Abstract
“True” cobras (genus Naja) are among the venomous snakes most frequently involved in snakebite accidents in Africa and Asia. The Cape cobra (Naja nivea) is one of the African cobras of highest medical importance, but much remains to be learned about its venom. Here, we used a shotgun proteomics approach to better understand the qualitative composition of N. nivea venom and tested its cytotoxicity and protease activity as well as its effect on intracellular Ca2+ release and NO synthesis. We identified 156 venom components representing 17 protein families, with the dominant ones being three-finger toxins, mostly of the short-chain type. Two-thirds of the three-finger toxin entries identified were assigned as cytotoxins, while the remainder were categorized as neurotoxins, including short-chain, long-chain, and ancestral three-finger toxins. We also identified snake venom metalloproteinases and members of CRISP, l-amino acid oxidase, and other families. Protease activity and its effect on intracellular Ca2+ release and NO synthesis were low. Phospholipase A2 activity was surprisingly high, despite this toxin family being marginally recovered in the analyzed venom. Cytotoxicity was relevant only at higher venom concentrations, with macrophage and neuroblastoma cell lines showing the lowest viability. These results are in line with the predominantly neurotoxic envenomation symptoms caused by Cape cobra bites. The present overview of the qualitatively complex and functionally intriguing venom of N. nivea may provide insights into the pathobiochemistry of this species’ venom.
Keywords: snakebite, Elapidae, bioassays, proteomics, venomics, 3FTx, Africa, Uraeus
1. Introduction
Snakebites are a frequent cause of death and morbidity in developing countries especially in Africa and Asia, and the socioeconomic consequences can be devastating when young working men (the main breadwinners for most families) are envenomated [1,2,3,4]. The high incidence and significant impact of snakebites in many of the economically weakest societies in the world have promoted interest in the composition of snake venoms [3] in order to facilitate the development of more effective therapeutics and treatment protocols [5,6,7,8]. This area of research has recently been accelerated by the emergence of disruptive technologies from the biotechnological sector [9,10,11,12]. It is important to understand the molecular profile of snake venoms, including the activity of all their proteins and peptides [13,14]. Proteomics has therefore been used to investigate the venom composition of several medically significant snake species [15,16,17,18,19,20].
Approximately 400 venomous snake species are currently recognized as medically important by the World Health Organization [21]. Among these, “true” cobras of the genus Naja deserve special attention. They are arguably among the most famous venomous snakes in the world, and their distinctive hooding behavior has made them widely recognizable to the general public. The genus Naja belongs to the family Elapidae and features ~30 species distributed across Africa and Asia [22,23]. Together with the genera Echis and Daboia (both belonging to the family Viperidae), the genus Naja is one of the genera leading causes of snakebites in the Old World [2]. Naja venoms are of severe toxicity, and virtually all Naja species are able to cause lethal envenomation in humans. The principal lethal components of Naja venoms are neurotoxins, but they also contain cytotoxins that cause tissue damage [24]. Considering the high medical relevance of cobra venoms, their effects and properties have been widely investigated by means of different approaches [24,25,26,27,28,29,30,31,32]. Venom research has focused on species responsible for the highest incidence of snakebites in different regions, such as the Indian cobra (Naja naja) in Asia [2,26]. While a series of studies investigated the venom of the Cape cobra (Naja nivea), this species has overall received less attention than several other true cobras. This representative of the subgenus Uraeus, exclusively comprising African non-spitting cobras (e.g., Naja haje, Naja annulifera), occurs in Namibia, Botswana, South Africa, and Lesotho [15,20,21]. It is one of the smaller African cobras (maximum length 150 cm), although larger specimens have been recorded [33,34,35,36] and often features a distinctive yellowish coloration that makes it very popular in the pet trade [37] (see Figure 1). N. nivea bites cause potentially lethal neurotoxic symptoms, particularly areflexic flaccid paralysis requiring prolonged ventilation and antivenom therapy, usually commencing less than 3 h post bite [38]. Considering the mainly neurotoxic symptoms generally arising after N. nivea envenoming, the potential of this species’ venom for causing different effects has scarcely been explored. The N. nivea venom shows high potency against mammalian targets (e.g., LD50 = 0.41–0.75 µg/g; murine) [17,39].
Figure 1.
Phenotypic variation and hooding behavior in the Cape cobra (Naja nivea). The top row shows juveniles (A,B); adults are shown in middle and bottom rows (C–F). Specimens in (C,D) display the widespread yellowish to brown coloration, (E) shows a speckled specimen, and a dark brown variety is shown in (F). Images by Andries Cilliers.
The Cape cobra played an important role in the establishment of the first functional snake venom gland organoids, and several individual toxins have already been isolated from the venom of this species [40,41,42,43,44]. However, to date, only a few studies have investigated the composition of Cape cobra venom using proteomics methods [25,27,45,46]. The composition and biological activities of snake venom may vary at intraspecific levels depending on factors like sex of the analyzed individuals, locality of origin, and seasonality, so it is important to analyze different specimens to gain a full understanding of the dynamic and variable venom components [47,48,49]. In the present study, we thus provide a qualitative profile of N. nivea venom from South Africa using a shotgun proteomics approach and investigate its bioactivity profile. Our results provide novel data to further support global efforts toward the development of more effective therapeutic tools to treat envenomation caused by Cape cobras.
2. Results
2.1. Venom Profiling by 1D-SDS-PAGE
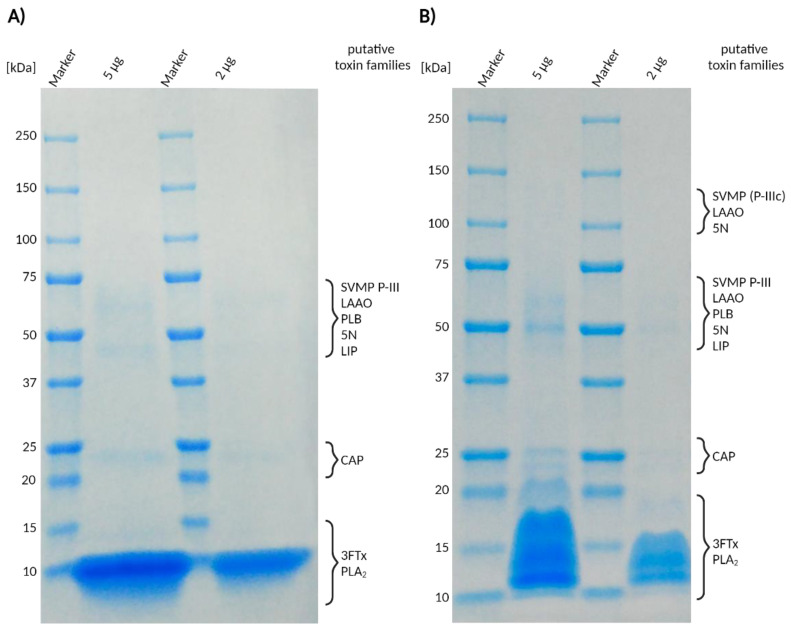
SDS-PAGE analysis was run under reducing and non-reducing conditions (see Figure 2). Under both conditions, the gels were characterized by the absence of intense bands at high molecular ranges. In contrast, the overwhelming majority of identified bands were present at lower molecular ranges (<25 kDa). The analysis under both SDS-PAGE conditions revealed similar banding patterns. Overall, the SDS-PAGE profiles suggest that the venom of N. nivea is strongly dominated by small venom proteins (<15 kDa). Bands at apparent masses for important snake venom toxin families known to occur in the genus Naja imply that a range of those are present, albeit in lower amounts. This includes snake venom metalloproteinases (svMPs), l-amino acid oxidase (LAAO), and cysteine-rich secretory proteins (CRISPs). However, in contrast to those, intense bands are retrieved under both reducing and non-reducing conditions (10 kDa reduced and 10–20 kDa in non-reduced), matching sizes for three-finger toxins (3FTxs) and phospholipase A2 (PLA2). These are often found in venoms of the Elapidae family, including members of the genus Naja [24,40,41,50,51,52]. The disappearance of the 10–20 kDa band (non-reduced) and formation of a 10 kDa band (reduced) suggest the dismantling of potential covalent oligomers, as they are known for only a few snake 3FTxs [53,54]. Since SDS-PAGE profiling provides only limited information on protein sizes within the sample, and any family assignments made are purely putative, additional mass spectrometric evidence is necessary to fully elucidate the venom profile of the analyzed venom.
Figure 2.
One-dimensional SDS-PAGE of Naja nivea venom under (A) reduced and (B) non-reduced conditions. Shown are stained protein bands of 5 µg and 2 µg venom. Putative toxin classes matching the molecular weight of the retrieved bands are assigned based on their known sizes. Abbreviations: svMP P-III = snake venom metalloproteinases of class P-III; LAAO = l-amino acid oxidase; PLA2 = phospholipase A2; PLB = phospholipase B; CE = carboxylesterase; 5N = 5′-nucleotidase; CRISP = cysteine-rich secretory proteins; 3FTx = three-finger toxins.
2.2. Shotgun Venom Proteomics
To support the protein assignments made via 1D-SDS-PAGE, we proceeded to perform a mass spectrometric validation of our findings. Using a shotgun proteomics strategy, we detected 168 proteins representing 23 distinct families (Table S1) across our entire sample. After removal of potential non-venom components from the analysis, our final dataset contained 156 venom components that could be assigned to 17 protein families. Most of the protein families were represented by more than one protein, and all had previously been found in snake venoms.
