Abstract
Enhancins are a group of proteins first identified in granuloviruses (GV) that have the ability to enhance nuclear polyhedrosis virus potency. We had previously identified an enhancin gene (E1) in the Lymantria dispar multinucleocapsid nucleopolyhedrovirus (LdMNPV) (D. S. Bischoff and J. M. Slavicek, J. Virol. 71:8133–8140, 1997). Inactivation of the E1 gene product within the viral genome lowered viral potency by an average of 2.9-fold. A second enhancin gene (E2) was identified when the entire genome of LdMNPV was sequenced (Kuzio et al., Virology 253:17–34, 1999). The E2 protein exhibits approximately 30% amino acid identity to the LdMNPV E1 protein as well as the enhancins from Trichoplusia ni GV, Pseudaletia unipuncta GV, Helicoverpa armigera GV, and Xestia c-nigrum GV. Northern analysis of viral RNA indicated that the E2 gene transcripts are expressed at late times postinfection from a consensus baculovirus late promoter. The effect of the enhancin proteins on viral potency was investigated through bioassay using two recombinant viruses, one with a deletion in the E2 gene (E2del) and a second with deletion mutations in both enhancin genes (E1delE2del). The enhancin gene viral constructs were verified by Southern analysis and shown not to produce enhancin gene transcripts by Northern analysis. The E2del virus exhibited an average decrease in viral potency of 1.8-fold compared to wild-type virus. In the same bioassays, the recombinant virus E1cat, which does not produce an E1 gene transcript, exhibited an average decrease in viral potency of 2.3-fold compared to control virus. The E1delE2del virus exhibited an average decrease in viral potency of 12-fold compared to wild-type virus. Collectively, these results suggest that both LdMNPV enhancin genes contribute to viral potency, that each enhancin protein can partially compensate for the lack of the other protein, and that both enhancin genes are necessary for wild-type viral potency.
Baculoviruses are viruses that are specific to insects and other invertebrates and include two genera, the nucleopolyhedroviruses (NPVs) and the granuloviruses (GVs). Both groups of viruses produce virions that are similar in both structure and pathology of infection, though NPVs have members that can package virions singularly or in groups and GVs can package virions only singularly (8, 10). Another distinction between these viruses is that NPVs produce occlusions that contain many viral particles and GV occlusions contain a single viral particle. Sequence analysis of NPV and GV genomes revealed that the NPVs and GVs contain many homologous genes (15, 16, 24).
Enhancins are proteins first found in GV occlusion bodies that have the ability to enhance the infection of some NPVs. Also referred to as virus-enhancing or synergistic factors, enhancins were first identified and isolated by Tanada (34, 37; see reference 35 for a review). The enhancin protein of Pseudaletia unipuncta GV (PuGV) was found to enhance the infection of P. unipuncta (PuNPV) only when both the PuGV protein and PuNPV were inoculated orally, indicating that the midgut is the site of the enhancin activity (38). The enhancin protein purified from the Trichoplusia ni GV (TnGV) enhanced Autographa californica multinucleocapsid NPV (AcMNPV) infection in Helicoverpa zea, Spodoptera exigua, P. unipuncta, and T. ni by 2- to 14-fold, depending on the host species (41). At present, enhancin genes have been sequenced in four GVs, Helicoverpa armigera GV (HaGV) (30), PuGV (30), TnGV (14), and Xestia c-nigrum GV (XcGV) (16). The first GV genome to be completely sequenced, XcGV, was found to have four different enhancin genes (16). In contrast, the Plutella xylostella GV genome lacks an enhancin gene (15).
Two functions have been proposed for enhancins, enhancement of virus-host cell fusion and disruption of the peritrophic membrane. The PuGV enhancin was thought to facilitate the uptake of virus particles by increasing the number of virus-membrane fusion events (23, 35, 36, 39). A specific binding site of the TnGV enhancin has been found in the midgut brush border membrane of P. unipuncta but not in the brush border membranes of T. ni, H. zea, or S. exigua, all of which demonstrate a response to the TnGV enhancin when infected with AcMNPV (41). The TnGV enhancin was found to damage the peritrophic membrane lining the larval midgut, exposing the gut wall to viral infection (7). Bioassay results of T. ni larvae infected with AcMNPV and various concentrations of the TnGV enhancin demonstrated that the major effect of enhancin appears to be an increase in infection efficiency caused by the disruption of the insect peritrophic membrane (11). This enhancin was later found to be a metalloproteinase, which degrades mucin, a major protein constituent of the peritrophic membrane (25, 40).
The Lymantria dispar MNPV (LdMNPV) is a baculovirus pathogenic to L. dispar, the gypsy moth, a forest and urban tree-defoliating pest in the northeastern and some midwestern states. The genome of LdMNPV is significantly larger than that of most other baculoviruses. The genome of the sequenced strain of LdMNPV is 161,046 bases (24), in contrast to 133,894 bases for AcMNPV (2), 128,413 for Bombyx mori NPV (12), 131,403 bases for H. armigera single nucleocapsid NPV (6), 131,990 bases for Orgyia pseudotsugata MNPV (1), and 135,611 bases for S. exigua MNPV (19). LdMNPV contains several unique open reading frames (ORFs) as well as ORFs with homology to GV ORFs. The first GV homolog found in LdMNPV was the enhancin 1 (E1) gene, which was also the first enhancin gene found in NPVs (5). The LdMNPV E1 gene product exhibits 29% amino acid identity to the sequenced enhancin genes of TnGV, PuGV, and HaGV and contains a conserved zinc-binding domain characteristic of metalloproteases. E1 gene transcripts are expressed at late times postinfection from a consensus baculovirus late promoter.
A viral isolate lacking a functional E1 gene was constructed to investigate enhancin function. Potency analysis revealed that viral strain without the enhancin 1 gene was approximately 2- to 3-fold less potent than wild-type viruses, suggesting that the LdMNPV enhancin affects viral potency (5). A second enhancin gene (E2) was recently identified in LdMNPV when the entire genome of isolate 5-6 was sequenced (24). Five other NPVs have been completely sequenced, but no enhancins have been found (AcMNPV [2], B. mori NPV [12], H. armigera NPV [6], O. pseudotsugata MNPV [1 ], and S. exigua MNPV [19]). In this study, we characterized the E2 gene and determined the effect of the enhancin genes both individually and collectively on viral potency.
MATERIALS AND METHODS
Cells, virus, and insects.
L. dispar 652Y cells were grown as monolayers at 27°C in Goodwin's IPL-52B medium (JRH Scientific) supplemented with 6.25 mM glutamine (Gibco-BRL) and 10% fetal bovine serum (Atlanta Biologicals). The LdMNPV isolate A21-MPV (31) served as the parental line for all subsequent viruses. A recombinant virus (E1cat), generated in a previous study (5), that contains the chloramphenicol acetyltransferase (cat) gene was also used in this study. L. dispar egg masses were obtained from the U.S. Department of Agriculture's Animal and Plant Health Inspection Service rearing facility at Otis Air Force Base (Otis, Mass.). Larvae were reared on gypsy moth diet (ICN) (3).
Northern blot analysis and primer extension mapping of transcripts.
