Abstract
The availability of fixed nitrogen limits overall agricultural crop production worldwide. The so-called modern “green revolution” catalyzed by the widespread application of nitrogenous fertilizer has propelled global population growth. It has led to imbalances in global biogeochemical nitrogen cycling, resulting in a “nitrogen problem” that is growing at a similar trajectory to the “carbon problem”. As a result of the increasing imbalances in nitrogen cycling and additional environmental problems such as soil acidification, there is renewed and increasing interest in increasing the contributions of biological nitrogen fixation to reduce the inputs of nitrogenous fertilizers in agriculture. Interestingly, biological nitrogen fixation, or life’s ability to convert atmospheric dinitrogen to ammonia, is restricted to microbial life and not associated with any known eukaryotes. It is not clear why plants never evolved the ability to fix nitrogen and rather form associations with nitrogen-fixing microorganisms. Perhaps it is because of the large energy demand of the process, the oxygen sensitivity of the enzymatic apparatus, or simply failure to encounter the appropriate selective pressure. Whatever the reason, it is clear that this ability of crop plants, especially cereals, would transform modern agriculture once again. Successfully engineering plants will require creating an oxygen-free niche that can supply ample energy in a tightly regulated manner to minimize energy waste and ensure the ammonia produced is assimilated. Nitrogen-fixing aerobic bacteria can perhaps provide a blueprint for engineering nitrogen-fixing plants. This short review discusses the key features of robust nitrogen fixation in the model nitrogen-fixing aerobe, gamma proteobacteria Azotobacter vinelandii, in the context of the basic requirements for engineering nitrogen-fixing plants.
Keywords: nitrogen fixation, NifLA, Fix, Rnf, plant engineering
1. Introduction
Nitrogen availability is the main limiting factor of agricultural productivity, so synthetic nitrogen fertilizers have been introduced to increase yields. These additions ensure food security but are costly and negatively affect the environment by damaging sensitive ecosystems, reducing air quality, and contributing to the nitrogen problem [1,2]. The addition of fertilizer increases the amount of reactive nitrogen in the soil. However, when rates of nitrogen addition and denitrification are unequal, the excess is transported to bodies of water, resulting in eutrophication [3,4]. This scenario is comparable to excess anthropogenic carbon dioxide production beyond the amount that can be used by plants or buried. The nitrogen problem has led to an increased interest in biological nitrogen fixation (BNF) in diazotrophs. Diazotrophs are responsible for an estimated 60% of all fixed nitrogen and are found free-living in soil, the rhizosphere of plants, or in the N-fixing nodules of legumes [5,6,7]. In the absence of other nitrogen sources, these organisms can convert nitrogen gas to ammonia enzymatically and release ammonia for use by crop plants. Increasing the productivity of BNF would help reduce the need for synthetic fertilizers and decrease the negative impacts of their use [8].
Nitrogen fixation is performed by many types of bacteria and archaea using nitrogenase [9,10]. Nitrogenase contains oxygen-sensitive iron-sulfur clusters and requires high amounts of ATP and low-potential electrons to reduce N2. Across diazotrophs, many mechanisms have been developed to ensure an oxygen-free environment for nitrogenase [11,12,13,14]. Some organisms create physical barriers to create compartments free of oxygen [15]. Obligate anaerobes and facultative anaerobes fix nitrogen in the absence of oxygen [16,17]. Photosynthetic diazotrophs fix nitrogen when photosynthesis is not being performed [12,16,18,19]. Aerobes have high respiration rates of consuming oxygen, which allows them to protect nitrogenase [17,20]. Though oxygen presence is detrimental to nitrogenase, oxygen is beneficial in aerobes, allowing greater ATP production [20]. The higher amounts of ATP produced in aerobes allow higher levels of N2 reduction than by anaerobic organisms.
Eukaryotic plants and algae have not evolved to have the ability to fix nitrogen; perhaps they have not had the right selective pressures to incorporate nitrogen fixation machinery [21]. There is significant interest in using synthetic biology and genetic engineering to transfer the ability to fix nitrogen from bacteria to plants or algae [22,23]. A potential solution is to engineer the mitochondria to express nitrogenase and cofactor biosynthesis proteins and so far has been carried out in yeast [24,25,26,27,28,29,30]. Like aerobic diazotrophs, the mitochondria produce high levels of ATP and consume oxygen. However, in addition to its complex environmental requirements, nitrogenase needs many components for assembly [31,32]. Regulatory systems would also be necessary to ensure that energy requirements and ammonia produced would be balanced with plant needs. Azotobacter vinelandii is an aerobic diazotroph that meets the needs of nitrogenase and can serve as a model for how to engineer the mitochondria to fix nitrogen effectively in the presence of oxygen. This short review outlines the features of the metabolism of A. vinelandii that could be employed as a blueprint for engineering nitrogen fixation into the mitochondria toward the goal of more sustainable agriculture. While the direct incorporation of these features into the mitochondria may be difficult, we hope to highlight the utility of these systems in A. vinelandii and that the role they play is vital to balancing nitrogen fixation with normal metabolic processes.
In order to engineer nitrogen fixation into the mitochondria, a number of essential requirements must be met [33]. One of the foremost requirements, which at first approximation seems unachievable in oxygenic phototrophs, is a niche free of oxygen. The second is meeting the high energy demand of nitrogen fixation since nitrogenase requires 16 equivalents of MgATP for each dinitrogen reduced. A third requirement is a source of low-potential electrons since eight electrons are required to reduce dinitrogen, and electrons with a reducing potential of NADH are inadequate for nitrogenase catalysis [34]. The fourth is high fidelity regulation that responds to energy status, carbon/nitrogen ratio, and oxygen presence [35]. A. vinelandii is a well-studied model diazotroph that has an inherent high rate of respiration, which is (1) an inspiration for the idea that one could successfully express active nitrogenase in the mitochondria and (2) the metabolic blueprint for implementing such a herculean feat.
2. A. vinelandii as a Blueprint
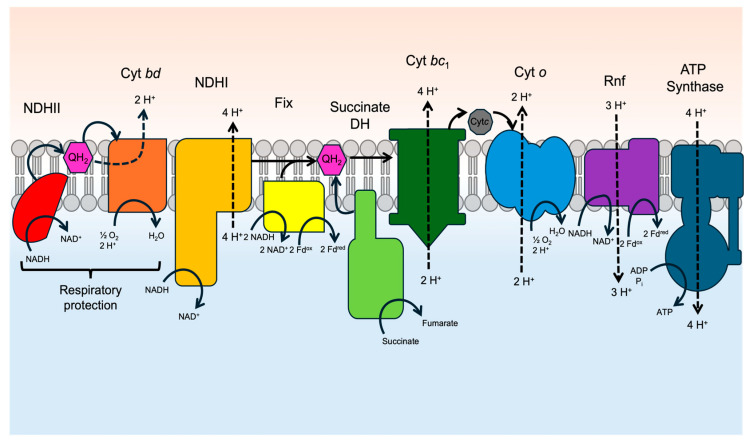
A. vinelandii possesses a fascinating mechanism that protects the nitrogen fixation apparatus from oxygen while enabling robust nitrogen fixation. The “respiratory protection mechanism”, as it is termed, takes advantage of alternative paths of electrons through the respiratory chain to modulate the rates of oxygen reduced with the amount of proton force available for ATP production [36]. Typically, electrons enter the electron transfer chain through respiratory complex I, where the oxidation of NADH is coupled to the reduction of quinone to quinol. Electrons are transferred in a similar manner through the remainder of the respiratory chain, where complex III oxidizes quinol and reduces cytochrome c, which is subsequently oxidized by complex IV coupled to the reduction of oxygen to water. The free energy release through the transfer of electrons down an electrochemical potential gradient in all three complexes (I, III, and IV) is coupled to the translocation of protons from the mitochondrial matrix of the microbial cell cytoplasm to the inner membrane space or microbial periplasm. The translocation of protons generates a proton gradient, or proton motive force, to produce ATP.