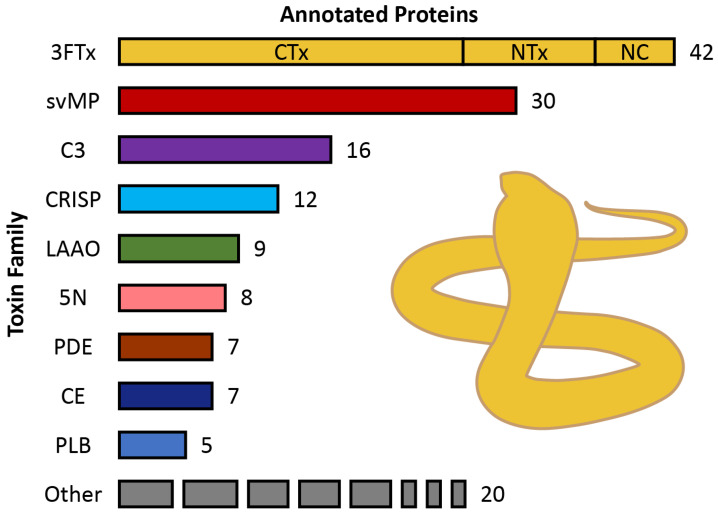
The qualitative composition of N. nivea venom is summarized in Figure 3. The most diverse family in the N. nivea venom proteome was the three-finger toxin (3FTx) family, with 42 members spanning three major subgroups (Table 1). Specifically, we identified six non-conventional (NC) neurotoxic 3FTxs. Most of these displayed similarities with other Naja NC 3FTxs, including N. nivea (P25680), N. haje (P01401), N. melanoleuca (P01400), and N. naja (P29181), but also with Bucandin (P81782), a presynaptic neurotoxin from the Malayan krait Bungarus candidus. We identified six short-chain and three long-chain neurotoxic 3FTxs, with the short neurotoxin 2 (P01423) and long neurotoxin 1 (P01390), both from Naja nivea, showing the highest MASCOT score for those subgroups, respectively. While most 3FTxs displayed similarity to known Naja toxins, one of the proteins was similar to a muscarinic m-1 toxin (P60234) from the Eastern green mamba Dendroaspis angusticeps.
Figure 3.
Bar chart depicting toxin diversity in Naja nivea venom. The numbers refer to the variety of venom proteins detected within each protein family, thus providing a snapshot of the toxin diversity within N. nivea venom. The most diverse component family was three-finger toxins (3FTxs), with cytotoxins (CTx, 26 entries), neurotoxins (NTx, 10 entries) including 1 muscarinic toxin, and non-conventional neurotoxins (NC, 6 entries). The second most diverse family was snake venom metalloproteinases (svMP), followed by venom complement C3 proteins, cysteine-rich secretory proteins (CRISP), l-amino acid oxidases (LAAO), 5′ nucleotidases (5N), carboxylesterases (CE), phosphodiesterases, and phospholipase B proteins (PLB). The less diverse families (<5 components) are grouped as “Other”.
Table 1.
Overview of the 3FTxs found in the analyzed Naja nivea venom. The identified 3FTxs were classified into the known subgroups and listed with the respective Uniprot ID, taxonomic origin, and MASCOT score for each hit identified via proteomics.
| Subgroup | Uniprot ID | Taxon | MASCOT Score | Subgroup | Uniprot ID | Taxon | MASCOT Score |
|---|---|---|---|---|---|---|---|
| Cytotoxins | P01453 | Naja annulifera | 39,445 | Cytotoxins (cont.) | P62390 | Naja annulifera | 1525 |
| P01456 | Naja nivea | 37,149 | P01446 | Naja kaouthia | 1314 | ||
| P01463 | Naja nivea | 25,422 | O93472 | Naja sputatrix | 1314 | ||
| P01462 | Naja annulifera | 24,579 | P01474 | Naja melanoleuca | 673 | ||
| P01461 | Naja annulifera | 10,362 | P60308 | Naja atra | 325 | ||
| P01458 | Naja nivea | 10,320 | Short Neurotoxins | P01423 | Naja nivea | 5245 | |
| Q98961 | Naja atra | 9746 | P01421 | Naja annulifera | 2871 | ||
| P60311 | Naja sputatrix | 9574 | P01422 | Naja annulifera | 2798 | ||
| O73857 | Naja sputatrix | 9574 | P01426 | Naja pallida | 1929 | ||
| P01451 | Naja oxiana | 9574 | P01424 | Naja melanoleuca | 1225 | ||
| O93473 | Naja sputatrix | 9572 | Q9W717 | Naja atra | 866 | ||
| A0A0U5ARS4 | Naja naja | 9572 | Long Neurotoxins | P01390 | Naja nivea | 47,565 | |
| A0A0U4W6H0 | Naja naja | 9572 | R4G2D8 | Acanthophis wellsi | 1388 | ||
| P01457 | Naja haje | 9446 | Q53B57 | Ophiophagus hannah | 84 | ||
| P01468 | Naja pallida | 9321 | Muscarinic Toxins | P60234 | Dendroaspis angusticeps | 669 | |
| P01469 | Naja mossambica | 9321 | Non-conventional | P25680 | Naja nivea | 7648 | |
| P0DSN1 | Naja nigricollis | 8467 | P01400 | Naja melanoleuca | 1554 | ||
| P86540 | Naja naja | 8281 | P01401 | Naja haje | 1539 | ||
| P83345 | Naja sagittifera | 8275 | P25677 | Naja annulifera | 902 | ||
| Q98956 | Naja atra | 8275 | P29181 | Naja naja | 596 | ||
| P14541 | Naja kaouthia | 1567 | P81782 | Bungarus candidus | 317 |
However, the most diverse subgroup was the cytotoxic 3FTxs (CTxs), which are also referred to as cardiotoxic 3FTxs with 26 members being homologs to toxins of spitting and non-spitting cobras, as N. nivea [55]. The six CTxs with the highest MASCOT score were CTxs 1, 2, and 3 (N. nivea; P01456, P01463, and P01458) and CTxs 2, 4, and 10 (N. annulifera; P01461, P01462, and P01453). In total, one third of the 3FTxs in N. nivea venom were therefore assigned as neurotoxins (16 members) and two thirds as cytotoxins (26 members), together accounting for 26.9% of all Cape cobra venom components (Table 1).
The second most diverse components were svMPs, which belong to the M12B family of metalloproteinases. We identified 30 svMPs, corresponding to 19.2% of the venom components. We also identified 16 venom complement C3 proteins (10.3%) and 12 CRISP members of the cysteine-rich secretory protein/antigen 5/pathogenesis-related protein (CAP) superfamily, representing 7.7% of the venom components. We detected nine LAAOs (5.8%), eight 5′ nucleotidases (5Ns, 5.1%), seven carboxylesterases (CEs, 4.5%), and seven phosphodiesterases (PDEs, 4.5%), as well as five phospholipase B proteins (PLBs, 3.2%).
The N. nivea venom sample analyzed also contained various less diverse toxin families with only 1–4 components, which are shown as “Other” in Figure 3. The most diverse of these were the aminopeptidases of the M1 family and endonuclease families, each represented by four members (2.6% of the protein diversity). The nerve growth factor β, snake venom serine protease (S1 family), and PLA2 were each represented by three members (1.9%). Two of the PLA2 proteins were basic and one acidic. One member was detected for the Kunitz, Ohanin/Vespryn, and vascular endothelial growth factor families, together accounting for 0.6% of the N. nivea venom analyzed.
2.3. Bioactivity Profiling
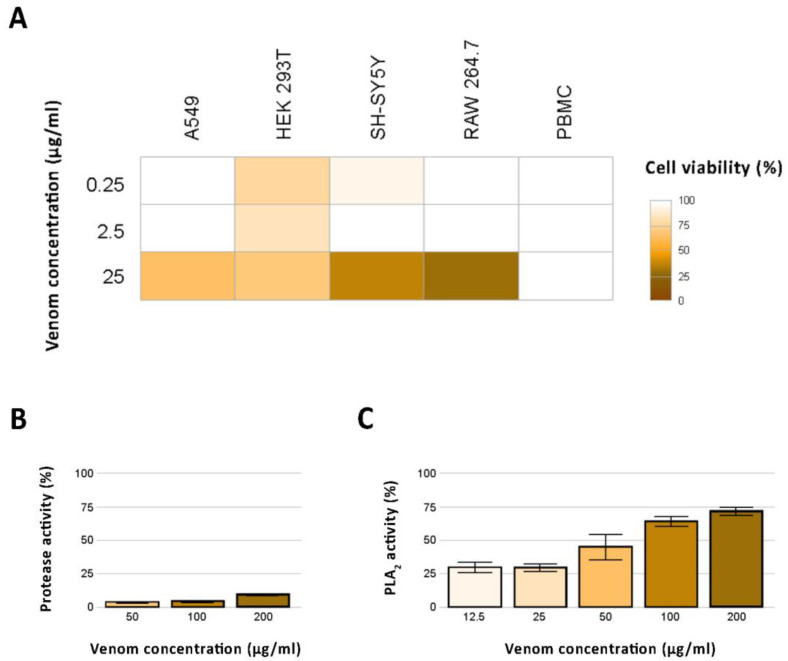
Considering the diversity of cytotoxins found in the produced N. nivea venom proteome, we first explored the cytotoxic effects of this species’ venom across four different mammalian cell lines (murine, RAW 264.7; human, A549, HEK 293T, and SH-SY5Y) and primary human peripheral blood mononuclear cells (PBMCs). Evident cytotoxicity was detectable almost exclusively at the highest venom concentration, with cell viability ranging from 29% (RAW 264.7) to 70% (HEK 293T) at 25 µg/mL. Virtually no cytotoxicity was detected at concentrations of 2.5 and 0.25 µg/mL across all tested cell lines but HEK 293T, for which viability ranged from 78% (0.25 µg/mL) to 70% (25 µg/mL). No cytotoxic effect was detected on PBMCs. Results of the cytotoxicity assays are presented in Figure 4A.
Figure 4.
In vitro effects of N. nivea venom. (A) The heatmap shows the cytotoxic effect of the analyzed N. nivea venom on the viability of the five tested cell lines (A549, HEK 293T, SH-SY5Y, RAW 264.7, PBMC). (B) Protease and (C) phospholipase A2 activity. The bar charts illustrate the photometrically determined protease and PLA2 activity of N. nivea venom at three concentrations relative to the trypsin control, and at five concentrations relative to the PLA2 stock solution provided with the assay kit, respectively.