Infected Ld652Y cells were harvested at various times postinfection (P.I.). Cytoplasmic RNA was isolated as described by Friesen and Miller (9). Poly(A) RNA was isolated from the cytoplasmic RNA by the PolyATtract mRNA isolation system (Promega), separated on a 1.2% agarose–formaldehyde gel, and then transferred to NitroPlus nitrocellulose transfer membrane (MSI). Northern blots were performed as described by Mahmoudi and Lin (26). Thirty-base oligonucleotides (complementary to bp 61158 to 61187 for the E1 gene or bp 156708 to 156737 for the E2 gene [24]) were end labeled with [γ-32P]ATP (NEN, Boston, Mass.) and used as strand-specific probes to detect the transcript of interest. Blots were imaged with the Storm 860 phosphorimager (Molecular Dynamics).
Primer extension reactions were performed using the primer extension system with avian myeloblastosis virus reverse transcriptase (Promega). Cytoplasmic RNA was isolated at 72 h postinfection from cells infected with A21-MPV or E2del. A 25-base oligonucleotide (complementary to positions 155841 to 155865 [24]) was used in the reactions after being end labeled with [γ-32P]ATP (NEN). Primer extension products were fractionated on 6% polyacrylamide–8 M urea gels, dried, and visualized by phosphorimager.
In vitro transcription and translation of E2 gene.
Plasmid pDB184 (see below) was digested with SstI, and a 2.8-kbp fragment containing the E2 gene was subcloned into the SstI site of pBluescript to generate pE2SstI. The E2 protein was expressed from pE2SstI by the TNT quick coupled transcription-translation system (Promega). The expressed protein was labeled with 35S-EasyTag express protein labeling mix (NEN). Reaction products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
Computer programs used for sequence comparison.
The amino acids of known enhancins available in GenBank were aligned using the Clustal W program (http://www2.ebi.ac.uk/clustalw/). Highlighted comparisons of proteins were produced using the Boxshade program (available at http://www.ch.embnet.org/software/BOX_form.html). An unrooted phylogenetic tree of known enhancin proteins was constructed using the PAUP 3.1 program (33). Bootstrap analysis with 100 replicates was performed to assess the integrity of the phylogeny generated.
Construction and verification of recombinant viruses.
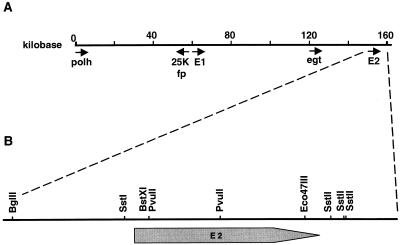
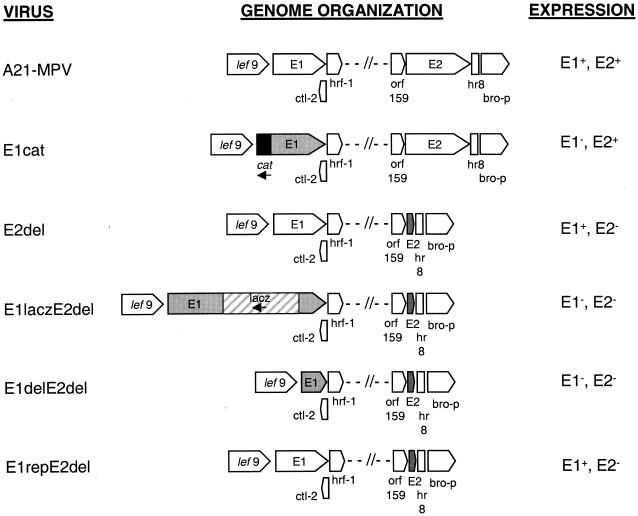
An E2 gene transplacement vector was constructed to generate an A21-MPV recombinant strain lacking the E2 gene. A vector containing the E2 gene was constructed by restricting A21-MPV cosmid 15 (4) with BglII and then religating the fragments. Because there is a single BglII restriction site within the SuperCos 1 vector (Stratagene), a plasmid was generated, pDB184, containing ca. 6.0 kbp of A21-MPV viral DNA (ca. bp 154.2 to 160, Fig. 1B). A second plasmid with a lacZ gene insertion in the E2 gene was made as follows. Plasmid pDB184 was restricted with BstXI. A 3.8-kbp XbaI-BamHI fragment containing the HSP70lacZ gene marker was isolated from plasmid p210.1 (provided by American Cyanamid, Princeton, N.J.). The ends of both of the fragments were end filled with T4 DNA polymerase, and the fragments were ligated together to form pDB186. To create a plasmid with a deletion in the E2 gene, plasmid pDB184 was then restricted with PvuII and BglII and with EcoR47III and EcoRI. The 1.7-kbp PvuII-BglII and 2.2-kbp EcoR47III-EcoRI fragments were isolated. These fragments were subcloned into the pBluescript vector (Stratagene) BamHI and EcoRI sites. The resulting plasmid, pDB185, has a 3.9-kbp insert missing 2.1 kbp (701 amino acids) within the E2 gene.
FIG. 1.
Genomic location of the two LdMNPV enhancin genes.E1 is located at bp 60268 to 62619, and E2 is located from bp 155812 to 158178 based on the sequence generated by Kuzio et al. (24). Several other LdMNPV genes are also indicated (A). The enlarged map of plasmid pDB184 indicates the Ca. 6 kb surrounding and encoding the E2 gene (kbp 155.8 to 158.2). The location of the E2 ORF and restriction sites used in subcloning of the gene are shown (B).
A recombinant virus, E2del, with a deletion in the E2 gene was created in a series of steps as follows. Viral DNA from A21-MPV (5) was cotransfected with plasmid pDB186 to make a virus with a lacZ gene insertion in the E2 gene, vE2lacz. Recombinant viruses were selected based on the presence of blue, occlusion-positive plaque phenotypes. E2lacz viral DNA was then cotransfected with plasmid pDB185. Resulting recombinant plaques had a white, occlusion-positive phenotype. Viral DNA was isolated by the method of O'Reilly et al. (28).
Using the E2del virus as a starting point, a virus, E1delE2del, was generated that lacked functional E1 and E2 genes. pDB126 (5) was cut with PstI, and a resulting 1.5-kbp band was isolated containing the C terminus of the E1 gene (ca. kb 59584 to 62609). This fragment was subcloned into the pBluescript vector PstI site to generate plasmid pDB140. pDB140 was restricted with NarI and end filled with T4 DNA polymerase. A 3.8-kbp XbaI-BamHI fragment containing the lacZ gene was restricted from plasmid p210.1, the ends were end filled, and the fragment was ligated to the pDB140 fragment to form pE1lacz. To create a plasmid with a deletion in the E1 gene, plasmid pDB126 (5) was restricted with NarI and BamHI and separately with HincII. The 5.3-kbp NarI-BamHI and 0.7-kbp HincII fragments were isolated, and the 5.3-kbp fragment was end filled. The fragments were then ligated together and the resulting plasmid, pE1del, had a 3.0-kbp insert missing 1.5 kbp (488 amino acids) and a frameshift within the E1 gene.
Virus E1laczE2del was generated by cotransfection of E2del viral DNA and pE1lacz plasmid DNA, and blue, occlusion-positive plaques were selected and plaque purified. DNA from the E1laczE2del recombinant virus was then cotransfected with pE1del plasmid DNA to generate the E1delE2del virus. Two different recombinant viruses of E1delE2del (AE2 and BF6) generated from different transfections and one isolate of E1laczE2del (5AA) were used for further characterization.
A recombinant virus in which the E1 gene was replaced in an enhancin gene double deletion virus was constructed by cotransfection of E1laczE2del viral DNA and plasmid pDB126 DNA. White occlusion-positive plaques were selected and plaque purified.
