Abstract
Recombinant hepatitis C virus (HCV) RNA-dependent RNA polymerase (RdRp) was reported to possess terminal transferase (TNTase) activity, the ability to add nontemplated nucleotides to the 3′ end of viral RNAs. However, this TNTase was later purported to be a cellular enzyme copurifying with the HCV RdRp. In this report, we present evidence that TNTase activity is an inherent function of HCV and bovine viral diarrhea virus RdRps highly purified from both prokaryotic and eukaryotic cells. A change of the highly conserved GDD catalytic motif in the HCV RdRp to GAA abolished both RNA synthesis and TNTase activity. Furthermore, the nucleotides added via this TNTase activity are strongly influenced by the sequence near the 3′ terminus of the viral template RNA, perhaps accounting for the previous discrepant observations between RdRp preparations. Last, the RdRp TNTase activity was shown to restore the ability to direct initiation of RNA synthesis in vitro on an initiation-defective RNA substrate, thereby implicating this activity in maintaining the integrity of the viral genome termini.
Replication of plus-strand RNA viruses requires a multisubunit enzyme, the replicase, which is composed of viral and cellular factors (6). Biochemical characterization of eukaryotic replicases is limited because of difficulty in obtaining sufficient quantities of purified replicase. Furthermore, the hepatitis C virus (HCV) replicase has not been reported to accept exogenously provided RNAs. These results have prompted studies of the recombinant HCV RNA-dependent RNA polymerase (RdRp), the subunit responsible for phosphoryl transfer (9, 16, 17, 26, 31, 38). While RdRps lack many properties of replicases, they are useful for characterizing some fundamental activities, such as the recognition of the initiation site and the kinetics of nucleotide polymerization (4, 18, 24).
The HCV RdRp has recently been demonstrated to initiate RNA synthesis preferentially from the 3′ terminus of the template RNA (16, 17, 26, 31). Initiation from the 3′ terminus raises a potential problem that viruses might encounter: cellular RNases that degrade even a few 3′ nucleotides could prevent the initiation of viral RNA replication. Several mechanisms have been proposed that might allow RNA viruses to preserve or restore the sequences at the termini of their genome. These include base-pairing-dependent and base-pairing-independent recombination (12), priming by oligonucleotides aborted during the initiation of RNA synthesis (29), telomerase-like addition of a repeated sequence (33), and nontemplated nucleotide addition (7, 12). Also, terminal adenylyl transferase activity was found to be associated with poliovirus polymerase 3Dpol (30), possibly causing restoration of infectivity of poliovirus RNAs lacking the wild-type poly(A) tail.
Recombinant HCV RdRp was reported to possess the ability to add nontemplated nucleotides to the 3′ end of viral RNAs (5). However, this terminal transferase (TNTase) activity was later purported to be a cellular enzyme copurifying with the HCV RdRp (25). In this report, we present evidence that TNTase activity is an inherent function of the HCV and bovine viral diarrhea virus (BVDV) RdRps. Furthermore, the nucleotides added via this TNTase activity are strongly influenced by the sequence near the 3′ terminus of the viral template RNA, thereby implicating this RdRp-associated activity in maintaining the integrity of the termini of the viral RNA genome.
MATERIALS AND METHODS
Cloning of recombinant RdRp NS5B.
The NS5B protein from BVDV (genotype 1b) was prepared as described by Zhong et al. (39). HCV genotype 1b isolate strain J4 (37) was the source to produce the HCV NS5B used in this study. cDNAs coding for full-length NS5B, a 21- or 51-residue C-terminally truncated proteins, were amplified using primers and PfuI polymerase (Stratagene Inc., San Diego, Calif.). C-terminal truncations were made to increase the solubility of the proteins, but these did not affect polymerase activity (9). All cDNAs were subcloned into pET21b for expression with a C-terminal polyhistidine tag and sequenced to confirm that the clone was correct. The sequence coding for a full-length HCV NS5B was also subcloned into pVL1392 (Invitrogen, San Diego, Calif.) to generate a recombinant baculovirus, BacFL. NS5B catalytic mutants were generated by site-directed mutagenesis of the GDD motif to GAA.
Cells and viruses.
Sf9 cells (Clontech) were grown in suspension in Grace's insect medium (Gibco-BRL) supplemented with 10% fetal calf serum (FCS). For protein expression, 108 cells were infected with recombinant baculovirus for 1 h at room temperature at a multiplicity of infection of 10 in a total volume of 20 ml of FCS-free medium. After infection, the cells were resuspended to 1 liter and incubated for 72 h at 28°C.
Generation of recombinant baculovirus and purification of full-length NS5B.
Cells (seeded at 106 Sf9 cells per 35-cm2 dish) were washed twice with 1.5 ml of FCS-free medium in preparation for transfection. A mixture of 1 μg of recombinant plasmid and 0.2 μg of linearized BaculoGold DNA (PharMingen, Inc.) was transfected according to the manufacturer's recommendations. The cells were incubated for 5 days at 28°C, and half of the supernatant was used for virus production on Sf9 cells. For plaque purification, the Sf9 cells were infected with serially diluted virus-containing FCS-free medium. Cell monolayers were overlaid with 0.5% SeaPlaque GTG agarose (FMC BioProducts, Olendorf, Germany) and incubated until plaques developed. Well-separated plaques were purified with a Pasteur pipette, and the eluted virus was amplified on Sf9 cells.
For NS5B purification from infected Sf9 cells, the cells were harvested by centrifugation at 1,000 rpm for 10 min and then frozen at −80°C until protein purification. NS5B was purified by previously reported methods (5, 25).
Purification of NS5B from E. coli.
NS5B was expressed from pET derivatives in Escherichia coli BL21(DE3)LysS. Bacteria were grown at 30°C in standard Luria-Bertani medium supplemented with ampicillin (final concentration, 50 μg/ml) and chloramphenicol (34 μg/ml) until the culture reached an optical density at 600 nm of 1.0. The culture temperature was then lowered to 25°C, and expression was induced for 4 h with 1 mM isopropylthiogalactoside. Cells were harvested after centrifugation at 3,000 rpm for 0.5 h. The purification steps were essentially as described by Behrens et al. (5), and the N termini of the expressed proteins were sequenced to confirm the correct translation of each protein. To quantify NS5B, serial dilutions were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) along with a series of bovine serum albumin (BSA) samples of known amounts (21). The gels were then stained with Coomassie brilliant blue, bands were quantified by densitometry scans, and the concentration of NS5B was derived from the BSA standards.
