Abstract
Carriers of certain human leukocyte antigen class I alleles show favorable prognosis of human immunodeficiency virus type 1 (HIV-1) infection, presumably due to effective CD8+ cytotoxic T-lymphocyte responses, but close relationships between class I variants mediating such responses to natural and to vaccine HIV-1 antigen have not been established. During 6 to 30 months of administration and follow-up in trials of ALVAC-HIV recombinant canarypox vaccines, cells from 42% of 291 HIV-1-negative vaccinated subjects typed at class I loci responded to an HIV-1 protein in a lytic bulk CD8+ cytotoxic T-lymphocyte assay. By 2 weeks after the second dose, higher proportions of vaccinees carrying one of two alleles consistently associated with slower progression of natural HIV-1 infection reacted at least once: B∗27 carriers reacted to Gag (64%; odds ratio [OR] = 10.3, P = 0.001) and Env (36%; OR = 4.6, P = 0.04), and B∗57 carriers reacted to Env (44%; OR = 6.6, P < 0.05). By 2 weeks after the third or fourth dose, B∗27 carriers had responded (two or more reactions) to Gag (33%; OR = 4.4, P < 0.05) and B∗57 carriers had responded to both Gag (39%; OR = 5.3, P = 0.013) and Env (39%; OR = 9.5, P = 0.002). Homozygosity at class I loci, although conferring an unfavorable prognosis following natural infection, showed no such disadvantage for vaccine response. Individual class I alleles have not previously demonstrated such clear and consistent relationship with both the clinical course of an infection and cellular immunity to a vaccine against the infectious agent. This proof of principle that class I an alleles modulate both processes has implications for development of HIV-1 and presumably other vaccines.
Cellular immune response systems, including those encoded by genes in the human leukocyte antigen (HLA) complex, influence the wide variation in outcome of human immunodeficiency virus type 1 (HIV-1) infection (8, 22, 26, 29, 30, 37, 52). Certain class I alleles have been consistently associated with slower (B∗27, B∗57) or faster (B∗35, B∗08) disease progression (8, 22, 26, 29, 30, 37), and diminished diversity (i.e., homozygosity) at class I loci confers substantial risk of accelerated infection (8, 30, 52). The mechanisms by which class I alleles differentially bind peptides, restrict the generation of CD8+ cytotoxic T lymphocytes (CTLs), and govern the clinical response to HIV-1 are under intense investigation (14b). Monkeys deficient in CD8+ T cells lack efficient control of viral replication and produce a strong CTL response to an effective vaccine (2, 24, 41, 50). In HIV-1-positive humans, CTLs presumably destroy infected cells (7, 12, 18, 33, 34, 46) by targeting HIV-1 peptides bound to surface-expressed class I molecules, establishing a dynamic equilibrium between a continuously mutating virus population and host-specific engagement with evolving virus.
In HIV-1-exposed but persistently seronegative individuals, the importance of observed CTL responses in protecting against infection is less clear (45, 49). The collective evidence that control of initial viral acquisition is regulated by specific HLA class I variants is also less convincing than for disease progression (5, 28, 36, 47).
Mechanisms of induced immunity to infection and natural immunity to disease progression almost surely differ. Ample CTL reactivity may be essential for protection by an HIV vaccine. Vaccine-generated CTL response mediates protection against simian immunodeficiency virus (1, 25, 51), but CTL-based efficacy has not yet been demonstrated for any human vaccine (43). Vaccines that control established infection may be less protective against initial virus acquisition.
Restriction of responses by polymorphic class I and/or class II HLA gene products has been suggested with vaccines against other viral pathogens (4, 21, 35), and early trials of HIV-vaccinia virus (16) hinted at such regulation. The unequivocal influence of class I polymorphism on the natural history of HIV infection compels evaluation of how this genetic variability may modulate HIV-1 vaccine response. Here we have observed HIV-1-specific CD8+ CTLs in significantly higher proportions of HIV-1-uninfected HIV vaccine recipients (3, 10, 13, 14, 39, 43) who carried class I alleles most consistently recognized as advantageous in infected individuals. To our knowledge, such a direct relationship has not previously been documented in humans for any infectious agent.
(This work was presented in part at the 7th Conference on Retroviruses and Opportunistic Infections, 29 January to 3 February 2000, San Francisco, Calif.)
MATERIALS AND METHODS
Subjects.
In compliance with federal guidelines and institutional review board policies, 291 volunteers received vaccine in four National Institute of Allergy and Infectious Diseases-sponsored AIDS Vaccine Evaluation Group (AVEG) randomized, double-blind trials summarized in Table 1 and below (3, 13, 14; L. Corey et al., Abstr. 12th Int. Conf. AIDS, abstr. 636, 1998; T. Evans et al., Abstr. 12th Int. Conf. AIDS, abstr. 277, 1998). The AVEG data coordinating center (The EMMES Corp, Potomac, Md.) provided epidemiologic, clinical, and laboratory data at enrollment and subsequent trial protocol and adherence data.