The respiratory chain of A. vinelandii includes the prototypical complexes but is supplemented with alternatives to complex I (NDHII) and complex IV (Cyt bd) (Figure 1) [37,38,39,40]. These alternative complexes are capable of oxidizing NADH and reducing O2 at higher rates than the standard complexes, but only the activity of Cyt bd translocates protons and contributes to the proton motive force (Figure 2). Modulating the expression and activity of the entire suite of respiratory complexes allows for high fidelity control of the rate of O2 reduction to protect nitrogenase from oxygen damage [37]. The respiratory protection mechanism is really the inspiration for engineering nitrogen fixation into the mitochondria of plants, where the large respiratory capacity of the organelle should be able to provide a similar reducing environment for maintaining active nitrogenase while at the same time regulating the production of ATP in concert.
Figure 1.
A. vinelandii respiratory protection, electron transport chain, and low-potential electron mechanisms. NDHII and Cyt bd are additional machinery used to increase A. vinelandii’s oxygen consumption. This occurs through the oxidation of NADH and subsequent reduction of quinone. The electrons are then transferred to Cyt bd where 2 protons are released across the membrane and 2 protons are used to form water. The typical electron transport chain consists of NDHI, Cyt bc1, Cyt c, and Cyt o. NDHI oxidizes NADH and transfers electrons to quinone while pumping out 4 protons. Quinone then passes electrons to Cyt bc1, then Cyt c, then Cyt o, where oxygen is reduced. Fix and Rnf reduce ferredoxin or flavodoxin using electrons from NADH. Solid lines indicate the movement of electrons, and dashed lines show the proton movement.
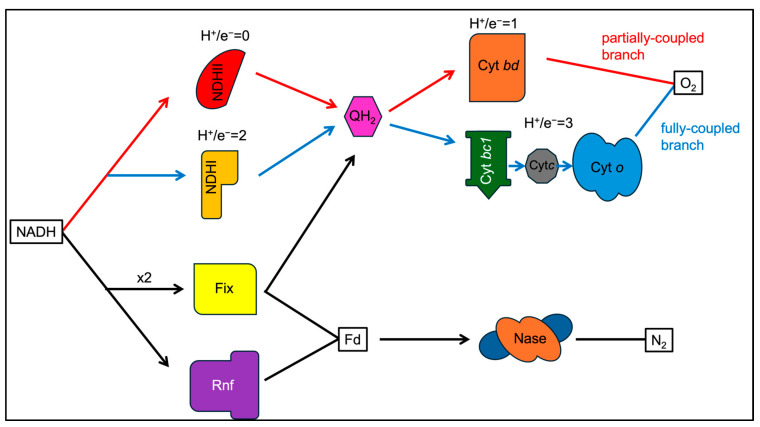
Figure 2.
Electron transport paths. Electrons from NADH can go through one of 4 paths. The first branch (shown in red) is the partially coupled branch involving NDHII and Cyt bd, where electron transport is only coupled to proton translocation in Cyt bd. The fully coupled branch (shown in blue) utilizes the typical electron transport chain machinery to pump protons out. The other two paths involve Fix and Rnf (shown in black), which generate low-potential electrons using electron bifurcation or proton motive force, respectively.
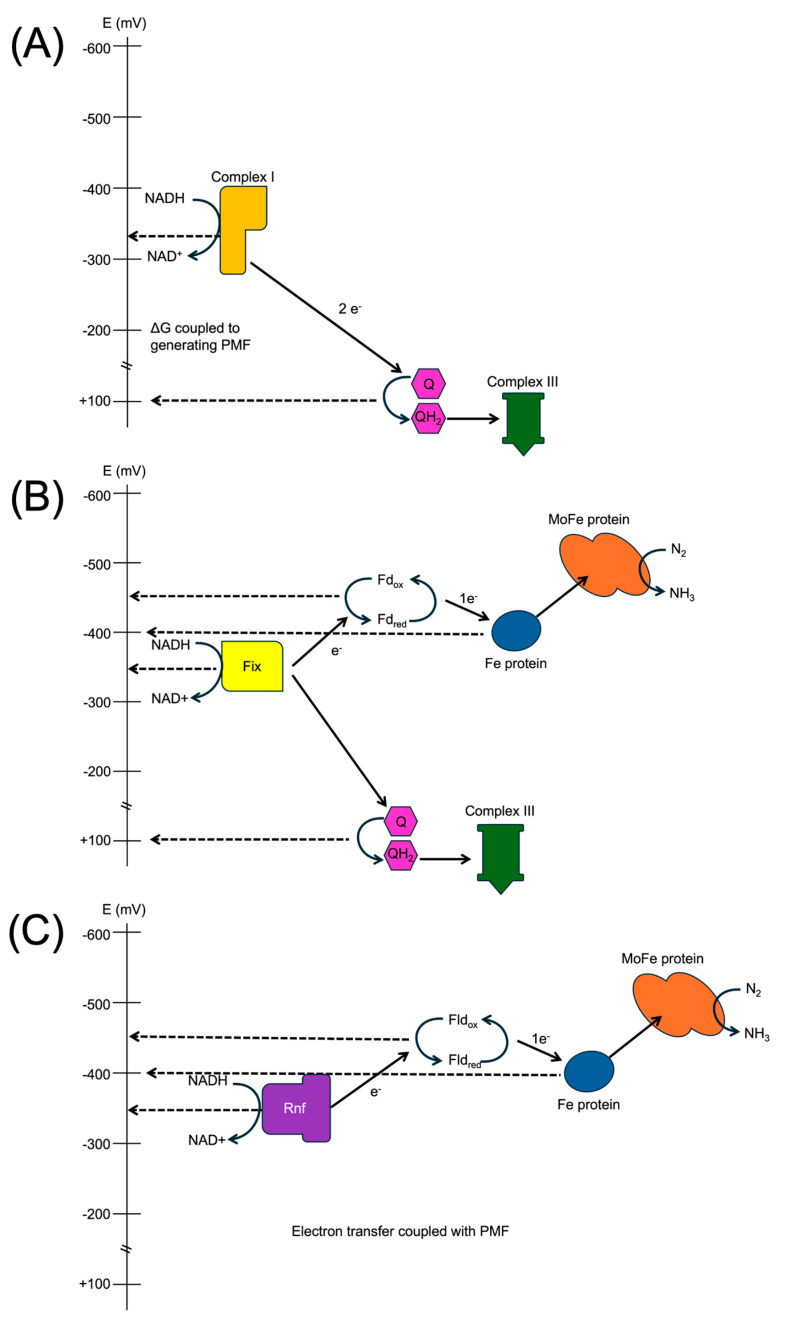
Low-potential electrons are required for biological nitrogen fixation, and the standard currency of electrons in the mitochondria and A. vinelandii is NADH. NADH has an insufficient reduction potential for nitrogenase catalysis even at very high ratios of NADH to NAD+ where the reduction potential approaches ~−400 mV. Reduction potentials approaching −500 mV are required and provided by low-potential ferredoxin and flavodoxin [41]. NADH is the primary metabolic currency of reducing equivalents in A. vinelandii; however, there are two elegant mechanisms intimately coupled to the respiratory chain and under the control of nitrogen-fixation-specific gene expression (Figure 3) [41,42,43]. The first mechanism is encoded by the Fix gene locus, which is thus termed because it has a function related to nitrogen fixation but is not formally part of the nif gene loci [44]. The FixABCX complex is a membrane-associated, heterotetrameric complex with multiple redox cofactors. This complex catalyzes an electron bifurcation reaction where the oxidation of NADH can result in the reduction of the more negative potential ferredoxin or flavodoxin when the reaction is stoichiometrically coupled to the reduction of the more positive potential quinone [43,45,46]. In this manner, half of an electron pair is diverted from the respiratory chain oxidation of NADH to the reduction of ferredoxin or flavodoxin, thereby impacting the flux of electrons to oxygen reduction and the generation of proton motive force. The second mechanism also diverts electrons from NADH away from oxygen reduction and the generation of proton motive force and simultaneously results in the consumption of proton motive force. The enzyme responsible for this activity is encoded by the Rnf genes, which are so called because their activity was implicated to be related to nitrogen fixation function [47,48]. The enzyme complex catalyzes the oxidation of NADH and the reduction of ferredoxin coupled to proton translocation from the periplasm to the cytoplasm. This exergonic translocation of protons provides the necessary free energy to drive the endergonic reduction of ferredoxin by NADH [49]. Deletion mutant analysis has shown that A. vinelandii can tolerate the deletion of either the FixABCX or the Rnf complex with limited effects under growth conditions [50]. However, the deletion of both mechanisms results in a nif-phenotype of A. vinelandii strains incapable of diazotrophic growth, indicating the requirement of these systems for low-potential electron generation for nitrogen fixation [43].