BVDV NS5B was purified as follows. Cell pellets (4 g) were thawed on ice, suspended in 15 ml of nickel-nitrilotriacetic acid (Ni-NTA) buffer A (20 mM Tris-HCl [pH 7.5], 500 mM NaCl, 10 mM MgCl2, 10 mM imidazole, 0.5% Triton X-100, 12.5% [vol/vol] glycerol, and a mixture of protease inhibitors [7 nM leupeptin, 42 nM pepstatin, and 220 μM phenylmethylsulfonyl fluoride]), and then lysed by passage thrice through a French press at 1,000 lb/in2. The lysate was clarified by centrifugation at 16,000 × g for 24 min, and the supernatant was loaded at 1 ml/min onto a 1-ml HiTrap nickel-chelating fast protein liquid chromatography (FPLC) column (Amersham Pharmacia) prepared per the manufacturer's instructions. The column was washed with 10 column volumes (CV) of buffer A before eluting NS5B with Ni-NTA buffer B (20 mM Tris-HCl [pH 7.5], 500 mM NaCl, 10 mM MgCl2, 350 mM imidazole, 0.5% Triton X-100, 12.5% [vol/vol] glycerol, and the mixture of protease inhibitors described above).
Additional purification of BVDV NS5B was performed with a 1-ml HiTrap sulphopropyl (SP) column (Amersham Pharmacia). One half of the pooled eluate containing NS5B was diluted sixfold with buffer A, loaded thrice (1 ml/min) onto a 1-ml SP column. The SP column was then washed with 5 CV of buffer A prior to elution with 15 CV containing a linear gradient from 0 to 70% buffer B (20 mM Tris-HCl [pH 7.5], 1 M NaCl, 5% glycerol, and protease inhibitors).
RdRp activity and TNTase assays.
DNAs were synthesized by Operon Inc. (Alameda, Calif.). RNAs were chemically synthesized by Dharmacon Inc. (Boulder, Colo.), purified by denaturing gel electrophoresis, checked for quality by toluidine blue staining, and quantified by spectrophotometry. Standard RdRp assays consisted of 2.5 pmol of template (unless stated otherwise) with 300 ng of NS5B in a 20-μl reaction containing 20 mM sodium glutamate (pH 8.2), 4 mM MgCl2, 12.5 mM dithiothreitol, 0.5% (vol/vol) Triton X-100, 1 mM MnCl2, 200 μM each ATP and UTP, 500 μM GTP, and 250 nM [α -32P]CTP (Amersham). TNTase assays were performed in the same buffer with the specified nucleoside triphosphates (NTPs). Both TNTase and RNA synthesis reactions were incubated at 25°C for 60 min and stopped by phenol-chloroform extraction followed by ethanol precipitation in the presence of 5 μg of glycogen and 0.5 M ammonium acetate. Products were separated by electrophoresis on denaturing 7 M urea–20% polyacrylamide gels. Gels were wrapped in plastic and exposed to film at −80°C. Quantification of radiolabeled bands was performed using a PhosphorImager (Molecular Dynamics).
RESULTS
Recombinant RdRp has TNTase activity.
RdRp-directed RNA synthesis from minimum-length templates often produces products longer than template length (15, 17). Several activities could contribute to the formation of these products, including the addition of nontemplated terminal nucleotides on either the newly synthesized RNA or the input template. We performed studies to determine whether highly purified (>95%; Fig. 1A) recombinant HCV RdRp possesses TNTase activity.
FIG. 1.
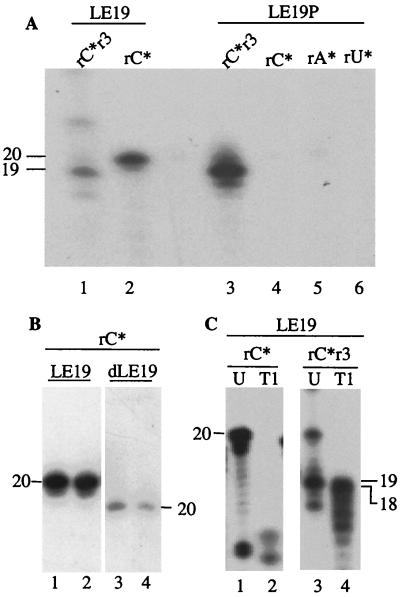
HCV NS5B has TNTase activity. (A) SDS-PAGE profile of different RdRps used in this study. BacFL is a full-length protein expressed using baculovirus. Proteins HΔ21 and HΔ51 are HCV NS5B with 21- and 51-amino-acid deletions from the C terminus. mΔ21 has a change of the GDD motif to GAA. Lane M, molecular size markers. (B) Predicted secondary structure of template LE19. (C) RNA synthesis assay (lanes 1 and 8) and TNTase assay (lanes 2 to 7 and 9 to 11) performed with RNA LE19. The nucleotide substrates used are listed above the autoradiogram. The asterisk (∗) indicates that the nucleotide is radiolabeled with α-32P. r3 denotes the presence of GTP, ATP, and UTP. The reactions in lanes 1 to 7 were performed with HΔ21, and those in 8 to 11 were performed with mΔ21. Sizes of reaction products are shown (in nt) on the side of the autoradiogram. (D) RNA synthesis (lane 1) and TNTase (lanes 2 to 7) assays performed with BacFL using template LE19. (E) RNA synthesis (lanes 1 and 2) and TNTase (lanes 3 and 4) assays performed with Δ51 using template LE19. The sizes of the RdRp products in this and other experiments were assigned by comparison with a series of BVDV RdRp products that varied by one nucleotide (M. Kim and C. Kao, data not shown).
The polymerase assays primarily used a 19-nucleotide (nt) RNA, LE19, derived from the 3′ end of the negative strand of the BVDV genome. RNA LE19 is predicted by the mfold program (14) to form an intramolecular fold consisting of a 5-bp stem-loop flanked by 3-nt single-stranded sequences at both the 5′ and 3′ ends (14) (Fig. 1B). The 3′ sequence contains a cytidylate that could be used as an initiation nucleotide. LE19 is a simplified version of an RNA that has been shown to direct de novo initiation by the HCV NS5B (18; C. T. Ranjith-Kumar, unpublished results).
