 cells. RNA transcribed from I377/NS3-3′X, carrying a frameshift mutation in the NS5A coding sequence, served as a negative control. To determine the effect of the mutation in 1bneo/delS on viral replication, we determined the levels of viral RNA 24, 48, and 72 h after transfection (Fig. 3A). This transient assay is based on the previously published observation that adapted replicons can amplify plus strand RNA more efficiently after transfection than the wild type (3, 11). While the BM4-5 replicon exhibited only moderately enhanced RNA levels compared with the wild-type levels, 1bneo/delS showed a much more dramatic increase compared with HCV1bneo (Fig. 3). Hence, these results indicated that adaptation of HCV-N-based replicons can occur in a fashion similar to that observed with I377/NS3-3′.
FIG. 3.
Transient replication of adapted mutants in Huh7 cells. (A) Viral plus strand RNA (vRNA) present in cells transfected with I377/NS3-3′X (lanes 1 to 3), I377/NS3-3′ (lanes 4 to 6), BM4-5 (lanes 7 to 9), HCV1bneo (lanes 10 to 12), and 1bneo/delS (lanes 13 to 15). RNA was extracted from transfected Huh7 cells 24 (lanes 1, 4, 7, 10, and 13), 48 (lanes 2, 5, 8, 11, and 14), and 72 h (lanes 3, 6, 9, 12, and 15) after the transfection. rRNA (28S) served as a control for the amount of RNA loaded per lane. (B) Levels of plus strand RNA in cells transfected with the indicated subgenomes 24, 48, and 72 h after the transfection as determined with a phosphorimager. PSL, arbitrary unit.
IFN response in Huh7 cells.
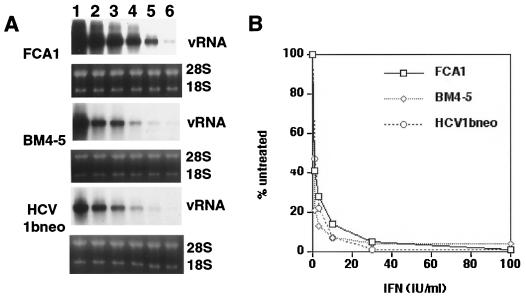
A major question in HCV biology is whether IFN-α-based antiviral therapy leads to the selection of IFN-α-resistant viral mutants. To address this question, we investigated whether the replication of HCV subgenomes is affected by IFN-α. Initial experiments with cell line FCA1 revealed that IFN-α reduced the levels of HCV RNA in Huh7 cells in a dose-dependent manner (Fig. 4). Inhibition was observed with IFN-α doses as low as 1 IU/ml, and 10- to 20-fold inhibition was obtained with 25 IU/ml. Of particular interest was the question of whether NS5A had any influence on the IFN response, as has been suggested previously. Because the I377/NS3-3′-based replicon encodes an NS5A protein that is expected to confer IFN resistance, in contrast to the HCV-N-based genomes, we directly compared pools of cells transfected with each replicon for their sensitivities to the antiviral action of the cytokine. Our results showed that both replicons responded in an equal fashion to IFN-α, suggesting that under our selected conditions IFN sensitivity is not influenced by variations in NS5A.
FIG. 4.
Antiviral activity of IFN-α. (A) Viral RNA (vRNA) levels present in cells treated with 0, 1, 3, 10, 30, and 100 IU of IFN-α/ml (lanes 1 to 6) for 72 h were determined by Northern blot analysis with a plus strand-specific RNA probe. rRNA (28S and 18S) served as a control for the amount of RNA loaded per lane. Examined were cell line FCA1 and G418-resistant pooled cells transfected with BM4-5 and HCV1bneo, respectively. (B) Signal intensities were measured with a phosphorimager and blotted as percentages of the values obtained with untreated cells (A, lane 1).