Figure 3.
NADH utilization comparison. Solid arrows indicate electron flow. (A) Complex I oxidizes NADH and transfers electrons to reduce quinone, coupling with proton transport. (B) Fix utilizes electron bifurcation to transfer one electron to high-potential quinone and one electron to low-potential ferredoxin. (C) Rnf couples electron transfer with proton motive force to reduce flavodoxin.
Respiratory protection and the activities of FixABCX and Rnf in generating low-potential electrons are two key factors that make nitrogen fixation possible in obligate aerobes. However, these additions add to the energy burden of metabolism. When you layer on the requirement of 16 ATP to reduce one N2 to two NH3 and H2, it is clear that there needs to be a highly coordinated and high-fidelity regulation system. This system must ensure that the amount of fixed nitrogen needed is balanced with oxygen reduction, generation and utilization of proton motive force, production of low-potential reducing equivalents, and production and utilization/hydrolysis of ATP. Looking forward to using these features of A. vinelandii to engineer mitochondria of crops plants for biological nitrogen fixation, it is clear without the appropriate coordinate regulation, the gene transfers would simply result in very sick plants and low crop yields [21,51].
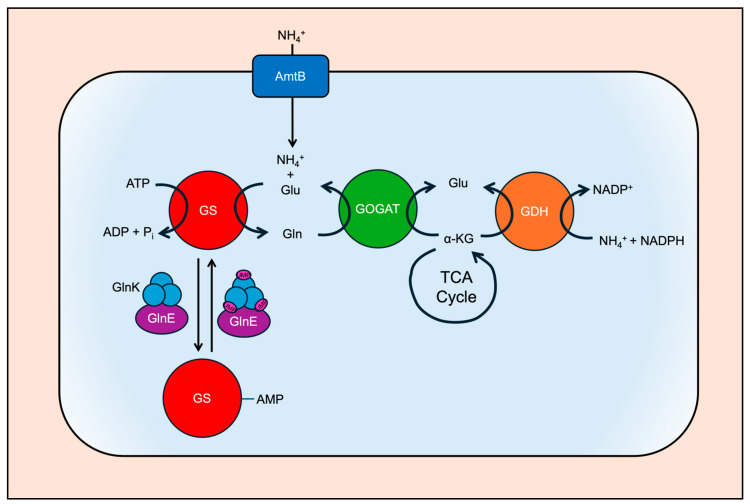
The expression of nitrogen fixation (nif) genes is controlled in response to several factors and is intimately linked to the regulation of nitrogen assimilation. Fixed nitrogen, usually ammonium ions or ammonia available in the cell’s environment, is then assimilated into amino acids. Many organisms, including bacteria and plants, use common machinery for the incorporation of fixed nitrogen, or nitrogen assimilation (Figure 4). Once imported by an Amt transporter, ammonium ions can then be assimilated by one of two different enzymatic paths [52]. The first path proceeds via the coupled reactions of glutamine synthase and glutamate synthase (also known as glutamine oxoglutarate aminotransferase), often termed GS-GOGAT. GS catalyzes the ATP-dependent incorporation of ammonium ions into glutamate to form glutamine. Then in sequence, GOGAT catalyzes the transfer of an amino group from resulting glutamine to α-ketoglutarate to form two glutamates [53,54,55]. The second enzymatic path is catalyzed by a single enzyme, glutamate dehydrogenase (GDH). GDH catalyzes the incorporation of ammonium ions onto α-ketoglutarate to form glutamate in a redox-dependent manner resulting in the oxidation of NADPH [56,57]. Though A. vinelandii contains a gene annotated as GDH, the GS-GOGAT has been shown to be the primary pathway for nitrogen assimilation [58]. Plants contain isoforms of GS, GOGAT, and GDH based on the cell location and their role in one of three assimilation pathways: primary assimilation, reassimilation of photorespiratory ammonia, and reassimilation of recycled nitrogen. In general, GS-GOGAT enzymes are used primarily due to GDH’s low affinity for ammonia [59,60,61].
Figure 4.
Nitrogen assimilation pathways in bacteria. Ammonium ions are transported into cells through Amt B, and it can then be utilized by either glutamine synthetase (GS) or glutamate dehydrogenase (GDH) to produce glutamine or glutamate, respectively. Glutamine can be utilized as a nitrogen donor to α-ketoglutarate (α-KG) to produce glutamate. Glutamine synthetase is reversibly modified by GlnE based on GlnK’s uridylyation status. In plants, ammonia is assimilated in the cytoplasm or chloroplast by GS. Ammonia can be uptaken by the roots and, in some species, produced in the mitochondria from photorespiration and proposed to passively move to the cytoplasm.
The expression of nitrogen assimilation genes is regulated by the NtrBC two component regulatory system. NtrB is a sensor histidine kinase and responds to nitrogen status through the activity of the PII protein, GlnB. Under nitrogen-limiting conditions, Gln B interactions stimulate NtrB phosphatase activity resulting in autophosphorylation on the conserved histidine in an ATP-dependent reaction where one monomer phosphorylates the other [62,63]. This process activates and phosphorylates NtrC, a σ54-dependent response regulator [64,65,66]. Under nitrogen-sufficient conditions, NtrB inactivates NtrC through dephosphorylation.
NtrC is a σ54-dependent response regulator with three domains [67,68,69,70]. The N-terminal domain acts as the receiver domain containing a conserved aspartate phosphorylation site. The central domain contains the nucleotide binding site, and the C-terminal domain contains a helix-turn-helix motif for DNA binding. When the aspartate residue on the N-terminal domain is phosphorylated, NtrC is competent to activate transcription; this induces oligomerization at upstream activator sequences and ATPase activity [71,72]. The ATPase activity, in combination with the interaction of the central domain with the sigma factor-containing RNA polymerase, activates transcription [73,74,75]. The helix-turn-helix motif of the C-terminal domain mediates DNA binding of the upstream activator sequence where the RNA polymerase binds [76,77]. NtrC’s DNA binding properties allow it to act as a transcriptional repressor when not phosphorylated.
In some nitrogen-fixing organisms, NtrBC is directly involved in the regulation of nitrogen fixation (nif) gene expression, but A. vinelandii has another layer of regulation of nitrogen fixation specific regulation that has some features in common with the NtrBC regulatory mechanisms but also some interesting twists of the two-component regulatory paradigm. The NifLA two-component regulatory system is constitutively expressed, and it regulates nitrogen fixation gene expression in response to redox status (oxygen), energy status (ATP/ADP ratios), and nitrogen status through crosstalk with the nitrogen assimilation apparatus [11].