TABLE 1.
Schedule of vaccine administration and assays for CTL response
| Protocol | Schedule at wka:
|
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 12 | 13 | 14 | 22 | 24 | 26 | 33 | 39 | 41 | 52 | 54 | 67 | 78 | 104 | 130 | |
| 022 | V | V | c | (V) | c | c | V | c | (V) | c | (V) | c | c | c | c | c | ||||||||
| 022A | V | V | c | c | c | c | (V) | c | V | c | c | (V) | c | (V) | c | c | c | c | ||||||
| 026 | V | V | c | (V) | c | V | c | (V) | c | c | c | c | ||||||||||||
| 029 | V | V | V, c | V, c | c | c | c | c | ||||||||||||||||
Weeks following enrollment according to protocols for four studies of ALVAC-HIV recombinant canarypox vaccine. V, ALVAC-HIV canarypox vaccine was given; (V), in certain arms of protocols 022, 022A, and 026, vaccination was not scheduled; c, CD8+ CTL lytic assay was scheduled.
Vaccine constructs and administration schedule.
Protocols 022, 022A, and 029 (3, 13; L. Corey, unpublished data; T. Evans, unpublished data) tested the recombinant canarypox ALVAC-HIV vCP205 vector (Aventis Pasteur, Inc., Marcy l'Etoile, France) encoding HIV-1 proteins gp120MN, the membrane-spanning domain of gp41LAI, GagLAI, and proteaseLAI. Protocol 026 utilized the similar ALVAC-HIV vCP300 vector (Aventis Pasteur, Inc.) encoding the HIV-1 proteins found in vCP205 plus CTL epitope-rich regions from nefBRU and polLAI. Certain individuals were given boosters of recombinant HIV-1 envelope glycoprotein subunit rgp120SF2 (Chiron Vaccines, Inc., Emeryville, Calif.). Protocols 022, 022A, and 026 scheduled three or four immunizations within the first 6 months with vCP205 (protocols 022 and 022A) or vCP300 (protocol 026) alone or in combination with rgp120SF2 (Table 1). Protocol 029 scheduled four immunizations within the first 21 days with vCP205 and a booster at days 28 and 84 with rgp120SF2.
CTL assays.
Schedules for the lytic assay for four viral proteins (Gag, Pol, Env, and Nef) according to standard AVEG methods (13, 14, 39) differed by protocol (Table 1). Individual recipient B lymphoblastoid cell lines (BLCLs) served as targets. We stimulated peripheral blood mononuclear cells (PBMCs) with recombinant HIV-1–vaccinia virus-infected autologous PBMCs cultured in RPMI 1640 (GIBCO BRL, Grand Island, N.Y.) with human antisera and interleukin 7 (R&D Systems, Minneapolis, Minn.). On day 7, we incubated cells with interleukin 2 (Amgen, Thousand Oaks, Calif.). On day 14 we assayed cytotoxicity using Na2CrO4-labeled BLCLs infected with vaccinia virus control (vP1170) or vaccinia virus recombinants expressing HIV-1 MN gp160 (vP1174), HIV-1 HXB2 Gag (vDK1), or HIV-1 GagLAI (vP1287). Vaccinia virus vP1288-IIIB or vP1218-MN was used for Pol or Nef BLCL targets, respectively. Virogenetics (Troy, N.Y.) provided all vectors except vDK1 (D. Kuritzkes, University of Colorado Health Sciences Center). Inclusion of unlabeled infected targets in excess reduced vaccinia virus background. Generally we tested effector-to-target ratios of 25:1 and 50:1 in triplicate with 5,000 targets/well. HIV-1-specific lysis of ≥10% over controls defined a positive result. A >50% loss of HIV-1-specific activity following selective depletion by anti-CD8-bearing magnetic beads (Dynabeads; Dynal, Great Neck, N.Y.) represented CD8+-cell-specific lysis. Positivity required ≥10% specific lysis for at least two separate effector-to-target ratios.
HLA typing.
We identified HLA-A, -B, and -C alleles in DNA extracted from banked PBMCs with the QIAmp DNA mini-kit (Qiagen Inc., Valencia, Calif.). Typing was performed with sequence-based typing and sequence-specific primer (SSP) methods, which resolved most of the ambiguous and apparent homozygous alleles. For nested sequence-based typing, we performed a primary amplification encompassing exons 2 and 3 of each locus (9, 53). Sequencing of their secondary amplification products employed either the AutoLoad kit or the Sequenase v.2 kit on an ALFexpress automated DNA sequencer (Amersham-Pharmacia Biotech, Inc., Piscataway, N.J.) or M-280 streptavidin Dynabeads (Dynal Inc., Lake Success, N.Y.). HLA SequiTyper v.2.0 software (Pharmacia) assigned alleles. For SSP typing, class I Unitray ABC SSP kits (Pel-Freez Clinical Systems, Brown Deer, Wis.) were used.