All viruses were verified by restriction enzyme analysis, and those with insertions or deletions in either enhancin gene were further verified by Southern blot analysis. Viral DNA restriction enzyme digests were electrophoresed on a 0.7% agarose–Tris–borate–EDTA gel, blotted on Nytran membrane (MSI), and probed with DNA fragments labeled with the nick translation kit (Bethesda Research Laboratories) and [α-32P]dCTP (NEN). For viruses with a deletion or insertion in the E1 gene, the blots were probed using a 735-bp BstXI fragment (bp 60981 to 61761 [24]) that was restricted and then gene cleaned (Q-Bio) from pDB126 (5). Blots containing DNA from recombinant E2 gene deletion viruses were probed with the 2.1-kbp BstXI-EcoR47III fragment deleted from these viruses.
Bioassay analysis of viruses.
Fourth-instar L. dispar larvae were infected by injection of 5 μl of budded virus for each test virus (titer of about 5 × 106 tissue culture-infective doses per ml). Larvae were placed on fresh food and allowed to feed until death. Polyhedra were then isolated from insects. The concentration of occluded virus required to kill 50% of the test larvae (LC50) and the mean time to kill 50% of the test larvae (LT50) for all viruses were determined by the droplet feeding method developed by Hughes et al. (18) as described by Slavicek et al. (32) on first-day, second-instar L. dispar larvae. Larvae were maintained at 27 ± 1°C at a photoperiod of 14 h light–10 h dark during the bioassay. LC50 values were calculated using Polo-PC (29), and LT50 values were determined by the ViStat 2.1 program (17). Statistical comparisons of bioassay data were performed using the StatView program from Abacus Concepts.
RESULTS AND DISCUSSION
Characterization of E2 gene.
The two enhancin genes in LdMNPV are located at kbp 60.3 to 62.6 (E1) and kbp 155.8 to 158.2 (E2) in the genome (Fig. 1A) (24). Analysis of the sequence upstream of the E2 gene revealed a potential baculovirus late promoter sequence, TTAAG, beginning 28 bp upstream of the enhancin gene start codon. Transcription of the E1 gene in LdMNPV (5) and of the HaGV enhancin (30) begins within consensus baculovirus late promoters (TTAAG and TTAAG, respectively, at the underlined nucleotides). All of the other enhancin genes identified to date in TnGV, PuGV, and XcGV also contain baculovirus late promoters. To determine the transcriptional start site of the E2 gene transcript, primer extension reactions were performed with a 25-base oligonucleotide. Transcription initiated at the first T residue within the consensus baculovirus late promoter sequence TTAAG (data not shown). No polyadenylation signal was identified immediately downstream of the E2 gene. Two consensus polyadenylation sites (AATAAA) are located 280 and 294 bp downstream of the stop codon in the E2 gene. These sites lie between the end of homologous repeat 8 and the bro-p gene. Recent studies on the mechanism of 3′-end formation of baculovirus late transcripts, using an in vitro assay, indicate that 3′ ends are formed by termination after transcription of a T-rich region, as opposed to a cleavage-polyadenylation mechanism using a consensus polyadenylation site (21). An earlier study of polyadenylation sites of late transcripts indicated that the sites occurred after a T-rich region a variable distance downstream of consensus polyadenylation sites (42), which may suggest that the consensus site has a role in the formation of late-transcript 3′ ends.
Identification and temporal analysis of E2 gene transcripts.
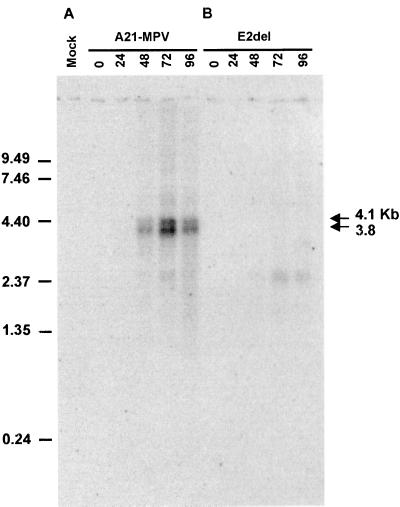
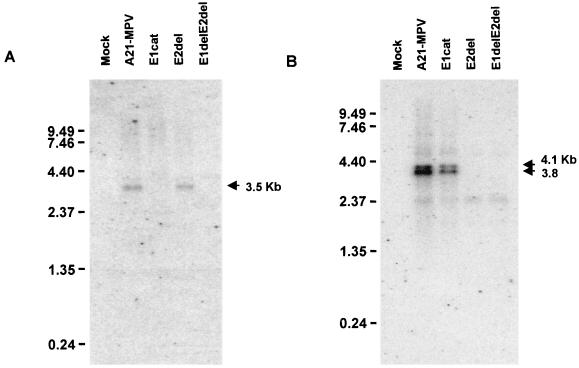
The temporal expression of the E2 gene transcript was investigated in Ld652Y cells infected with isolate A21-MPV. A primary transcript of approximately 3.8 kb and a secondary transcript of approximately 4.1 kb were found using a strand-specific oligonucleotide probe at late times postinfection 48, 72, and 96 h p.i. (Fig. 2A). No transcripts were evident at the end of the 1-h infection period or at 24 h p.i. The 4.1-kb transcript is more clearly seen in Fig. 8. The lengths of E2 gene transcripts are longer than expected if the most proximal polyadenylation sites are used. With these sites, transcripts of approximately 2.9 kb would be generated. Further downstream, past the bro-p gene, are two consensus polyadenylation sites, and downstream from these sites are several T-rich areas. An E2 gene transcript terminating near these sites would be approximately 3.9 kb in length, which is the approximate transcript length observed.
FIG. 2.
Temporal analysis of the enhancin 2 gene transcripts. Cells were infected with A21-MPV or E2del, and cytoplasmic RNA was isolated at the indicated hours postinfection. Poly(A) RNA was isolated from the cytoplasmic RNA, and 1 μg was separated by formaldehyde-agarose gel electrophoresis, blotted, and probed with a strand-specific oligonucleotide complementary to positions 156708 to 156737 in the E2 ORF. RNA from uninfected cells was used as a mock infection sample. RNA size standards (in kilobases) are indicated on the left.
The E1 gene also generated a transcript significantly longer than expected based on the size of the gene (5). The E1 gene ORF is 2.3 kbp in length, whereas a 3.5-kb transcript is generated. It was speculated that the E1 gene transcript proceeds through the hrf-1 gene, which is immediately downstream, and terminates downstream of polyadenylation sites close to the hrf-1 gene (5). Polyadenylation signal sequences are also absent between the enhancin gene and the immediate downstream gene in TnGV, PuGV, and HaGV (14, 30). In addition, analysis of HaGV enhancin RNAs indicated that the HaGV enhancin gene and the downstream ORF1 gene may be part of the same bicistronic message (30), and therefore similar to LdMNPV E1 and E2 gene transcription. A faint signal for a transcript of approximately 2.4 kb was also found at 72 and 96 h p.i. (Fig. 2A). However, this transcript was also found in cells infected with the E2del virus (Fig. 2B) and not in the mock-infected cells, which suggests that the probe is detecting a viral transcript originating from a different gene.
Characteristics of E2 protein and comparison to other enhancins.