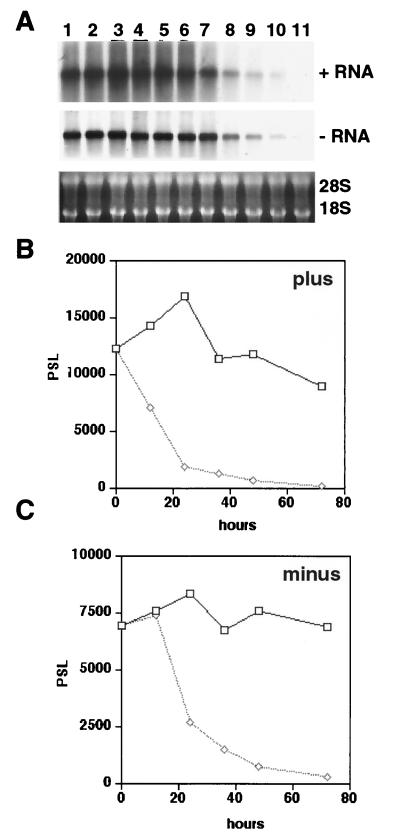
Cells supporting HCV replication are thought to harbor plus strand RNA base paired with minus strand RNA in replication complexes as well as excess plus strand RNA derived from the RNA amplification reaction. To examine whether IFN-α treatment affected only free plus strand RNA or replication complexes or both, we measured the levels of plus and minus strand RNA in cells treated with 100 IU of IFN-α/ml. Our results showed that both RNA strands were reduced with a half-life of about 12 h (Fig. 5). However, the decline of minus strand RNA appeared to be delayed by approximately 12 h, which could reflect differences in the mechanism of RNA degradation between free and complexed RNA.
FIG. 5.
Time course of the IFN-α response. (A) FCA1 cells were incubated with 100 IU of IFN-α/ml (lanes 7 to 11) or without IFN-α (lanes 1 to 6) for 12, 24, 36, 48, and 72 h (lanes 2 to 5 and 7 to 11, respectively). Plus and minus strand RNA was analyzed by Northern blotting with strand-specific RNA probes. (B and C) Levels of plus and minus strand RNA as determined with a phosphorimager. PSL, arbitrary unit; diamonds, IFN-α-treated cells; squares, untreated cells.
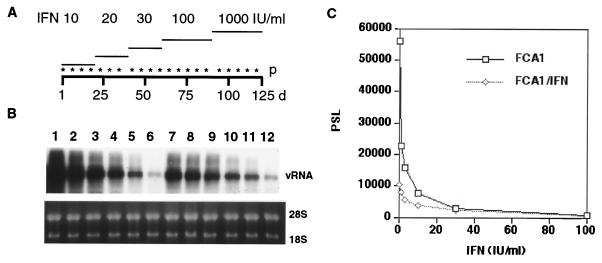
To examine whether IFN-α treatment permitted the selection of IFN-resistant mutants, we maintained cell line FCA1 for over 100 days in the presence of G418 and increasing concentrations of IFN-α to a final concentration of 1,000 IU/ml (Fig. 6). Although we noticed that these conditions reduced the growth rate of the cells, we did not observe massive cell death, which occurs during the selection of G418-resistant colonies with subgenomic replicons. Consistent with this observation, viral RNA continued to replicate in IFN-α-treated cultures, albeit at approximately 10-fold-reduced levels compared to those for untreated cells (results not shown). We examined whether IFN-α-resistant replicons were present in the FCA1 cells after treatment with increasing doses of the cytokine up to 1,000 IU/ml for 4 months. These cells were incubated for 6 days without the cytokine followed by an incubation with 1 to 100 IU of IFN-α/ml for 72 h before viral RNA levels were determined. The results showed that IFN-α-treated cells (FCA1/IFN) increased their viral RNA levels to 20% of the RNA level present in the parental FCA1 cell line when IFN-α was removed from the culture medium. Furthermore, the FCA1/IFN cells exhibited a slightly attenuated dose-response pattern compared with FCA cells (Fig. 6). Whether this difference reflects changes in the response of the cells to IFN-α or alterations in the virus is not yet known. Based on the observation that the two cell lines did not differ in terms of the maximal reduction of viral replication (100 IU/ml), we favor the former over the latter possibility.
FIG. 6.
Long-term treatment of FCA1 cells with IFN-α. (A) FCA1 cells were incubated for the indicated number of days (d) with increasing doses of IFN-α in the presence of G418 (1 mg/ml). The cells were passaged (p) at a ratio of 1:6 during the incubation with 10 to 30 IU of IFN/ml and at a ratio of 1:10 with 100 and 1,000 IU of IFN/ml at the indicated (asterisks) time points. (B) Control FCA cells (FCA1; lanes 1 to 6) and IFN-treated cells (FCA1/IFN; lanes 7 to 12) were incubated for 6 days without IFN and G418 and then with 0, 1, 3, 10, 30, or 100 IU of IFN-α/ml for 72 h (lanes 1 to 6 and 7 to 12, respectively). Viral RNA (vRNA) levels were determined by Northern blot analysis with a plus strand-specific probe. (C) Levels of viral RNA determined with a phosphorimager in FCA1 (squares) and FCA1/IFN (diamonds) cells treated with the indicated amounts of IFN-α. PSL, arbitrary unit.