Recombinant HCV NS5B, named HΔ21, purified from E. coli produced a 19-nt RNA from LE19 when all rNTPs were present (Fig. 1C, lane 1). In addition, two low-abundance higher-molecular-weight bands of ∼20 and 24 nt were observed. To discern whether these bands were produced by terminal nucleotide addition on the template RNA, HΔ21 was incubated with α-32P-radiolabeled rCTP, rATP, or rUTP in RdRp reaction buffer. Products of 20 to 22 nt were observed in reactions containing [α-32P]-rATP and [α-32P]-rCTP, whereas reactions containing [α-32P]-rUTP yielded only minor products from HΔ21 (Fig. 1C, lanes 2 to 4). The 19-nt newly synthesized RNA was not observed, as expected, since its synthesis should not occur in reactions containing only one nucleotide. Also, the 20- and 24-nt RNAs that were quite prominent in the reaction containing only [α-32P]-CTP were less obvious in reactions containing all four nucleotides (including [α-32P]-CTP), suggesting that the presence of unlabeled nucleotides decreased the terminal labeling of the RNAs.
Next, radiolabeled dCTP, dATP, and dTTP were evaluated as substrates for the putative TNTase activity. Labeled products were apparent only in the reactions containing dCTP, but not dATP or dTTP. The migration of this product was altered slightly in comparison to the reaction with rCTP (Fig. 1C, lanes 2 and 5).
To determine whether the TNTase activity is an inherent property of the HCV NS5B or due to a contaminant in the HΔ21 preparation, the highly conserved GDD sequence in motif C was changed to GAA, resulting in mΔ21. Protein mΔ21 was unable to initiate RNA synthesis from template LE19 or to add terminal nucleotides to LE19 using either radiolabeled rNTPs or dNTPs (Fig. 1C, lanes 8 to 11, and data not shown). Hence, TNTase activity requires a catalytically active polymerase.
Two additional HCV RdRps were generated to eliminate the possibility that an E. coli protein(s) responsible for TNTase activity copurified with HΔ21 but not mΔ21. First, a version lacking the C-terminal 51 amino acids, HΔ51, was produced in E. coli. Second, a full-length HCV NS5B, BacFL, was produced using a recombinant baculovirus. Both proteins, HΔ51 and BacFL, were as pure as HΔ21 (Fig. 1A) and directed RNA synthesis using a number of templates, including RNA LE19 (Fig. 1D and 1E). HΔ51 and BacFL possess high levels of TNTase activity with LE19, generating labeled RNAs of 20 to 21 nt with radiolabeled rATP, rCTP, and dCTP more efficiently than with rUTP and dATP (Fig. 1D, lanes 2 to 7). These results are consistent with those from HΔ21, suggesting that highly purified HCV NS5B produced from either prokaryotic or eukaryotic cells possesses TNTase activity with similar substrate preferences.
To further demonstrate that the HCV RdRp has TNTase activity, we used a compound reported to inhibit HCV RdRp activity (32), 3-[5-(5-benzo[b]thiophen-2-yl-furan-2-ylmethylene)-4-oxo-2-thioxo-thiazolidin-3-yl]-propionic acid, whose structure is shown in Table 1. To maintain the inhibitor in solution, a final concentration of 5% dimethyl sulfoxide was added to the RNA synthesis assay. At this concentration, RNA synthesis was not significantly affected. However, RNA synthesis and TNTase activity were both decreased in a manner dependent on the concentration of the inhibitor (Table 1). These data provide additional evidence that the HCV RdRp indeed possesses TNTase activity.
TABLE 1.
Effect of HCV RdRp-specific inhibitor (structure shown in inset) on RNA synthesis and TNTase activitya
Values are percentages of the values with no inhibitor, which were set at 100%.
Recombinant BVDV RdRp also has TNTase activity.
To determine whether TNTase activity is a property of another RdRp from the Flaviviridae, recombinant BVDV NS5B produced in E. coli was tested. BΔ23, the BVDV NS5B lacking the C-terminal 23 residues, was more robust in directing RNA synthesis from template LE19 than HΔ21 (Fig. 2A, lanes 1 and 3). Similar to the products from HΔ21, several bands longer than 19 nt and a presumed prematurely terminated RNA of ∼17 nt were observed with BΔ23. At least a portion of the 20-nt RNA is likely due to TNTase activity, since RNA of this length was produced by BΔ23 in reactions that contained [α-32P]-rCTP or [α-32P]-rATP as the only nucleotide in the reaction (Fig. 2A, lanes 4 and 5). Other prominent, higher-molecular-weight bands may be due to nontemplated nucleotide addition on the newly synthesized RNA, a known property of the BVDV RdRp (15, 18), or by the terminal addition of multiple nucleotides at the 3′ terminus of LE19.
FIG. 2.
BVDV NS5B has TNTase activity that cofractionates with RdRp activity. (A) RNA synthesis and TNTase assays performed on RNA LE19. The nucleotide substrates used are shown above the autoradiogram. The asterisk corresponds to the radiolabeled nucleotide. r3 denotes the presence of GTP, ATP, and UTP. Sizes of the reaction products (in nt) are given on the side of the autoradiogram. Lanes 1 and 2 contain results from RNA synthesis and TNTase assays, respectively, performed using HΔ21, while lanes 3 to 8 were performed with BΔ23. (B) Silver-stained SDS-PAGE profile depicting the various SP-Sepharose column fractions. Fraction numbers are at the bottom of the gel. (C) Quantification of relative RNA synthesis and TNTase activity of the BΔ23 eluted from an SP-Sepharose column. RNA synthesis and TNTase activities in fraction 22 were set at 100%, and other fractions were normalized to these values.
In comparison to HΔ21, the TNTase activity of BΔ23 produced fewer labeled products relative to template-directed RNA synthesis (compare Fig. 2A, lanes 3 and 4, with Fig. 1C, lanes 1 and 2). The ratio of template-directed RNA synthesis to terminally labeled RNA was 0.3 and 0.8 with CMP and AMP incorporation, respectively, after adjusting for specific activity. The ratio of newly synthesized RNA to terminally labeled RNA with the HCV RdRp was approximately 3 and 4 with [α-32P]-CTP and [α-32P]ATP, respectively. Furthermore, another difference from the HΔ21 was that BΔ21 was unable to label RNA LE19 with dCTP (Fig. 2, lane 7).