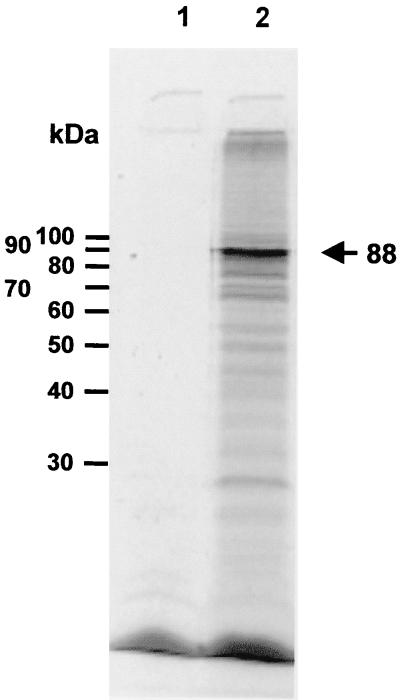
The E2 gene could encode a 788-amino-acid protein with a predicted molecular mass of 88,408 Da. A rabbit reticulocyte-coupled transcription-translation system was used to express E2 to determine if the gene did encode an expressed protein of the predicted size. Plasmid E2SstI, which contained the E2 gene under the control of the T7 promoter, was used to express the E2 gene, and Bluescript Sk+ was used as a negative control. A number of bands were produced by pE2SstI, though a major band was seen at 88 kDa, which corresponds to the expected size of the E2 protein (Fig. 3, lane 2). The E2 protein is similar in size to the LdMNPV E1 protein, which contains 783 amino acids. Both LdMNPV enhancin proteins are considerably smaller than the known GV enhancins, most of which range in size from 856 to 902 amino acids. The E1 protein in XcGV is of intermediate size, with a length of 824 amino acids.
FIG. 3.
Analysis of expression of the enhancin 2 gene. The enhancin 2 gene was expressed in a rabbit reticulocyte transcription-translation system, and the proteins were separated by SDS-PAGE. Lane 1, Bluescript SK+; lane 2, pE2Sstl, expressing E2. Positions of molecular mass standards are indicated to the left, and the position of the enhancin protein is indicated to the right.
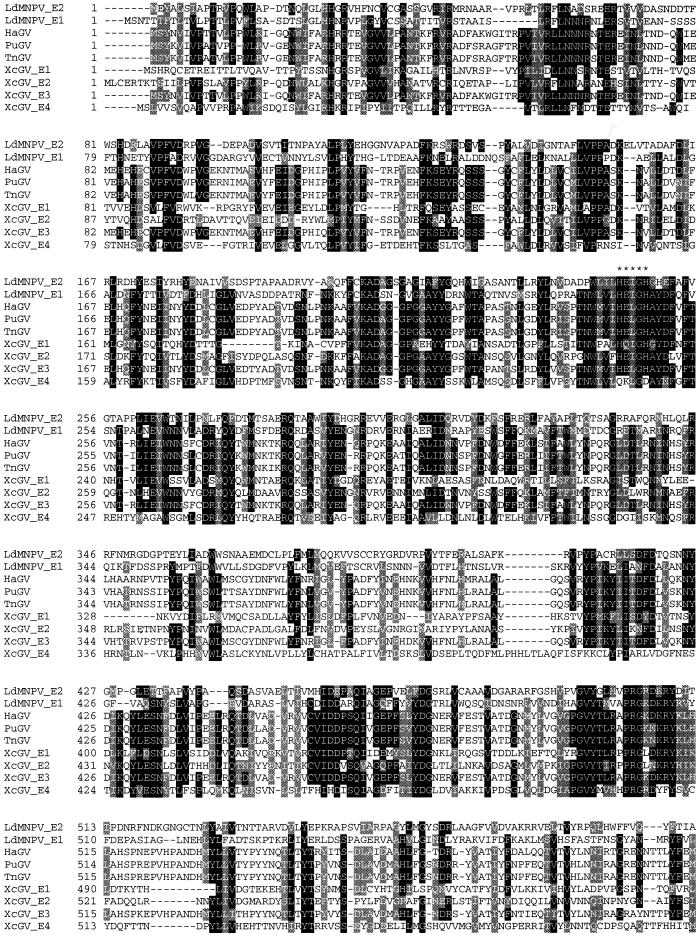
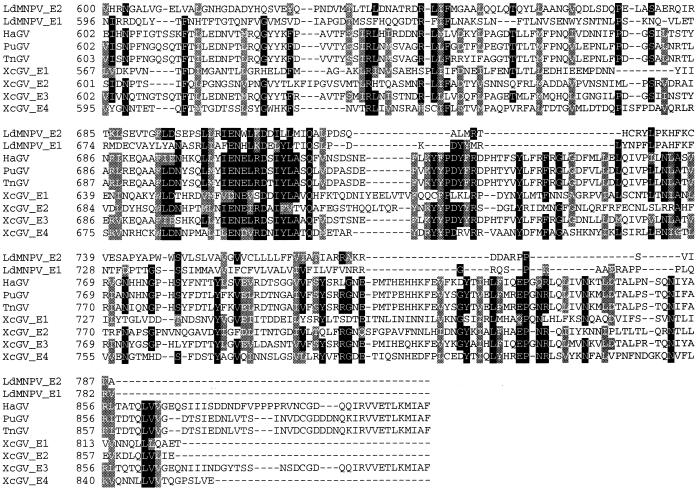
Alignment of the known GV and LdMNPV enhancins revealed that the proteins exhibit the greatest homologies in the area bracketed by amino acid positions 200 and 300 (Fig. 4). The region from approximately amino acid positions 440 to 510 is another area of high homology among the enhancins. The LdMNPV E2 enhancin exhibits amino acid identities of about 28, 28, 26, 24, 27, 26, and 26% with the XcGV 1 to 4, HaGV, PuGV, and TnGV enhancins, respectively. There is an overall amino acid identity of about 6% among all the enhancin genes compared in Fig. 4. The most heterogeneous region of the enhancins is at the C-terminal end. The LdMNPV E2 protein exhibits an amino acid identity of only approximately 7% with the GV enhancins in the region from amino acid positions 704 to 788. The basis for the difference in the length of the LdMNPV enhancins compared to the GV proteins is within the C-terminal end. The LdMNPV enhancins lack two blocks of amino acids present in the GV proteins within the amino acid regions from approximately 724 to 758 and 807 to 902, relative to the HaGV enhancin. The LdMNPV E1 and E2 enhancins exhibit an amino acid identity of about 30%. The proteins are most conserved within the region from amino acids 1 to 509 (36% amino acid identity), and least conserved in the region from 510 to 788 (21%). The three regions from 203 to 269 (48.5%), 444 to 509 (41.5%), and 700 to 778 (41%) of the LdMNPV enhancins exhibit higher levels of amino acid identity than the other areas of the proteins.
FIG. 4.
Aligned amino acid sequences of known baculovirus enhancin genes. Black and gray areas indicate positions with 70% or greater sequence identity or similarity, respectively, among all enhancins. The enhancin sequences aligned include those of LdMNPV, HaGV, PuGV, TnGV, and the four from XcGV. Asterisks indicate the area containing the HEXXH conserved zinc-binding domain of metalloproteases.
