Abstract
Although p48 is the most conserved subunit of mammalian DNA polymerase α-primase (pol-prim), the polypeptide is the major species-specific factor for mouse polyomavirus (PyV) DNA replication. Human and murine p48 contain two regions (A and B) that show significantly lower homology than the rest of the protein. Chimerical human-murine p48 was prepared and coexpressed with three wild-type subunits of pol-prim, and four subunit protein complexes were purified. All enzyme complexes synthesized DNA on single-stranded (ss) DNA and replicated simian virus 40 DNA. Although the recombinant protein complexes physically interacted with PyV T antigen (Tag), we determined that the murine region A mediates the species specificity of PyV DNA replication in vitro. More precisely, the nonconserved phenylalanine 262 of mouse p48 is crucial for this activity, and pol-prim with mutant p48, h-S262F, supports PyV DNA replication in vitro. DNA synthesis on RPA-bound ssDNA revealed that amino acid (aa) 262, aa 266, and aa 273 to 288 are involved in the functional cooperation of RPA, pol-prim, and PyV Tag.
The small DNA tumor viruses simian virus 40 (SV40) and mouse polyomavirus (PyV) have provided excellent model systems to study the mechanisms and regulation of eukaryotic DNA replication (reviewed in references 7, 9, 55, and 67). Their DNA replication in vivo and in vitro largely depends on the replication apparatus of their hosts, as only one protein, namely, the multifunctional viral large T antigen (Tag), is supplied by the virus (20, 26, 49).
The establishment of cell-free SV40 and PyV DNA replication systems was a major step towards the purification and characterization of essential host replication factors (67). These studies allowed the understanding of the molecular mechanisms involved in the assembly and progression of replication forks (7, 28, 67). Along with genetic and other biochemical approaches, these DNA replication systems provided insights into the processes of initiation and elongation (67). Interestingly, several parallels between eukaryotic and prokaryotic DNA duplication emerged which show that central processes of DNA metabolism are in part conserved in all kingdoms (33, 60, 61). The studies of the polyomaviral systems have allowed the development of a general model for eukaryotic DNA replication. In an initial step, Tag recognizes the replication origin (ori) and forms a double hexameric complex, which causes specific distortions of the double-stranded (ds) DNA (4, 15, 20, 37, 49). Subsequently, dsDNA is unwound, and multiprotein complexes are assembled by Tag and host replication proteins, such as eukaryotic single-stranded (ss) DNA-binding protein (replication protein A [RPA]), topoisomerase I (topo I), and DNA polymerase α-primase (pol-prim), are recruited (10, 11, 17, 23, 38, 53, 54). After binding and distortion of the dsDNA, the helicase function of Tag melts and unwinds the ds ori DNA (4, 59). For this function, Tag depends on RPA as a cofactor to stabilize the ssDNA resulting from the helicase activity of Tag (7, 29, 72). The unwinding of dsDNA requires an additional cofactor, topo I, to relax the supercoiled DNA and to avoid the introduction of torsional stress into the DNA (24, 67). During the initiation of leading-strand replication, the primase activity of pol-prim synthesizes the first RNA primer at the ori (8, 28, 51, 69). This RNA molecule is directly transferred to the DNA polymerase subunit of pol-prim and elongated to 30 to 40 nucleotides. The multiprotein complex RFC (replication factor C) catalyzes the DNA polymerase switch to a leading-strand synthesis complex consisting of PCNA (proliferating cell nuclear antigen) and DNA polymerase δ, which carries out processive DNA synthesis (8, 28, 36, 39, 67, 73). In cooperation with Tag and RPA, pol-prim multiply initiates discontinuous Okazaki fragment DNA synthesis on the lagging strand in synchrony with the leading-strand synthesis (16, 19, 38, 41, 70, 71). Again after a polymerase switch, these RNA-DNA primers are elongated by DNA polymerase δ or ɛ in a complex with PCNA (8, 28, 67, 73). The preinitiation, initiation, and elongation steps of viral DNA synthesis at the origin of DNA replication require specific physical contacts between these protein complexes in addition to the DNA binding and enzymatic activities of the replication factors (16, 64). This model, which originated from work with cell-free polyomavirus DNA replication systems, also illustrates basic mechanisms of chromosomal DNA replication (28, 67).
Although PyV and SV40 depend on the same mechanisms for their DNA replication and their Tags perform identical replication functions, each virus has a specific host range resulting from host-dependent DNA duplication (55). PyV propagates in mouse cells, whereas SV40 multiplies in primate cells. This species specificity can be reproduced in cell-free DNA replication systems, and the cellular pol-prim is the major species-specific factor for PyV and SV40 DNA replication (6, 18, 40, 42, 58). Recently, it was shown that a central region of murine p48 is required for the initiation and elongation steps of PyV DNA replication (31). To determine the cellular requirements for the host-specific DNA replication of PyV and whether one or more regions of p48 are involved in the regulation of PyV DNA replication, chimerical polypeptides were created by exchanging various stretches of mouse and human p48. In addition, single amino acids were mutated in human p48. The mutant and wild-type polypeptides were coexpressed with the p180, p68, and p58 subunits of pol-prim and purified as enzyme complexes. The functional tests showed that the least conserved region A and especially the phenylalanine 262 of mouse p48 can induce an otherwise-human pol-prim to support PyV DNA replication. The residues at positions 262 and 266, as well as amino acids (aa) 273 to 288, are crucial for the replication mediator function of PyV Tag on RPA-bound templates.
MATERIALS AND METHODS
Computer analysis of proteins.
Sequence analyses were performed by using the Geneworks program (IntelliGenetics, Brussels, Belgium) for protein alignments, the jpred2 program for secondary-structure predictions (14; http://jura.ebi.ac.uk:8888/), and the bioinbgu software for predicting three-dimensional protein structures (21, 22; http://www.cs.bgu.ac.il/≈bioinbgu/).
Production of the mutant p48 proteins.
To obtain the chimerical human-mouse p48 subunits (Fig. 1B), cDNAs were produced by overlap extension PCR as described previously (66). The double mutants were constructed by successive repetition of this method. These DNAs were ligated into pVL1392-hp48. After in vivo recombination, baculoviruses expressing recombinant proteins were created using Baculo-Gold DNA and standard procedures (Pharmingen, Hamburg, Germany) (47).
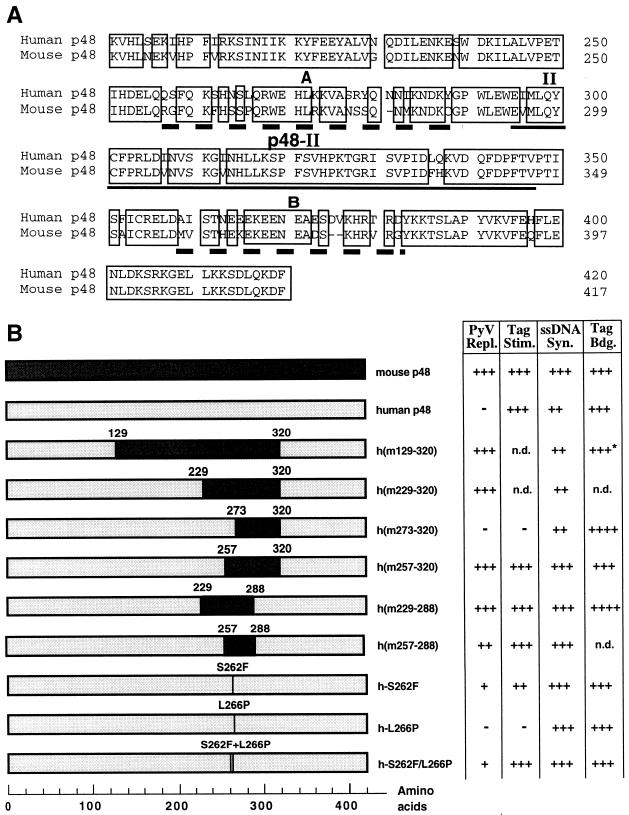
FIG. 1.