Furthermore, we assessed the protease and PLA2 activity of the analyzed N. nivea venom. The venom showed low levels of protease activity across all tested concentrations, ranging from 5% to 10% in a concentration-dependent manner (Figure 4B). Also, the PLA2 activity varied in a positive concentration-dependent manner but was considerably more remarkable, ranging from 31% to 73% (Figure 4C). Finally, aiming to test whether N. nivea venom impacts cell–cell communication and inflammation, we investigated its effect on intracellular Ca2+ release in HEK 293T and SH-SY5Y cells, as well as on NO levels in RAW 264.7 cells. Our analysis revealed no apparent effect of N. nivea venom on either of the tested targets. Additional information on the performed bioassays is provided in Tables S2–S7.
3. Discussion
Through the application of a shotgun proteomics approach paired with one-dimensional SDS-PAGE profiling, we determined the qualitative profile of N. nivea venom components (Figure 2 and Figure 3). This provided deeper insights into the putative biological diversity of venom proteins and the medical consequences of envenomation by Cape cobras, which is characterized by the rapid onset of neurotoxic effects. In some verified bites, life-threatening flaccid paralysis developed within 3 h, requiring a combination of prolonged ventilation and antivenom therapy [38]. Untreated bites are often fatal, but even after prompt treatment, it may still be necessary for patients to remain in-hospital for several days [38]. We have identified several components that may be responsible for the extreme and prolonged neurotoxicity of N. nivea venom.
Fatalities caused by elapid snakebites often reflect the activity of 3FTxs and PLA2 [56], two of the largest protein families found in elapid snake venoms [57]. The 3FTxs contain the characteristic, three-finger fold motif, comprising three loops that protrude from the protein core to form a finger-like structure [56]. Plesiotypic 3FTxs bind to neuromuscular α1 nicotinic acetylcholine receptors. Such ancestral toxins are often described as “weak neurotoxins” or non-conventional due to their low affinity for human receptors and the consequently mild neurotoxic symptoms they typically elicit in humans [55,56]. However, their toxicity in reptiles and birds is significantly higher, and the term “weak neurotoxins” is therefore likely inadequate [58]. Elapids have also evolved a large repertoire of apotypic 3FTxs through the loss of functionally constraining cysteine residues [56,59]. Our exploratory examination of the N. nivea venom profile by means of SDS-PAGE under reducing- and non-reducing conditions showed that Cape cobra venom is predominantly composed of smaller-sized proteins below 15 kDa. This banding pattern is to be expected by venoms predominantly composed of 3FTxs such as those of the genus Naja.
Furthermore, we conducted a shotgun proteomics investigation of the N. nivea venom (Table 1, Figure 3, and Supplementary Table S1). This analysis supported our initial observations and revealed that 3FTxs are indeed the principal components of N. nivea venom. Of these, two thirds were identified as cytotoxic 3FTxs, with the remainder classified instead as putative neurotoxins. Seven of the seemingly neurotoxic 3FTxs are α-neurotoxins that potently interfere with human α1 nicotinic acetylcholine receptors and contribute to the lethality as in other cobra venoms [60]. We also found six NC 3FTxs, which can cause mild neurotoxic effects in humans [61]. While the overwhelming majority of identified 3FTxs with putative neurotoxic effects displayed similarities with known toxins previously described from Naja venoms (e.g., P25680, P01390, P01423), some of these resembled non-Naja elapid toxins. These include one toxin similar to bucandin from the Malayan krait B. candidus and one similar to an uncharacterized toxin from the death adder Acanthophis wellsi (R4G2D8). Remarkably, one of the short-chain 3FTxs was found to be similar to a muscarinic m-1 toxin (P60234) from the Eastern green mamba D. angusticeps, which binds irreversibly and with high specificity to M1 (CHRM1) muscarinic acetylcholine receptors [62]. This prevents the binding of receptor antagonists and downstream signaling for extended time periods, potentially explaining the long-lasting neurotoxic effects of N. nivea envenomation.
Despite the medical importance of the Cape cobra, the composition of its venom has only recently been assessed using proteomics [25,27,45,46]. The available proteomics studies on N. nivea venom have been analyzed based on different workflows and (semi)-quantification (see Table 2). Some differences in the results are therefore anticipated, due to discrepancies in the experimental designs applied, such as databases used for annotation and technological strengths and weaknesses of the different mass spectrometry protocols [63]. Nonetheless, consistent with previous works, we identified 3FTxs as the most abundant and diverse components of N. nivea venom. Cytotoxins were generally more diverse than neurotoxins [25,27,45,46], which were identified as a lower abundant fraction of the 3FTxs, consistent with the venom compositions of some other Naja species [25,27,45]. The lower diversity of neurotoxic compared to cytotoxic 3FTxs may reflect the potency and target redundancy of the neurotoxins. A handful of highly potent neurotoxic components are sufficient to ensure the rapid onset of paralysis, and an extension of this chemical arsenal would therefore likely be unnecessary in a scenario where venom is primarily used for prey subjugation.
Table 2.
Venom diversity comparison of five studies, including the dataset presented here, on N. nivea venom proteomes. Toxin families: three-finger toxins (3FTx), cytotoxins (CTx), neurotoxins (NTx), non-conventional 3FTx (NC), muscarinic toxins (MTx), snake venom metalloproteinases (svMP), snake venom serine proteases (svSP), aminopeptidases (AP), cysteine-rich secretory protein (CRISP), l-amino acid oxidase (LAAO), 5′-nucleotidase (5N), phosphodiesterase (PDE), carboxylesterase (CE), phospholipase A2 (PLA2), phospholipase B (PLB), nerve growth factor (NGF), Kunitz-type inhibitor (KUN), vascular endothelial growth factors (VEGF), hyaluronidases (HYAL), cystatin (CYS), C-type lectins and lectin-related proteins (CTL), and disintegrins (DI). Abbr.: mass spectrometry (MS), area under the curve of the extracted ion chromatograms (AUC XIC), label-free quantification (LFQ), mean spectral intensity (MSI), and normalized spectral abundance factor (NASF).
| Toxin Family and Subgroups | This Study | Kazandjian et al., 2021 |
Nguyen et al., 2022 |
Tan et al., 2022 |
McFarlane et al., 2024 |
|---|---|---|---|---|---|
| 3FTx | ✓ | ✓ | ✓ | ✓ | ✓ |
|
✓ | ✓ | ✓ | ✓ | ✓ |
|
✓ | ✓ | ✓ | ✓ | ✓ |
|
✓ | ✓ | ✓ | ✓ | ✓ |
|
✓ | ✓ | ✓ | ✓ | ✓ |
|
✓ | ✓ | |||
|
✓ | ||||
| svMP | ✓ | ✓ | ✓ | ✓ | |
| svSP | ✓ | ✓ | ✓ | ||
| AP and other peptidases | ✓ | ✓ | |||
| Venom Complement C3 | ✓ | ✓ | ✓ | ✓ | |
| CRISP | ✓ | ✓ | ✓ | ✓ | ✓ |
| LAAO | ✓ | ✓ | ✓ | ✓ | |
| 5N | ✓ | ✓ | ✓ | ✓ | |
| PDE | ✓ | ✓ | ✓ | ✓ | |
| CE and other esterases | ✓ | ✓ | ✓ | ✓ | |
| PLA2 | ✓ | ✓ | ✓ | ✓ | |
| PLB | ✓ | ✓ | |||
| Endonuclease | ✓ | ||||
| NGF | ✓ | ✓ | ✓ | ✓ | ✓ |
| KUN | ✓ | ✓ | ✓ | ✓ | ✓ |
| Vespryn | ✓ | ✓ | ✓ | ✓ | |
| VEGF | ✓ | ✓ | |||
| HYAL | ✓ | ✓ | |||
| CYS | ✓ | ||||
| CTL | ✓ | ||||
| DI | ✓ | ||||
| Other proteins and peptides | ✓ | ✓ | |||
| Proteomics workflow | Shotgun | Top-down | Shotgun | Bottom-up | Bottom-up |
| (Semi)-quantification | No. of toxins | MS (AUC XIC) | MS (LFQ) | RP-HPLC + MS (MSI) | MS (NASF) |
The appearance of a band around 10 kDa that is smaller under reducing conditions and the disappearance of a band >15 kDa that is larger (non-reduced) indicate that structuring disulfide bonds might have been cleaved and monomers were formed. Even if 3FTxs are mostly known as monomeric venom compounds, a few dimeric 3FTxs were discovered in different snake venoms recently. While Haditoxin (Ophiophagus hannah) and fulditoxin (Micrurus fulvis) are non-covalent homodimeric 3FTxs, covalent 3FTx complexes have been identified in the venom of the closely related Naja kauthia (α-cobratoxin homodimer and heterodimers), as well as two rear-fanged snakes Bioga irregularis (irditoxin) and Spilotes sulphureus (sulditoxin) [53,54,64,65]. McFarlane et al. (2024) hypothesized about the existence of covalent 3FTxs in the venom of N. nivea. By using a different venom pool, they showed a comparable banding pattern in reduced and non-reduced venom SDS PAGEs [46]. By LC-MS intact mass profiling, the authors listed over 20 masses in the range of potential NTxs and CTxs, which could mathematically form dimers of 12 to 16 kDa, corresponding to the reported bands. The 3FTxs with the highest MASCOT score we identified in our N. nivea venom sample were identical to the study of McFarlane et al., where they also formed the highest abundancies by NSAF quantification. This includes the cytotoxins 1, 2, and 3 (P01456, P01463, P01458); the long neurotoxin 1 (P01390); and cytotoxins 2 and 10 (P01462, P01453) of Naja annulifera, which belongs to the same subgenus Uraeus, leading to assumptions that the covalent 3FTxs of N. nivea can be cytotoxic, neurotoxic, or even a combination of cyto- and neurotoxic 3FTxs.