The typing scheme achieved resolution to four digits at the A and B loci and to two or four digits at the C locus in the current HLA nomenclature (6). Full resolution was achieved for all but 3% of A and B alleles, which failed to amplify in three attempts or remained ambiguous after both techniques were applied. We defined homozygosity as identity at the four-digit level and assigned Bw4 specificity according to standard nomenclature (38).
Statistical analysis.
We defined a meaningful CTL response using two different criteria, either one or two positive measurements during the study interval, and calculated proportions of individuals with a CTL response using each criterion. For successive intervals following vaccination responses were examined separately for each viral protein (Gag, Pol, Env, and Nef). Data on “Any,” Gag, and Env responses are presented; the sparseness of Pol and Nef data precluded meaningful analysis. “Any” refers to aggregate analysis of one or more proteins in any combination. Analyses requiring presence of both Gag and Env or requiring exclusively Gag or Env did not add enough information about differential HLA effects to present separately.
Analyses for protocols 022, 022A, and 026 focused on cumulative CTL response during three intervals: (i) from initial dose to 2 weeks after the second dose (criterion = one positive measurement), (ii) from initial dose to 2 weeks after the third or fourth dose (criterion = two positives), and (iii) from 2 to 76 weeks after last dose (criterion = two positives). These corresponded, respectively, to the 6-week, 26-week, and 2.5-year intervals from first inoculation. For protocol 029 we considered CTL measurements by 2 weeks after the fourth consecutive weekly dose (6 weeks after the first inoculation) along with the other three protocols at both 6 and 26 weeks. However, because protocol 029 subjects had also received 4 doses by week 6, analysis of their CTL measurements began with the post-third- or -fourth-dose, 26-week interval. Results for CTLs during the 26-week interval (criterion = two positives) are presented in greater detail; cumulative effects during shorter and longer intervals were quite consistent with those shown.
The four protocols differed in the scheduled numbers of CTL measurements for each individual (Table 1), but the number of CTL responses closely paralleled the number of measurements, i.e., measurement and response frequencies were closely proportional. We formally excluded this potential protocol bias. We also analyzed each putative association with an allele in the presence and again in the absence of any other that might have explained the finding.
Although typing resolved alleles to four digits in nearly all cases, the majority represented the principal allele in the two-digit serogroup. Relatively few four-digit alleles occurred at high enough frequency for separate statistical analysis, but when possible, we searched for such separate effects. Since no effect seen using aggregated two-digit allele designation was better captured by four-digit stratification, we have presented only data on aggregated alleles.
Differences in allele frequencies and CTL responses by age, gender, or race, by vaccine study site, or by protocol were not large enough to require adjustment. About half of protocol 022A participants were at somewhat higher risk of exposure to HIV-1 prior to vaccination and could have been selectively primed for a CTL response; however, there was a negligible difference in response between the subjects in the two risk categories, the HLA allele frequencies of the two groups were comparable, and differential exposure was not a factor in the HLA relationships.
All significance testing was based on Fisher's exact test. We have reported unadjusted P values because this was not an exploratory search but a test of involvement of four HLA alleles for which there is ample a priori evidence.
RESULTS
Study population characteristics and overall response.
Vaccine recipients consisted primarily of Caucasians (88%) and males (68%); the mean (standard deviation) age was 34.3 (9.9), with a range of 18 to 60 years. The distributions of class I allele and phenotype frequencies in the Caucasian vaccinees resembled those in other North American populations (Table 2). No significant deviation from Hardy-Weinberg equilibrium was observed for Caucasians or the small number of African-Americans. HLA phenotype distributions and proportions of homozygotes at each locus were comparable among the four protocols (data not shown).
TABLE 2.