Two additional approaches were used to further confirm that BVDV NS5B possesses TNTase activity. First, we fractionated the BΔ23 on a cation-exchange SP-Sepharose column. Fractions were assessed for total proteins by SDS-PAGE and staining with silver (Fig. 2B) and quantified for RNA synthesis and TNTase activity with rCTP. Fractions 18 to 26 contained the majority of the BΔ23, with the peak abundance in fractions 22 and 23. The highest activity for both RdRp and TNTase was also found in fractions 22 and 23 (Fig. 2C). The second approach used a panel of mutant BΔ23 proteins that had been tested previously for RNA synthesis (22). Mutant proteins R295A, D448A, D449A, C497A, NΔ120, and CΔ155, which were unable to perform template-dependent RNA synthesis, were found to be incapable of terminal nucleotide addition, while RNA synthesis-competent mutant proteins such as K282A, Y289A, D350A, and S405A retained TNTase activity (22; C. T. Ranjith-Kumar, data not shown). We note that D448A and D449A have mutations in the highly conserved GDD motif of the BVDV NS5B. Activities of the mutant BVDV NS5B and the coelution of RNA synthesis and TNTase activities strongly support our assertion that BΔ23 possesses TNTase activity.
Template requirements for TNTase activity.
We first determined whether the TNTase activity does add nucleotides to the 3′ terminus of the acceptor RNA, as would be expected. An RNA containing a puromycin 3′ of the initiation cytidylate (the first template nucleotide used for nucleotide polymerization) has previously been shown to retain the ability to initiate RNA synthesis by the HCV RdRp (17) (Fig. 3A, lanes 1 to 3). However, a puromycin containing a 3′ moiety attached via an amide linkage should abrogate terminal nucleotide addition. RNA LE19 containing a 3′-terminal puromycin, LE19P, was tested as a template for RNA synthesis and TNTase assays. While HΔ21 produced RNA products from LE19P, this template was not radiolabeled in a TNTase reaction in the presence of rCTP, rATP, or rUTP (Fig. 3A, lanes 4 to 6). A single-stranded DNA version of LE19, dLE19, could be radiolabeled in the TNTase assay, but at only 5% in comparison to RNA LE19 (Fig. 3B, compare lanes 3 and 4 to 1 and 2). We note that radiolabeled LE19 RNA and dLE19 reproducibly have slightly different mobilities. Taken together, these results indicate that one or more riboses in RNA LE19 contribute to efficient TNTase activity, while the 3′-terminal ribose hydroxyl was absolutely required.
FIG. 3.
Template requirements for TNTase activity. RdRp and TNTase assays performed with HΔ21. The nucleotide substrates used are shown above the autoradiogram. Asterisk (∗) identifies the radiolabeled nucleotide. r3 denotes the presence of GTP, ATP, and UTP in the reactions. The lengths of the reaction products (in nucleotides) are indicated on the side of the autoradiogram. (A) Reactions performed with RNA LE19 are in lanes 1 and 2, while those performed with the puromycin-modified LE19P are in lanes 3 to 6. (B) Comparison of TNTase activity performed with HΔ21, [α-32P]rCTP, and either an RNA or DNA version of LE19. (C) Autoradiogram of the products from TNTase (lanes 1 and 2) and RNA synthesis (lanes 3 and 4) reactions, untreated (U) or treated with RNase T1 (T1).
To confirm that nucleotides are added to the 3′ end of the template, RNase T1 digestions were performed on products of TNTase and RNA synthesis reactions. RNase T1 cleaves the phosphodiester bond 3′ of a guanylate. Digestion of a terminally labeled LE19 RNA should yield an RNA of 6 nt or longer, depending on whether one or more nucleotides were added to the 3′ terminus. We observed that RNase T1 digestion of RNA LE19 labeled in the TNTase assay resulted in bands of about 6 nt (Fig. 3C, lanes 1 and 2). RNA synthesized by HΔ21 should result in an 18-nt RNA when treated with RNase T1. Indeed, this was readily observed (Fig. 3C, lanes 3 and 4).
Reaction condition required for TNTase activity.
Mn2+ has been shown to enhance RNA synthesis by the HCV RdRp (9). To determine whether TNTase activity also required Mn2+, reactions were performed with [α-32P]rCTP with and without 1 mM Mn2+ (Fig. 4A) using both HΔ21 and BΔ23. For the HΔ21, both RNA synthesis and TNTase activity were decreased by about fourfold in the absence of Mn2+ (Fig. 4A, lanes 1 to 8). We note that significant TNTase activity remained in the absence of Mn2+, and thus it is not necessary for the TNTase activity of HΔ21. The absence of Mn2+ had more of a differential effect on RNA synthesis and TNTase activity with BΔ23. RNA synthesis was reduced to 30% and TNTase activity to 5% (Fig. 4B, lanes 1 to 8). Mn2+ may also affect nontemplated nucleotide addition on the newly synthesized RNA by the BΔ23, as evidenced by the abundance of RdRp products longer than 19 nt in the RNA synthesis reactions (Fig. 4B, lanes 9 and 10) (M. Kaganovich, unpublished results).
FIG. 4.
Effect of Mn2+ and KCl on TNTase activity. (A) RdRp and TNTase assays using HΔ21. Both assays were performed with enzyme HΔ21 and RNA LE19. The nucleotides used in the reactions are listed above the autoradiogram. Asterisk (∗) identifies the radiolabeled nucleotide. r3 denotes the presence of GTP, ATP, and UTP in the reactions. Lengths of the reaction products (in nucleotides) are given between the two autoradiograms. Lanes + and −, reactions performed in the presence and absence of Mn2+, respectively. (B) RdRp and TNTase assays for BΔ23. The layout of the figure is identical to that in panel A. (C) Effects of increasing KCl concentration on RNA synthesis and TNTase activity of HΔ21. (D) Effects of increasing KCl concentration on RNA synthesis and TNTase activity of BΔ23.