Amino acid alignments and construction of chimerical human-murine p48. (A) Human and murine p48 subunits (aa 200 to 420 and 417, respectively) were aligned using the Geneworks program; identical amino acids are boxed. The highly conserved region p48-II (marked by a single line) was named according to the system in reference 57. Two regions, A and B (marked by a thick, dashed line), have a significantly lower degree of amino acid identity. (B) All chimerical p48 subunits containing human and murine sequences used in these studies. They were named according to their murine amino acids. The chimerical protein h(m273–320) consists of murine aa 273 to 320, whereas the remaining residues are coded by the human p48 cDNA. In the proteins h-S262F, h-L266P, and h-S262F/L266P, the serine at position 262 of human p48 was changed to phenylalanine, leucine 266 became a proline, or both amino acids were mutated. The activities of these polypeptides are summarized on the right. The binding of pol-prim to PyV Tag (Tag Bdg.) (see Fig. 4) is summarized. The interaction of chimerical HHHh(m129-320) with PyV Tag was determined earlier (31) and is marked with an asterisk. Stimulation on RPA-bound ssDNA (Tag Stim.) is indicated as follows: +++, 2.0–2.5; ++, 1.5–1.9; -, no stimulation [<1.1]. PyV DNA replication (PyV Repl.) and DNA synthesis on ssDNA (ssDNA Syn.) are indicated as follows: +++, 80 to 100%; ++, 40 to 70%; +, 5 to 30% of that of MMMM; -, no replication).
Proteins.
PyV Tag and DNA pol-prim (consisting of subunits p180, p68, p58, and p48) were purified from baculovirus-infected insect cells as described previously (6, 44, 57). RPA was bacterially expressed and purified as outlined before (27, 45). The monoclonal antibodies SJK237-71, SJK287-38 (63), and F5 (48), specific for DNA pol-prim and PyV Tag, respectively, were purified by affinity chromatography (25).
Protein manipulations and immunological techniques.
Protein concentrations were determined according to the method of Bradford (5) using a commercial reagent with bovine immunoglobulin G (Bio-Rad) as a standard. Sodium dodecyl sulfate (SDS) gel electrophoresis was carried out as described previously (34) with 10-kDa ladders (Life Technologies) as molecular mass markers. After polyacrylamide gel electrophoresis, proteins were visualized by staining them with Coomassie brilliant blue. For quantitative comparison, the dried gels were scanned and the intensities of staining were quantified with the Image Quant program (Amersham Pharmacia Biotech, Freiburg, Germany).
Modified enzyme-linked immunosorbent assays were carried out as described previously (17, 31).
DNA synthesis on ΦX174 ssDNA.
The DNA replication of ΦX174 ssDNA was carried out in a reaction mixture containing 66 ng of ΦX174 ssDNA (New England Biolabs); 20 mM Tris-acetate (pH 7.3); 5 mM magnesium acetate; 20 mM potassium acetate; 1 mM dithiothreitol; 0.1 mg of bovine serum albumin (BSA)/ml; 1 mM ATP; 0.1 mM (each) CTP, GTP, UTP, dATP, dCTP, dGTP, and TTP; and 0.1 mM [α-32P]dCTP (Amersham Pharmacia Biotech) (43). Comparisons between different pol-prim preparations were made by adding 0.2 U of primase per assay. The incorporation of dCMP was determined by acid precipitation of DNA on GF52 filters (Schleicher & Schüll, Dassel, Germany) and scintillation counting.
DNA synthesis on ΦX174 ssDNA with RPA and Tag.
DNA synthesis was carried out in an assay mixture (40 μl) containing 30 mM HEPES-KOH (pH 7.8); 7 mM MgAc; 0.1 mM EGTA; 1 mM dithiothreitol; 0.25 mg of BSA/ml; 0.01 mg of creatine kinase/ml; 40 mM creatine phosphate; 4 mM ATP; 0.2 mM (each) CTP, GTP, and UTP; 0.1 mM (each) dATP, dGTP, and dTTP; 0.002 mM dCTP; 1 μl of [α32P]dCTP (specific activity, 3,000 Ci/mmol; 10 μCi/μl; Amersham Pharmacia Biotech); 250 ng (0.76 nmol of nucleotides) of ΦX174 ssDNA; 1 μg of PyV Tag; and 0.69 μg of RPA as described earlier (70, 71). The reaction mixture was assembled on ice, and the reaction was started by the addition of pol-prim. After incubation for 1 h at 37°C, the reaction mixture was spotted onto GF52 filters and precipitated in 10% trichloroacetic acid. The amount of DNA synthesis was measured by liquid scintillation counting.
Preparation of S100 extracts and replication of viral DNA in vitro.
S100 extracts were prepared from logarithmically growing human 293S and mouse FM3A cells as described before (46, 58). The replication of PyV DNA in vitro was performed according to methods described previously (6, 57). Briefly, the assay contained 1.2 μg of PyV Tag, 250 ng of pUC-Py1 DNA (PyV origin DNA [52]), and 200 μg of S100 or depleted S100 extract in 30 mM HEPES–KOH (pH 7.8); 1 mM dithiothreitol; 7 mM magnesium acetate; 1 mM EGTA (pH 7.8); 4 mM ATP; 0.3 mM (each) CTP, GTP, and UTP; 0.1 mM (each) dATP and dGTP; 0.05 mM (each) dCTP and dTTP; 40 mM creatine phosphate; 80 μg of creatine kinase/ml, and 5 μCi each of [α32P]dCTP and [α32P]dTTP (3,000 Ci/mmol; Amersham Pharmacia Biotech); pol-prim was added as indicated. The incorporation of radioactive dNMP was measured by acid precipitation of DNA and scintillation counting. The total radioactivity was determined by liquid scintillation counting after 5 μl of a 200-fold dilution of the replication assay mixture was spotted onto GF52 filters.
The replication of SV40 DNA in vitro was performed as described above with minor modifications (46, 52). The SV40 assay mixture (60 μl) contained 0.6 μg of SV40 Tag, 0.25 μg of SV40 origin DNA (pUC-HS), 0.6 μg of human RPA, and 300 μg of S100 or depleted S100 extract from FM3A cells. All other conditions were as described above.
Nucleotide sequence accession numbers.
The Protein Data Bank accession codes of domains referred to in this study were as follows: DNA binding domain 3 of mouse c-Myb, 1idy; a dimerization domain of thermolysin protease, 1trla; and mouse c-Myb DNA binding domain 2, 1msec.
RESULTS
Purification of DNA pol-prim containing chimerical human-murine p48-primase.
The murine p48 subunit and specific regions therein control the replication of PyV DNA in vitro (6, 18, 31). The catalytic subunit p48 of primase is highly conserved between mice and humans (>90% identical amino acids). However, mammalian p48 contains two regions, A and B (murine aa 257 to 288 and 358 to 379), which are significantly less conserved (65 and 59% amino acid identity, respectively) (Fig. 1A). Outside these regions, p48 has 92 (aa 288 to 357), 95 (N terminus; aa 1 to 256), and 97% (C terminus; aa 380 to 417) identical amino acids.