LdMNPV enhancin 2 contains a signature pattern characteristic of a zinc-binding domain found within metalloproteases (22, 27). All of the enhancins sequenced to date except enhancin 4 of XcGV (16) have the characteristic HEXXH zinc-binding site of metalloproteinases. In metalloproteases, the zinc ion is chelated by the two histidine residues in the HEXXH site and by a third residue, typically a histidine, cysteine, or aspartic or glutamic acid residue, located anywhere from 20 to 120 amino acids downstream of the binding site (see references 13 and 20 for reviews). A glutamic acid residue in position 299 (51 amino acids downstream of the binding site) of the LdMNPV E2 gene is conserved among all enhancin proteins (Fig. 4). In addition, most enhancins contain conserved aspartic or glutamic residues at the LdMNPV enhancin 2 positions 291, 307, and 313, which are 43, 59, and 65 amino acids downstream of the binding site, respectively. It is interesting that the XcGV E4 lacks the conserved HEXXH zinc-binding site of metalloproteinases present in the other enhancins (HEI/LGH), and of all the enhancins, this protein exhibits the least homology to LdMNPV E2. The amino acid residues before and after the binding site are well conserved in comparison to the other enhancins, while the binding site (QKLGD) contains only the conserved isoleucine/leucine and glycine variable residues found in the baculovirus enhancins. This may suggest that the XcGV E4 is not functional and hence would no longer be under evolutionary pressure to conserve amino acids within the protein.
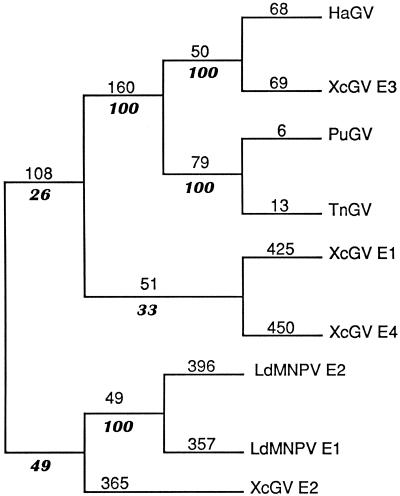
To address the relationships of the LdMNPV and GV enhancins, a phylogenetic analysis was performed with the enhancin protein sequences shown in Fig. 4. An unrooted parsimonious tree was calculated with the PAUP heuristic search algorithm, and bootstrap analysis was performed to assess variability of the phylogeny (Fig. 5). XcGV E1, E3, and E4 and HaGV, TnGV, and PuGV enhancins formed one clade (clade 1), and the LdMNPV E1 and E2 and XcGV E2 enhancins formed another clade (clade 2). Clade 1 contained two subgroups, one consisting of HaGV, XcGV E3, PuGV, and TnGV, and the other of XcGV E1 and XcGV E4. The first subgroup of clade 1 formed two further subgroups of HaGV and XcGV E3 and of PuGV and TnGV. The first subgroup of clade 1 and the two subgroups contained within are well supported by bootstrap analysis. A subgroup consisting of LdMNPV E1 and LdMNPV E2 formed within clade 2, which was well supported by bootstrap analysis. In contrast, the positions of XcGV E1, E2, and E4 within the phylogeny are not well supported by bootstrap analysis. This finding could suggest either that these enhancins originated from a distant ancestor or that each has a different ancestor. It is interesting that while the LdMNPV enhancins exhibit branch lengths comparable to those of XcGV E1, E2, and E4, the subgroup that they form is well supported by bootstrap analysis. The phylogenetic analysis indicates that the HaGV, PuGV, TnGV, and XcGV E3 enhancins originated from a common ancestor and the LdMNPV enhancins came from a common ancestor. To delineate the phylogenetic relationship of the LdMNPV and GV enhancins as well as the relationship of the XcGV E1, E2, and E4 enhancins within the GVs will require identification of and analysis with additional enhancins from NPVs and GVs.
FIG. 5.
Phylogenetic tree of known baculovirus enhancins. The most parsimonious tree for known baculovirus enhancins was constructed with the heuristic search algorithm of PAUP. Numbers above the lines in roman type indicate phylogenetic distance, and italic numbers below the lines indicate the frequency of a given cluster after bootstrap analysis (100 replicates).
Construction and verification of recombinant viruses.
To address the role of the LdMNPV E2 in viral potency, a recombinant virus was constructed that lacked the E2 gene. In addition, to determine if one enhancin gene could compensate for the lack of the other, a virus lacking both enhancin genes was generated. The E2 gene (2,364 nucleotides in length) was subcloned from the genome into a plasmid, pDB184 (Fig. 1B). A lacZ gene was inserted into the BstXI site of pDB184, at nucleotide 66 within the E2 gene ORF, and used to construct the intermediate virus, E2lacz. This virus was then recombined with a plasmid containing a deletion of E2 gene nucleotides 66 to 2168 to produce the virus E2del (Fig. 6). A lacZ gene was inserted into the NarI sites of the E1 gene (2,349 nucleotides in length) at nucleotides 1309 and 1492, and the resulting plasmid, pE1lacz, was recombined with the E2del virus to generate the virus E1laczE2del. This virus was recombined with a plasmid containing a deletion of E1 gene nucleotides 26 to 1492 to produce the virus E1delE2del. To confirm the effect of deletion of both enhancin genes, a virus was constructed in which the E1 gene was replaced. Plasmid pDB126, which contains the E1 gene, was recombined with E1laczE2del5AA to give E1repE2del (Fig. 6). The identity of the recombinant viruses was verified by restriction enzyme and Southern blot analysis (data not shown).
FIG. 6.
Schematic representation of A21-MPV and enhancin recombinant viruses. Arrows show the location and direction of ORFs in the regions of the E1 and E2 genes (24). The shaded ORFs are those where the enhancin gene has been either deleted or disrupted. The cat gene is indicated by a black box, and the lacZ gene is indicated by a striped box. Expression of the enhancin gene products is indicated for each virus on the right.
Northern analysis was used to verify that the enhancin transcripts were not being produced in the double deletion recombinant viruses as well as the single enhancin gene deletion viruses (Fig. 7). Poly(A) RNA isolated at 72 h p.i. from mock, A21-MPV, E1cat, E2del, and E1delE2del AE2 viral infections was probed with strand-specific probes for E1 gene (Fig. 7A) or E2 gene (Fig. 7B) transcripts. Oligonucleotide sequences that contained little homology were selected in order to differentiate between the enhancin gene transcripts. The 3.5-kb E1 gene transcript is missing in lanes containing RNA isolated from cells infected with E1cat and E1delE2del AE2 viruses, and the 3.8- and 4.1-kb E2 gene transcripts are missing in lanes containing RNA isolated from cells infected with E2del and E1delE2del AE2 viruses, as expected, confirming that the E1 and E2 genes have been removed.
FIG. 7.
Northern blot analysis of enhancin gene transcripts from cells infected with recombinant viruses. Cells were infected with A21-MPV, E1cat, E2del, and E1delE2del AE2 viruses. Poly(A) RNA was isolated from the cytoplasmic RNA, and 1μg was separated by formaldehyde-agarose gel electrophoresis, blotted, and probed with a strand-specific oligonucleotide for the enhancin 1 gene (A) or enhancin 2 gene (B). RNA from uninfected cells was used as a control (lane mock). Positions of RNA size standards (in kilobases) are indicated on the left.
Bioassay analysis.