Number and proportion of ALVAC HIV vaccine recipients with HLA-A, -B, and -C alleles
| No. of chromosomesa | Alleleb | Frequency | %c |
|---|---|---|---|
| 546 | A∗01 | 73 | 13.3 |
| A∗0101 | 55 | ||
| A∗02 | 157 | 28.8 | |
| A∗0201 | 129 | ||
| A∗03 | 76 | 13.9 | |
| A∗0301 | 69 | ||
| A∗11 | 27 | 4.9 | |
| A∗1101 | 14 | ||
| A∗2301 | 17 | 3.1 | |
| A∗24 | 54 | 10.0 | |
| A∗2402 | 46 | ||
| A∗2501 | 9 | 1.6 | |
| A∗26 | 14 | 2.6 | |
| A∗2601 | 11 | ||
| A∗29 | 21 | 3.9 | |
| A∗2902 | 17 | ||
| A∗30 | 14 | 2.6 | |
| A∗31 | 15 | 2.7 | |
| A∗3101 | 13 | ||
| A∗32 | 20 | 3.7 | |
| A∗3201 | 19 | ||
| A∗3301 | 5 | 1.4 | |
| A∗3402 | 1 | 0.2 | |
| A∗3601 | 12 | 2.2 | |
| A∗4301 | 1 | 0.2 | |
| A∗6601 | 3 | 0.5 | |
| A∗68 | 24 | 4.4 | |
| A∗6801 | 13 | ||
| A∗6901 | 2 | 0.4 | |
| A∗74 | 2 | 0.4 | |
| 570 | B∗07 | 80 | 14.0 |
| B∗0702 | 75 | ||
| B∗08 | 64 | 11.2 | |
| B∗0801 | 62 | ||
| B∗13 | 8 | 1.4 | |
| B∗14 | 16 | 2.7 | |
| B∗1402 | 10 | ||
| B∗15 | 43 | 7.5 | |
| B∗1501 | 28 | ||
| B∗18 | 23 | 4.0 | |
| B∗1801 | 20 | ||
| B∗27 | 24 | 4.2 | |
| B∗2705 | 11 | ||
| B∗35 | 38 | 6.7 | |
| B∗3501 | 18 | ||
| B∗37 | 9 | 1.6 | |
| B∗38 | 6 | 1.1 | |
| B∗39 | 15 | 2.6 | |
| B∗3901 | 10 | ||
| B∗40 | 35 | 6.1 | |
| B∗4001 | 31 | ||
| B∗41 | 5 | 0.8 | |
| B∗42 | 6 | 1.1 | |
| B∗44 | 71 | 12.5 | |
| B∗4402 | 16 | ||
| B∗4403 | 19 | ||
| B∗4405 | 11 | ||
| B∗45 | 13 | 2.3 | |
| B∗46 | 1 | 0.2 | |
| B∗47 | 2 | 0.4 | |
| B∗48 | 2 | 0.4 | |
| B∗4901 | 13 | 2.3 | |
| B∗50 | 6 | 1.1 | |
| B∗51 | 33 | 5.8 | |
| B∗5101 | 29 | ||
| B∗5201 | 5 | 0.9 | |
| B∗5301 | 10 | 1.8 | |
| B∗55 | 10 | 1.8 | |
| B∗5601 | 2 | 0.4 | |
| B∗5701 | 19 | 3.3 | |
| B∗58 | 6 | 1.1 | |
| B∗8101 | 1 | 0.2 | |
| 572 | Cw∗01 | 24 | 4.2 |
| Cw∗02 | 21 | 3.7 | |
| Cw∗03 | 68 | 11.8 | |
| Cw∗04 | 62 | 10.8 | |
| Cw∗05 | 46 | 8.0 | |
| Cw∗06 | 59 | 10.3 | |
| Cw∗07 | 192 | 33.6 | |
| Cw∗08 | 19 | 3.3 | |
| Cw∗12 | 22 | 3.8 | |
| Cw∗14 | 8 | 1.4 | |
| Cw∗15 | 15 | 2.6 | |
| Cw∗16 | 26 | 4.5 | |
| Cw∗17 | 9 | 1.6 | |
| Cw∗18 | 1 | 0.2 |
The number of chromosomes is double the number of subjects with typing data (n = 273 at A, 285 at B, and 286 at C).
Alleles were typed to high resolution for A and B loci and to medium or high resolution for the C locus; frequencies of four digit A and B subtypes are shown for subjects with no other subtype or those carried by ≥10 subjects
Percent of all alleles that the indicated allele represents.
Among 291 vaccine recipients with CD8+ CTL lytic assay data, a CD8+ CTL response to Any, Gag, and Env occurred in 12, 19, and 11% (one positive assay) by 2 weeks after the first dose of vaccine, in 11, 13, and 9% (two positive assays) by 2 weeks after the third or fourth dose, and in 27, 24, and 13% (two positive assays) during the entire 2.5-year follow-up.
HLA-A alleles.