The sequence comparison and the role of the central region of mouse p48 comprising aa 129 to 320 (31) suggest that region A might mediate the species-specific functions to support cell-free PyV DNA replication. To address this question, chimerical p48 subunits were produced and coexpressed with human p180, p68, and p58 (Fig. 1B). The recombinant protein complexes were purified to near homogeneity by phosphocellulose and immunoaffinity chromatography and contained all four subunits (Fig. 2). The purified enzyme complexes had DNA polymerase and primase activity, with specific enzyme activities varying from 2,120 to 19,700 and from 160 to 840 U/mg, respectively (Table 1). Some preparations of human pol-prim also had low specific enzyme activities in the range of 2,000 to 5,000 U/mg of DNA polymerase α and 350 to 540 U/mg of primase (data not shown) (31). Due to these variations, we used the same amounts of active enzyme for functional assays as indicated in the figures. In the experiments shown, a human pol-prim with 19,700 and 840 U/mg was preferentially used, but other enzyme preparations were also studied and gave the same results.
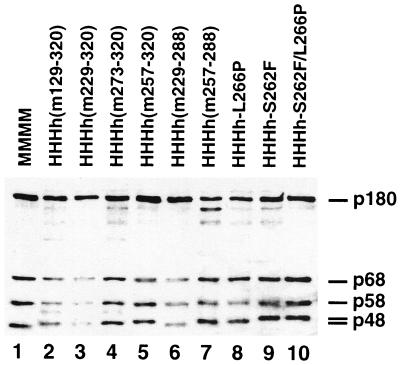
FIG. 2.
Production of DNA pol-prim complexes. The purified pol-prim complexes were analyzed by SDS gel electrophoresis and stained with Coomassie brilliant blue. Lane 1, murine pol-prim (MMMM); lanes 2–10, hybrid pol-prim (HHHx) with human p180, p68, and p58 and mutant p48. In lanes 2 and 4, the protein between p48 and p58 with a molecular mass of about 56 kDa is most likely the heavy chain of the immunoglobulin G bleeding from the affinity resin used for purification.
TABLE 1.
Enzyme activities of recombinant DNA pol-prim complexes
| Enzyme complexa | Protein concn (mg/ml) | Specific DNA polymerase activity (U/mg) | Specific primase activity (U/mg) |
|---|---|---|---|
| MMMM | 0.23 | 3,560 | 260 |
| HHHh(m129–320) | 0.45 | 3,130 | 170 |
| HHHh(m229–320) | 0.47 | 2,610 | 210 |
| HHHh(m273–320) | 0.34 | 3,750 | 380 |
| HHHh(m257–320) | 1.18 | 2,420 | 500 |
| HHHh(m229–288) | 1.40 | 2,120 | 370 |
| HHHh(m257–288) | 1.42 | 2,390 | 240 |
| HHHh-L266P | 1.37 | 1,970 | 160 |
| HHHh-S262F | 0.72 | 2,870 | 250 |
| HHHh-S262F/L266P | 1.13 | 2,900 | 230 |
| HHHH | 0.13 | 19,690 | 840 |
MMMM, murine pol-prim; HHHH, human pol-prim; HHHx, hybrid pol-prim containing mutant p48 subunit plus human subunits p180, p68, and p58.
The purified protein complexes were further analyzed by SDS gel electrophoresis. The Coomassie brilliant blue-stained gel (Fig. 2) was scanned, and the intensities were quantified. The average subunit ratio (p180:p68:p58:p48) was about 1:1.3:1.6:2.2, which is similar to that of published chimerical pol-prim complexes (1:1.5:2.2:2.6 [58]). However, the complex HHHh(m229–320) had a subunit distribution of 1:0.4:0.2:0.3. The replication-competent enzyme complex HMMM, which consists of human p180 and three murine subunits, also assembles in a suboptimal way and had a subunit compositon of 1:1.8:1:2.3 (58). Importantly, its primase polypeptides were only present after the stringency of the washing conditions was reduced; otherwise, both subunits were quantitatively lost (56). Despite the low abundance of the small subunits, the specific primase activity of HHHh(m229–320) was only slightly below average and was comparable to that of the mouse enzyme complex and higher than that of other enzyme complexes (Table 1).
The polyacrylamide gel electrophoresis analysis showed that the protein complex HHHh(m257–288) had a p180 subunit which was significantly more degraded than that of the other purified complexes (Fig. 2, lane 7). The ratio of p180 to the proteolysis products p160 and p140 was about 44:39:17. Two additional enzyme complexes, HHHh(m273–320) and HHHh-S262F, showed a high proteolytical degradation of p180 (ratios of proteolysis products, 68:25:7 and 71:20:9, respectively) (Fig. 2, lanes 4 and 9). The high proteolysis rate of p180 in specific preparations could be due to a high protease activity during purification, although the same protease inhibitor cocktail was used in all purifications. The p180 subunits of HHHh(m129–320) and HHHh-L266P were proteolytically degraded, but not significantly above that of an average pol-prim preparation (Fig. 2, lanes 2 and 8) (6, 13, 57, 58).
DNA synthesis by chimerical DNA pol-prim on natural ssDNA.
Several of the chimerical pol-prim complexes contained lower levels of primase or proteolytically degraded subunits. Therefore, we wanted to determine whether these enzymes initiate and elongate DNA synthesis on natural ssDNA templates. Human, murine, and chimerical pol-prim complexes synthesized dsDNA (Fig. 3A). Although we consistently observed differences in their efficiencies of DNA synthesis, we could not determine any correlation between the level of incorporated dNMPs and protein degradation or specific activities (compare Fig. 2 and 3A, as well as Table 1).
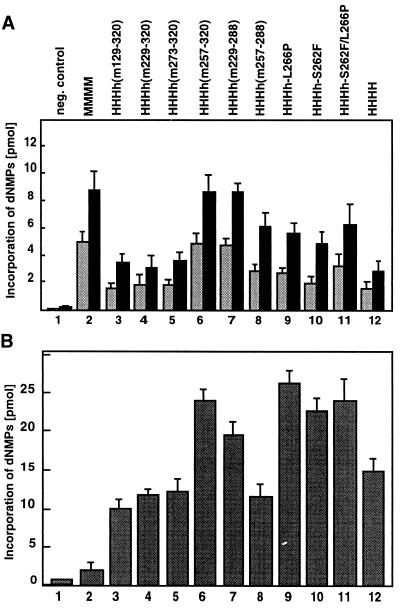
FIG. 3.
DNA synthesis on ssDNA and SV40 DNA replication by chimerical DNA pol-prim. (A) Mutant (bars 3 to 11) and wild-type (bars 2 and 12) pol-prim complexes were used to synthesize nucleic acids on ssDNA in the presence of NTPs, dNTPs, and Mg2+ for 30 (shaded bars) and 60 (solid bars) min. (B) These enzyme complexes were also used in an SV40 DNA replication system by using mouse cell extracts depleted of endogenous pol-prim. Bars 3 to 11, mutant pol-prim with human p180, p68, p58, and mutant p48; bars 2 and 12, murine (MMMM) and human (HHHH) pol-prim. Bars 1, negative control without pol-prim. The assays were repeated twice, and the mean values are presented. The error bars indicate standard deviations.
SV40 DNA replication by chimerical DNA pol-prim in vitro.
To test the functional cooperation of the various enzyme complexes with other replication factors, we used the SV40 DNA replication system, since we recently showed that chimerical p48 containing human N- and C-terminal amino acids supports cell-free replication of SV40 DNA (31). As expected, mouse pol-prim did not replicate SV40 DNA, whereas all chimerical enzyme complexes studied here, as well as the human enzyme, did so (Fig. 3B, bars 2 to 12). The enzyme complexes containing h(m129–320), h(m229–320), h(m273–320), h(m257–288), and human p48 showed similar activities in the SV40 DNA replication system (Fig. 3B, bars 3, 4, 5, 8, and 12). The pol-prim complexes containing p48 h(m257–320), h(m229-288), h-L266P, h-S262F, and h-S262F/L266P showed a slightly higher replication activity than the human enzyme (Fig. 3B, bars 6, 7, 9, 10, 11, and 12, respectively). These data indicated that all chimerical proteins efficiently supported cell-free DNA replication.