KCl can modulate RdRp-template interactions (24). The effects of increasing salt concentration on RNA synthesis and TNTase activities of HΔ21 and BΔ23 were evaluated (Fig. 4C). Increasing the KCl concentration from 10 to 250 mM caused a corresponding decrease in both enzymatic activities. However, for the HCV RdRp, TNTase activity proved more sensitive to salt than RNA synthesis. At 75 mM KCl, TNTase activity was decreased by at least 100-fold, while RNA synthesis remained at approximately 20% of the standard reaction value. For BΔ23, increasing the salt concentration did not have significantly different effects on RNA synthesis and TNTase activity. At 75 mM KCl, RNA synthesis and TNTase activities were 10 and 5%, respectively (Fig. 4D). In summary of this section, RdRps from closely related viruses have different requirements for the TNTase activity.
3′-Terminal cytidylate is not required for TNTase activity.
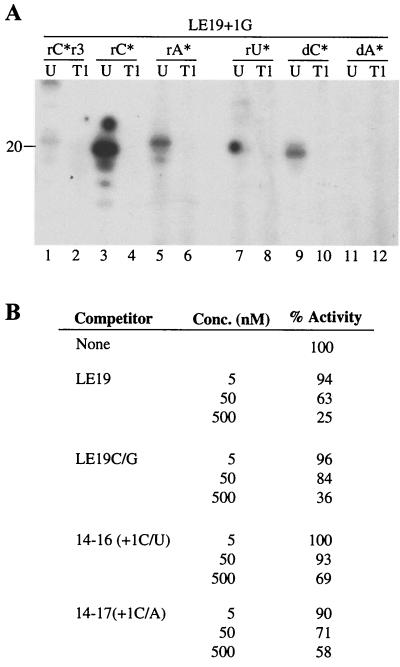
The 3′-terminal cytidylate is preferred for the de novo initiation of RNA synthesis by the HCV RdRp (17). Whether the 3′C was required for TNTase activity was examined using LE19+1G, a modified version of RNA LE19 containing a 3′ guanylate. As expected, LE19+1G was severely reduced in the ability to direct the synthesis of the product RNA (Fig. 5A, lane 1). However, LE19+1G was readily radiolabeled with rCTP, rATP, and dCTP (Fig. 5A, lanes 3, 5, and 9), indicating that the TNTase activity can be observed with different 3′-terminal nucleotides. When treated with RNase T1, terminally labeled LE19+1G would be cleaved after the 3′-terminal G, resulting in an 18-nt RNA that is not radiolabeled and a 2-nt product that could not be resolved from the unincorporated label in our gels. We observed that RNase T1 treatment of reactions performed with LE19+1G abolished the radiolabeled bands (Fig. 5A, lanes 2, 4, 6, 8, 10, and 12). This is consistent with the hypothesis that terminally added radiolabeled nucleotides are present at the 3′ end of the template. In contrast to RNA LE19, which incorporated [α-32P]ATP preferentially over [α-32P]CTP, terminal labeling with LE19+1G preferentially used [α-32P]CTP. This result suggests that the nucleotide(s) incorporated by the TNTase at the 3′ end of the template may depend on the template sequence.
FIG. 5.
TNTase activity does not require the initiation nucleotide. (A) RNA synthesis (lanes 1 and 2) and TNTase (lanes 3 to 12) assays performed with HΔ21 using LE19+1G. The nucleotide substrates used in each reaction are listed above the autoradiogram. Asterisk (∗) identifies the radiolabeled nucleotide. r3 denotes the presence of GTP, ATP, and UTP in the reaction. The length of the reaction products (in nucleotides) is on the side of the autoradiogram. T1, reactions treated with RNase T1, U, untreated control. (B) LE19+1G retains interaction with HΔ21. This assay measures the synthesis from template SLD3 as affected by the presence of a second RNA in the reaction. SLD3 has the sequence 5′-GGGCUUGCAUAGCAAGUCUGAGACC-3′ (17). RNAs 14–16 and 14–17 have sequences 5′-AAAUCCUCUGAUAU-3′ and 5′-AAAUCCUCUGAUAA-3′, respectively.
Since the HCV RdRp could utilize LE19+1G for terminal nucleotide addition, either LE19+1G must retain the ability to interact with NS5B or else the TNTase activity is not associated with RdRp. There is precedent in that an initiation-incompetent stem-loop RNA has previously been demonstrated to bind the HCV NS5B (17). To experimentally distinguish these two possibilities, we assessed whether LE19+1G could interact with HΔ21 in a template competition assay (Fig. 5B). In these reactions, an initiation-competent RNA, SLD3, was incubated with increasing concentrations of RNA LE19 and LE19+1G. The amount of synthesis from SLD3 was quantified to assess the inhibitory effects of the competitors. Both LE19 and LE19+1G were more effective competitors than two 14-nt RNAs, 14-16 and 14-17 (Fig. 5B). These results are consistent with our claim that LE19+1G retained the ability to interact with HΔ21 and that the TNTase activity is a property of the RdRp.
Restoration of initiation site by terminal nucleotide addition.
Since the initiation nucleotide is not necessary for TNTase activity, it is possible that the TNTase activity could add nucleotides that are used to initiate RNA synthesis. To test this possibility, we used the scheme summarized in Fig. 6A. Briefly, RNA LE19 was incubated for 1 h with HΔ21, and 100 μM unlabeled rATP, rCTP, or dCTP and then treated with alkaline phosphatase to render the free nucleotides incapable of participating in RNA polymerization, followed by phenol extraction and ethanol precipitation. Products of the TNTase reactions were then used as the template for RNA synthesis with HΔ21. To discourage additional TNTase activity, syntheses were performed in the presence of 75 mM KCl, a concentration that effectively reduced TNTase activity (Fig. 6B, compare lanes 1 and 2). RNA LE19, which incorporated 3′-terminal rCMP or dCMP, directed the synthesis of 20- and 21-nt RNAs (Fig. 6B, lanes 5 and 6). However, LE19 terminally modified with AMP directed primarily a 19-nt RNA that likely initiated from the original 3′-terminal cytidylate (Fig. 6B, lane 4 and 5).
FIG. 6.
TNTase activity can restore de novo initiation on an initiation-incompetent RNA. (A) Schematic of the treatments used in the experiments shown in panels B and C. APase, alkaline phosphatase. (B) RNA synthesis and TNTase assays performed with HΔ21, RNA LE19, and the nucleotide used in the TNTase assay (T-NTPs) indicated above the autoradiogram. The asterisk (∗) denotes that the nucleotide is radiolabeled. RNA synthesis reaction in lanes 3 to 6 used [α-32P]CTP and unlabeled ATP, GTP, and UTP. Lengths of the reaction products (in nucleotides) are to the right of the autoradiogram. Lanes + and −, reactions performed in the presence and absence of 75 mM KCl, respectively. (C) TNTase and RdRp reactions performed using 14-1, 14–16, and 14–17 as the templates. Nucleotides used in the TNTase reactions are identified in the column labeled T-NTP. The absence and presence of 75 mM KCl is indicated (− and +, respectively). RNA synthesis reactions contained [α-32P]CTP and unlabeled ATP, GTP, and UTP.