DISCUSSION
In this report we demonstrated that a subgenomic replicon based on an infectious HCV1b isolate can replicate in tissue culture cells with an approximately 1,000-fold-higher colony formation efficiency than the previously reported I377/NS3-3′ replicon (Table 2) (3, 14). In agreement with others, we found that the I377/NS3-3′ replicon undergoes adaptation as a consequence of replication in Huh7 cells (3, 12). Moreover, the transfer of adapted mutations can also enhance the RNA replication of related replicons, as shown with the 1bneo/delS replicon (Fig. 3). Whether adaptive mutations occur in cells transfected with wild-type HCV-N-based replicons has not yet been determined. However the results obtained with 1bneo/delS provide support for this possibility. It is worthy of note that serine residue 1176, which is deleted in the products of replicons BM4-5 and 1bneo/delS, maps to one of three proposed hyperphosphorylation sites in NS5A and coincides with residues associated with other adaptive mutations that were described previously (3, 11, 20). Nevertheless, our observations have opened the possibility of analyzing HCV isolates from different patient sera for their biological properties in transfected cells. For example, clones from patients taken before and after therapy with IFN and ribavirin can now be isolated and tested for their biological behavior including drug resistance in tissue culture cells. Although the correlation between the expression of the NS proteins from subgenomic RNA templates in vitro and viral replication in cells is incomplete, it could be used as one predictor for the ability of a particular construct to replicate in Huh7 cells (Fig. 2C). However, our results showed that other unknown conditions have to be met for the establishment of additional HCV replicons in transfected tissue culture cells.
Our observation that the highest levels of HCV replication occurred in growing, subconfluent cells is consistent with a report by Pietschmann et al. (16) but is in direct contradiction with similar studies performed previously with hepatitis B virus (HBV) (19a). With HBV the accumulation of viral DNA occurs primarily in confluent or resting cells, which reflects the natural situation, where the majority of hepatocytes in the liver are in G0. Although our observation could reflect a property specific for subgenomic replicons and not for complete HCV genomes, it suggests that dividing hepatocytes might produce a higher level of virus than resting cells in an infected liver. Furthermore, it predicts that virus production increases as a consequence of liver disease characterized by the death and regeneration of hepatocytes. Finally, the observation that replication appears to be dependent on the cell cycle or cell density or both points to the presence of host factors that could play a major role in HCV replication. Evidence for the role of such factors comes from studies indicating that IRES-mediated translation is activated during the G2/M phase of the cell cycle, during which cap-dependent translation is suppressed (8, 18).
Antiviral therapy with IFN-α and ribavirin is ineffective in about 50% of treated patients (1, 4, 15). Thus, a major question concerns the mechanism responsible for the failure of IFN-α to inhibit viral replication in a large fraction of HCV patients. Our results showed that subgenomic replicons based on the I377/NS3-3′ construct are sensitive to the antiviral program induced by IFN-α in Huh7 cells (Fig. 4 to 6). IFN reduced genomic (plus strand) RNA levels with a half-life of approximately 12 h. Interestingly, inhibition of minus strands was delayed by about 12 h, which could suggest that replication complexes are more resistant to degradation by the cellular antiviral response than free plus strand RNA. Hence, a major unresolved question concerns the mechanism responsible for the antiviral activity of IFN-α.
Recently PKR has been invoked as an important IFN-induced pathway in the HCV system because it can bind to NS5A proteins derived from some IFN-resistant HCV isolates (3, 5, 6). Our results showed that I377/NS3-3 and HCV-N-based replicon HCV1bneo behaved almost identically in terms of their response to the cytokine (Fig. 4). These observations are surprising considering that I377/NS3-3, in contrast to HCV1bneo, encodes a form of NS5A believed to confer IFN-α resistance but are consistent with results reported previously by Blight et al. and Enomoto et al. (3, 5, 6). It should be noted that the mutations in NS5A identified in cell lines FCA1 and FCA4 (Table 1) are outside of the IFN sensitivity-determining region of NS5A (3, 4). Thus, under physiological conditions NS5A might be either sequestered in replication complexes or expressed at lower levels and, either way, does not seem to act as an inhibitor of PKR (7). Alternatively, it is possible that PKR is not activated in the Huh7 culture system. However, it is known that the IFN response is multifactorial, and perhaps as a consequence there is, so far, no known virus that has adopted an IFN-resistant phenotype. Our results obtained from the long-term incubation of FCA1 cells with IFN-α did not reveal any evidence for a selection of such variants (Fig. 6). The most likely explanation lies in the mechanism by which the IFN response is regulated through the activation of inhibitors of IFN signal transduction pathways (19). Our observation that IFN-treated cells were able to maintain low levels of subgenomic replicons is in good agreement with models predicting the long-term suppression of the IFN response after a relatively short activation period. Nevertheless, our observations should not be interpreted to mean that IFN-α-resistant HCV variants cannot be identified in Huh7 cells. For example, their isolation might depend on alternative selection strategies, i.e., the incubation of cell lines such as FCA1 with very high initial doses of IFN-α.