Physical interaction of PyV Tag and chimerical DNA pol-prim.
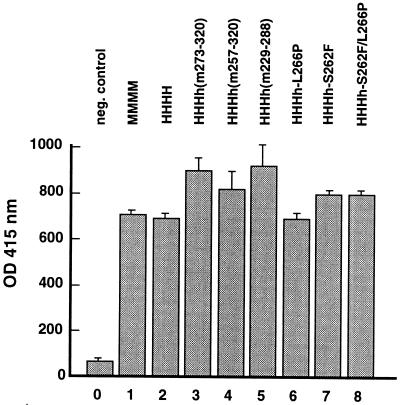
To test whether the physical interactions of chimerical, mouse and human pol-prim complexes with PyV Tag are species specific, modified enzyme-linked immunosorbent assays were carried out. PyV Tag bound to human, mouse, and chimerical pol-prims with nearly the same efficiency. The chimerical complexes HHHh(m273–320) and HHHh(m229–288) showed the strongest binding to PyV Tag (Fig. 4). Thus, the direct physical contacts between PyV Tag and various pol-prim complexes are not host specific.
FIG. 4.
Interaction of PyV T antigen with recombinant DNA pol-prim. Pol-prim and PyV Tag physically interact (6, 31). BSA (1 μg; bar 0) or purified, recombinant pol-prim complexes (1 μg; bars 1–8) as indicated were immobilized. After incubation with 1 μg of PyV Tag for 1 h at room temperature, Tag–pol-prim complexes were determined with Tag-specific antibody F5 and a mouse-specific peroxidase-coupled secondary antibody. The mean values and standard deviations of three experiments are presented in optical density (OD) units.
Region A is required for species-specific replication of PyV DNA in vitro.
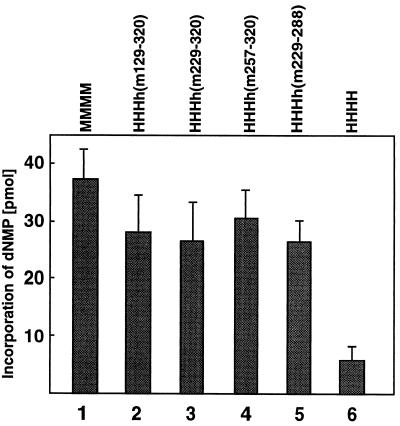
PyV DNA replication in vitro requires the mouse p48 subunit during the initiation of leading-strand DNA synthesis (6, 31). To determine the amino acids controlling PyV DNA replication, the chimerical p48 subunits were tested in the replication of PyV DNA. As expected, mouse pol-prim supported in vitro PyV DNA replication, and its replication activity was used as a positive control (Fig. 5, bar 1). The human enzyme was inactive in the replication assay and served as a negative control (Fig. 5, bar 6). Chimerical p48 containing mouse residues spanning aa 129 to 320, 229 to 320, 257 to 320, and 229 to 288 supported DNA synthesis with plasmids which have a PyV origin of replication.
FIG. 5.
Species specificity of PyV DNA replication in vitro is mediated by the least conserved region A of mouse p48. DNA polymerase of recombinant pol-prim (0.5 U) was used to replicate PyV DNA in human extracts depleted of pol-prim. The indicated enzyme complexes were added to the human 293S cell extracts supplemented with PyV Tag and a PyV origin-containing plasmid as described in Materials and Methods. The data are the mean values and standard deviations of four DNA replication assays.
Exchange of serine at position 262 of human p48 with phenylalanine allows PyV DNA replication in vitro.
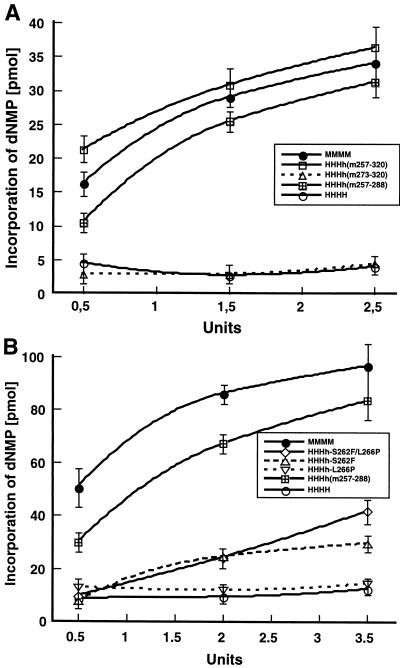
The above-mentioned data suggested that the least conserved region A, aa 257 to 288, probably controls the species specificity of PyV DNA replication in vitro. Although the applied enzyme concentrations were equivalent to those present in crude extracts (0.4 to 0.5 U per replication reaction), we wondered whether raising the level of human pol-prim and chimerical enzymes would influence their capability to support PyV DNA replication (Fig. 6). The addition of mouse pol-prim allowed the incorporation of radioactive dNMPs in a concentration-dependent manner (Fig. 6A). Human pol-prim HHHH and HHHh(m273–320) did not support PyV DNA replication above background, even at the highest concentrations, and the acid-insoluble radioactivity was between 2 and 4 pmol (Fig. 6A). The pol-prim complex HHHh(m257–288) containing murine region A supported PyV DNA replication, and the incorporation of radioactive dNMPs rose in a concentration-dependent manner; 0.5, 1.5, and 2.5 U had 66, 88, and 92%, respectively, of the replication activity of murine pol-prim. For comparison, the chimerical complex HHHh(m257–320), which contains region A plus C-terminal flanking sequences, was also studied. Interestingly, it was slightly more active than the mouse enzyme (Fig. 6A). These results indicate that high pol-prim levels do not suppress species specificity in the PyV replication system.
FIG. 6.
Phenylalanine at position 262 of mouse p48 allows cell-free PyV DNA replication. Increasing enzyme concentrations of pol-prim complexes were tested for their ability to support PyV DNA replication in vitro in human extracts depleted of pol-prim. (A) the activities of hybrid enzyme complexes HHHh(m273–320), HHHh(m257–320), and HHHh(m257–288) were compared with the replication activities of murine pol-prim (MMMM) and human pol-prim (HHHH). (B) The abilities of HHHh(m257–288), HHHh-L266P, HHHh-S262F, and HHHh-S262F/L266P to replicate PyV DNA were measured. For comparison, replication assays with the replication-competent murine pol-prim (MMMM) and replication-inactive human pol-prim (HHHH) were performed in parallel. The data are the mean values and standard deviations of three DNA replication assays.
The failure of HHHh(m273–320) to replicate PyV DNA suggests that the region aa 257 to 273 contains essential residues which mediate the species specificity of PyV DNA replication. The comparison of amino acid sequences (Fig. 1) and amino acid structures suggested that F262 and P266 might be crucial for the function of p48. To examine this more precisely, we produced the mutants h-S262F, h-L266P, and h-S262F/L266P and investigated their activities (Fig. 2 and 6B; Table 1). Purified chimerical enzyme HHHh-L266P (0.5 U) had an activity (13 pmol) which was slightly above the background determined with human pol-prim (8 pmol). However, incorporation of radioactive dNMPs did not increase in a concentration-dependent manner (Fig. 6B). Characterization of the second point mutant revealed that low levels of HHHh-S262F (0.5 U) hardly supported PyV DNA replication above that of the negative control HHHH. However, increasing the enzyme level to 2 and 3.5 U allowed replication of PyV DNA in a concentration-dependent manner (Fig. 6B). The maximal incorporation was 30 pmol, or 31% of that of mouse pol-prim. The complex HHHh-S262F/L266P containing human p48 with two mouse residues was more efficient in PyV DNA replication in vitro than HHHh-S262F. In the presence of 3.5 U, 42 pmol of dNMPs was incorporated into PyV DNA, which corresponds to 44% of that of murine pol-prim (Fig. 6B). For comparison, the pol-prim complexes HHHH and HHHh(m257–288) were analyzed in parallel. The human enzyme was inactive, whereas the replication activity of HHHh(m257–288) was between that of HHHh-S262F/L266P and mouse pol-prim. These data suggested that phenylalanine 262 of murine p48 has a role in PyV DNA replication and that additional amino acids, such as proline 266 and those within and near region A, stimulate PyV DNA replication in human extracts.