To confirm that terminal nucleotide addition could give an initiation-incompetent RNA the ability to direct RNA synthesis, we used templates 14–16 and 14–17, which contain a 3′ U and a 3′ A, respectively. Relative to 14-1, which has a 3′ C, 14–16 and 14–17 directed RNA synthesis at 10 and 2%, respectively (C. T. Ranjith-Kumar, data not shown). Also, the presence of 75 mM KCl in the reactions decreased terminal nucleotide addition (Fig. 6C, lanes 2, 6, and 10). RNAs 14-1, 14–16, and 14–17 were used for terminal nucleotide addition in three independent reactions, one which lacked all NTPs, one containing only rATP, and one containing only rCTP. After TNTase assays, these templates were used for RNA synthesis in the presence of all four NTPs and 75 mM KCl. The predominant product formed after mock treatment or AMP addition to template 14-1 was a 14-mer, while no products were seen with RNAs 14–16 and 14–17 (Fig. 6C, lanes 2, 3, 6, 7, 10, and 11). Quite dramatically, terminal addition of CMP(s) prior to the RdRp reaction allowed both 14–16 and 14–17 to initiate RNA synthesis, producing RNAs of 15 and 16 nt (Fig. 6C, lanes 8 and 12). With 14-1, 14–16, and 14–17, multiple bands were seen. These could have resulted from one or more CMPs added to the 3′ terminus of these templates.
Identity of terminally added nucleotide(s) is affected by template sequence.
The observation that RNA LE19 and LE19+1G accepted different terminal nucleotides suggests that nucleotide selection and incorporation are affected by the RNA sequence. To examine this further, a series of 14-nt RNAs with changes concentrated at the 5′ and 3′ ends were tested. Quantification of the TNTase assays using radiolabeled rATP and rCTP is shown in Table 2. In contrast to RNA LE19, the majority of the RNAs in this series incorporated rCMP more efficiently than rAMP, although substantial variation for the incorporation was observed. For example, threefold more CMP than AMP was incorporated into RNA 14-1, while 40-fold more AMP than CMP was incorporated into RNA 14–18 (Table 2). Hence, changes in the template sequence and/or structure could affect the efficiency of terminal nucleotide addition.
TABLE 2.
Nucleotide(s) added by the TNTase activity of HΔ21 are affected by template sequence
| Template | Sequencea (5′ to 3′) | Quantification of TNTase activityb
|
|||
|---|---|---|---|---|---|
| Expt 1
|
Expt 2 (ratio, CMP/ AMP) | ||||
| CMP | AMP | Ratio, CMP/ AMP | |||
| 14-1 | AAAUCCUCUGAUAC | 8,915 | 2,972 | 3.0 | 4.2 |
| 14-2 | AACCUUACUGAUAC | 10,309 | 5,102 | 2.0 | 2.8 |
| 14-3 | CCAUUCUCUGAUAC | 10,410 | 3,097 | 4.4 | 3.5 |
| 14-4 | GCAUUCUCUGAUAC | 10,465 | 2,377 | 3.4 | |
| 14-5 | UUAUUCUCUGAUAC | 8,127 | 4,702 | 1.7 | |
| 14-6 | AUAUUCUCUGAUAC | 8,444 | 2,390 | 3.5 | |
| 14-14 | AAAGCCUCUGAUAC | 9,127 | 3,661 | 2.5 | |
| 14-15 | AAACGCUCUGAUAC | 9,805 | 2,995 | 3.3 | |
| 14-16 | AAAUCCUCUGAUAU | 8,919 | 5,223 | 1.7 | |
| 14-17 | AAAUCCUCUGAUAA | 1,216 | 989 | 1.2 | 1.2 |
| 14-18 | AAAUCCUCUGAUAG | 140 | 6,087 | 0.0 | 0.0 |
| 14-19 | AAAUCCUCUGAUUC | 1,986 | 5,734 | 0.4 | 0.1 |
| 14-20 | AAAUCCUCUGAAAC | 8,927 | 5,988 | 1.5 | |
| 14-21 | AAAUCCUCUGUUAC | 6,332 | 8,253 | 0.9 | 0.5 |
| 14-22 | AAAUCCUCUCAUAC | 91 | 1,154 | 0.1 | 0.3 |
Nucleotides that are different from those in 14-1 are in boldface type.
The values in the CMP and AMP columns were quantified by a PhosphorImager and used to calculate the CMP/AMP ratio in experiment 1. The results in experiment 2 were from an independent set of assays. Blanks in this column indicate that the templates were not assayed.
Next, we examined whether the positions of the nucleotide substitutions in RNAs 14-2 to 14–15 (Table 2) correlate with preference for the incorporation of rCMP or rAMP. Templates with single or multiple changes in the 5′ portion of the RNAs favored CMP addition in a manner similar to 14-1. However, changes near the 3′ portion more readily impacted the efficiency of CMP or AMP incorporation (Table 2). Notably, a change of the 3′ C to a G in template 14–18 and a change of a C to a G five nucleotides from the 3′ end (RNA 14–22) significantly favored AMP addition. These results are consistent with our previous observation that a change of the 3′-terminal nucleotide of RNA LE19 altered the preference of the terminally incorporated nucleotide (compare Fig. 5, lanes 3 and 5, to Fig. 1, lanes 2 and 3). In total, the sequence of the template, especially 3′-terminal nucleotides, can determine the identity and the overall efficiency of terminally incorporated nucleotides.
DISCUSSION
TNTase activity associated with the HCV RdRp was reported by Behrens et al. (5), but was claimed to be a contaminating activity by Lohmann et al. (25). We present several independent lines of evidence that viral RdRps possess TNTase activity. (i) Several RdRps, including those purified from prokaryotic or eukaryotic cells, have TNTase activity. (ii) Mutations in the highly conserved GDD motif of the HCV and BVDV RdRps, which are not predicted to change the folding of the polypeptide, inactivated both polymerase and TNTase activity. (iii) An inhibitor specific for the HCV RdRp also inhibited TNTase activity. (iv) TNTase activity in BVDV RdRp cofractionated with the enzyme and with RNA synthesis. We also demonstrate that the identity of the nucleotide incorporated at the 3′ terminus of the template is affected by the template sequence. Last, TNTase activity was shown to restore the ability to direct initiation of RNA synthesis in vitro using an RNA that was unable to direct the initiation of RNA synthesis.