The work of Nguyen et al. (2022) [22] presents a higher protein family diversity, potentially due to their use of an in-house venom gland transcriptome as a database for protein identification. Additional retrieved venom components included hyaluronidases (HYALs), cystatin (CYS), C-type lectins and lectin-related proteins (CTL), and disintegrins (DIs), each accounting for less than 0.01% of the produced N. nivea venom proteome. Regarding the other toxin families, exclusively the study of Tan et al. (2022) [45] reported the absence of PLA2 in Cape cobra venom. This discrepancy underpins the importance of investigating different samples of a species using different methods, in order to avoid generalized assumptions stemming from limited sample sizes and analytical platforms. The N. nivea venom investigated by Kazandjian et al. (2021) showed only five toxin families in total, most likely as a consequence of their purely top-down-based workflow, known for its limitations when it comes to the analysis of low-abundance analytes with high molecular mass (e.g., svMP, LAAO, PDE, 5N) [27,66,67]. That said, it cannot be excluded that venom variation is at play in Cape cobras. This phenomenon has been reported widely across the snake kingdom and it is well known that venom profiles can differ tremendously between individuals, based on factors such as locality, sex, and life history stages [68]. That said, this subject has not been studied in Cape cobras sufficiently and we recommend that future studies investigate it. Interestingly, the analysis of coloration phenotypes in the context of venom variation has recently started to gain traction, but the results are inconclusive [69,70,71]. Facing the large phenotypic variability of Cape cobras (see Figure 1), it would be interesting to also investigate this particularity in the future.
Interestingly, all studies aiming to characterize N. nivea venom (including the present one) detected the lowest amounts of PLA2, in contrast to the venoms of other African and Asian members of the genus Naja [25,27,45]. Although proteomic studies suggest that PLA2 proteins are underrepresented in non-spitting cobras of the subgenus Uraeus, insights from bioactivity-guided experiments partially contradict these findings. For example, our PLA2 activity assay detected considerable concentration-dependent effects, between 30.95% (lowest concentration) and 72.56% (highest concentration) in comparison to crude bee venom used as a positive control. Likewise, PLA2 activities of 1–30 U/mg were reported in 11 individual specimens of N. nivea, as well as 65 U/mg in N. haje, 62 U/mg in N. melanoleuca (African non-spitting cobras), 125 U/mg in N. nigricollis (African spitting cobra), and 70 U/mg in Hemachatus haemachatus (African spitting elapid, but not a “true” cobra) [72]. Although these data agree with the proteomics-based theory of the higher relevance of PLA2 in spitting cobra venoms, they also show that the extreme variability recently reported in several animal venoms may play an important role, and that several non-spitting cobra venoms may feature ~25% (N. nivea) or even ~50% (e.g., N. haje and N. melanoleuca) of the PLA2 activity present in spitting cobras (N. nigricollis) [72]. This highlights the need to investigate different populations from each venomous species and generate multiple datasets to account for the phenomenon of intraspecific venom variability in snakes and other venomous taxa.
In light of the detected diversity of components in the analyzed N. nivea venom, we investigated its function by testing its protease activity and role in intracellular Ca2+ release and NO synthesis. Furthermore, considering the high diversity of cytotoxic 3FTxs observed, we assessed the venom’s cytotoxic activity on a diverse array of mammalian cells. Despite finding minuscule effects for the protease activity and Ca2+ and NO assays (Figure 4B and Tables S4–S6), we observed the presence of cytotoxic activity. Indeed, at the highest concentration (i.e., 25 µg/mL), N. nivea decreased the viability of most tested cells (Figure 4A), whereas lower concentrations had virtually no cytotoxic effect, and the PBMCs were completely unaffected. The highest venom concentration caused a decrease in cell viability of approximately 30% in the epithelial-like kidney cells (HEK 293T) and epithelial lung cells (A549). These findings appear concordant with a previous cell viability assay performed by Nguyen and colleagues on human N/TERT keratinocytes, and suggest that N. nivea venom generally exerts a weak cytotoxic effect on epithelial cells [25]. Nonetheless, our bioassays recovered a considerable cytotoxicity of N. nivea venom at a concentration of 25 µg/mL on SH-SY5Y neuroblastoma cells, reducing their viability to approximately 37%. An even more marked decrease in viability was detected in the macrophage cell line RAW 264.7 (29% viability at venom concentration of 25 µg/mL). As a decrease in macrophage number may play a role in impairing the inflammatory response and delaying wound healing [73,74], it may increase the risk of secondary infection, and should thus be considered when treating N. nivea snakebites. Although infection following bites of most snakes is not particularly common, snake venoms have recently been shown to contain complex microbiomes, and thus the aspect of microbial infection post envenoming should be taken into account [75,76]. In such a scenario, the decreased viability of macrophages may impact patient recovery. Overall, the tested venom exerted some effects against cancerous cell lines; hence, it would be interesting to investigate the effects of individual toxins to potentially derive novel anti-cancer agents.
While our study provided some novel insights into the composition and activity of Naja nivea venom, several limitations need to be considered. Firstly, our analysis is based on a shotgun approach, which is a powerful tool to unveil the molecular diversity of venoms. However, unfortunately, this method does not allow a quantitative analysis and, hence, abundances of toxins cannot be provided. Second, we worked with a commercially purchased pooled venom sample; hence, we were unable to further investigate the effects of venom variation. Lastly, our selection of cell lines for functional analyses is limited. In order to understand better the effects of Naja nivea venom on cytotoxicity, local effects, and long-term consequences, a broader set of cell lines and animal models should be employed [77,78]. We recommend that future studies on Naja nivea venom incorporate these considerations to gain deeper insights into the matter.
4. Conclusions
Snakebite envenomation is as a neglected tropical disease as well as a global medical burden. Detailed analysis of the venom systems of medically important snakes is needed to develop effective antivenoms and other treatments. We used a shotgun proteomics approach and 1D-SDS-PAGE profiling to increase the current understanding on the molecular diversity of Cape cobra venom, which is among the most potent venoms of the African “true” cobras (genus Naja). We identified a total of 156 venom components belonging to 17 distinct protein families and showed that the venom profile of N. nivea is rich in proteins of lower molecular weight. The major venom components were 3FTxs, one-third of which were neurotoxins and two-thirds cytotoxins. Several of the neurotoxic 3FTxs were α-neurotoxins, which are highly toxic in humans. Additional toxins such as members of CRISP, LAAO, and svMP were also identified. In line with previous studies and with what has been reported for some other non-spitting Naja species, few PLA2 proteins were found in our dataset. However, the results of our bioactivity profiling suggest that N. nivea venoms can display considerable PLA2 activity. Future work should therefore evaluate multiple populations of these species to account for dynamic intraspecific variability in venom composition.
5. Materials and Methods
5.1. Origin of Sample Material
Crude N. nivea venom was obtained from a commercial supplier (Latoxan, Portes lès Valence, France). The venom was collected from their captive stock animals stemming from a South African population (unknown locality and pool size) and lyophilized before shipment. The sample was stored at −20 °C before use.
5.2. One-Dimensional SDS-PAGE
To generate insight into the overall protein landscape within the N. nivea venom, we applied a one-dimensional sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) as described previously [79]. Therefore, we dissolved the venom in ddH2O and Laemmli buffer toward final concentrations of 5 µg and 2 µg. For SDS-PAGE under reducing conditions, the mix contained 5% of 2-mercaptoethanol. Analysis under non-reducing conditions was lacking 2-mercaptoethanol. Next, samples were incubated at 95 °C for 5 min and then loaded on 4–20% Mini-PROTEAN® TGX™ Precast Protein Gels (Bio-Rad, Hercules, CA, USA). Electrophoresis of the samples and the used protein size marker (Precision Plus Protein All Blue Standard 10–250 kDa, Bio-Rad) was carried out at 150 V for 60 min in a Mini-PROTEAN Tetra Vertical Electrophoresis Cell (Bio-Rad). Gels were stained by ROTI BLUE quick solution (Carl Roth, Karlsruhe, Germany) and destained with ddH2O.
5.3. Tryptic Digestion
The shotgun proteomics strategy was based on a protocol established for other animal venoms [80,81]. Briefly, 10 μg of venom was dissolved in 25 mM ammonium bicarbonate solution containing 0.6 µM ProteaseMax (Promega, Madison, WI, USA). Disulfide bonds were reduced by 5 mM dithiothreitol during 30 min of incubation at 50 °C. Next, free cysteines were alkylated via 10 mM iodoacetamide for 30 min at 24 °C. The addition of excess cysteine was used to quench the reaction. Afterwards, trypsin was added (50:1) and the sample was digested overnight at 37 °C. The reaction was then inhibited through the addition of 1% trifluoroacetic acid. Purification of the sample was carried out through a C18-ZipTip system (Millipore, Burlington, MA, USA), and the generated tryptic peptides were vacuum-dried and subsequently redissolved in 10 μL of 0.1% trifluoroacetic acid.
5.4. Mass Spectrometry
Following digestion and purification, the tryptic peptides were subjected to high-performance liquid chromatography on an UltiMate 3000RSLCnano device (Thermo Fisher Scientific, Waltham, MA, USA) for separation and decomplexation. We therefore injected 1 μg of the peptide solution into a 50 cm μPAC C18 column (Pharma Fluidics, Bath, UK) dissolved in 0.1% formic acid at 35 °C. A linear gradient of 3–44% acetonitrile over 240 min was used and column washing was performed with 72% acetonitrile with the flow set to 300 nL/min. Eluting peptides were analyzed in an Orbitrap Eclipse Tribrid MS (Thermo Fisher Scientific) linked to an Advion TriVersa NanoMate electrospray ionization system (Advion BioSciences, Ithaca, NY, USA) running in positive ionization mode with the spray voltage set to 1.5 kV at 250 °C. Scanning was performed in data-independent acquisition mode with a scanning time of 3 s and mass-to-charge ratio (m/z) range of 375–1500 with a resolution of 120,000. Auto-gain control was set to standard and a maximum injection time of 50 ms was applied. Most intense ions of each cycle with a threshold count >50,000 and charge states of 2–7 were selected with an isolation window of 1.6 m/z for higher-energy collisional dissociation (normalized collision energy = 30%). Fragment spectra were acquired in the linear ion trap using a rapid scan rate and normal mass range. The maximum injection time was set to 100 ms and selected precursor ions were excluded for 15 s following fragmentation.