Cooperation of DNA pol-prim with RPA and PyV Tag during DNA synthesis.
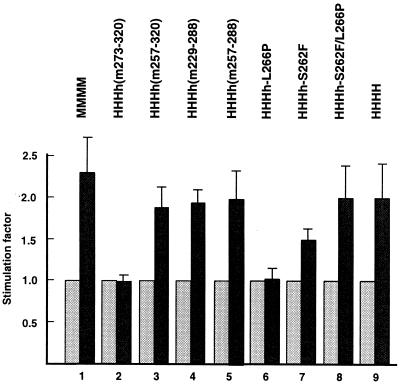
On natural ssDNA, low concentrations of RPA inhibit primer synthesis by pol-prim, and SV40 Tag relieves this inhibition (10, 38, 52, 68, 70). To synthesize RNA on ssDNA bound by RPA, human pol-prim requires replication mediator proteins such as SV40 Tag for optimal activity (3, 10, 38, 52, 68, 70). Since the physical interaction of chimerical pol-prim and PyV Tag, as well as the priming activity of chimerical pol-prim on artifical and natural DNA, is not or is hardly influenced by these amino acid exchanges (Fig. 3A and 4; Table 1), we used stimulation of primer synthesis on RPA-bound ssDNA as a simple model system to study the functional cooperation among PyV Tag, RPA, and various pol-prim complexes. In addition, we wanted to know whether this assay shows species specificity in the PyV system. PyV Tag stimulated DNA synthesis by human and murine pol-prim on RPA-bound ΦX174 ssDNA (Fig. 7 [summarized in Fig. 1B]). The stimulation of murine pol-prim by PyV Tag is only slightly higher than that of human pol-prim (Fig. 7, bars 1 and 9). These results showed that the stimulation of pol-prim by PyV Tag in the presence of RPA is not species specific and that PyV Tag matches SV40 Tag in this assay (52).
FIG. 7.
DNA synthesis of DNA pol-prim on natural ssDNA in the presence of RPA and PyV Tag. The cooperation of PyV Tag with mouse, human, and mutant pol-prims was tested on natural ssDNA bound by human RPA. In the reaction, ΦX174 ssDNA, RPA, and pol-prim complexes as indicated were incubated in the absence (shaded bars) or in the presence (solid bars) of PyV Tag. The incorporation of acid-insoluble dNMPs in the absence of Tag was arbitrarily set to 1. The presented data are the mean values and standard deviations of three experiments.
In the presence of RPA, PyV Tag also stimulated the complexes HHHh(m257–320), HHHh(m229–288), and HHHh(m257–288) (Fig. 7, bars 3, 4, and 5, respectively). However, the chimerical HHHh(m273–320), which was inactive in PyV DNA replication in cell extracts and in the PyV origin-dependent initiation (Fig. 6A and data not shown), did not respond to the addition of PyV Tag in this assay (Fig. 7, bars 2). In addition, the introduction of a proline at position 266 of human p48, which does not allow the replication of PyV DNA (Fig. 6B), also diminished the ability of PyV Tag to stimulate pol-prim (Fig. 7, bars 6). Importantly, the inability of Tag to cooperate with these enzyme complexes was not dependent on their ability to synthesize DNA de novo on ssDNA templates, since both complexes incorporated dNMPs as well as the human enzyme did (Fig. 3A). The negative effect of P266 on p48 could be fully rescued by introducing a second mutation, S262F, and the complex HHHh-S262F/L266P, which supports cell-free PyV DNA replication (Fig. 6B), was stimulated to an extent comparable to human pol-prim (Fig. 7, bars 8). PyV Tag stimulated HHHh-S262F, which had some PyV DNA replication activity (Fig. 6B), with a reduced efficiency (Fig. 7, bars 7).
Comparison of the stimulatory effect of PyV Tag on the chimerical protein complexes with that on human pol-prim showed that aa 262, 266, and 273 to 288 are important for the function of the primase to initiate DNA synthesis in the presence of RPA and PyV Tag. Furthermore, the amino acid pairs S262 and L266 of human p48, as well as F262 and L266 of mouse p48, are required for optimal initiation activity. Two other important sites for the cooperation of these initiation proteins are the regions aa 257 to 272 and 273 to 288, since the human aa 257 to 272 together with murine aa 273 to 288 do not allow optimal cooperation of pol-prim with Tag and RPA. However, exchanging the mouse sequence aa 273 to 320 with the human one (human p48) or the human sequence aa 257 to 272 with that from mouse h(m257–320) rescues this impaired function of h(m273–320).
DISCUSSION
The cell-free polyomavirus DNA replication systems have enabled the elucidation of the functions and activities of purified eukaryotic replication proteins (for a review, see references 55 and 67 and references therein). The cellular pol-prim is required for the initiation of leading- and lagging-strand DNA synthesis and is involved in the control of PyV and SV40 DNA replication (6, 18, 50, 65–67).
p48 region A controls PyV DNA replication.
The PyV DNA replication system revealed that mouse p48 controls viral DNA replication (6). Recent results suggested that the central part of mouse p48 mediates the species specificity of PyV DNA replication in vitro (31). Exchanging human and mouse amino acids of p48 showed that the less conserved region A of p48 controls the species specificity of cell-free PyV DNA replication. The characterization of the additional mutant polypeptides revealed that a single amino acid, F262 within region A, is sufficient to allow PyV DNA replication in an otherwise-human setting (Fig. 1 and 6). The efficiency of the enzyme complex is quite low and reaches only about 31% of that of the mouse enzyme at extremely high concentrations. Since murine amino acids neighboring F262 stimulated the replication activity, these data suggest that the host specificity of PyV DNA replication is rather complex and that multiple functions are involved to achieve full replication activity.
The interpretation that multiple activities of pol-prim control optimal PyV DNA replication is supported by the PyV Tag-mediated stimulation of mutant pol-prim on RPA-bound ssDNA. This assay has been used to study the functional cooperation of SV40 Tag, RPA, and pol-prim (10, 38, 52, 68, 70). The stimulation assay with SV40 Tag requires protein-protein and protein-DNA interactions as well as catalytic steps, but it is independent of the unwinding of dsDNA (70, 71). The origin-dependent primer synthesis and that on RPA-bound ssDNA involve similar interactions of SV40 Tag, RPA, and pol-prim, since the same monoclonal antibodies abrogate both reactions and interfere with the physical binding of these initiation proteins (70, 71). The adaptation of the assay to the PyV system gave important hints concerning the functional cooperation of RPA, PyV Tag, and pol-prim, especially of the primase region A (Fig. 7). Although the assay is not species specific, it showed that aa 262 and 266, as well as the residues aa 257 to 272 and 273 to 320 of p48, have major relevance during the initiation reaction on RPA-bound ssDNA and interfere with the stimulatory activity of PyV Tag. The data suggest that these residues of p48 control the replication mediator function of PyV Tag (summarized in Fig. 1B) (3). Since this activity of PyV Tag requires simultaneous physical interactions of Tag with RPA and pol-prim on an ssDNA template (3, 52, 68, 70, 71), the results show that specific positions of region A are involved in the cooperation of these initiation factors during primer synthesis. As the same mutations of primase interfered with its ability to support PyV DNA replication in vitro as well as with its functional cooperation in the stimulation assay (summarized in Fig. 1B), we anticipate that such cooperation also occurs during the origin-dependent initiation of viral DNA replication in vitro.