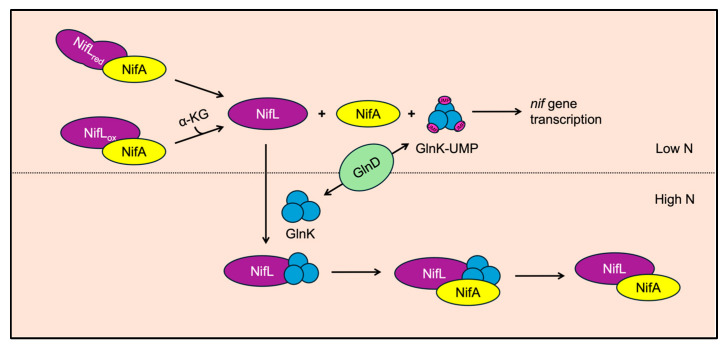
In Proteobacteria, including A. vinelandii, nitrogen fixation (nif) gene expression is controlled by NifLA, which is an interesting variation of the two-component regulatory system paradigm [35,78,79]. NifA is a σ54-dependent transcriptional activator that stimulates the expression of nif genes [80,81,82,83,84]. NifL is homologous to sensor histidine kinases (SHKs) like NtrB. However, NifL does not hydrolyze ATP and does not function as a kinase/phosphatase like NtrB. It is also not subject to phosphorylation in its role in modulating NifA activity [84]. NifL undergoes conformational changes in response to cellular signals of oxygen, energy, and fixed nitrogen status and modulates NifA activity by binding or releasing NifA (Figure 5) [85].
Figure 5.
NifLA system mechanism. To activate nif gene expression, NifA cannot be interacting with NifL. This complex dissociates when NifL is reduced or when NifA is saturated by α-ketoglutarate (a-KG). When nitrogen levels in the cell are low, GlnK is uridylylated by GlnD and unable to bind NifL. When nitrogen levels are high, GlnK is de-uridylyated and binds NifL, stimulating the formation of the NifLA-GlnK complex.
NifL senses these cellular cues and propagates the signal via its four discrete domains, similar to class I SHKs [84,86,87,88]. The amino-terminal (N-terminal) portion of NifL contains tandem PAS domains, PAS1 and PAS2. The PAS1 domain includes a solvent-accessible FAD cofactor that is readily oxidized by intracellular oxygen and is the only part of NifL with a known structure [89,90,91]. Oxidation of the PAS1 FAD causes reorganization of hydrogen bonds within the FAD-binding pocket. These changes lead to a reorientation of the non-FAD-containing PAS2 domain to stimulate NifL binding to NifA, inhibiting nitrogenase expression in oxidizing conditions [92,93,94,95]. The carboxy-terminal (C-terminal) kinase-like DH and GHKL domains perceive energy and fixed nitrogen signals [96]. The NifL DH domain is homologous to the dimerization and histidine phosphotransfer (DHp) domain that contains the conserved His in SHKs, and the GHKL domain closely resembles the catalytic domain of canonical SHKs [96,97]. However, NifL does not function either as a kinase or a phosphatase. Rather than hydrolyzing ATP to catalyze phosphorylation, the GHKL domain binds adenosine nucleotides ADP and ATP, sensing the cellular energy status via the ADP/ATP ratio and assuming a NifA-binding conformation when bound to ADP. NifL exhibits a 10-fold higher affinity for ADP than ATP, which ensures that nitrogenase is only expressed in energy-rich conditions that can support the energetic demands of nitrogenase [88]. Similar to the sensor kinase domain of NtrB, NifL also perceives nitrogen status through interactions with the PII protein GlnK [98,99,100].
As a member of the PII protein family, GlnK is post-translationally modified by GlnD based on the α-ketoglutarate/glutamine ratio in the cell [100]. The uridylylation state of GlnK modulates interactions with NifL; GlnK is reversibly uridylylated by GlnD. In low-nitrogen conditions, GlnK is uridylylated and it is unable to interact with NifL. Once deuridylylated in high-nitrogen conditions, GlnK can interact with NifL and stimulate the formation of the NifLA complex [98]. Though the interactions between NifL and GlnK and the role of alpha-ketoglutarate and nucleotides in these interactions are not clearly understood, it is clear that GlnK plays an important role in communicating information about the cell’s fixed nitrogen status.
NifLA’s regulation is robust and can respond to elements needed to identify optimal conditions for nitrogen fixation in bacteria, which suggests that it should be effective at responding to these elements in plants to ensure the process is balanced with the plant metabolism. It is unclear whether there will be compatibility with the plant’s nitrogen assimilation machinery and NifLA’s regulation; however, NifLA’s mechanism for sensing carbon/nitrogen status will compensate for incompatibility. The role of GlnE in nitrogen assimilation is to promote the inactivation of glutamine synthetase (GS) based on the uridylylation status of GlnK. In the absence of GlnE, GS is always active, and glutamine is continuously produced in the presence of ammonium ions. Previous work in microbial systems that do not fix nitrogen has shown that the deletion of GlnE results in growth limitations in the presence of excess ammonium ions [101,102]. This has been interpreted to result from the absence of GS regulation and the consumption of α-ketoglutarate, resulting in a stall in carbon metabolism. In our previous work, we have shown that when GlnE is deleted in A. vinelandii, an analogous phenotype is observed when grown under nitrogen-replete conditions, but growth in diazotrophic conditions overcomes this deleterious growth phenotype [103]. We attributed this growth phenotype to previous observations that NifA can bind α-ketoglutarate and that binding regulates NifA’s ability to promote the transcription of nif genes. These results indicate that the NifLA system has its own high-fidelity sensor of carbon/nitrogen status and is not dependent on the crosstalk with the nitrogen assimilation regulatory machinery.
3. Conclusions
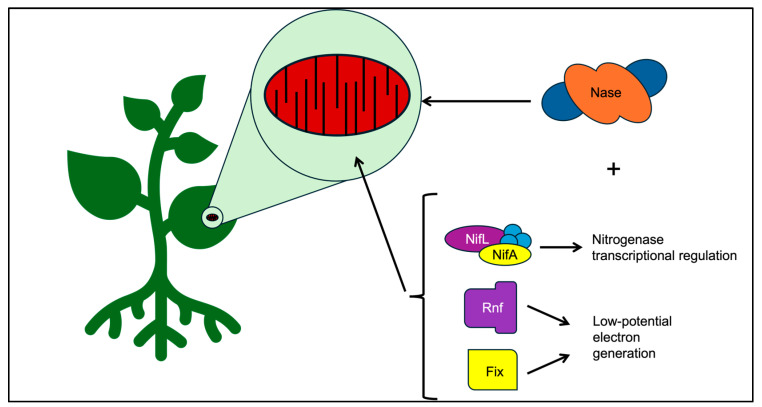
The quest for engineering nitrogen fixation plants has several perceived barriers, and probably some that still need to be realized. The perceived barriers include providing an appropriate niche in a plant where the nitrogen fixation enzymatic machinery can be protected from oxygen and where there are ample energy reserves to accomplish energy-intensive nitrogen reduction. The mitochondrion is a niche that can satisfy these requirements, and nitrogenase and cofactor synthesis proteins have been successfully expressed and targeted for localization in the mitochondria of yeast [29,30]. Still, several modifications are needed to convert it to a nitrogen-fixing organelle in a manner that does not simply result in severe growth limitations. A. vinelandii utilizes a high respiration rate, membrane-associated complexes for low-potential electron generation, and robust transcriptional regulation to fix nitrogen efficiently in the air, making it an ideal blueprint for several features (Figure 6).
Figure 6.
Summary of proposed components based on A. vinelandii as a blueprint. A. vinelandii contains NifLA, Rnf, and Fix systems that ensure the needs of nitrogenase are met. NifLA regulates transcription of nitrogenase based on cellular energy levels, oxygen presence, and nitrogen levels. Rnf and Fix produce low-potential electrons for use by nitrogenase.