5.5. Data Analysis
Data acquisition and analysis were carried out in Xcalibur v4.3.73.11 and Proteome Discoverer v2.4.0.305 (Thermo Fisher Scientific). Protein identification using MASCOT v2.6.2 and the UniProt “Serpentes” database (3 June 2021; 117,084,901 residues; 274,620 sequences) served as a repository for peptide searches with the following settings: precursor ion mass tolerance = 10 ppm, carbamidomethylation as global modification, methionine oxidation as variable modification, and one missed cleavage allowed. The fragment ion mass tolerance during linear ion trap MS2 detection was 0.8 Da, and the false discovery rate was limited to 0.01 in a decoy database. For qualitative analysis, we only considered proteins that were identified with a MASCOT score of at least 50, a false discovery rate (FDR) confidence of “high”, and at least two verified peptides. A comprehensive list of all identified venom components, their characteristics, and annotations is provided in Table S1. The raw proteomic data were uploaded to PRIDE under the ID PXD036965.
5.6. Cytotoxicity Assays
Cytotoxicity assays on venom from N. nivea were performed using several cell lines. We initially tested the human alveolar basal epithelial cell line A549 (CLS Cell Lines Service, Eppelheim, Germany) [82]. Briefly, cells were placed in 96-well plates and grown until reaching confluence. Venom was diluted in water, ionomycin (7.74 mg/mL stock, Cayman Chemicals, Ann Arbor, USA) was uptaken in DMSO, and cells were treated with either venom (0.25, 2.5, 25 µg/mL), ddH2O (negative control), or ionomycin (100 µM, positive control). Next, they were incubated at 37 °C under 5% CO2 atmosphere. After 48 h, cell viability was determined by photometrically quantifying the ATP content via the CellTiter-Glo Luminescent Cell Viability assay (Promega) following the manufacturer’s instructions.
Additional cytotoxicity assays were undertaken on human embryonic kidney cells (HEK 293T), neuroblastoma cells (SH-SY5Y), and RAW 264.7 macrophages and peripheral blood mononuclear cells (PBMCs), following the protocol described by Erkoc and colleagues [83,84]. Cell viability was determined using the OranguTM assay (Cell Guidance Systems, Cambridge, United Kingdom). Therefore, 2 × 105 cells from each tested cell line were placed in 96-well plates and exposed to different venom concentrations or ddH2O as a control for 24 h. Subsequently, 10 μL of OranguTM cell-counting solution was added, followed by 60 min of incubation. Absorbance was measured at λ = 450 nm with a wavelength of λ = 650 nm using an EnSpire 2300 Multimode Plate Reader (Perkin Elmer, Waltham, MA, USA). Celecoxib (100 µM) and ddH2O were applied as the positive control as negative control, respectively. Readings were normalized to the vehicle (ddH2O) and set to 100% cell viability. For every assay, 1 μL of treatment (i.e., venom, positive control, or negative control) was added to 99 μL of medium present in each well of the 96-well plates used. All experiments were carried out in triplicates.
5.7. Protease Activity Assay
To understand the proteolytic potential of the tested venom, we employed a photometric general protease activity assay. Protease activity was determined via a Protease Assay Kit (539125, Sigma-Aldrich, St. Louis, MO, USA), as described earlier [85]. Venom at three different concentrations (50, 100, 200 μg/mL) was tested against trypsin and ddH2O controls. In a 96-well plate, 25 μL of fluorescein thiocarbamoyl-casein derivatives (FTC-casein) was mixed with 25 μL of incubation buffer and 10 μL of either venom or control and incubated (37 °C, 220 rpm for 2 h). Then, 120 μL of 5% trichloroacetic acid solution (Carl Roth) was added prior to incubation for another 20 min. This was followed by 15 min of centrifugation at 4 °C and 500× g. Next, 40 μL of supernatant was transferred to a flat-bottom 96-well plate and then mixed with 160 μL of assay buffer. Absorbance was detected at λ = 492 nm on a BioTek Eon microplate reader using the Gen v2.09 software. The absorbance values from each treatment were normalized to positive controls (set at 100%), after subtracting the values from negative controls (set at 0%). All experiments were carried out in triplicates.
5.8. Phospholipase A2 Activity Assay
Phospholipase A2 (PLA2) activity was measured via the EnzChek™ Phospholipase A2 Assay Kit (E10217, Thermo Fisher), as described earlier [79]. We prepared the Lipid Mix by adding 30 μL of 10 mM dioleoylphosphatidylcholine (DOPC) to 30 μL of 10 mM dioleoylphosphatidylglycerol (DOPG), and 30 μL of 1 mM PLA2 substrate. We then added 5 mL of 1 × reaction buffer. To prepare 5 mL of substrate-liposome, we slowly injected 50 μL of Lipid Mix into the side of the vortex in the beaker using a pipettor fitted with a narrow orifice gel-loading tip. Subsequently, 50 μL of the controls and 50 μL of each venom sample at different concentrations (3.125, 6.25, 12.5, 25, and 50 μg/mL) were injected into wells of a black 96-well plate, and 50 μL of the substrate-liposome mix was added. The plate was in the dark incubated at room temperature for 50 min. Fluorescence was measured on a Synergy H4 microplate reader (BioTek) equipped for excitation at λ = 470 nm and fluorescence emission at λ = 515 nm. Measurements were normalized against the positive control (100%) after subtraction of the negative control (0%). All experiments were carried out in triplicates.
5.9. Assessment of Intracellular Calcium (Ca2+) Levels
We measured Ca2+ release within cells as described previously [83,84]. Briefly, 2 × 104 HEK 293T cells were placed in 96-well poly-D-lysine-coated plates and incubated (37 °C, 24 h). Then, they were incubated with 4.19 µg/mL of Fluo-8-AM in Hanks’ balanced salt solution (HBSS) for another 1 h at 37 °C. Next, the Fluo-8/HBSS was replaced with 100 µL of fresh HBSS. Images were acquired using an ImageXpress Micro confocal high-content imaging system (Molecular Devices, Workingham, United Kingdom) with five frames per second. For the induction assay, cells were treated with a vehicle (negative control), N. nivea venom at three different concentrations (0.25, 2.5, 25 µg/mL), or 5 µM ionomycin (positive control). In the inhibition assay, 5 µM ionomycin was added to the venom-treated cells after 30 min. For both assays, images were periodically taken every second for 20 s. Analysis of data was performed with the software MetaXpress v. 3.1.0.65, using a fluorescence intensity threshold established from pre-treatment cells. Cells exceeding this threshold were quantified. In the induction assay, the number of cells surpassing the threshold in venom-treated samples was compared to untreated samples. For the inhibition assay, venom-treated samples were compared to ionomycin-treated cells. Data were normalized to the positive control (100%) for the induction assay, and to the vehicle/negative control (100%) for the inhibition assay. All experiments were carried out in triplicates.
5.10. Assessment of Nitric Oxide (NO) Levels
Effects of N. nivea venom on the synthesis of NO were assessed following the method described by Erkoc and colleagues [83,84]. RAW 264.7 macrophages were seeded at a density of 2 × 104 cells in a 96-well plate and incubated at 37 °C for 24 h. Then, N. nivea venom at varying concentrations (0.25, 2.5, 25 µg/mL), vehicle (negative control), or 0.1 µg/mL of lipopolysaccharide (LPS, positive control) was added. Alternatively, cells were pre-treated with venom or vehicle for 30 min before being exposed to 0.1 µg/mL of LPS to evaluate the inhibition of NO production. After 24 h of incubation, supernatants were collected and stored at −80 °C. A standard curve was established with sodium nitrite concentrations ranging from 0 to 3.45 µg/mL. For the assay, 80 µL of either cell supernatant or standard was mixed with 20 µL of sulfanilamide solution (40 mg/mL in 1 M HCl) and 20 µL of naphthalene diamine solution (60 mg/mL of N-(1-naphthyl)ethylenediamine dihydrochloride in water), followed by a 15 min incubation. Absorbance was quantified at 540 nm using an EnSpire Plate Reader (Perkin Elmer). Data from the induction and inhibition assays were normalized to 100% based on the positive control and vehicle/negative control, respectively. All experiments were carried out in triplicates.
Acknowledgments
We thank Richard M. Twymann for language proofreading of the manuscript. Thomas Lindner supported this study intellectually, and Andries Cilliers provided the image used for Figure 1. All figures were assembled using BioRender or GraphPad Prism (version 9.2.0 (332)) and modified using Adobe Photoshop CC 2019 (release 20.0.0).
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/toxins16100438/s1. Figure S1: Additional SDS-PAGE gel with increased venom concentrations; Table S1: Full results of the proteomic analysis performed on the analyzed Naja nivea venom; Table S2: Raw data of the cytotoxicity assays performed on A549 cells; Table S3: Raw data of the cytotoxicity assays performed on PBMC, RAW 264.7, HEK 293T, and SH-SY5Y cells; Table S4: Raw data of the protease activity assay; Table S5: Raw data of PLA2 activity assay; Table S6: Raw data of assessment of intracellular Ca2+ release; Table S7: Raw data of assessment of NO levels.
Author Contributions
T.L., L.S., I.A., S.S., J.E., K.H., M.H., T.T. and G.L. conducted the experiments. T.L. contributed venom samples and, together with T.T., M.D. and G.L., analyzed the data. T.L. and A.V. designed the study. T.L., I.A., M.D., S.S., K.H. and A.V. obtained the funding. T.L., I.A., A.V. and M.D. wrote the manuscript with comments from the other authors. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Proteomic raw data files have been uploaded to the open access PRIDE database (PXD03696). All identified venom components are described in Table S1. Crude venom samples are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
Key Contribution
The venom composition of the Cape cobra, a species of high medical concern, is presented, and its functional profile is investigated, with a focus on potential cytotoxic effects. Novel bioactivity data for enzymatic activities and in vitro cell viability are presented alongside a new proteomic dataset for the species.