Putative functions of p48 region A and flanking residues.
The results presented here and other biochemical data can now be discussed in light of the recently published structure of a distantly related archaeal primase (1, 2). Partial protease digests of human p48 suggested that regions A and p48-II form a proteolytically stable domain (data not shown). In good agreement with the three-dimensional structure of the archeal primase, programs (14, 21, 22) suggested that aa 240 to 340 of mouse p48 consists of three α-helices (data not shown). These predicted secondary structures were compared with published domain structures (21, 22). The comparisons revealed homologies of human p48 with DNA binding domain 3 of mouse c-Myb (homology score z, 9.2) and a dimerization domain of thermolysin protease (z = 8.6). These values are both above the confidence threshold of the method (z = 4.8 ±1 [21]). Mouse p48, h-S262F, h-L266P, and h-S262F/L266P showed structural homologies to mouse c-Myb DNA binding domain 2 in complex with DNA (z = 5.4, 6.5, 12.5, and 10.4, respectively). Interestingly, the third helix of mouse c-Myb DNA binding domain 2 interacts with dsDNA, suggesting that a helix spanning aa 290 to 303 of mouse p48 (aa 290 to 304 of human p48), which overlaps with the highly conserved region p48-II (Fig. 1A), might bind to ds nucleic acids. This hypothesis is in agreement with earlier results showing that R304 of human p48, which corresponds to R303 of mouse p48, is involved in the recognition of the nucleoside triphosphate (NTP) that will become the 5′-terminal nucleotide of the primer during the synthesis of the first dinucleotide and that it possibly binds to a phosphate of the primer strand during the elongation step (1, 32). In close proximity, mouse amino acid D305 (D306 in human p48) has essential catalytic functions, since together with D109 and D111, it chelates divalent metal ions (1, 2, 12).
According to its hydrophilic character and in agreement with the crystal structure of the archeal primase, region aa 251 to 290 is most likely located at the surface of the protein, where it may interact with other proteins, such as PyV Tag and RPA. By combining structural and various biochemical data, the species specificity can be explained in two ways (Fig. 1B) (1, 2, 6, 12, 31, 32). First, PyV Tag binds to the region aa 251 to 290 and supports the initiation reaction on a template by stabilizing the DNA-NTP-NTP-primase complex during the synthesis of the first dinucleotide. This assumption would explain the fact that priming at an origin and on RPA-bound ssDNA behave differently with the wild-type human and murine proteins. In the ssDNA–NTP–NTP–pol-prim–Tag–RPA complex, the DNA is still relatively flexible, whereas at an origin of replication such a complex is structurally more restricted, and the equivalent reaction at an origin becomes species specific. The failure of PyV Tag to stimulate pol-prim containing mutant p48 h-L266P and h(m273–320) can be explained by the assumption that these mutant p48 forms do not allow the formation of a functional initiation complex. It is important to mention here that the formation of the DNA-NTP-NTP-primase complex or the following step during the synthesis of the first dinucleotide is the rate-limiting step in primer synthesis (1).
In a second explanation, RPA binds to region aa 250 to 290 and perhaps also to aa 290 to 303. These primase-RPA interactions prevent the correct formation of an ssDNA-NTP-NTP-primase complex and inhibit RNA synthesis by p48. This assumption is supported by the previous finding that inhibition of primase on ssDNA by a low concentration of RPA requires physical interactions involving RPA (70). For RNA synthesis by primase, PyV Tag would displace the inactive primase-RPA complex and allow p48 to bind to DNA and to form a functional ssDNA–NTP–NTP–pol-prim–Tag complex. This reaction probably requires the cooperation of several components and perhaps other pol-prim subunits such as p58, e.g., to stabilize the PyV-primase complex versus the RPA-primase complex. The explanation that RPA is involved in the species specificity of viral DNA replication is supported by earlier findings that mammalian and insect RPAs support SV40 DNA replication whereas yeast RPA does not (30, 38, 56, 58, 68). In both models, protein-protein interactions and conformational restrictions of the protein-template-primer complex contribute to the species specificity of the origin-dependent initiation reaction.
PyV DNA replication in transformed cell lines.
The biochemical data, which define pol-prim and region A of p48 as a host-specific modulator (Fig. 5 and 6) (6, 18, 31, 40, 58), seem to contradict previous findings that PyV DNA is replicated in some human transformed cell lines whereas their untransformed parental cell lines do not support the viral replication (62). This apparent contradiction can be resolved by various assumptions. Transformed cells could become permissive by a single-amino-acid exchange in the primase gene product, e.g., F262. An alternative explanation is that during the transformation process one or several factors are overexpressed and suppress the phenotype of normal human cells. Pol-prim is most likely not one of these factors, because raising the concentration of human pol-prim and HHHh(m273–320), which were both active in cell-free SV40 DNA replication (Fig. 3B) (31, 58), did not allow PyV DNA replication in replication-competent human 293S extracts (Fig. 6). On the other hand, the failure of these extracts to support PyV DNA replication could be caused by depletion or inactivation of the suppressor during extract preparation. Sverdrup et al. (62) speculated that a kinase or phosphatase might be responsible for the permissiveness of the transformed 293S and C33A. Interestingly, a canonical consensus phosphorylation site (-SPQR-) of cyclin-dependent kinases (Cdks), which is recognized by cyclin E-Cdk2 in vitro (data not shown), is present in region A of murine but not human p48. HeLa cells, which do not support PyV DNA replication in vivo and in vitro, contain a truncated form of HPV-18 E1 which inhibits activation of papillomavirus DNA replication. This inhibition can be overcome by ectopically expressing cyclin E-Cdk2 (35). However, the introduction of the SPQR motif of mouse p48 into human p48 did not allow replication of PyV DNA in 293S cell extracts, and it only allowed a slightly more efficient PyV replication together with the F262 mutation in human p48. Therefore, the role of cyclin E-Cdk2 in the species specificity of PyV DNA replication is still open.
Our findings suggest that the control of PyV DNA replication by pol-prim is a multistep process. By exchanging various polypeptide stretches and amino acids, we determined that amino acid F262 of mouse p48 plays a crucial role in PyV DNA replication. In addition, P266, as well as other residues within murine regions 257 to 288, are involved in the cooperation of primase, RPA, and PyV Tag on a template. Furthermore, the other subunits of pol-prim, especially murine p58, can enhance cell-free PyV DNA replication (6, 31).
ACKNOWLEDGMENTS
We thank D. Willbold, M. Görlach, G. Palm, M. Augustin, and R. Hilgenfeld for discussions of protein structures, R. Smith for critical reading of the manuscript, and A. Willitzer for technical support.
The work was financially supported by the Deutsche Forschungsgemeinschaft (Na 190/8, Na 190/10, and Na 190/12) and the EC (CT970125). The IMB is a Gottfried-Wilhelm-Leibniz-Institut and is financially supported by the federal government and the Land Thüringen.