Author Contributions
Conceptualization, S.B., F.M. and J.W.P.; writing—review and editing, S.B., F.M. and J.W.P. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research was funded by US DOE, Office of Science, Office of Basic Energy Sciences, grant number DE-SC0024311.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Borlaug N.E. Feeding a World of 10 Billion People: The Miracle Ahead. Biotechnol. Biotechnol. Equip. 1997;11:3–13. doi: 10.1080/13102818.1997.10818934. [DOI] [Google Scholar]
- 2.Erisman J.W., Galloway J., Dise N., Bleeker A., Grizzetti B., Leach A., Vries W. Nitrogen: Too Much of a Vital Resource. WWF; Zeist, The Netherlands: 2015. [Google Scholar]
- 3.Ahmed M., Rauf M., Mukhtar Z., Saeed N.A. Excessive Use of Nitrogenous Fertilizers: An Unawareness Causing Serious Threats to Environment and Human Health. Environ. Sci. Pollut. Res. 2017;24:26983–26987. doi: 10.1007/s11356-017-0589-7. [DOI] [PubMed] [Google Scholar]
- 4.Gruber N., Galloway J.N. An Earth-System Perspective of the Global Nitrogen Cycle. Nature. 2008;451:293–296. doi: 10.1038/nature06592. [DOI] [PubMed] [Google Scholar]
- 5.Newton W.E. Chapter 8—Physiology, Biochemistry, and Molecular Biology of Nitrogen Fixation. In: Bothe H., Ferguson S.J., Newton W.E., editors. Biology of the Nitrogen Cycle. Elsevier; Amsterdam, The Netherlands: 2007. pp. 109–129. [Google Scholar]
- 6.Rubio L.M., Ludden P.W. Biosynthesis of the Iron-Molybdenum Cofactor of Nitrogenase. Annu. Rev. Microbiol. 2008;62:93–111. doi: 10.1146/annurev.micro.62.081307.162737. [DOI] [PubMed] [Google Scholar]
- 7.Fowler D., Coyle M., Skiba U., Sutton M.A., Cape J.N., Reis S., Sheppard L.J., Jenkins A., Grizzetti B., Galloway J.N., et al. The Global Nitrogen Cycle in the Twenty-First Century. Philos. Trans. R. Soc. B Biol. Sci. 2013;368:20130164. doi: 10.1098/rstb.2013.0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Udvardi M., Below F.E., Castellano M.J., Eagle A.J., Giller K.E., Ladha J.K., Liu X., Maaz T.M., Nova-Franco B., Raghuram N., et al. A Research Road Map for Responsible Use of Agricultural Nitrogen. Front. Sustain. Food Syst. 2021;5:660155. doi: 10.3389/fsufs.2021.660155. [DOI] [Google Scholar]
- 9.Hoffman B.M., Lukoyanov D., Yang Z.-Y., Dean D.R., Seefeldt L.C. Mechanism of Nitrogen Fixation by Nitrogenase: The Next Stage. Chem. Rev. 2014;114:4041–4062. doi: 10.1021/cr400641x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyd E.S., Hamilton T.L., Peters J.W. An Alternative Path for the Evolution of Biological Nitrogen Fixation. Front. Microbiol. 2011;2:205. doi: 10.3389/fmicb.2011.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixon R., Kahn D. Genetic Regulation of Biological Nitrogen Fixation. Nat. Rev. Microbiol. 2004;2:621–631. doi: 10.1038/nrmicro954. [DOI] [PubMed] [Google Scholar]
- 12.Boyd E., Peters J.W. New Insights into the Evolutionary History of Biological Nitrogen Fixation. Front. Microbiol. 2013;4:201. doi: 10.3389/fmicb.2013.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dos Santos P.C., Fang Z., Mason S.W., Setubal J.C., Dixon R. Distribution of Nitrogen Fixation and Nitrogenase-like Sequences amongst Microbial Genomes. BMC Genom. 2012;13:162. doi: 10.1186/1471-2164-13-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyd E.S., Anbar A.D., Miller S., Hamilton T.L., Lavin M., Peters J.W. A Late Methanogen Origin for Molybdenum-Dependent Nitrogenase. Geobiology. 2011;9:221–232. doi: 10.1111/j.1472-4669.2011.00278.x. [DOI] [PubMed] [Google Scholar]
- 15.Minchin F.R., James E.K., Becana M. Oxygen Diffusion, Production Of Reactive Oxygen And Nitrogen Species, And Antioxidants In Legume Nodules. In: Dilworth M.J., James E.K., Sprent J.I., Newton W.E., editors. Nitrogen-Fixing Leguminous Symbioses. Springer; Dordrecht, The Netherlands: 2008. pp. 321–362. [Google Scholar]
- 16.Gallon J.R. The Oxygen Sensitivity of Nitrogenase: A Problem for Biochemists and Micro-Organisms. Trends Biochem. Sci. 1981;6:19–23. doi: 10.1016/0968-0004(81)90008-6. [DOI] [Google Scholar]
- 17.Yates M.G., Jones C.W. Respiration and Nitrogen Fixation in Azotobacter. In: Rose A.H., Tempest D.W., editors. Advances in Microbial Physiology. Volume 11. Academic Press; Cambridge, MA, USA: 1974. pp. 97–135. [Google Scholar]
- 18.Berman-Frank I., Lundgren P., Falkowski P. Nitrogen Fixation and Photosynthetic Oxygen Evolution in Cyanobacteria. Res. Microbiol. 2003;154:157–164. doi: 10.1016/S0923-2508(03)00029-9. [DOI] [PubMed] [Google Scholar]
- 19.Fay P. Oxygen Relations of Nitrogen Fixation in Cyanobacteria. Microbiol. Rev. 1992;56:340–373. doi: 10.1128/mr.56.2.340-373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poole R.K., Hill S. Respiratory Protection of Nitrogenase Activity in Azotobacter Vinelandii—Roles of the Terminal Oxidases. Biosci. Rep. 1997;17:303–317. doi: 10.1023/A:1027336712748. [DOI] [PubMed] [Google Scholar]
- 21.Hutson M. Will Plants Ever Fertilize Themselves? The New Yorker. Feb 6, 2024.