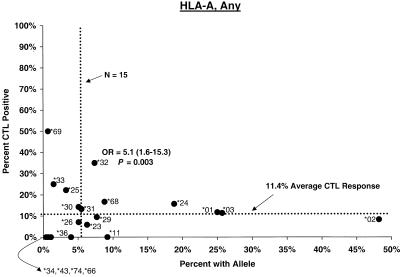
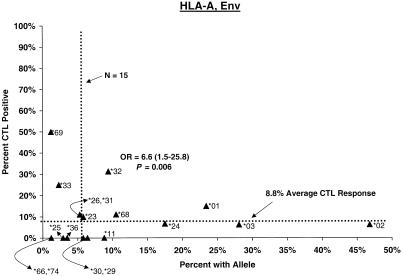
Compared with individuals carrying other A alleles, a significantly higher proportion of A∗32 carriers showed CTL responses (Any, 35%; odds ratio [OR] = 5.1, P = 0.003; Env, 31%; OR = 6.6, P = 0.006) (Fig. 1). The A∗32 effect on response to Env was already apparent by 2 weeks after the second dose as well (OR = 5.3, P < 0.05), and it remained strong throughout the postvaccination period, even when vaccinees with B∗27 and B∗57 were excluded (see below). A∗32 has been associated with relatively slow disease progression in two related population studies (26, 30), and a trend toward protection was seen in a small group of transfusion recipients with long-standing infection (15). For A∗0201, the most common allele and one prominently featured in experimental protocols, during the interval through the third or fourth dose, positive CTL responses occurred in somewhat lower proportions (Any and Gag) than for other A∗02 or other A allele carriers (data not shown).
FIG. 1.
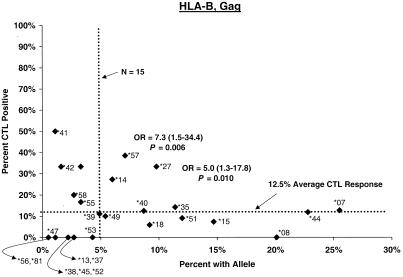
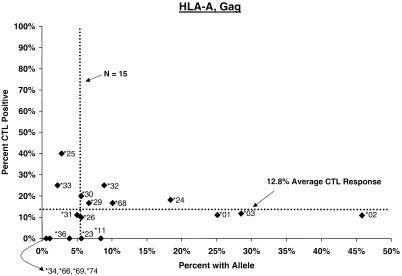
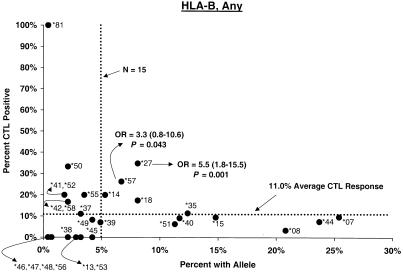
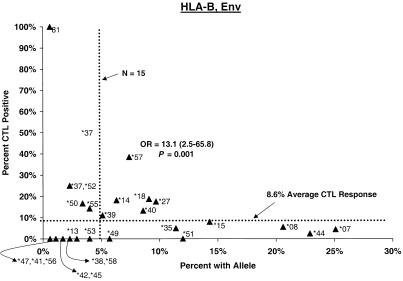
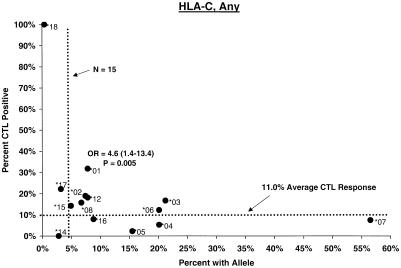
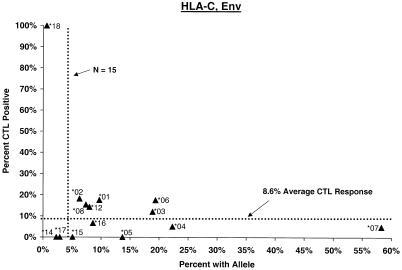
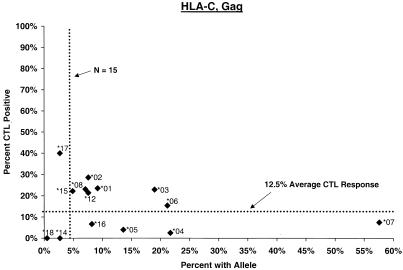
Proportion of vaccinees carrying HLA class I allele with CTL response. Class I allele phenotype in ALVAC-HIV recombinant canarypox vaccine recipients and cumulative proportions with bulk CD8+ CTL responses to major HIV protein components by 2 weeks after the third or fourth dose (26 weeks after enrollment) are shown. The dotted line labeled “N = 15” signifies the threshold below which proportions with CTL are less meaningful; the horizontal dotted line signifies the average proportion with a CTL response to the HIV protein for all alleles at the locus.
HLA-B alleles.