Funding Statement
T.L., I.A., M.D. and L.S. are funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), refs. 505696476 (T.L. and L.S.), 545040837 (I.A.), and 540833593 (M.D.). K.H. was funded by the Bundesministerium für Bildung und Forschung (BMBF, Federal Ministry of Education and Research), grant number 01KI2024. This work was further financially supported by the LOEWE Centre for Translational Biodiversity Genomics (LOEWE-TBG) granted to A.V. and S.S. via the Landesoffensive zur Förderung der Wissenschaftlich-Ökonomischen Exzellenz (LOEWE) of the Hessian Ministry of Higher Education, Research and the Arts (HMWK). I.A. gratefully acknowledges the financial support received from the European Cooperation in Science and Technology (COST) through the Action CA19144 EUVEN (ref. E-COST-GRANT-CA19144-9da371bb). K.H. received funding from the Federal Ministry of Science and Education via the project “ASCRIBE” (ID = 01KI2024).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Kasturiratne A., Wickremasinghe A.R., De Silva N., Gunawardena N.K., Pathmeswaran A., Premaratna R., Savioli L., Lalloo D.G., De Silva H.J. The Global Burden of Snakebite: A Literature Analysis and Modelling Based on Regional Estimates of Envenoming and Deaths. PLoS Med. 2008;5:e218. doi: 10.1371/journal.pmed.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gutiérrez J.M., Calvete J.J., Habib A.G., Harrison R.A., Williams D.J., Warrell D.A. Snakebite Envenoming. Nat. Rev. Dis. Primers. 2017;3:17063. doi: 10.1038/nrdp.2017.63. [DOI] [PubMed] [Google Scholar]
- 3.Harrison R.A., Casewell N.R., Ainsworth S.A., Lalloo D.G. The Time Is Now: A Call for Action to Translate Recent Momentum on Tackling Tropical Snakebite into Sustained Benefit for Victims. Trans. R. Soc. Trop. Med. Hyg. 2019;113:835–838. doi: 10.1093/trstmh/try134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fry B.G. Snakebite: When the Human Touch Becomes a Bad Touch. Toxins. 2018;10:170. doi: 10.3390/toxins10040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calvete J.J. Proteomic Tools against the Neglected Pathology of Snake Bite Envenoming. Expert Rev. Proteom. 2011;8:739–758. doi: 10.1586/epr.11.61. [DOI] [PubMed] [Google Scholar]
- 6.Xie C., Slagboom J., Albulescu L.O., Somsen G.W., Vonk F.J., Casewell N.R., Kool J. Neutralising Effects of Small Molecule Toxin Inhibitors on Nanofractionated Coagulopathic Crotalinae Snake Venoms. Acta Pharm. Sin. B. 2020;10:1835–1845. doi: 10.1016/j.apsb.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knudsen C., Ledsgaard L., Dehli R.I., Ahmadi S., Sørensen C.V., Laustsen A.H. Engineering and Design Considerations for Next-Generation Snakebite Antivenoms. Toxicon. 2019;167:67–75. doi: 10.1016/j.toxicon.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Knudsen C., Laustsen A.H. Recent Advances in Next Generation Snakebite Antivenoms. Trop. Med. Infect. Dis. 2018;3:42. doi: 10.3390/tropicalmed3020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khalek I.S., Senji Laxme R.R., Ngyuen Y.T.K., Khochare S., Patel R.N., Woehl J., Smith J.M.M., Saye-Francisco K., Kim Y.J., Mindrebo L.M., et al. Synthetic development of a broadly neutralizing antibody against snake venom long-chain a-neurotoxins. Sci. Transl. Med. 2024;16:735. doi: 10.1126/scitranslmed.adk1867. [DOI] [PubMed] [Google Scholar]
- 10.Laustsen A.H. Recombinant snake antivenoms get closer to the clinic. Trends Immunol. 2024;45:P225–P227. doi: 10.1016/j.it.2024.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivera-de-Torre E., Rimbault C., Jenkins T.P., Sørensen C.V., Damsbo A., Saez N.J., Duhoo Y., Hackney C.M., Ellgaard L., Laustsen A.H. Strategies for Heterologous Expression, Synthesis, and Purification of Animal Venom Toxins. Front. Bioeng. Biotechnol. 2022;9:811905. doi: 10.3389/fbioe.2021.811905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lüddecke T., Paas A., Harris R.J., Talmann L., Kirchhoff K.N., Billion A., Hardes K., Steinbrink A., Gerlach D., Fry B.G., et al. Venom Biotechnology: Casting Light on Nature’s Deadliest Weapons Using Synthetic Biology. Front. Bioeng. Biotechnol. 2023;11:1166601. doi: 10.3389/fbioe.2023.1166601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calvete J.J. Next-Generation Snake Venomics: Protein-Locus Resolution through Venom Proteome Decomplexation. Expert Rev. Proteom. 2014;11:315–329. doi: 10.1586/14789450.2014.900447. [DOI] [PubMed] [Google Scholar]
- 14.Lüddecke T., Blank S. Animal Toxins: Biodiscovery, Mechanistic Insights and Translational Potential. Toxins. 2024;16:130. doi: 10.3390/toxins16030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petras D., Heiss P., Harrison R.A., Süssmuth R.D., Calvete J.J. Top-down Venomics of the East African Green Mamba, Dendroaspis angusticeps, and the Black Mamba, Dendroaspis Polylepis, Highlight the Complexity of Their Toxin Arsenals. J. Proteom. 2016;146:148–164. doi: 10.1016/j.jprot.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 16.Alape-Girón A., Flores-Díaz M., Sanz L., Madrigal M., Escolano J., Sasa M., Calvete J.J. Studies on the Venom Proteome of Bothrops Asper: Perspectives and Applications. Toxicon. 2009;54:938–948. doi: 10.1016/j.toxicon.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 17.Whiteley G., Casewell N.R., Pla D., Quesada-Bernat S., Logan R.A.E., Bolton F.M.S., Wagstaff S.C., Gutiérrez J.M., Calvete J.J., Harrison R.A. Defining the Pathogenic Threat of Envenoming by South African Shield-Nosed and Coral Snakes (Genus Aspidelaps), and Revealing the Likely Efficacy of Available Antivenom. J. Proteom. 2019;198:186–198. doi: 10.1016/j.jprot.2018.09.019. [DOI] [PubMed] [Google Scholar]
- 18.Sunagar K., Undheim E.A.B., Scheib H., Gren E.C.K., Cochran C., Person C.E., Koludarov I., Kelln W., Hayes W.K., King G.F., et al. Intraspecific Venom Variation in the Medically Significant Southern Pacific Rattlesnake (Crotalus Oreganus Helleri): Biodiscovery, Clinical and Evolutionary Implications. J. Proteom. 2014;99:68–83. doi: 10.1016/j.jprot.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 19.Petras D., Heiss P., Süssmuth R.D., Calvete J.J. Venom Proteomics of Indonesian King Cobra, Ophiophagus Hannah: Integrating Top-down and Bottom-up Approaches. J. Proteome Res. 2015;14:2539–2556. doi: 10.1021/acs.jproteome.5b00305. [DOI] [PubMed] [Google Scholar]
- 20.Pla D., Bande B.W., Welton R.E., Paiva O.K., Sanz L., Segura Á., Wright C.E., Calvete J.J., Gutiérrez J.M., Williams D.J. Proteomics and Antivenomics of Papuan Black Snake (Pseudechis papuanus) Venom with Analysis of Its Toxicological Profile and the Preclinical Efficacy of Australian Antivenoms. J. Proteom. 2017;150:201–215. doi: 10.1016/j.jprot.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 21.Snakebite Information and Data Platform 2020. [(accessed on 20 June 2024)]. Available online: https://www.who.int/teams/control-of-neglected-tropical-diseases/snakebite-envenoming/snakebite-information-and-data-platform.