ACKNOWLEDGMENTS
We thank Devron R. Averett, Kerry Campbell, Rene Daniel, and Bill Mason for helpful comments on the manuscript and acknowledge services provided by the FCCC nucleotide sequencing facility. We are especially grateful to Charlie Rice for the communication of unpublished results about adapted variants. We thank Stan Lemon and Charlie Rice for providing HCV clones HCV-N and I377/NS3-3′.
This work was supported by grants from the National Institutes of Health and by an appropriation from the Commonwealth of Pennsylvania.
REFERENCES
- 1.Alter H J, Seeff L B. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin Liver Dis. 2000;20:17–35. doi: 10.1055/s-2000-9505. [DOI] [PubMed] [Google Scholar]
- 2.Beard M R, Abell G, Honda M, Carroll A, Gartland M, Clarke B, Suzuki K, Lanford R, Sangar D V, Lemon S M. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology. 1999;30:316–324. doi: 10.1002/hep.510300137. [DOI] [PubMed] [Google Scholar]
- 3.Blight K J, Kolykhalov A A, Rice C M. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1975. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 4.Davis G L, Esteban-Mur R, Rustgi V, Hoefs J, Gordon S C, Trepo C, Shiffman M L, Zeuzem S, Craxi A, Ling M H, Albrecht J. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. International Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1493–1499. doi: 10.1056/NEJM199811193392102. [DOI] [PubMed] [Google Scholar]
- 5.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J Clin Investig. 1995;96:224–230. doi: 10.1172/JCI118025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 7.Gale M J, Jr, Korth M J, Katze M G. Repression of the PKR protein kinase by the hepatitis C virus NS5A protein: a potential mechanism of interferon resistance. Clin Diagn Virol. 1998;10:157–162. doi: 10.1016/s0928-0197(98)00034-8. [DOI] [PubMed] [Google Scholar]
- 8.Honda M, Kaneko S, Matsushita E, Kobayashi K, Abell G A, Lemon S M. Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology. 2000;118:152–162. doi: 10.1016/s0016-5085(00)70424-0. [DOI] [PubMed] [Google Scholar]
- 9.Kedzierski W, Porter J C. A novel non-enzymatic procedure for removing DNA template from RNA transcription mixtures. BioTechniques. 1991;10:210–214. [PubMed] [Google Scholar]
- 10.Kolykhalov A A, Agapov E V, Blight K J, Mihalik K, Feinstone S M, Rice C M. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 11.Krieger N, Lohmann V, Bartenschlager R. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J Virol. 2001;75:4614–4624. doi: 10.1128/JVI.75.10.4614-4624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lohmann V, Korner F, Dobierzewska A, Bartenschlager R. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J Virol. 2001;75:1437–1449. doi: 10.1128/JVI.75.3.1437-1449.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 14.Lohmann V, Overton H, Bartenschlager R. Selective stimulation of hepatitis C virus and pestivirus NS5B RNA polymerase activity by GTP. J Biol Chem. 1999;274:10807–10815. doi: 10.1074/jbc.274.16.10807. [DOI] [PubMed] [Google Scholar]
- 15.McHutchison J G, Gordon S C, Schiff E R, Shiffman M L, Lee W M, Rustgi V K, Goodman Z D, Ling M H, Cort S, Albrecht J K. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- 16.Pietschmann T, Lohmann V, Rutter G, Kurpanek K, Bartenschlager R. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J Virol. 2001;75:1252–1264. doi: 10.1128/JVI.75.3.1252-1264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reed K E, Rice C M. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr Top Microbiol Immunol. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- 18.Sachs A B. Cell cycle-dependent translation initiation: IRES elements prevail. Cell. 2000;101:243–245. doi: 10.1016/s0092-8674(00)80834-x. [DOI] [PubMed] [Google Scholar]
- 19.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 19a.Sureau C, Romet-Lemonne J-L, Mullins J I, Essex M. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with a cloned circular HBV DNA. Cell. 1986;47:37–47. doi: 10.1016/0092-8674(86)90364-8. [DOI] [PubMed] [Google Scholar]
- 20.Tanji Y, Kaneko T, Satoh S, Shimotohno K. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol. 1995;69:3980–3986. doi: 10.1128/jvi.69.7.3980-3986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]