Distributions of CTL responses in vaccine recipients with B alleles relatively consistently associated elsewhere (8, 11, 22, 23, 26, 27, 29, 30, 37, 40, 42, 52) with faster or slower HIV disease progression are summarized in Table 3. In both the post-second-dose and the post-third- or -fourth-dose intervals, vaccine recipients carrying B∗27 or B∗57, the two alleles most widely observed to retard disease progression (11, 22, 30, 37, 40, 42), differed significantly from those with other alleles in the cumulative proportion showing CTL positivity (Fig. 1). Proportions of B∗27 and B∗57 carrier vaccinees who responded to Any, Gag, and Env during the second interval were 35, 33, and 18% and 26, 39, and 39%, respectively. For B∗27 the differences were clear during the earliest interval for Any (OR = 5.4, P = 0.002), Gag (OR = 10.3, P = 0.001), and Env (OR = 4.6, P = 0.04); big differences persisted through the second interval for Any (OR = 5.5, P = 0.001) and Gag (OR = 5.0, P = 0.01) but not for Env. The Gag effect was also evident for the entire 2.5-year study period. For B∗57, a higher response frequency was seen early for Env (OR = 6.6, P = 0.018) and became stronger after the third or fourth dose for Any (OR = 3.3, P = 0.043), Gag (OR = 5.3, P = 0.013), and Env (OR = 9.5, P < 0.002). During this 26-week interval, the CTL assay was positive at least once in 72% of B∗57 carriers (OR = 7.9, P < 5 × 10−4). The effects of these two individual alleles persisted in analyses excluding volunteers with the B∗27/B∗57 genotype or other apparently protective alleles.
TABLE 3.
Association of HLA class I alleles with CD8+ CTL response to ALVAC-HIV canarypox vaccinea
| Allele consistently associated with altered HIV disease progressionb | HIV protein | 6 wksc
|
26 wksd
|
2.5 yrse
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. (%) of persons with CTL response
|
ORf | No. (%) of persons with CTL response
|
ORf | No. (%) of persons with CTL response
|
ORf | |||||
| With allele | Without allele | With allele | Without allele | With allele | Without allele | |||||
| B∗08 (F) | Any | 9 (16) | 27 (12) | 1.3 | 2 (3) | 29 (13) | 0.2g | 15 (25) | 60 (27) | 0.9 |
| Gag | 4 (13) | 20 (21) | 0.6 | 0 (0) | 23 (16) | 0.0h | 8 (15) | 51 (25) | 0.5 | |
| Env | 6 (19) | 11 (11) | 1.9 | 2 (6) | 13 (9) | 0.6 | 7 (13) | 28 (14) | 0.9 | |
| B∗27 (S) | Any | 8 (40) | 28 (11) | 5.4i | 8 (35) | 23 (9) | 5.5i | 14 (61) | 61 (23) | 5.2i |
| Gag | 7 (64) | 17 (15) | 10.3i | 6 (33) | 17 (10) | 4.4g | 12 (55) | 47 (20) | 4.9i | |
| Env | 4 (36) | 13 (11) | 4.6g | 3 (18) | 12 (8) | 2.6 | 4 (18) | 31 (13) | 1.5 | |
| B∗35 (F) | Any | 3 (9) | 33 (14) | 0.6 | 4 (11) | 27 (11) | 1.1 | 8 (22) | 67 (27) | 0.8 |
| Gag | 2 (13) | 22 (19) | 0.6 | 3 (14) | 20 (12) | 1.2 | 6 (18) | 53 (23) | 0.7 | |
| Env | 1 (7) | 16 (14) | 0.4 | 1 (5) | 14 (9) | 0.5 | 3 (9) | 32 (14) | 0.6 | |
| B∗57 (S) | Any | 4 (21) | 32 (12) | 1.9 | 5 (26) | 26 (10) | 3.3g | 7 (37) | 68 (25) | 1.7 |
| Gag | 2 (20) | 22 (19) | 1.1 | 5 (39) | 18 (11) | 5.3g | 7 (39) | 52 (21) | 2.3 | |
| Env | 4 (44) | 13 (11) | 6.6g | 5 (39) | 10 (6) | 9.5i | 7 (41) | 28 (12) | 5.3i | |
Cumulative responses: one positive CTL measurement by 2 weeks after second dose and two positive CTL measurements by 2 weeks after third or fourth dose and by 1.5 years after third through sixth doses. The numbers of subjects with typing and CTL measurements varied by protocol and interval.
References 8, 11, 21, 22, 25, 26, 28, 29, 35, 38, 40, and 50. Previous association with progression: F, faster; S, slower.
Two weeks after second dose.
Two weeks after third or fourth dose (postvaccination interval relative to third or fourth dose in protocols 022, 022A, and 026 and relative to fourth dose in protocol 029; see Table 1 and the text).