- 22.Mus F., Crook M.B., Garcia K., Garcia Costas A., Geddes B.A., Kouri E.D., Paramasivan P., Ryu M.-H., Oldroyd G.E.D., Poole P.S., et al. Symbiotic Nitrogen Fixation and the Challenges to Its Extension to Nonlegumes. Appl. Environ. Microbiol. 2016;82:3698–3710. doi: 10.1128/AEM.01055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogers C., Oldroyd G.E.D. Synthetic Biology Approaches to Engineering the Nitrogen Symbiosis in Cereals. J. Exp. Bot. 2014;65:1939–1946. doi: 10.1093/jxb/eru098. [DOI] [PubMed] [Google Scholar]
- 24.Pankievicz V.C.S., Irving T.B., Maia L.G.S., Ané J.-M. Are We There yet? The Long Walk towards the Development of Efficient Symbiotic Associations between Nitrogen-Fixing Bacteria and Non-Leguminous Crops. BMC Biol. 2019;17:99. doi: 10.1186/s12915-019-0710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burén S., Rubio L.M. State of the Art in Eukaryotic Nitrogenase Engineering. FEMS Microbiol. Lett. 2018;365:fnx274. doi: 10.1093/femsle/fnx274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beatty P.H., Good A.G. Future Prospects for Cereals That Fix Nitrogen. Science. 2011;333:416–417. doi: 10.1126/science.1209467. [DOI] [PubMed] [Google Scholar]
- 27.Curatti L., Rubio L.M. Challenges to Develop Nitrogen-Fixing Cereals by Direct Nif-Gene Transfer. Plant Sci. 2014;225:130–137. doi: 10.1016/j.plantsci.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Guo K., Yang J., Yu N., Luo L., Wang E. Biological Nitrogen Fixation in Cereal Crops: Progress, Strategies, and Perspectives. Plant Commun. 2023;4:100499. doi: 10.1016/j.xplc.2022.100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dobrzyńska K., Pérez-González A., Echavarri-Erasun C., Coroian D., Salinero-Lanzarote A., Veldhuizen M., Dean D.R., Burén S., Rubio L.M. Nitrogenase Cofactor Biosynthesis Using Proteins Produced in Mitochondria of Saccharomyces Cerevisiae. mBio. 2023;15:e03088-23. doi: 10.1128/mbio.03088-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burén S., Young E.M., Sweeny E.A., Lopez-Torrejón G., Veldhuizen M., Voigt C.A., Rubio L.M. Formation of Nitrogenase NifDK Tetramers in the Mitochondria of Saccharomyces Cerevisiae. ACS Synth. Biol. 2017;6:1043–1055. doi: 10.1021/acssynbio.6b00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burén S., Jiménez-Vicente E., Echavarri-Erasun C., Rubio L.M. Biosynthesis of Nitrogenase Cofactors. Chem. Rev. 2020;120:4921–4968. doi: 10.1021/acs.chemrev.9b00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lill R., Mühlenhoff U. Maturation of Iron-Sulfur Proteins in Eukaryotes: Mechanisms, Connected Processes, and Diseases. Annu. Rev. Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 33.Seefeldt L.C., Hoffman B.M., Dean D.R. Mechanism of Mo-Dependent Nitrogenase. Annu. Rev. Biochem. 2009;78:701–722. doi: 10.1146/annurev.biochem.78.070907.103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robson R.L., Postgate J.R. Oxygen and Hydrogen in Biological Nitrogen Fixation. Annu. Rev. Microbiol. 1980;34:183–207. doi: 10.1146/annurev.mi.34.100180.001151. [DOI] [PubMed] [Google Scholar]
- 35.Bueno Batista M., Dixon R. Manipulating Nitrogen Regulation in Diazotrophic Bacteria for Agronomic Benefit. Biochem. Soc. Trans. 2019;47:603–614. doi: 10.1042/BST20180342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oelze J. Respiratory Protection of Nitrogenase in Azotobacter Species: Is a Widely Held Hypothesis Unequivocally Supported by Experimental Evidence? FEMS Microbiol. Rev. 2000;24:321–333. doi: 10.1111/j.1574-6976.2000.tb00545.x. [DOI] [PubMed] [Google Scholar]
- 37.D’mello R., Purchase D., Poole R.K., Hill S. Expression and Content of Terminal Oxidases in Azotobacter Vinelandii Grown with Excess NH4+ Are Modulated by O2 Supply. Microbiology. 1997;143:231–237. doi: 10.1099/00221287-143-1-231. [DOI] [PubMed] [Google Scholar]
- 38.D’Mello R., Hill S., Poole R.K. Determination of the Oxygen Affinities of Terminal Oxidases in Azotobacter Vinelandii Using the Deoxygenation of Oxyleghaemoglobin and Oxymyoglobin: Cytochrome Bd Is a Low-Affinity Oxidase. Microbiology. 1994;140:1395–1402. doi: 10.1099/00221287-140-6-1395. [DOI] [Google Scholar]
- 39.Rey L., Maier R.J. Cytochrome c Terminal Oxidase Pathways of Azotobacter Vinelandii: Analysis of Cytochrome C4 and C5 Mutants and up-Regulation of Cytochrome c-Dependent Pathways with N2 Fixation. J. Bacteriol. 1997;179:7191–7196. doi: 10.1128/jb.179.22.7191-7196.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moshiri F., Chawla A., Maier R.J. Cloning, Characterization, and Expression in Escherichia coli of the Genes Encoding the Cytochrome d Oxidase Complex from Azotobacter Vinelandii. J. Bacteriol. 1991;173:6230–6241. doi: 10.1128/jb.173.19.6230-6241.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alleman A.B., Peters J.W. Mechanisms for Generating Low Potential Electrons across the Metabolic Diversity of Nitrogen-Fixing Bacteria. Appl. Environ. Microbiol. 2023;89:e00378-23. doi: 10.1128/aem.00378-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyd E.S., Costas A.M.G., Hamilton T.L., Mus F., Peters J.W. Evolution of Molybdenum Nitrogenase during the Transition from Anaerobic to Aerobic Metabolism. J. Bacteriol. 2015;197:1690–1699. doi: 10.1128/JB.02611-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ledbetter R.N., Garcia Costas A.M., Lubner C.E., Mulder D.W., Tokmina-Lukaszewska M., Artz J.H., Patterson A., Magnuson T.S., Jay Z.J., Duan H.D., et al. The Electron Bifurcating FixABCX Protein Complex from Azotobacter Vinelandii: Generation of Low-Potential Reducing Equivalents for Nitrogenase Catalysis. Biochemistry. 2017;56:4177–4190. doi: 10.1021/acs.biochem.7b00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pühler A., Aguilar M.O., Hynes M., Müller P., Klipp W., Priefer U., Simon R., Weber G. Advances in the Genetics of Free-Living and Symbiotic Nitrogen Fixing Bacteria. In: Veeger C., Newton W.E., editors. Advances in Nitrogen Fixation Research: Proceedings of the 5th International Symposium on Nitrogen Fixation, Noordwijkerhout, The Netherlands, 28 August–3 September 1983. Springer; Dordrecht, The Netherlands: 1984. pp. 609–619. [Google Scholar]
- 45.Peters J.W., Miller A.-F., Jones A.K., King P.W., Adams M.W. Electron Bifurcation. Curr. Opin. Chem. Biol. 2016;31:146–152. doi: 10.1016/j.cbpa.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 46.Peters J.W., Beratan D.N., Bothner B., Dyer R.B., Harwood C.S., Heiden Z.M., Hille R., Jones A.K., King P.W., Lu Y., et al. A New Era for Electron Bifurcation. Curr. Opin. Chem. Biol. 2018;47:32–38. doi: 10.1016/j.cbpa.2018.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saeki K., Tokuda K., Fujiwara T., Matsubara H. Nucleotide Sequence and Genetic Analysis of the Region Essential for Functional Expression of the Gene for Ferredoxin I, fdxN, in Rhodobacter Capsulatus: Sharing of One Upstream Activator Sequence in Opposite Directions by Two Operons Related to Nitrogen Fixation. Plant Cell Physiol. 1993;34:185–199. doi: 10.1093/oxfordjournals.pcp.a078406. [DOI] [PubMed] [Google Scholar]
- 48.Schmehl M., Jahn A., Meyer zu Vilsendorf A., Hennecke S., Masepohl B., Schuppler M., Marxer M., Oelze J., Klipp W. Identification of a New Class of Nitrogen Fixation Genes in Rhodobacter Capsalatus: A Putative Membrane Complex Involved in Electron Transport to Nitrogenase. Mol. Gen. Genet. MGG. 1993;241:602–615. doi: 10.1007/BF00279903. [DOI] [PubMed] [Google Scholar]