REFERENCES
- 1.Arezi B, Kuchta R D. Eukaryotic DNA primase. Trends Biochem Sci. 2000;25:572–576. doi: 10.1016/s0968-0004(00)01680-7. [DOI] [PubMed] [Google Scholar]
- 2.Augustin M A, Huber R, Kaiser J T. Crystal structure of a DNA-dependent RNA polymerase (DNA primase) Nat Struct Biol. 2001;8:57–61. doi: 10.1038/83060. [DOI] [PubMed] [Google Scholar]
- 3.Beernink H T, Morrical S W. RMPs: recombination/replication mediator proteins. Trends Biochem Sci. 1999;24:385–389. doi: 10.1016/s0968-0004(99)01451-6. [DOI] [PubMed] [Google Scholar]
- 4.Borowiec J A. DNA helicases. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 545–574. [Google Scholar]
- 5.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Brückner A, Stadlbauer F, Guarino L A, Brunahl A, Schneider C, Rehfuess C, Prives C, Fanning E, Nasheuer H P. The mouse DNA polymerase α-primase subunit p48 mediates species-specific replication of polyomavirus DNA in vitro. Mol Cell Biol. 1995;15:1716–1724. doi: 10.1128/mcb.15.3.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brush G S, Kelly T J. Mechanisms for replicating DNA. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 1–43. [Google Scholar]
- 8.Burgers P M. Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma. 1998;107:218–227. doi: 10.1007/s004120050300. [DOI] [PubMed] [Google Scholar]
- 9.Challberg M, Kelly T J. Animal virus DNA replication. Annu Rev Biochem. 1989;58:671–717. doi: 10.1146/annurev.bi.58.070189.003323. [DOI] [PubMed] [Google Scholar]
- 10.Collins K L, Kelly T J. The effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase α-primase. Mol Cell Biol. 1991;11:2108–2115. doi: 10.1128/mcb.11.4.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins K L, Russo A A R, Tseng B Y, Kelly T J. The role of the 70kDa subunit of human DNA polymerase a in DNA replication. EMBO J. 1993;12:4555–4566. doi: 10.1002/j.1460-2075.1993.tb06144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Copeland W C, Tan X. Active site mapping of the catalytic mouse primase subunit by alanine scanning mutagenesis. J Biol Chem. 1995;270:3905–3913. doi: 10.1074/jbc.270.8.3905. [DOI] [PubMed] [Google Scholar]
- 13.Copeland W C, Wang T S. Catalytic subunit of human DNA polymerase alpha overproduced from baculovirus-infected insect cells. Structural and enzymological characterization. J Biol Chem. 1991;266:22739–22748. [PubMed] [Google Scholar]
- 14.Cuff J A, Barton G J. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 15.Dean F B, Borowiec J A, Eki T, Hurwitz J. The simian virus 40 T antigen double hexamer assembles around the DNA at the replication origin. J Biol Chem. 1992;267:14129–14137. [PubMed] [Google Scholar]
- 16.Dornreiter I, Erdile L F, Gilbert I U, von Winkler D, Kelly T J, Fanning E. Interaction of DNA polymerase α-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dornreiter I, Höss A, Arthur A K, Fanning E. SV40 T antigen binds directly to the catalytic subunit of DNA polymerase alpha. EMBO J. 1990;9:3329–3336. doi: 10.1002/j.1460-2075.1990.tb07533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eki T, Enomoto T, Masutani C, Miyajima A, Takada R, Murakami Y, Ohno T, Hanaoka F, Ui M. Mouse DNA primase plays the principal role in determination of permissiveness for polyomavirus DNA replication. J Virol. 1991;65:4874–4881. doi: 10.1128/jvi.65.9.4874-4881.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falaschi A. Eukaryotic DNA replication: a model for a fixed double replisome. Trends Genet. 2000;16:88–92. doi: 10.1016/s0168-9525(99)01917-4. [DOI] [PubMed] [Google Scholar]
- 20.Fanning E, Knippers R. Structure and function of simian virus 40 large T antigen. Annu Rev Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- 21.Fischer D, Eisenberg D. Protein fold recognition using sequence-derived predictions. Protein Sci. 1996;5:947–955. doi: 10.1002/pro.5560050516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer D, Eisenberg D. Assigning folds to the proteins encoded by the genome of Mycoplasma genitalium. Proc Natl Acad Sci USA. 1997;94:11929–11934. doi: 10.1073/pnas.94.22.11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gannon J V, Lane D P. Interactions between SV40 T antigen and DNA polymerase α. New Biol. 1990;2:84–92. [PubMed] [Google Scholar]
- 24.Hangaard A, Bendixen C, Westergaard O. DNA topoisomerases. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 587–617. [Google Scholar]
- 25.Harlow E, Lane D P. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 26.Hassell J A, Brinton B T. SV40 and polyomavirus DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 639–677. [Google Scholar]
- 27.Henricksen L A, Umbricht C B, Wold M S. Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 28.Hübscher U, Nasheuer H P, Syväoja J. Eukaryotic DNA polymerases, a growing family. Trends Biochem Sci. 2000;25:143–147. doi: 10.1016/s0968-0004(99)01523-6. [DOI] [PubMed] [Google Scholar]
- 29.Iftode C, Daniely Y, Borowiec J A. Replication protein A (RPA): the eukaryotic SSB. Crit Rev Biochem Mol Biol. 1999;24:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- 30.Kamakaka R T, Kaufman P D, Stillman B, Mitsis P G, Kadonaga J T. Simian virus 40 origin- and T-antigen-dependent DNA replication with Drosophila factors in vitro. Mol Cell Biol. 1994;14:5114–5122. doi: 10.1128/mcb.14.8.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kautz A R, Schneider A, Weisshart K, Geiger C, Nasheuer H P. Different regions of primase subunit p48 control mouse polyomavirus and simian virus 40 DNA replication in vitro. J Virol. 2001;75:1751–1760. doi: 10.1128/JVI.75.4.1751-1760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirk B W, Kuchta R D. Arg304 of human DNA primase is a key contributor to catalysis and NTP binding: primase and the family X polymerases share significant sequence homology. Biochemistry. 1999;38:7727–7736. doi: 10.1021/bi990247c. [DOI] [PubMed] [Google Scholar]
- 33.Kornberg A, Baker T. DNA replication. 2nd ed. New York, N.Y: W. H. Freeman & Company; 1992. [Google Scholar]
- 34.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Lin B Y, Ma T, Liu J S, Kuo S R, Jin G, Broker T R, Harper J W, Chow L T. HeLa cells are phenotypically limiting in cyclin E/CDK2 for efficient human papillomavirus DNA replication. J Biol Chem. 2000;275:6167–6174. doi: 10.1074/jbc.275.9.6167. [DOI] [PubMed] [Google Scholar]
- 36.Maga G, Stucki M, Spadari S, Hüubscher U. DNA polymerase switching: I. Replication factor C displaces DNA polymerase alpha prior to PCNA loading. J Mol Biol. 2000;295:791–801. doi: 10.1006/jmbi.1999.3394. [DOI] [PubMed] [Google Scholar]
- 37.Mastrangelo I A, Hough P V C, Wall J S, Dodson M, Dean F B, Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature. 1989;338:658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- 38.Melendy T, Stillman B. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J Biol Chem. 1993;268:3389–3395. [PubMed] [Google Scholar]
- 39.Mossi R, Keller R C, Ferrari E, Hübscher U. DNA polymerase switching: II. Replication factor C abrogates primer synthesis by DNA polymerase alpha at a critical length. J Mol Biol. 2000;295:803–814. doi: 10.1006/jmbi.1999.3395. [DOI] [PubMed] [Google Scholar]