Two to 78 weeks after third through sixth doses.
Associations remain comparably significant when subjects carrying additional alleles associated with increased CTL are excluded from analysis and whether subjects in protocol 029 are included in the analysis of response 2 weeks after the second dose.
P < 0.05.
P < 0.01.
P < 0.05.
Of the B alleles most consistently associated with rapid progression (8, 22, 23, 26, 27, 29, 30, 37), no B∗08-positive vaccinee met the criterion for response to Gag during the second postconversion interval (OR, indeterminate; P = 0.006), and the proportion of B∗08-positive subjects responding during the other intervals was 40 to 50% lower than for other B alleles (Table 3). For B∗35 there were nonsignificant 30 to 50% reductions in proportions responding at the different intervals. With the CTL assay performed on bulk populations of PBMCs rather than allele-specific CTL clones, reduced responses in individuals with those two alleles could have been masked by a compensatory contribution from companion A or B alleles mediating an increased response; however, we found no indication of such compensation (data not shown).
Higher proportions of volunteers carrying certain B alleles (B∗1501 and B∗4901) made more transient CTL responses, measured either as a single positive assay or during a single interval. These relationships do not correspond to known effects of these markers in natural history studies. In the case of B∗14, carriers occasionally found to have more favorable outcomes (22, 37) showed a trend here toward increased response during the interval following the third or fourth dose (ORs = 2.1 to 2.9; P = 0.14 to 0.24). Although tightly linked with Cw∗08, the B∗14 allele shows a slightly stronger effect. Various other alleles that have been reported less consistently in association with disease progression did not show corresponding relationships with CTL response here.
A- and B-locus alleles displaying the “public” antigen encoded by the Bw4 sequence (38) have been associated with favorable prognosis in natural infection (14a, 26, 29). Here, certain of those Bw4 alleles showed a weak association with prominent CTL response to Gag, but this trend did not persist when the three Bw4-bearing alleles (A∗32, B∗27, and B∗57) previously found to be protective were excluded from analysis.
HLA-C alleles.
No C allele by itself appeared to be associated with significantly stronger or weaker CTL response. A significantly increased frequency of response in Cw∗01 carriers (OR = 4.6, P = 0.005) (Fig. 1) was readily explained by its strong disequilbrium with B∗27. Similarly, although the proportion of Cw∗07-positive subjects who mounted a CTL response to Gag and Env was low (ORs = 0.3; P = 0.02 to 0.06), the effect of the allele could not be separated from that of a Cw∗07-bearing haplotype (see below).
Haplotypes.
None of the associations observed with apparent two-locus (A-C and B-C) haplotypes surpassed those of their component A or B alleles alone in magnitude. For the apparent three-locus haplotype A∗02-Cw∗07-B∗07, during the second and third intervals, lower proportions of the haplotype carriers mounted CTL responses to both major proteins (ORs = 0.3 to 0.8, P = 0.08 to 0.10). For individual carriers of A∗02, Cw∗07, or B∗07 alone, proportions with CTL responses were less diminished.
Homozygosity.
Identity of alleles at the A, B, or C locus to the four-digit level did not reduce the cumulative proportion of vaccinees showing a CTL response to Any, Gag, or Env in the 6-week, the 26-week, or the 2.5-year interval; the data are shown for the 26-week interval (Table 4). The six vaccinees who were homozygous at the C locus and who were positive for CTL against Env at 6 weeks actually experienced a somewhat higher response frequency, and two carried specific A or B markers shown to account for that higher frequency. Allele identity at two loci (six to eight subjects) and three loci (four subjects) was likewise not associated with relative unresponsiveness.
TABLE 4.
Association of homozygosity at HLA class I loci with CD8+ CTL response to ALVAC-HIV canarypox vaccinea
| Locusb | HIV proteinc | No. (%) of persons at 2 wks after 3rd or 4th dose (wk 26)
|
ORd | |
|---|---|---|---|---|
| Homozygous | Heterozygous | |||
| HLA-A | Any | 3 (9) | 31 (13) | 0.7 |
| Gag | 3 (16) | 16 (15) | 1.3 | |
| Env | 2 (10) | 18 (17) | 1.2 | |
| HLA-B | Any | 3 (12) | 22 (10) | 1.1 |
| Gag | 3 (18) | 14 (13) | 1.6 | |
| Env | 1 (6) | 16 (14) | 0.6 | |
See text for typing details; a total of 273 and 285 were unambiguously typed to the four-digit level at the A and B loci, respectively.
Vaccine-specific viral protein against which CTL response was measured.
All P values were >0.15.
Effect of sexual exposure.