The biological activity of the recombinant enhancin viruses was determined through bioassay of L. dispar larvae. A previously constructed recombinant virus (5) containing a cat gene insertion in the E1 gene was also used in the bioassays. The potency of the E2del virus was found to be consistently less than that of the control virus in three separate bioassays (Table 1). The decrease in potency ranged from 1.2-to 2.9-fold. In agreement with our previous findings, the E1cat recombinant virus exhibited a drop in potency compared to the control virus (Table 1). In this study, the decrease ranged from 1.6- to 3.8-fold, which is in good agreement with the range of 1.4-to 4.0-fold reported in the previous study (5). These results indicate that enhancin 2 had an effect on viral potency and that both enhancin genes encode functional proteins that increase viral potency. Analysis of the time-mortality response showed similar killing speeds for the E2del, E1cat, and A21-MPV viruses (Table 1). Taken together, these results indicate that the enhancins are functioning at the level of viral entry into the insect host and do not affect systemic transmission. E2del, E1cat, and A21-MPV exhibited essentially identical rates of budded virus formation (data not shown), which indicates that the enhancin genes are not involved in viral replication. In addition, this finding indicates that a random mutation in a gene involved in DNA replication did not occur in the recombinant viruses. In contrast to the present findings, a decrease in the survival time of 50% of larvae (ST50) was noted in bioassays of mid-fourth-instar T. ni larvae infected with AcMNPV in combination with the TnGV enhancin protein (11). Larvae that showed the same mortality with virus and with virus and enhancin had a lower ST50 in the larvae infected with virus and enhancin. Perhaps a dual activity is possible for some enhancin proteins, or the impact on ST50 was a function of the experimental conditions employed.
TABLE 1.
Dose-response of second-instar L. dispar larvae infected per os by droplet feeding with A21-MPV, E1cat, and E2 del
| Assay no. | Virus | LC50
|
LT50
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PIBa/ml | 95% fiducial limits
|
Slope | Het.b | Difference vs. A21-MPV(fold) | LT50 (day) ± SE | Slope ± SE | % Mortality | |||
| Lower | Upper | |||||||||
| 1 | A21-MPV | 4.8 × 104 | 1.8 × 103 | 4.2 × 105 | 0.5 ± 0.1 | 2.4 | 7.4 ± 0.3 | 10.5 ± 2.7 | 50.0 | |
| E1cat | 7.8 × 104 | 2.4 × 103 | 7.8 × 105 | 0.5 ± 0.1 | 3.1 | 1.6 | 7.8 ± 1.1 | 13.7 ± 3.9 | 48.2 | |
| E2del | 1.4 × 105 | 8.3 × 103 | 1.3 × 106 | 0.6 ± 0.1 | 3.2 | 2.9 | 7.3 ± 0.2 | 26.9 ± 8.1 | 42.3 | |
| 2 | A21-MPV | 3.4 × 105 | 5.2 × 104 | 1.2 × 106 | 0.9 ± 0.1 | 1.1 | 8.1 ± 0.3 | 13.3 ± 4.5 | 64.7 | |
| E1cat | 5.3 × 105 | 1.1 × 105 | 1.6 × 106 | 0.7 ± 0.1 | 0.4 | 1.6 | 8.4 ± 1.1 | 11.6 ± 3.2 | 60.0 | |
| E2del | 4.0 × 105 | 1.2 × 105 | 1.0 × 106 | 0.7 ± 0.1 | 0.5 | 1.2 | 8.1 ± 0.2 | 12.6 ± 3.7 | 78.8 | |
| 3 | A21-MPV | 2.4 × 105 | 4.9 × 104 | 8.5 × 105 | 0.9 ± 0.1 | 1.7 | 7.9 ± 0.3 | 8.5 ± 1.6 | 74.3 | |
| E1cat | 9.1 × 105 | 3.8 × 105 | 1.9 × 106 | 0.8 ± 0.1 | 0.1 | 3.8 | 7.6 ± 0.1 | 25.2 ± 5.4 | 68.9 | |
| E2del | 3.1 × 105 | 1.3 × 105 | 6.2 × 105 | 0.9 ± 0.1 | 0.8 | 1.2 | 8.1 ± 0.2 | 18.9 ± 3.6 | 76.5 | |
PIB, polyhedron inclusion body.
Het., heterogeneity.
To determine whether each LdMNPV enhancin protein contributes to viral potency in an additive manner, the contribution of both enhancin genes to viral potency was determined through bioassay of three separately generated viruses with the E2 gene deleted and the E1 gene deleted or inactivated by insertion of the lacZ gene. In two bioassays, all of the E1/E2 gene recombinant viruses exhibited decreased potency compared to isolate A21-MPV (Table 2). The decrease in potency ranged from 3.6- to 24.7-fold and averaged 12.7-fold. The drop in potency in these bioassays was analyzed by the unpaired t test (P < 0.0003) and by analysis of variance (P < 0.0007) and found to be significant. As expected, no significant difference in the LT50s was seen for any of the enhancin gene deletion viruses in comparison with A21-MPV (Table 3). This finding corroborates the LT50 results of the single enhancin gene deletion viruses and supports the conclusion that no inadvertent mutations occurred in genes involved in viral replication during the construction of the recombinant viruses. In addition, comparison of the kinetics of budded-virus synthesis of the enhancin gene recombinant viruses with wild-type virus revealed no differences (data not shown). Furthermore, since the same result was obtained with three separately generated viruses with the E2 gene deleted and the E1 gene deleted or inactivated, it is unlikely that a random mutation affecting viral potency would have occurred in all three constructs.
TABLE 2.
Dose-mortality response of second-instar L. dispar larvae infected per os by droplet feeding with A21-MPV and enhancin deletion virusesa
| Bioassay | Virus | LC 50 PIB/ml | 95% fiducial limits
|
Slope | Het. | Difference vs. A21-MPV (fold) | |
|---|---|---|---|---|---|---|---|
| Lower | Upper | ||||||
| 1 | A21-MPV | 1.1 × 106 | 5.5 × 105 | 2.0 × 106 | 1.2 ± 0.2 | 1.00 | |
| E1delE2del AE2 | 1.2 × 107 | 6.1 × 106 | 2.6 × 107 | 1.0 ± 0.2 | 0.39 | 10.9 | |
| E1delE2del BF6 | 4.0 × 106 | 1.8 × 106 | 9.8 × 106 | 0.7 ± 0.1 | 0.73 | 3.6 | |
| E1laczE2del 5AA | 1.3 × 107 | 2.7 × 106 | 8.9 × 107 | 0.9 ± 0.2 | 1.24 | 11.8 | |
| 2 | A21-MPV | 7.3 × 104 | 2.3 × 104 | 1.7 × 105 | 1.0 ± 0.2 | 0.49 | |
| E1delE2del AE2 | 1.0 × 106 | 1.6 × 105 | 3.4 × 106 | 0.6 ± 0.1 | 0.55 | 13.7 | |
| E1delE2del BF6 | 8.3 × 105 | 2.4 × 105 | 2.1 × 106 | 0.8 ± 0.1 | 0.25 | 11.4 | |
| E1laczE2del 5AA | 1.8 × 106 | 2.2 × 105 | 7.7 × 106 | 0.9 ± 0.1 | 1.24 | 24.7 | |
See Table 1, footnotes a and b.
TABLE 3.