- 22.Wüster W., Crookes S., Ineich I., Mané Y., Pook C.E., Trape J.F., Broadley D.G. The Phylogeny of Cobras Inferred from Mitochondrial DNA Sequences: Evolution of Venom Spitting and the Phylogeography of the African Spitting Cobras (Serpentes: Elapidae: Naja nigricollis Complex) Mol. Phylogenet. Evol. 2007;45:437–453. doi: 10.1016/j.ympev.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 23.Kazemi E., Nazarizadeh M., Fatemizadeh F., Khani A., Kaboli M. The Phylogeny, Phylogeography, and Diversification History of the Westernmost Asian Cobra (Serpentes: Elapidae: Naja oxiana) in the Trans-Caspian Region. Ecol. Evol. 2020;11:2024–2039. doi: 10.1002/ece3.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panagides N., Jackson T.N.W., Ikonomopoulou M.P., Arbuckle K., Pretzler R., Yang D.C., Ali S.A., Koludarov I., Dobson J., Sanker B., et al. How the Cobra Got Its Flesh-Eating Venom: Cytotoxicity as a Defensive Innovation and Its Co-Evolution with Hooding, Aposematic Marking, and Spitting. Toxins. 2017;9:103. doi: 10.3390/toxins9030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen G.T.T., O’Brien C., Wouters Y., Seneci L., Gallissà-Calzado A., Campos-Pinto I., Ahmadi S., Laustsen A.H., Ljungars A. High-Throughput Proteomics and in Vitro Functional Characterization of the 26 Medically Most Important Elapids and Vipers from Sub-Saharan Africa. GigaScience. 2022;11:giac121. doi: 10.1093/gigascience/giac121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Avella I., Wüster W., Luiselli L., Martínez-Freiría F. Toxic Habits: An Analysis of General Trends and Biases in Snake Venom Research. Toxins. 2022;14:884. doi: 10.3390/toxins14120884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kazandjian T.D., Petras D., Robinson S.D., van Thiel J., Greene H.W., Arbuckle K., Barlow A., Carter D.A., Wouters R.M., Whiteley G., et al. Convergent Evolution of Pain-Inducing Defensive Venom Components in Spitting Cobras. Science. 2021;371:386–390. doi: 10.1126/science.abb9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris R.J., Nekaris K.A.I., Fry B.G. Monkeying around with Venom: An Increased Resistance to α-Neurotoxins Supports an Evolutionary Arms Race between Afro-Asian Primates and Sympatric Cobras. BMC Biol. 2021;19:253. doi: 10.1186/s12915-021-01195-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avella I., Barajas-Ledesma E., Casewell N.R., Harrison R.A., Rowley P.D., Crittenden E., Wüster W., Castiglia R., Holland C., van der Meijden A. Unexpected Lack of Specialisation in the Flow Properties of Spitting Cobra Venom. J. Exp. Biol. 2021;224:jeb229229. doi: 10.1242/jeb.229229. [DOI] [PubMed] [Google Scholar]
- 30.Du T.Y., Hall S.R., Chung F., Kurdyukov S., Crittenden E., Patel K., Dawson C.A., Westhorpe A.P., Bartlett K.E., Rasmussen S.A., et al. Molecular Dissection of Cobra Venom Highlights Heparinoids as an Antidote for Spitting Cobra Envenoming. Sci. Transl. Med. 2024;16:eadk4802. doi: 10.1126/scitranslmed.adk4802. [DOI] [PubMed] [Google Scholar]
- 31.Kakati H., Patra A., Mukherjee A.K. Composition, Pharmacology, and Pathophysiology of the Venom of Monocled Cobra (Naja kaouthia)—A Medically Crucial Venomous Snake of Southeast Asia: An Updated Review. Toxicon. 2024;249:108056. doi: 10.1016/j.toxicon.2024.108056. [DOI] [PubMed] [Google Scholar]
- 32.Wang C.R., Zenaidee M.A., Snel M.F., Pukala T.L. Exploring Top-Down Mass Spectrometric Approaches to Probe Forest Cobra (Naja melanoleuca) Venom Proteoforms. J. Proteome Res. 2024;23:4601–4613. doi: 10.1021/acs.jproteome.4c00486. [DOI] [PubMed] [Google Scholar]
- 33.Wallach V., Wüster W., Broadley D.G. In Praise of Subgenera: Taxonomic Status of Cobras of the Genus Naja Laurenti (Serpentes: Elapidae) Zootaxa. 2009;2236:26–36. doi: 10.11646/zootaxa.2236.1.2. [DOI] [Google Scholar]
- 34.Uetz P., Freed P., Aguilar R., Reyes F., Hošek J. The Reptile Database 2023. [(accessed on 20 June 2024)]. Available online: http://www.reptile-database.org.
- 35.Phelps T. Cape Cobra: Maximum Size. Afr. Herp News. 2007;42:22–23. [Google Scholar]
- 36.Spawls S., Branch B. The Dangerous Snakes of Africa. Bloomsbury; London, UK: 2022. [Google Scholar]
- 37.Trutnau L. Giftschlangen. 4th ed. Ulmer; Stuttgart, Germany: 1998. [Google Scholar]
- 38.Blaylock R., Lichtman A., Potgieter P. Clinical Manifestations of Cape Cobra (Naja nivea) Bites. A Report of 2 Cases. S. Afr. Med. J. 1985;68:342–344. [PubMed] [Google Scholar]
- 39.Ramos-Cerrillo B., de Roodt A.R., Chippaux J.-P., Olguín L., Casasola A., Guzmán G., Paniagua-Solís J., Alagón A., Stock R.P. Characterization of a New Polyvalent Antivenom (Antivipmyn® Africa) against African Vipers and Elapids. Toxicon. 2008;52:881–888. doi: 10.1016/j.toxicon.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 40.Mebs D. Myotoxic Activity of Phospholipases A2 Isolated from Cobra Venoms: Neutralization by Polyvalent Antivenoms. Toxicon Off. J. Int. Soc. Toxinol. 1986;24:1001–1008. doi: 10.1016/0041-0101(86)90006-1. [DOI] [PubMed] [Google Scholar]
- 41.Botes D.P., Strydom D.J., Anderson C.G., Christensen P.A. Snake Venom Toxins. Purification and Properties of Three Toxins from Naja nivea (Linnaeus) (Cape Cobra) Venom and the Amino Acid Sequence of Toxin Delta. J. Biol. Chem. 1971;246:3132–3139. doi: 10.1016/S0021-9258(18)62205-7. [DOI] [PubMed] [Google Scholar]
- 42.Hokama Y., Iwanaga S., Tatsuki T., Suzuki T. Snake Venom Proteinase Inhibitors: III. Isolation of Five Polypeptide Inhibitors from the Venoms of Hemachatus haemachatus (Ringhal’s Corbra) and Naja nivea (Cape Cobra) and the Complete Amino Acid Sequences of Two of Them. J. Biochem. 1976;79:559–578. doi: 10.1093/oxfordjournals.jbchem.a131100. [DOI] [PubMed] [Google Scholar]
- 43.Botes D.P., Viljoen C.C. The Amino Acid Sequence of Three Non-Curarimimetictoxins from Naja nivea Venom. Biochim. Biophys. Acta BBA Protein Struct. 1976;446:1–9. doi: 10.1016/0005-2795(76)90091-X. [DOI] [PubMed] [Google Scholar]
- 44.Post Y., Puschhof J., Beumer J., Kerkkamp H.M., de Bakker M.A.G., Slagboom J., de Barbanson B., Wevers N.R., Spijkers X.M., Olivier T., et al. Snake Venom Gland Organoids. Cell. 2020;180:233–247. doi: 10.1016/j.cell.2019.11.038. [DOI] [PubMed] [Google Scholar]
- 45.Tan C.H., Wong K.Y., Huang L.-K., Tan K.Y., Tan N.H., Wu W.-G. Snake Venomics and Antivenomics of Cape Cobra (Naja nivea) from South Africa: Insights into Venom Toxicity and Cross-Neutralization Activity. Toxins. 2022;14:860. doi: 10.3390/toxins14120860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McFarlane L.O., Pukala T.L. Proteomic Investigation of Cape Cobra (Naja nivea) Venom Reveals First Evidence of Quaternary Protein Structures. Toxins. 2024;16:63. doi: 10.3390/toxins16020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chippaux J.P., Williams V., White J. Snake Venom Variability: Methods of Study, Results and Interpretation. Toxicon. 1991;29:1279–1303. doi: 10.1016/0041-0101(91)90116-9. [DOI] [PubMed] [Google Scholar]
- 48.Casewell N.R., Wagstaff S.C., Wüster W., Cook D.A.N., Bolton F.M.S., King S.I., Pla D., Sanz L., Calvete J.J., Harrison R.A. Medically Important Differences in Snake Venom Composition Are Dictated by Distinct Postgenomic Mechanisms. Proc. Natl. Acad. Sci. USA. 2014;111:9205–9210. doi: 10.1073/pnas.1405484111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Casewell N.R., Jackson T.N.W., Laustsen A.H., Sunagar K. Causes and Consequences of Snake Venom Variation. Trends Pharmacol. Sci. 2020;41:570–581. doi: 10.1016/j.tips.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chanda A., Mukherjee A.K. Quantitative Proteomics to Reveal the Composition of Southern India Spectacled Cobra (Naja Naja) Venom and Its Immunological Cross-Reactivity towards Commercial Antivenom. Int. J. Biol. Macromol. 2020;160:224–232. doi: 10.1016/j.ijbiomac.2020.05.106. [DOI] [PubMed] [Google Scholar]
- 51.Kakati H., Patra A., Kalita B., Chanda A., Rapole S., Mukherjee A.K. A Comparison of Two Different Analytical Workflows to Determine the Venom Proteome Composition of Naja Kaouthia from North-East India and Immunological Profiling of Venom against Commercial Antivenoms. Int. J. Biol. Macromol. 2022;208:275–287. doi: 10.1016/j.ijbiomac.2022.03.095. [DOI] [PubMed] [Google Scholar]
- 52.Vanuopadath M., Raveendran D., Nair B.G., Nair S.S. Venomics and Antivenomics of Indian Spectacled Cobra (Naja naja) from the Western Ghats. Acta Trop. 2022;228:106324. doi: 10.1016/j.actatropica.2022.106324. [DOI] [PubMed] [Google Scholar]
- 53.Pawlak J., Mackessy S.P., Sixberry N.M., Stura E.A., Le Du M.H., Ménez R., Foo C.S., Ménez A., Nirthanan S., Kini R.M. Irditoxin, a Novel Covalently Linked Heterodimeric Three-Finger Toxin with High Taxon-Specific Neurotoxicity. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2009;23:534–545. doi: 10.1096/fj.08-113555. [DOI] [PubMed] [Google Scholar]
- 54.Osipov A.V., Kasheverov I.E., Makarova Y.V., Starkov V.G., Vorontsova O.V., Ziganshin R.K., Andreeva T.V., Serebryakova M.V., Benoit A., Hogg R.C., et al. Naturally Occurring Disulfide-Bound Dimers of Three-Fingered Toxins: A Paradigm for Biological Activity Diversification. J. Biol. Chem. 2008;283:14571–14580. doi: 10.1074/jbc.M802085200. [DOI] [PubMed] [Google Scholar]
- 55.Mackessy S.P. Handbook of Venoms and Toxins of Reptiles. 2nd ed. CRC Press; Boca Raton, FL, USA: 2021. [Google Scholar]