While all evidence is in agreement that TNTase activity is a property of RdRps that requires the RdRp catalytic site, it can have distinct requirements even between RdRps from closely related viruses. We had previously seen that the BVDV and HCV RdRps have different requirements with regard to recognition of the initiation cytidylate (17). It is possible that one or a few amino acid differences in the RdRp may alter the specificity for a biochemical activity. It remains to be determined whether specific mutations within an RdRp will differentially affect the two activities.
TNTase activity may be a common property of RNA polymerases. DNA-dependent RNA polymerases are known to add nucleotides to the ends of newly synthesized RNAs (19, 28). The replicases of BMV and turnip crinkle virus have also been documented to add terminal nucleotides to either the template or product RNA (12, 34). The poliovirus RdRp could also add nontemplated nucleotides to the blunt end of the RNA primer-template (2). Poliovirus 3Dpol was reported to bind the primer-template complex in two possible orientations, one that leads to product RNA synthesis and another that results in TNTase activity. This result is consistent with our observation that TNTase and RNA synthesis activity have different requirements.
Our results suggest that TNTase activity can often be masked by the reaction conditions. In reactions containing unlabeled NTPs, the detection of labeled TNTase products in autoradiograms can be significantly decreased. The incorporation of the terminal nucleotide could be significantly affected by the template sequence, especially the nucleotides near the 3′ terminus of the RNA (Table 2). These results may explain why TNTase activity was inconsistently observed by independent groups (1, 25, 36).
RdRp's TNTase activity may affect the initiation of RNA synthesis by adding nucleotides to the 3′ terminus of template RNAs. For most plus-strand RNA viruses in the Flaviviridae family and the alphavirus superfamily, initiation of RNA synthesis occurs by a de novo mechanism from a nucleotide at or near the 3′ terminus of the template RNA. It is unlikely that TNTase activity would abolish RNA synthesis, since initiation has been shown to occur on templates containing additional nucleotides (3, 27). The HCV RdRp can also initiate RNA synthesis from an initiation nucleotide that is not present at the very 3′ terminus of the template (17, 31).
Additionally, TNTase activity may be advantageous to a virus under certain circumstances. RNA synthesis by the cucumber mosaic virus replicase requires a nontemplated nucleotide at the 3′ terminus of the template RNA (35). Indeed, initiation from an internal nucleotide may be an advantageous feature for some replicases. The Tacaribe arenavirus, Hantaan virus, and Respiratory syncytial virus have evolved a mechanism to initiate RNA synthesis from an internal template nucleotide and to realign the nascent oligonucleotide RNA to the end of the template, thus circumventing the difficult task of initiating at the very 3′-terminal nucleotide (10, 11, 20). Finally, the ends of viral genomes (the sequences that contain the initiation site) may be damaged by exonucleases, which are widely distributed in intracellular compartments (13). TNTase activity with some specificity for the incorporated nucleotide could potentially be used by RNA viruses as a mechanism to restore the 3′ initiation site. Such an activity was proposed for the turnip crinkle virus RNA replicase (8), and results consistent with this activity were recently observed (12). The data herein show that the HCV RdRp could restore an initiation nucleotide and then use it for RNA synthesis in vitro, consistent with the demonstration of polymerase-dependent TNTase activity. TNTase activity associated with viral RdRps and possibly RNA replicases could be at least partially responsible for the reported telomerase-like activity (33).
ACKNOWLEDGMENTS
We thank the IU cereal killers for helpful discussions. We thank Michael Darcy and Dash Dhanak for providing the HCV RdRp-specific inhibitor.
C. Kao acknowledges a fellowship from the Linda and Jack Gill Foundation. We thank the USDA and the NSF for funding the Kao laboratory.
REFERENCES
- 1.Al R H, Xie Y, Wang Y, Hagedorn C H. Expression of recombinant hepatitis C virus non-structural protein 5B in Escherichia coli. Virus Res. 1998;53:141–149. doi: 10.1016/s0168-1702(97)00147-0. [DOI] [PubMed] [Google Scholar]
- 2.Arnold J J, Ghosh S K B, Cameron C E. Poliovirus RNA-dependent RNA polymerase (3Dpol) J Biol Chem. 1999;274:37060–37069. doi: 10.1074/jbc.274.52.37060. [DOI] [PubMed] [Google Scholar]
- 3.Ball L A. Requirements for the self-directed replication of flock house virus RNA 1. J Virol. 1995;69:720–727. doi: 10.1128/jvi.69.2.720-727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beckman M T, Kirkegaard K. Site size of cooperative single-stranded RNA binding by poliovirus RNA-dependent RNA polymerase. J Biol Chem. 1998;273:6724–6730. doi: 10.1074/jbc.273.12.6724. [DOI] [PubMed] [Google Scholar]
- 5.Behrens S-E, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 6.Buck K W. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgyan J, Garcia-Arenal F. Template-independent repair of the 3′ end of cucumber mosaic virus satellite RNA controlled by RNAs 1 and 2 of helper virus. J Virol. 1998;72:5061–5066. doi: 10.1128/jvi.72.6.5061-5066.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpenter C D, Simon A E. In vivo restoration of biologically active 3′ ends of virus-associated RNAs by nonhomologous RNA recombination and replacement of a terminal motif. J Virol. 1996;70:478–486. doi: 10.1128/jvi.70.1.478-486.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrari E, Wright-Minogue J, Fang J W S, Baroudy B M, Lau J Y N, Hong Z. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J Virol. 1999;73:1649–1654. doi: 10.1128/jvi.73.2.1649-1654.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcin D, Kolakofsky D. Tacaribe arenavirus RNA synthesis in vitro is primer dependent and suggests an unusual model for the initiation of genome replication. J Virol. 1992;66:1370–1376. doi: 10.1128/jvi.66.3.1370-1376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcin D, Lezzi M, Dobbs M, Elliott R M, Schmaljohn C, Kang C Y, Kolakofsky D. The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J Virol. 1995;69:5754–5762. doi: 10.1128/jvi.69.9.5754-5762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan H, Simon A E. Polymerization of nontemplate bases before transcription initiation at the 3′ ends of templates by an RNA-dependent RNA polymerase: an activity involved in 3′ end repair of viral RNAs. Proc Natl Acad Sci USA. 2000;97:12451–12456. doi: 10.1073/pnas.97.23.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irie M. Structure-function relationships of acid ribonucleases: lysosomal, vacuolar, and periplasmic enzymes. Pharmacol Ther. 1999;81:77–89. doi: 10.1016/s0163-7258(98)00035-7. [DOI] [PubMed] [Google Scholar]