Time-mortality response of second-instar L. dispar larvae infected per os by droplet feeding with A21-MPV and enhancin deletion viruses
| Bioassay | Virus | Mortality of ∼50%
|
Mortality of ∼80%
|
||||
|---|---|---|---|---|---|---|---|
| LT50 (h) ± SE | Slope ± SE | % Mortality | LT50 (h) ± SE | Slope ± SE | % Mortality | ||
| 1 | A21-MPV | 9.0 ± 0.4 | 10.1 ± 2.4 | 46.9 | 8.2 ± 0.2 | 16.1 ± 3.1 | 93.6 |
| E1delE2del BF6 | 8.7 ± 0.4 | 10.6 ± 1.9 | 40.0 | 8.9 ± 0.4 | 8.0 ± 1.6 | 78.6 | |
| E1delE2delAE2 | 9.4 ± 0.5 | 8.9 ± 1.9 | 48.5 | 8.2 ± 0.3 | 11.9 ± 2.6 | 80.0 | |
| E1laczE2del 5AA | 8.2 ± 0.3 | 13.1 ± 3.6 | 51.5 | 8.6 ± 0.3 | 11.1 ± 2.2 | 75.8 | |
| 2 | A21-MPV | 7.3 ± 0.2 | 19.9 ± 4.6 | 60.0 | 7.7 ± 0.1 | 19.7 ± 3.8 | 81.3 |
| E1delE2del AE2 | 7.7 ± 0.2 | 16.6 ± 4.1 | 53.3 | 7.5 ± 0.2 | 14.7 ± 3.2 | 79.2 | |
| E1delE2del BF6 | 7.8 ± 0.2 | 19.4 ± 4.7 | 56.7 | 7.3 ± 0.1 | 24.0 ± 5.3 | 80.8 | |
| E1laczE2del 5AA | 7.8 ± 0.1 | 25.0 ± 6.3 | 47.1 | 7.5 ± 0.1 | 18.1 ± 3.3 | 82.4 | |
To confirm that the large decrease in viral potency was due to deletion of both enhancin genes, a virus was constructed in which the E1 gene was replaced in an E1laczE2del recombinant (E1repE2del) and bioassayed. E1repE2del exhibited greater potency than E1laczE2del and essentially the same potency as E2del (Table 4). In this bioassay, E2del and E1delE2del AE2 exhibited decreases in potency of 1.4-and 10-fold, respectively, which is in good agreement with earlier bioassays (Tables 1 and 2). The finding that the replacement of the E1 gene restores viral potency to the level of E2del indicates that the drop in potency in E1delE2del was due to the deletion of both enhancin genes, as opposed to a mutation in another gene that affects viral potency.
TABLE 4.
Dose-mortality response of second-instar L. dispar larvae infected per os by droplet feeding with A21-MPV and enhancin deletion and replacement virusesa
| Virus | LC50 (PIB/ml) | 95% fiducial limits
|
Slope | Het. | Difference vs. A21-MPV (fold) | |
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| A21-MPV | 1.2 × 105 | 6.5 × 104 | 2.1 × 105 | 1.2 ± 0.2 | 0.18 | |
| E2del | 1.7 × 105 | 3.2 × 104 | 6.6 × 105 | 1.0 ± 0.1 | 1.66 | 1.4 |
| E1delE2del AE2 | 1.6 × 106 | 3.0 × 105 | 9.8 × 106 | 0.7 ± 0.1 | 2.23 | 10.0 |
| E1repE2del | 2.7 × 105 | 1.3 × 105 | 5.2 × 105 | 0.8 ± 0.1 | 0.60 | 2.3 |
See Table 1, footnotes a and b.
Taken together, the bioassay results indicate that the E1 and E2 gene products can partially compensate for the loss of one of the genes, since deletion of the E1 or E2 gene decreased viral potency by approximately 2-fold, whereas deletion of both enhancin genes decreased potency by approximately 12-fold. The contribution to viral potency by the enhancin proteins does not appear to be additive, since each protein alone is capable of increasing viral potency approximately 10-fold (relative to a virus with no enhancin genes), whereas when both proteins are present, an increase in potency of 12-fold is observed. This suggests that there is a limit to the degree of potency enhancement that the enhancin proteins can effect. Since the total contribution to viral potency of both enhancin proteins together is greater than that of each individually, the presence of both genes makes the virus more competitive than a strain that contains only one. The potency results obtained in this study provide a basis for the presence of two enhancin genes in LdMNPV and possibly for the finding of multiple enhancin genes in XcGV. The mechanism by which the LdMNPV enhancin genes increase viral potency, their site of action, and the means of conveyance to the action site are under investigation.
ACKNOWLEDGMENTS
We thank Jennifer Koch and Suzanne Thiem for critical review of the manuscript.
This work was supported by the U.S. Department of Agriculture, Forest Service, Northeastern Research Station.
REFERENCES
- 1.Ahrens C H, Russell R L Q, Funk C J, Evans J T, Harwood S H, Rohrmann G F. The sequence of the Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus genome. Virology. 1997;229:381–399. doi: 10.1006/viro.1997.8448. [DOI] [PubMed] [Google Scholar]
- 2.Ayres M D, Howard S C, Kuzio J, Lopez-Ferber M, Possee R D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 3.Bell L C, Owens C D, Shapiro M. Development of mass rearing technology. In: Doane C C, McManus M L, editors. The gypsy moth: research toward integrated pest management. Forest Service Technology Bulletin no. 1584. U.S. Washington, D.C.: Department of Agriculture; 1981. p. 608. [Google Scholar]
- 4.Bischoff D S, Slavicek J M. Characterization of the Lymantria dispar nucleopolyhedrovirus 25K FP gene. J Gen Virol. 1996;77:1913–1923. doi: 10.1099/0022-1317-77-8-1913. [DOI] [PubMed] [Google Scholar]
- 5.Bischoff D S, Slavicek J M. Molecular analysis of an enhancin gene in the Lymantria dispar nuclear polyhedrosis virus. J Virol. 1997;71:8133–8140. doi: 10.1128/jvi.71.11.8133-8140.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, Ijkel W F J, Tarchini R, Sun X, Sandbrink H, Wang H, Peters S, Zuidema D, Lankhorst R K, Vlak J M, Hu Z. The sequence of the Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus genome. J Gen Virol. 2001;82:241–257. doi: 10.1099/0022-1317-82-1-241. [DOI] [PubMed] [Google Scholar]
- 7.Derksen A C G, Granados R. Alteration of a lepidopteran peritrophic membrane by baculoviruses and enhancement of viral infectivity. Virology. 1988;167:242–250. doi: 10.1016/0042-6822(88)90074-8. [DOI] [PubMed] [Google Scholar]
- 8.Federici B A. Baculovirus pathogenesis. In: Miller L K, editor. The baculoviruses. New York, N.Y: Plenum Press; 1997. pp. 33–59. [Google Scholar]
- 9.Friesen P D, Miller L K. Temporal regulation of baculovirus RNA: overlapping early and late transcripts. J Virol. 1985;54:392–400. doi: 10.1128/jvi.54.2.392-400.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Funk C J, Braunagel S C, Rohrmann G F. Baculovirus structure. In: Miller L K, editor. The baculoviruses. New York, N.Y: Plenum Press; 1997. pp. 7–32. [Google Scholar]
- 11.Gallo L G, Corsaro B G, Hughes P R, Granados R R. In vivo enhancement of baculovirus infection by the viral enhancing factor of a granulosis virus of the cabbage looper, Trichoplusia ni (Lepidoptera: Noctuidae) J Invertebr Pathol. 1991;58:203–210. [Google Scholar]
- 12.Gomi S, Majima K, Maeda S. Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J Gen Virol. 1999;80:1323–1337. doi: 10.1099/0022-1317-80-5-1323. [DOI] [PubMed] [Google Scholar]