- 56.Utkin Y., Sunagar K., Jackson T.N.W., Reeks T., Fry B.G. Chapter 8: Three Finger Toxins (3FTx) In: Fry B.G., editor. Venomous Reptiles: Evolution, Pathophysiology and Biodiscovery. Oxford University Press; Oxford, UK: 2015. [Google Scholar]
- 57.Tasoulis T., Isbister G.K. A Review and Database of Snake Venom Proteomes. Toxins. 2017;9:290. doi: 10.3390/toxins9090290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fry B.G., Lumsden N.G., Wüster W., Wickramaratna J.C., Hodgson W.C., Manjunatha Kini R. Isolation of a Neurotoxin (α-Colubritoxin) from a Nonvenomous Colubrid: Evidence for Early Origin of Venom in Snakes. J. Mol. Evol. 2003;57:446–452. doi: 10.1007/s00239-003-2497-3. [DOI] [PubMed] [Google Scholar]
- 59.Sunagar K., Jackson T.N.W., Undheim E.A.B., Ali S.A., Antunes A., Fry B.G. Three-Fingered RAVERs: Rapid Accumulation of Variations in Exposed Residues of Snake Venom Toxins. Toxins. 2013;5:2172–2208. doi: 10.3390/toxins5112172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bekbossynova A., Zharylgap A., Filchakova O. Venom-Derived Neurotoxins Targeting Nicotinic Acetylcholine Receptors. Molecules. 2021;26:3373. doi: 10.3390/molecules26113373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Utkin Y.N., Kukhtina V.V., Kryukova E.V., Chiodini F., Bertrand D., Methfessel C., Tsetlin V.I. “Weak Toxin” from Naja kaouthia Is a Nontoxic Antagonist of Alpha 7 and Muscle-Type Nicotinic Acetylcholine Receptors. J. Biol. Chem. 2001;276:15810–15815. doi: 10.1074/jbc.M100788200. [DOI] [PubMed] [Google Scholar]
- 62.Carsi J.M., Potter L.T. M1-Toxin Isotoxins from the Green Mamba (Dendroaspis angusticeps) That Selectively Block M1 Muscarinic Receptors. Toxicon Off. J. Int. Soc. Toxinol. 2000;38:187–198. doi: 10.1016/S0041-0101(99)00141-5. [DOI] [PubMed] [Google Scholar]
- 63.Von Reumont B.M., Anderluh G., Antunes A., Ayvazyan N., Beis D., Caliskan F., Crnković A., Damm M., Dutertre S., Ellgaard L., et al. Modern Venomics—Current Insights, Novel Methods, and Future Perspectives in Biological and Applied Animal Venom Research. GigaScience. 2022;11:giac048. doi: 10.1093/GIGASCIENCE/GIAC048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Foo C.S., Jobichen C., Hassan-Puttaswamy V., Dekan Z., Tae H.-S., Bertrand D., Adams D.J., Alewood P.F., Sivaraman J., Nirthanan S., et al. Fulditoxin, Representing a New Class of Dimeric Snake Toxins, Defines Novel Pharmacology at Nicotinic ACh Receptors. Br. J. Pharmacol. 2020;177:1822–1840. doi: 10.1111/bph.14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Modahl C.M., Mrinalini, Frietze S., Mackessy S.P. Adaptive Evolution of Distinct Prey-Specific Toxin Genes in Rear-Fanged Snake Venom. Proc. Biol. Sci. 2018;285:20181003. doi: 10.1098/rspb.2018.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Melani R.D., Nogueira F.C.S., Domont G.B. It Is Time for Top-down Venomics. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017;23:44. doi: 10.1186/s40409-017-0135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walker A.A., Robinson S.D., Hamilton B.F., Undheim E.A.B., King G.F. Deadly Proteomes: A Practical Guide to Proteotranscriptomics of Animal Venoms. Proteomics. 2020;20:1900324. doi: 10.1002/pmic.201900324. [DOI] [PubMed] [Google Scholar]
- 68.Smith C.F., Nikolakis Z.L., Ivey K., Perry B.W., Schield D.R., Balchan N.R., Parker J., Hansen K.C., Saviola A.J., Castoe T.A., et al. Snakes on a plain: Biotic and abiotic factors determine venom compositional variation in a wide-ranging generalist rattlesnake. BMC Biol. 2023;21:136. doi: 10.1186/s12915-023-01626-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schulte L., Uhrig L., Eichberg J., Schwartze M., Auth I., Schulz M., Lindner T., Hien P., Hardes K., Vilcinskas A., et al. Comparative Venom Analysis between Melanistic and Normally-Colored Phenotypes of the Common Adder (Vipera berus) R. Soc. Open Sci. 2024;11:241268. doi: 10.1098/rsos.241268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Avella I., Damm M., Freitas I., Wüster W., Lucchini N., Zuazo Ó., Süssmuth R.D., Martínez-Freiría F. One Size Fits All—Venomics of the Iberian Adder (Vipera Seoanei, Lataste 1878) Reveals Low Levels of Venom Variation across Its Distributional Range. Toxins. 2023;15:371. doi: 10.3390/toxins15060371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Silva R.J.D., Fontes M.R.M., Rodrigues R.R., Bruder E.M., Stein M.F.B., Sipoli G.P.M., Pinhão R., Lopes C.A.D.M. A report on a case of melanism in a specimen of Crotalus durissus terrificus (Laurenti, 1768) J. Venom. Anim. Toxins. 1999;5:91–97. doi: 10.1590/S0104-79301999000100008. [DOI] [Google Scholar]
- 72.Schwick G., Dickgiesser F. Probleme Der Antigen- Und Fermentanalyse Im Zusammenhang Mit Der Herstellung Polyvalenter Schlangenseren. In: Behringwerke A.G., editor. Giftschlangen der Erde. Elwert Universitäts- und Verlags-Buchhandlung; Marburg, Germany: 1963. p. 34. [Google Scholar]
- 73.Ross E.A., Devitt A., Johnson J.R. Macrophages: The Good, the Bad, and the Gluttony. Front. Immunol. 2021;12:708186. doi: 10.3389/fimmu.2021.708186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maruyama K., Asai J., Ii M., Thorne T., Losordo D.W., D’Amore P.A. Decreased Macrophage Number and Activation Lead to Reduced Lymphatic Vessel Formation and Contribute to Impaired Diabetic Wound Healing. Am. J. Pathol. 2007;170:1178–1191. doi: 10.2353/ajpath.2007.060018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Esmaeilishirazifard E., Usher L., Trim C., Denise H., Sangal V., Tyson G.H., Barlow A., Redway K.F., Taylor J.D., Kremyda-Vlachou M., et al. Bacterial Adaptation to Venom in Snakes and Arachnida. Microbiol. Spectr. 2022;10:e0240821. doi: 10.1128/spectrum.02408-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ul-Hasan S., Rodríguez-Román E., Reitzel A.M., Adams R.M.M., Herzig V., Nobile C.J., Saviola A.J., Trim S.A., Stiers E.E., Moschos S.A., et al. The Emerging Field of Venom-Microbiomics for Exploring Venom as a Microenvironment, and the Corresponding Initiative for Venom Associated Microbes and Parasites (iVAMP) Toxicon X. 2019;4:100016. doi: 10.1016/j.toxcx.2019.100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bartlett K.E., Hall S.R., Rasmussen S.A., Crittenden E., Dawson C.A., Albulescu L.-O., Laprade W., Harrison R.A., Saviola A.J., Modahl C.M., et al. Dermonecrosis Caused by a Spitting Cobra Snakebite Results from Toxin Potentiation and Is Prevented by the Repurposed Drug Varespladib. Proc. Natl. Acad. Sci. USA. 2024;121:e2315597121. doi: 10.1073/pnas.2315597121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bin Haidar H., Almeida J.R., Williams J., Guo B., Bigot A., Senthilkumaran S., Vaiyapuri S., Patel K. Differential Effects of the Venoms of Russell’s Viper and Indian Cobra on Human Myoblasts. Sci. Rep. 2024;14:3184. doi: 10.1038/s41598-024-53366-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paas A., Dresler J., Talmann L., Vilcinskas A., Lüddecke T. Venom Ex Machina? Exploring the Potential of Cell-Free Protein Production for Venom Biodiscovery. Int. J. Mol. Sci. 2024;25:8286. doi: 10.3390/ijms25158286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hurka S., Brinkrolf K., Özbek R., Förster F., Billion A., Heep J., Timm T., Lochnit G., Vilcinskas A., Lüddecke T. Venomics of the Central European Myrmicine Ants Myrmica Rubra and Myrmica Ruginodis. Toxins. 2022;14:358. doi: 10.3390/toxins14050358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Avella I., Schulte L., Hurka S., Damm M., Eichberg J., Schiffmann S., Henke M., Timm T., Lochnit G., Hardes K., et al. Proteogenomics-Guided Functional Venomics Resolves the Toxin Arsenal and Activity of Deinagkistrodon Acutus Venom. Int J. Biol. Macromol. 2024;278:135041. doi: 10.1016/j.ijbiomac.2024.135041. [DOI] [PubMed] [Google Scholar]
- 82.Hurka S., Lüddecke T., Paas A., Dersch L., Schulte L., Eichberg J., Hardes K., Brinkrolf K., Vilcinskas A. Bioactivity Profiling of In Silico Predicted Linear Toxins from the Ants Myrmica Rubra and Myrmica Ruginodis. Toxins. 2022;14:846. doi: 10.3390/toxins14120846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Erkoc P., von Reumont B.M., Lüddecke T., Henke M., Ulshöfer T., Vilcinskas A., Fürst R., Schiffmann S. The Pharmacological Potential of Novel Melittin Variants from the Honeybee and Solitary Bees against Inflammation and Cancer. Toxins. 2022;14:818. doi: 10.3390/toxins14120818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Erkoc P., Schiffmann S., Ulshöfer T., Henke M., Marner M., Krämer J., Predel R., Schäberle T.F., Hurka S., Dersch L., et al. Determining the Pharmacological Potential and Biological Role of Linear Pseudoscorpion Toxins via Functional Profiling. iScience. 2024;27:110209. doi: 10.1016/j.isci.2024.110209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schulte L., Damm M., Avella I., Uhrig L., Erkoc P., Schiffmann S., Fürst R., Timm T., Lochnit G., Vilcinskas A., et al. Venomics of the Milos Viper (Macrovipera Schweizeri) Unveils Patterns of Venom Composition and Exochemistry across Blunt-Nosed Viper Venoms. Front. Mol. Biosci. 2023;10:1254058. doi: 10.3389/fmolb.2023.1254058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomic raw data files have been uploaded to the open access PRIDE database (PXD03696). All identified venom components are described in Table S1. Crude venom samples are available from the corresponding author upon reasonable request.