- 40.Murakami Y, Eki T, Yamada M, Prives C, Hurwitz J. Species-specific in vitro synthesis of DNA containing the polyoma virus origin of replication. Proc Natl Acad Sci USA. 1986;83:6347–6351. doi: 10.1073/pnas.83.17.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakami Y, Hurwitz J. DNA polymerase α stimulates the ATP-dependent binding of simian virus tumor T antigen to the SV40 origin of replication. J Biol Chem. 1993;268:11018–11027. [PubMed] [Google Scholar]
- 42.Murakami Y, Wobbe R, Weissbach C L, Dean F B, Hurwitz J. Role of DNA polymerase α and DNA primase in simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA. 1986;83:2869–2873. doi: 10.1073/pnas.83.9.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nasheuer H P, Grosse F. DNA polymerase α-primase from calf thymus. Determination of the polypeptide responsible for primase activity. J Biol Chem. 1988;263:8981–8988. [PubMed] [Google Scholar]
- 44.Nasheuer H P, Grosse F. Immunoaffinity-purified DNA polymerase α displays novel properties. Biochemistry. 1987;26:8458–8466. doi: 10.1021/bi00399a064. [DOI] [PubMed] [Google Scholar]
- 45.Nasheuer H P, von Winkler D, Schneider C, Dornreiter I, Gilbert I, Fanning E. Purification and functional characterization of bovine RP-A in an in vitro SV40 DNA replication system. Chromosoma. 1992;102:S52–S59. doi: 10.1007/BF02451786. [DOI] [PubMed] [Google Scholar]
- 46.Nesper J, Smith R W P, Kautz A R, Sock E, Wegner M, Grummt F, Nasheuer H P. A cell-free replication system for human polyomavirus JC DNA. J Virol. 1997;71:7421–7428. doi: 10.1128/jvi.71.10.7421-7428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors—a laboratory manual. W. H. New York, N.Y: Freeman and Company; 1992. [Google Scholar]
- 48.Pallas D C, Schley C, Mahoney M, Harlow E, Schaffhausen B S, Roberts T M. Polyomavirus small T antigen: overproduction in bacteria, purification, and utilization for monoclonal and polyclonal antibody production. J Virol. 1986;60:1075–1084. doi: 10.1128/jvi.60.3.1075-1084.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng Y C, Acheson N H. Polyomavirus and simian virus 40 large T antigens: a review. Recent Res Dev Virol. 1999;1:655–682. [Google Scholar]
- 50.Reynisdottir I, Bhattacharyya S, Zhang D, Prives C. The retinoblastoma protein alters the phosphorylation state of polyomavirus large T antigen in murine cell extracts and inhibits polyomavirus origin DNA replication. J Virol. 1999;73:3004–3013. doi: 10.1128/jvi.73.4.3004-3013.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salas M, Miller J T, Leis J, DePamphilis M L. Mechanisms for priming DNA synthesis. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 131–176. [Google Scholar]
- 52.Schneider C, Weisshart K, Guarino L A, Dornreiter I, Fanning E. Species-specific functional interactions of DNA polymerase α-primase with simian virus 40 (SV40) T antigen require SV40 origin DNA. Mol Cell Biol. 1994;14:3176–3185. doi: 10.1128/mcb.14.5.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simmons D T, Melendy T, Usher D, Stillman B. Simian virus 40 large T antigen binds to topoisomerase I. Virology. 1996;222:365–374. doi: 10.1006/viro.1996.0433. [DOI] [PubMed] [Google Scholar]
- 54.Smale S T, Tjian R. T-antigen-DNA polymerase α complex implicated in simian virus 40 DNA replication. Mol Cell Biol. 1986;6:4077–4087. doi: 10.1128/mcb.6.11.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith R W P, Nasheuer H P. Control of papovaviral DNA replication. In: Pandalai S G, editor. Recent research development in virology. 2. Transworld Research Network. India: Trivandrum; 2000. pp. 67–92. [Google Scholar]
- 56.Stadlbauer F. Molekulare Charakterisierung der Wirtsspezifität der DNA-Replikation von Papovaviren. Ph.D. dissertation. Munich, Germany: LMU München; 1995. [Google Scholar]
- 57.Stadlbauer F, Brueckner A, Rehfuess C, Eckerskorn C, Lottspeich F, Förster V, Tseng B Y, Nasheuer H P. DNA replication in vitro by recombinant DNA-polymerase-α-primase. Eur J Biochem. 1994;222:781–793. doi: 10.1111/j.1432-1033.1994.tb18925.x. [DOI] [PubMed] [Google Scholar]
- 58.Stadlbauer F, Voitenleitner C, Brückner A, Fanning E, Nasheuer H P. Species-specific replication of simian virus 40 DNA in vitro requires the p180 subunit of human DNA polymerase α-primase. Mol Cell Biol. 1996;16:94–104. doi: 10.1128/mcb.16.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stahl H, Dröge P, Knippers R. DNA helicase activity of SV40 large tumor antigen. EMBO J. 1986;5:1939–1944. doi: 10.1002/j.1460-2075.1986.tb04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stillman B. Initiation of chromosomal DNA replication in eukaryotes. Lessons from lambda. J Biol Chem. 1994;269:7047–7050. [PubMed] [Google Scholar]
- 61.Stillman B. Comparison of DNA replication in cells from prokarya and eukarya. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 435–460. [Google Scholar]
- 62.Sverdrup F, Schaffhausen B S, Androphy E J. Polyomavirus large T can support DNA replication in human cells. Virology. 1998;240:50–56. doi: 10.1006/viro.1997.8865. [DOI] [PubMed] [Google Scholar]
- 63.Tanaka S, Hu S-Z, Wang T S-F, Korn D. Preparation and preliminary characterization of monoclonal antibodies against human DNA polymerase α. J Biol Chem. 1982;257:8386–8390. [PubMed] [Google Scholar]
- 64.Trowbridge P W, Roy R, Simmons D T. Human topoisomerase I promotes initiation of simian virus 40 DNA replication in vitro. Mol Cell Biol. 1999;19:1686–1694. doi: 10.1128/mcb.19.3.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Voitenleitner C, Fanning E, Nasheuer H P. Phosphorylation of DNA polymerase α-primase by cyclin A-dependent kinases regulates initiation of DNA replication in vitro. Oncogene. 1997;14:1611–1615. doi: 10.1038/sj.onc.1200975. [DOI] [PubMed] [Google Scholar]
- 66.Voitenleitner C, Rehfuess C, Hilmes M, Rear O, Liao P C, Gage D A, Ott R, Nasheuer H P, Fanning E. Cell cycle-dependent regulation of human DNA polymerase α-primase activity by phosphorylation. Mol Cell Biol. 1999;19:646–656. doi: 10.1128/mcb.19.1.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 68.Wang M, Park J S, Ishiai M, Hurwitz J, Lee S H. Species specificity of human RPA in simian virus 40 DNA replication lies in T-antigen-dependent RNA primer synthesis. Nucleic Acids Res. 2000;28:4742–4749. doi: 10.1093/nar/28.23.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang T S-F. Cellular DNA polymerases. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 461–493. [Google Scholar]
- 70.Weisshart K, Förster H, Kremmer E, Schlott B, Grosse F, Nasheuer H P. Protein-protein interactions of the primase subunits p58 and p48 with simian virus 40 T antigen are required for efficient primer synthesis in a cell-free system. J Biol Chem. 2000;275:17328–17337. doi: 10.1074/jbc.M000717200. [DOI] [PubMed] [Google Scholar]
- 71.Weisshart K, Taneja P, Fanning E. The replication protein A binding site in simian virus 40 (SV40) T antigen and its role in the initial steps of SV40 DNA replication. J Virol. 1998;72:9771–9781. doi: 10.1128/jvi.72.12.9771-9781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wold M S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 73.Yuzhakov A, Kelman Z, Hurwitz J, O'Donnell M. Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. EMBO J. 1999;18:6189–6199. doi: 10.1093/emboj/18.21.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]