- 13.Häse C C, Finkelstein R A. Bacterial extracellular zinc-containing metalloproteases. Microbiol Rev. 1993;57:823–837. doi: 10.1128/mr.57.4.823-837.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashimoto Y, Corsaro B G, Granados R R. Location and nucleotide sequence of the gene encoding the viral enhancing factor of the Trichoplusia ni granulosis virus. J Gen Virol. 1991;72:2645–2651. doi: 10.1099/0022-1317-72-11-2645. [DOI] [PubMed] [Google Scholar]
- 15.Hashimoto Y, Hayakawa T, Ueno Y, Fujita T, Sano Y, Matsumoto T. Sequence analysis of the Plutella xylostella granulovirus genome. Virology. 2000;275:358–372. doi: 10.1006/viro.2000.0530. [DOI] [PubMed] [Google Scholar]
- 16.Hayakawa T, Ko R, Okano K, Seong S, Goto C, Maeda S. Sequence analysis of the Xestia c-nigrum granulovirus genome. Virology. 1999;262:277–297. doi: 10.1006/viro.1999.9894. [DOI] [PubMed] [Google Scholar]
- 17.Hughes P R. ViStat: statistical package for the analysis of baculovirus bioassay data. Ithaca, N.Y: Boyce Thompson Institute at Cornell University; 1990. [Google Scholar]
- 18.Hughes P R, van Beek N A M, Wood H A. A modified droplet feeding method for rapid assay of Bacillus thuringiensis and baculoviruses in noctuid larvae. J Invertebr Pathol. 1986;48:187–192. [Google Scholar]
- 19.Ijkel W F J, van Strien E A, Heldens J G M, Broer R, Zuidema D, Goldbach R W, Vlak J M. Sequence and organization of the Spodoptera exigua multicapsid nucleopolyhedrovirus. J Gen Virol. 1999;80:3289–3304. doi: 10.1099/0022-1317-80-12-3289. [DOI] [PubMed] [Google Scholar]
- 20.Jiang W, Bond J S. Families of metalloendopeptidases and their relationships. FEBS Lett. 1992;312:11–114. doi: 10.1016/0014-5793(92)80916-5. [DOI] [PubMed] [Google Scholar]
- 21.Jin J, Guarino L A. 3′-End formation of baculovirus late RNAs. J Virol. 2000;74:8930–8937. doi: 10.1128/jvi.74.19.8930-8937.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jongeneel C V, Bouvier J, Bairoch A. A unique signature identifies a family of zinc-dependent metallopeptidases. FEBS Lett. 1989;242:211–214. doi: 10.1016/0014-5793(89)80471-5. [DOI] [PubMed] [Google Scholar]
- 23.Kozuma K, Hukuhara T. Fusion characteristics of a nuclear polyhedrosis virus in cultured cells: time course and effect of a synergistic factor and pH. J Invertebr Pathol. 1994;63:63–67. [Google Scholar]
- 24.Kuzio J, Pearson M N, Harwood S H, Funk C J, Evans J R, Slavicek J M, Rohrmann G F. Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology. 1999;253:17–34. doi: 10.1006/viro.1998.9469. [DOI] [PubMed] [Google Scholar]
- 25.Lepore L S, Roelvink P R, Granados R R. Enhancin, the granulosis virus protein that facilitates nucleopolyhedrovirus (NPV) infections, is a metalloprotease. J Invertebr Pathol. 1996;68:131–140. doi: 10.1006/jipa.1996.0070. [DOI] [PubMed] [Google Scholar]
- 26.Mahoudi M, Lin V K. Comparison of two different hybridization systems in Northern transfer analysis. BioTechniques. 1989;7:331–333. [PubMed] [Google Scholar]
- 27.Murphy G J P, Murphy G, Reynolds J J. The origin of matrix metalloproteinases and their familial relationships. FEBS Lett. 1991;289:4–7. doi: 10.1016/0014-5793(91)80895-a. [DOI] [PubMed] [Google Scholar]
- 28.O'Reilly D R, Miller L K, Lucknow V A. Baculovirus expression vectors: a laboratory manual. New York, N.Y: Freeman; 1992. [Google Scholar]
- 29.Robertson J L, Preisler M K. Pesticide bioassays with arthropods. Boca Raton, Fla: CRC Press; 1992. [Google Scholar]
- 30.Roelvink P W, Corsaro B G, Granados R R. Characterization of the Helicoverpa armigera and Pseudaletia unipuncta granulovirus enhancin genes. J Gen Virol. 1995;76:2693–2705. doi: 10.1099/0022-1317-76-11-2693. [DOI] [PubMed] [Google Scholar]
- 31.Slavicek J M, Mercer M J, Kelly M E, Hayes-Plazolles N. Isolation of a baculovirus variant that exhibits enhanced polyhedra production stability during serial passage in cell culture. J Invertebr Pathol. 1996;67:153–160. [Google Scholar]
- 32.Slavicek J M, Popham H J R, Riegel C I. Deletion of the Lymantria dispar multicapsid nucleopolyhedrovirus ecdysteroid UDP-glucosyl transferase gene enhances viral killing speed in the last instar of the gypsy moth. Biol Control. 1999;16:91–103. [Google Scholar]
- 33.Swofford P L. PAUP: phylogenetic analysis using parsimony, version 3.1. Washington, D.C.: Smithsonian Institution; 1993. [Google Scholar]
- 34.Tanada Y. Synergism between two viruses of the armyworm, Pseudaletia unipuncta (Haworth) (Lepidoptera, Noctuidae) J Insect Pathol. 1959;1:215–231. [Google Scholar]
- 35.Tanada Y. A synopsis of studies on the synergistic property of an insect baculovirus: a tribute to Edward A. Steinhaus. J Invertebr Pathol. 1985;45:125–138. [Google Scholar]
- 36.Tanada Y, Hess R T, Omi E M. Invasion of a nuclear polyhedrosis virus in midgut of the armyworm, Pseudaletia unipuncta, and the enhancement of a synergistic enzyme. J Invertebr Pathol. 1975;26:99–104. doi: 10.1016/0022-2011(75)90174-3. [DOI] [PubMed] [Google Scholar]
- 37.Tanada Y, Himeno M, Omi E M. Isolation of a factor, from the capsule of a granulosis virus, synergistic for a nuclear-polyhedrosis virus of the armyworm. J Invertebr Pathol. 1973;21:31–90. doi: 10.1016/0022-2011(73)90110-9. [DOI] [PubMed] [Google Scholar]
- 38.Tanada Y, Inoue H, Hess R T, Omi E M. Site of action of a synergistic factor of a granulosis virus of the armyworm, Pseudaletia unipuncta. J Invertebr Pathol. 1980;34:249–255. [Google Scholar]
- 39.Uchima K, Harvey J P, Omi E M, Tanada Y. Binding sites on the midgut cell membrane for the synergistic factor of a granulosis virus of the armyworm (Pseudaletia unipuncta) Insect Biochem. 1988;18:645–650. [Google Scholar]
- 40.Wang P, Granados R R. An intestinal mucin is the target substrate for a baculovirus enhancin. Proc Natl Acad Sci USA. 1997;94:6977–6982. doi: 10.1073/pnas.94.13.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang P, Hammer D A, Granados R R. Interaction of Trichoplusia ni granulosis virus-encoded enhancin with the midgut epithelium and peritrophic membrane of four lepidopteran insects. J Gen Virol. 1994;75:1961–1967. doi: 10.1099/0022-1317-75-8-1961. [DOI] [PubMed] [Google Scholar]
- 42.Westwood J A, Jones I M, Bishop D H L. Analysis of alternative poly(A) signals for use in baculovirus expression vectors. Virology. 1993;195:90–99. doi: 10.1006/viro.1993.1349. [DOI] [PubMed] [Google Scholar]