- 49.Bertsova Y.V., Serebryakova M.V., Baykov A.A., Bogachev A.V. The Flavin Transferase ApbE Flavinylates the Ferredoxin:NAD+-Oxidoreductase Rnf Required for N2 Fixation in Azotobacter Vinelandii. FEMS Microbiol. Lett. 2021;368:fnab130. doi: 10.1093/femsle/fnab130. [DOI] [PubMed] [Google Scholar]
- 50.Alleman A.B., Garcia Costas A., Mus F., Peters J.W. Rnf and Fix Have Specific Roles during Aerobic Nitrogen Fixation in Azotobacter Vinelandii. Appl. Environ. Microbiol. 2022;88:e01049-22. doi: 10.1128/aem.01049-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang W., Chen Y., Huang K., Wang F., Mei Z. Molecular Mechanism and Agricultural Application of the NifA–NifL System for Nitrogen Fixation. Int. J. Mol. Sci. 2023;24:907. doi: 10.3390/ijms24020907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Heeswijk W.C., Westerhoff H.V., Boogerd F.C. Nitrogen Assimilation in Escherichia coli: Putting Molecular Data into a Systems Perspective. Microbiol. Mol. Biol. Rev. 2013;77:628–695. doi: 10.1128/MMBR.00025-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alibhai M., Villafranca J.J. Kinetic and Mutagenic Studies of the Role of the Active Site Residues Asp-50 and Glu-327 of Escherichia coli Glutamine Synthetase. Biochemistry. 1994;33:682–686. doi: 10.1021/bi00169a008. [DOI] [PubMed] [Google Scholar]
- 54.Colanduoni J., Nissan R., Villafranca J.J. Studies of the Mechanism of Glutamine Synthetase Utilizing pH-Dependent Behavior in Catalysis and Binding. J. Biol. Chem. 1987;262:3037–3043. doi: 10.1016/S0021-9258(18)61465-6. [DOI] [PubMed] [Google Scholar]
- 55.Meek T.D., Villafranca J.J. Kinetic Mechanism of Escherichia coli Glutamine Synthetase. Biochemistry. 1980;19:5513–5519. doi: 10.1021/bi00565a008. [DOI] [PubMed] [Google Scholar]
- 56.Sakamoto N., Kotre A.M., Savageau M.A. Glutamate Dehydrogenase from Escherichia coli: Purification and Properties. J. Bacteriol. 1975;124:775–783. doi: 10.1128/jb.124.2.775-783.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharkey M.A., Engel P.C. Apparent Negative Co-Operativity and Substrate Inhibition in Overexpressed Glutamate Dehydrogenase from Escherichia coli. FEMS Microbiol. Lett. 2008;281:132–139. doi: 10.1111/j.1574-6968.2008.01086.x. [DOI] [PubMed] [Google Scholar]
- 58.Kennedy C., Toukdarian A. Genetics of Azotobacters:Applications to Nitrogen Fixation and Related Aspects of Metabolism. Annu. Rev. Microbiol. 1987;41:227–258. doi: 10.1146/annurev.mi.41.100187.001303. [DOI] [PubMed] [Google Scholar]
- 59.Howitt S.M., Udvardi M.K. Structure, Function and Regulation of Ammonium Transporters in Plants. Biochim. Biophys. Acta BBA—Biomembr. 2000;1465:152–170. doi: 10.1016/S0005-2736(00)00136-X. [DOI] [PubMed] [Google Scholar]
- 60.Lam H.-M., Coschigano K.T., Oliveira I.C., Melo-Oliveira R., Coruzzi G.M. THE MOLECULAR-GENETICS OF NITROGEN ASSIMILATION INTO AMINO ACIDS IN HIGHER PLANTS. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;47:569–593. doi: 10.1146/annurev.arplant.47.1.569. [DOI] [PubMed] [Google Scholar]
- 61.Schjoerring J.K., Husted S., Mäck G., Mattsson M. The Regulation of Ammonium Translocation in Plants. J. Exp. Bot. 2002;53:883–890. doi: 10.1093/jexbot/53.370.883. [DOI] [PubMed] [Google Scholar]
- 62.Ninfa A.J., Bennett R.L. Identification of the Site of Autophosphorylation of the Bacterial Protein Kinase/Phosphatase NRII. J. Biol. Chem. 1991;266:6888–6893. doi: 10.1016/S0021-9258(20)89585-4. [DOI] [PubMed] [Google Scholar]
- 63.Ninfa E.G., Atkinson M.R., Kamberov E.S., Ninfa A.J. Mechanism of Autophosphorylation of Escherichia coli Nitrogen Regulator II (NRII or NtrB): Trans-Phosphorylation between Subunits. J. Bacteriol. 1993;175:7024–7032. doi: 10.1128/jb.175.21.7024-7032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keener J., Kustu S. Protein Kinase and Phosphoprotein Phosphatase Activities of Nitrogen Regulatory Proteins NTRB and NTRC of Enteric Bacteria: Roles of the Conserved Amino-Terminal Domain of NTRC. Proc. Natl. Acad. Sci. USA. 1988;85:4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ninfa A.J., Magasanik B. Covalent Modification of the glnG Product, NRI, by the glnL Product, NRII, Regulates the Transcription of the glnALG Operon in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1986;83:5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weiss V., Magasanik B. Phosphorylation of Nitrogen Regulator I (NRI) of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1988;85:8919–8923. doi: 10.1073/pnas.85.23.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reitzer L.J., Magasanik B. Isolation of the Nitrogen Assimilation Regulator NR(I), the Product of the glnG Gene of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1983;80:5554–5558. doi: 10.1073/pnas.80.18.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morett E., Segovia L. The Sigma 54 Bacterial Enhancer-Binding Protein Family: Mechanism of Action and Phylogenetic Relationship of Their Functional Domains. J. Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kustu S., Santero E., Keener J., Popham D., Weiss D. Expression of Sigma 54 (ntrA)-Dependent Genes Is Probably United by a Common Mechanism. Microbiol. Rev. 1989;53:367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Drummond M., Whitty P., Wootton J. Sequence and Domain Relationships of ntrC and nifA from Klebsiella Pneumoniae: Homologies to Other Regulatory Proteins. EMBO J. 1986;5:441–447. doi: 10.1002/j.1460-2075.1986.tb04230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Makino K., Amemura M., Kim S.K., Nakata A., Shinagawa H. Role of the Sigma 70 Subunit of RNA Polymerase in Transcriptional Activation by Activator Protein PhoB in Escherichia coli. Genes Dev. 1993;7:149–160. doi: 10.1101/gad.7.1.149. [DOI] [PubMed] [Google Scholar]
- 72.Stewart V. Nitrate Regulation of Anaerobic Respiratory Gene Expression in Escherichia coli. Mol. Microbiol. 1993;9:425–434. doi: 10.1111/j.1365-2958.1993.tb01704.x. [DOI] [PubMed] [Google Scholar]
- 73.Weiss D.S., Batut J., Klose K.E., Keener J., Kustu S. The Phosphorylated Form of the Enhancer-Binding Protein NTRC Has an ATPase Activity That Is Essential for Activation of Transcription. Cell. 1991;67:155–167. doi: 10.1016/0092-8674(91)90579-N. [DOI] [PubMed] [Google Scholar]
- 74.Austin S., Dixon R. The Prokaryotic Enhancer Binding Protein NTRC Has an ATPase Activity Which Is Phosphorylation and DNA Dependent. EMBO J. 1992;11:2219–2228. doi: 10.1002/j.1460-2075.1992.tb05281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wedel A., Kustu S. The Bacterial Enhancer-Binding Protein NTRC Is a Molecular Machine: ATP Hydrolysis Is Coupled to Transcriptional Activation. Genes Dev. 1995;9:2042–2052. doi: 10.1101/gad.9.16.2042. [DOI] [PubMed] [Google Scholar]
- 76.Porter S.C., North A.K., Wedel A.B., Kustu S. Oligomerization of NTRC at the glnA Enhancer Is Required for Transcriptional Activation. Genes Dev. 1993;7:2258–2273. doi: 10.1101/gad.7.11.2258. [DOI] [PubMed] [Google Scholar]