- 14.Jaeger J A, Turner D H, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kao C C, Del Vecchio A M, Zhong W. De novo initiation of RNA synthesis by a recombinant Flaviviridae RNA-dependent RNA polymerase. Virology. 1999;253:1–7. doi: 10.1006/viro.1998.9517. [DOI] [PubMed] [Google Scholar]
- 16.Kao C C, Ecker D, Singh P. De novo initiation of viral RNA-dependent RNA synthesis. 2001. Virology, in press. [DOI] [PubMed] [Google Scholar]
- 17.Kao C C, Yang X, Kline A, Wang Q M, Barket D, Heinz B A. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J Virol. 2000;74:11121–11128. doi: 10.1128/jvi.74.23.11121-11128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim M-J, Zhong W, Hong Z, Kao C C. Template nucleotide moieties for de novo initiation of RNA synthesis by a recombinant RNA-dependent RNA polymerase. J Virol. 2000;74:10312–10322. doi: 10.1128/jvi.74.22.10312-10322.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krupp G. RNA synthesis: strategies for the use of bacteriophage RNA polymerases. Gene. 1988;72:75–89. doi: 10.1016/0378-1119(88)90129-1. [DOI] [PubMed] [Google Scholar]
- 20.Kuo L, Fearns R, Collins P L. Analysis of the gene start and gene end signals of human respiratory syncytial virus: quasi-templated initiation at position 1 of the encoded mRNA. J Virol. 1997;71:4944–4953. doi: 10.1128/jvi.71.7.4944-4953.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Lai V C H, Kao C C, Ferrari E, Park J, Uss A S, Wright-Minogue J, Hong Z, Lau J Y N. Mutational analysis of bovine viral diarrhea virus RNA-dependent RNA polymerase. J Virol. 1999;73:10129–10136. doi: 10.1128/jvi.73.12.10129-10136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohmann V, Roos A, Korner F, Koch J O, Bartenschlager R. Biochemical and kinetic analysis of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology. 1998;249:108–118. doi: 10.1006/viro.1998.9311. [DOI] [PubMed] [Google Scholar]
- 24.Lohmann V, Roos A, Korner F, Koch J O, Bartenschlager R. Biochemical and structural analysis of the NS5B RNA-dependent RNA polymerase of the hepatitis C virus. J Viral Hepat. 2000;7:167–174. doi: 10.1046/j.1365-2893.2000.00218.x. [DOI] [PubMed] [Google Scholar]
- 25.Lohmann V, Korner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo G, Hamatake R K, Mathis D M, Racela J, Rigat K L, Lemm J, Colonno R J. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J Virol. 2000;74:851–863. doi: 10.1128/jvi.74.2.851-863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller W A, Dreher T W, Hall T C. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (−)-sense genomic RNA. Nature. 1985;313:68–70. doi: 10.1038/313068a0. [DOI] [PubMed] [Google Scholar]
- 28.Milligan J F, Groebe D R, Witherell G W, Uhlenbeck O C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagy P D, Carpenter C D, Simon A E. A novel 3′-end repair mechanism in an RNA virus. Proc Acad Natl Sci USA. 1997;94:1113–1118. doi: 10.1073/pnas.94.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neufeld K L, Galarza J M, Richards O C, Summers D F, Ehrenfeld E. Identification of terminal adenylyl transferase activity of the poliovirus 3Dpol. J Virol. 1994;68:5811–5818. doi: 10.1128/jvi.68.9.5811-5818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh J-W, Sheu G-T, Lai M M C. Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J Biol Chem. 2000;275:17710–17717. doi: 10.1074/jbc.M908781199. [DOI] [PubMed] [Google Scholar]
- 32.Pevear, D. C., T. J. Nitz, and M. Seipel. Nov. 1998. Methods for preventing and treating pestivirus infection and associated diseases. Patent publication number WO 98/36752.
- 33.Rao A L N, Dreher T W, Marsh L E, Hall T C. Telomeric function of the tRNA-like structure of brome mosaic virus RNA. Proc Natl Acad Sci USA. 1989;86:5335–5339. doi: 10.1073/pnas.86.14.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siegel R W, Adkins S, Kao C C. Sequence-specific recognition of a subgenomic promoter by a viral RNA polymerase. Proc Natl Acad Sci USA. 1997;94:11238–11243. doi: 10.1073/pnas.94.21.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu G, Kaper J M. Requirement of 3′-terminal guanosine in (−)-stranded RNA for in vitro replication of cucumber mosaic virus satellite RNA by viral RNA-dependent RNA polymerase. J Mol Biol. 1994;238:655–657. doi: 10.1006/jmbi.1994.1326. [DOI] [PubMed] [Google Scholar]
- 36.Yamashita T, Kaneko S, Shirota Y, Qin W, Nomura T, Kobayashi K, Murakami S. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J Biol Chem. 1998;273:15479–15486. doi: 10.1074/jbc.273.25.15479. [DOI] [PubMed] [Google Scholar]
- 37.Yanagi M, St Claire M, Shapiro M, Emerson S U, Purcell R H, Bukh J. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology. 1998;244:161–172. doi: 10.1006/viro.1998.9092. [DOI] [PubMed] [Google Scholar]
- 38.Zhong W, Ferrari E, Lesburg C A, Maag D, Ghosh S K, Cameron C E, Lau J Y, Hong Z. Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase. J Virol. 2000;74:9134–9143. doi: 10.1128/jvi.74.19.9134-9143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong W, Gutshall I L, Del Vecchio A M. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of Bovine viral diarrhea virus. J Virol. 1998;72:9365–9369. doi: 10.1128/jvi.72.11.9365-9369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]