- 77.Klose K.E., North A.K., Stedman K.M., Kustu S. The Major Dimerization Determinants of the Nitrogen Regulatory Protein NTRC from Enteric Bacteria Lie in Its Carboxy-Terminal Domain. J. Mol. Biol. 1994;241:233–245. doi: 10.1006/jmbi.1994.1492. [DOI] [PubMed] [Google Scholar]
- 78.Martinez-Argudo I., Little R., Shearer N., Johnson P., Dixon R. The NifL-NifA System: A Multidomain Transcriptional Regulatory Complex That Integrates Environmental Signals. J. Bacteriol. 2004;186:601–610. doi: 10.1128/JB.186.3.601-610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dixon R. The Oxygen-Responsive NIFL-NIFA Complex: A Novel Two-Component Regulatory System Controlling Nitrogenase Synthesis in γ-Proteobacteria. Arch. Microbiol. 1998;169:371–380. doi: 10.1007/s002030050585. [DOI] [PubMed] [Google Scholar]
- 80.Poza-Carrión C., Jiménez-Vicente E., Navarro-Rodríguez M., Echavarri-Erasun C., Rubio L.M. Kinetics of Nif Gene Expression in a Nitrogen-Fixing Bacterium. J. Bacteriol. 2014;196:595–603. doi: 10.1128/JB.00942-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lei S., Pulakat L., Gavini N. Genetic Analysis of Nif Regulatory Genes by Utilizing the Yeast Two-Hybrid System Detected Formation of a NifL-NifA Complex That Is Implicated in Regulated Expression of Nif Genes. J. Bacteriol. 1999;181:6535–6539. doi: 10.1128/JB.181.20.6535-6539.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Little R., Dixon R. The Amino-Terminal GAF Domain of Azotobacter Vinelandii NifA Binds 2-Oxoglutarate to Resist Inhibition by NifL under Nitrogen-Limiting Conditions *. J. Biol. Chem. 2003;278:28711–28718. doi: 10.1074/jbc.M301992200. [DOI] [PubMed] [Google Scholar]
- 83.Martinez-Argudo I., Little R., Dixon R. Role of the Amino-Terminal GAF Domain of the NifA Activator in Controlling the Response to the Antiactivator Protein NifL. Mol. Microbiol. 2004;52:1731–1744. doi: 10.1111/j.1365-2958.2004.04089.x. [DOI] [PubMed] [Google Scholar]
- 84.Austin S., Buck M., Cannon W., Eydmann T., Dixon R. Purification and in Vitro Activities of the Native Nitrogen Fixation Control Proteins NifA and NifL. J. Bacteriol. 1994;176:3460–3465. doi: 10.1128/jb.176.12.3460-3465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boyer N.R., Tokmina-Lukaszewska M., Bueno Batista M., Mus F., Dixon R., Bothner B., Peters J.W. Structural Insights into Redox Signal Transduction Mechanisms in the Control of Nitrogen Fixation by the NifLA System. Proc. Natl. Acad. Sci. USA. 2023;120:e2302732120. doi: 10.1073/pnas.2302732120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martinez-Argudo I., Little R., Dixon R. A Crucial Arginine Residue Is Required for a Conformational Switch in NifL to Regulate Nitrogen Fixation in Azotobacter Vinelandii. Proc. Natl. Acad. Sci. USA. 2004;101:16316–16321. doi: 10.1073/pnas.0405312101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Little R., Reyes-Ramirez F., Zhang Y., van Heeswijk W.C., Dixon R. Signal Transduction to the Azotobacter Vinelandii NIFL–NIFA Regulatory System Is Influenced Directly by Interaction with 2-Oxoglutarate and the PII Regulatory Protein. EMBO J. 2000;19:6041–6050. doi: 10.1093/emboj/19.22.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Söderbäck E., Reyes-Ramirez F., Eydmann T., Austin S., Hill S., Dixon R. The Redox- and Fixed Nitrogen-Responsive Regulatory Protein NIFL from Azotobacter Vinelandii Comprises Discrete Flavin and Nucleotide-Binding Domains. Mol. Microbiol. 1998;28:179–192. doi: 10.1046/j.1365-2958.1998.00788.x. [DOI] [PubMed] [Google Scholar]
- 89.Key J., Hefti M., Purcell E.B., Moffat K. Structure of the Redox Sensor Domain of Azotobacter Vinelandii NifL at Atomic Resolution: Signaling, Dimerization, and Mechanism. Biochemistry. 2007;46:3614–3623. doi: 10.1021/bi0620407. [DOI] [PubMed] [Google Scholar]
- 90.Slavny P., Little R., Salinas P., Clarke T.A., Dixon R. Quaternary Structure Changes in a Second Per-Arnt-Sim Domain Mediate Intramolecular Redox Signal Relay in the NifL Regulatory Protein. Mol. Microbiol. 2010;75:61–75. doi: 10.1111/j.1365-2958.2009.06956.x. [DOI] [PubMed] [Google Scholar]
- 91.Little R., Salinas P., Slavny P., Clarke T.A., Dixon R. Substitutions in the Redox-Sensing PAS Domain of the NifL Regulatory Protein Define an Inter-Subunit Pathway for Redox Signal Transmission. Mol. Microbiol. 2011;82:222–235. doi: 10.1111/j.1365-2958.2011.07812.x. [DOI] [PubMed] [Google Scholar]
- 92.Little R., Martinez-Argudo I., Dixon R. Role of the Central Region of NifL in Conformational Switches That Regulate Nitrogen Fixation. Biochem. Soc. Trans. 2006;34:162–164. doi: 10.1042/BST0340162. [DOI] [PubMed] [Google Scholar]
- 93.Slavny P. Ph.D. Thesis. University of East Anglia; Norwich, UK: 2010. Role of the PAS2 Domain of the NifL Regulatory Protein in Redox Signal Transduction. [Google Scholar]
- 94.Macheroux P., Hill S., Austin S., Eydmann T., Jones T., Kim S.O., Poole R., Dixon R. Electron Donation to the Flavoprotein NifL, a Redox-Sensing Transcriptional Regulator. Biochem. J. 1998;332:413–419. doi: 10.1042/bj3320413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hill S., Austin S., Eydmann T., Jones T., Dixon R. Azotobacter Vinelandii NIFL Is a Flavoprotein That Modulates Transcriptional Activation of Nitrogen-Fixation Genes via a Redox-Sensitive Switch. Proc. Natl. Acad. Sci. USA. 1996;93:2143–2148. doi: 10.1073/pnas.93.5.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bhate M.P., Molnar K.S., Goulian M., DeGrado W.F. Signal Transduction in Histidine Kinases: Insights from New Structures. Structure. 2015;23:981–994. doi: 10.1016/j.str.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Möglich A. Signal Transduction in Photoreceptor Histidine Kinases. Protein Sci. 2019;28:1923–1946. doi: 10.1002/pro.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Little R., Colombo V., Leech A., Dixon R. Direct Interaction of the NifL Regulatory Protein with the GlnK Signal Transducer Enables the Azotobacter vinelandiiNifL-NifA Regulatory System to Respond to Conditions Replete for Nitrogen*. J. Biol. Chem. 2002;277:15472–15481. doi: 10.1074/jbc.M112262200. [DOI] [PubMed] [Google Scholar]
- 99.Arcondéguy T., Lawson D., Merrick M. Two Residues in the T-Loop of GlnK Determine NifL-Dependent Nitrogen Control of Nif Gene Expression *. J. Biol. Chem. 2000;275:38452–38456. doi: 10.1074/jbc.M001935200. [DOI] [PubMed] [Google Scholar]
- 100.Rudnick P., Kunz C., Gunatilaka M.K., Hines E.R., Kennedy C. Role of GlnK in NifL-Mediated Regulation of NifA Activity in Azotobacter Vinelandii. J. Bacteriol. 2002;184:812–820. doi: 10.1128/JB.184.3.812-820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Foor F., Janssen K.A., Magasanik B. Regulation of Synthesis of Glutamine Synthetase by Adenylylated Glutamine Synthetase. Proc. Natl. Acad. Sci. USA. 1975;72:4844–4848. doi: 10.1073/pnas.72.12.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kustu S., Hirschman J., Burton D., Jelesko J., Meeks J.C. Covalent Modification of Bacterial Glutamine Synthetase: Physiological Significance. Mol. Gen. Genet. MGG. 1984;197:309–317. doi: 10.1007/BF00330979. [DOI] [PubMed] [Google Scholar]
- 103.Mus F., Tseng A., Dixon R., Peters J.W. Diazotrophic Growth Allows Azotobacter Vinelandii To Overcome the Deleterious Effects of a glnE Deletion. Appl. Environ. Microbiol. 2017;83:e00808-17. doi: 10.1128/AEM.00808-17. [DOI] [PMC free article] [PubMed] [Google Scholar]