Among volunteers in protocol 022A, low-risk sexual exposure was reported by 60 (50.8%) and higher-risk exposure was reported by 58 (49.2%). Those at higher risk showed no appreciable difference in early CTL response. Cumulatively through the post-third- or -fourth-dose interval, very similar proportions of the 20 to 28 low-risk participants and the 18 to 28 higher-risk participants had produced CTL responses to Any, Gag, or Env viral proteins. None of the class I relationships described here showed a meaningful change when adjusted for exposure level (data not shown).
DISCUSSION
By 26 weeks after at least three doses of canarypox-HIV vaccine, 9 to 13% of recipients had mounted a meaningful response, as indicated by two positive CD8+ CTL lytic assays against Gag, Env, Pol, or Nef. The proportions responding to Gag or Env were significantly higher among those carrying HLA-B∗27 or -B∗57, the two B alleles repeatedly associated (11, 22, 37, 40, 42) with a more benign course of natural HIV-1 infection. For B∗57, the protection against disease progression has been persuasively documented not only in homosexual clade B virus-infected Caucasians (primarily B∗5701) (22, 37, 42) but also in heterosexual clade A virus-infected Africans (primarily B∗5703) (11). The favorable effect on CTL response for both alleles was apparent even after the second dose of vaccine (i.e., at 6 weeks) but was particularly strong among B∗27 carriers for both Gag and Env at this earlier time point. Although this distinction with B∗27 is based on small numbers, it invites speculation about whether that allele differs fundamentally in the breadth or specificity of its peptide binding repertoire (31) or to a rapid response (54).
Although a remarkable coincidence could explain these identical class I allele-specific associations with both vaccine response and outcome of natural infection, they more likely reflect the particular capacity of these HLA molecules to bind conserved HIV-1 peptides, transporting them to the cell surface or displaying them effectively to the T-cell receptor. For the two protective HLA-B alleles, dominant epitopes and other features favorable for CTL response have been well documented (17, 19, 31, 42), and predicted peptide binding motifs in clade B HIV-1 may be especially plentiful (44). Whatever the precise molecular explanation, the generalizability of the advantage for B∗57 across race, clade, and mode of transmission and the consistency of that advantage for response to both natural and vaccine-formulated HIV-1 antigens warrant close investigation.
As for the two class alleles previously observed in conjunction with poorer prognosis, B∗08 showed a lower CTL response to vaccine Gag and B∗35 showed a lower response to Env in all three intervals. However, neither of these reductions in response was as striking as the augmentation associated with the protective alleles. If that pattern represents a discrepancy between natural and vaccine-induced responses, several explanations are possible. First, the B∗08 association with rapid disease progression has not remained as consistent as for B∗35 in recent analyses (R. A. Kaslow, unpublished data). Second, compared with Gag and Env antigens produced by naturally replicating, mutating HIV-1 isolates, cloned HIV-1 genes in the tested vaccines may have expressed peptide motifs relatively more favorable for binding to or presentation by B∗08 or B∗35. Third, in contrast to the uniform findings thus far for different subtypes of B∗57, some B∗35 allele subtypes appear to mediate effective responses, and the lack of observed overall effect on CTL may reflect the heterogeneous influence of more effective and less effective subtypes intermingled (14b, 20, 48). Fourth, assessment of alleles accompanying B∗08 or B∗35 disclosed no masking of effects of these two by others mediating better responses to HIV canarypox vaccine motifs; however, compensation at the level of specific CD8+ CTL responses is still possible. The contrast between response to naturally and artificially presented peptides in the context of B∗35 deserves further evaluation.
A∗32, an allele inconsistently predisposing to slower disease progression (15, 30), merits further examination in light of its association with increased frequency of CTL response; it seems more difficult to justify further attention to the more transient associations of alleles not previously recognized as determinants (e.g., B∗1501 and B∗4901). Conversely, certain alleles occasionally associated with unfavorable or favorable prognosis elsewhere (e.g., A∗24) (8, 27, 30) did not show the predicted influence on CTL response. Nonspecific (i.e., bulk PBMC) response in the lytic assay and unstable estimation of ORs for less common alleles both here and in cohorts of infected subjects preclude any simple explanation for the discrepant impact of these class I alleles on natural and vaccine-induced responses.
No C-locus polymorphism showed a specific effect that was clearly independent of a similar or stronger effect from a B allele in linkage disequilibrium. Although certain associations of the C-locus or B-C haplotypes with rapid or slow HIV disease progression have been reported (8, 22, 30, 37), so far none of the effects of C alleles seem sufficiently consistent and independent of their linked B alleles to be regarded seriously in their own right.
In contrast to the clear negative effect of class I homozygosity on the course of natural infection (8, 30, 52), responsiveness to vaccine proteins was entirely indifferent to reduced diversity (i.e., homozygosity). The advantage of higher degrees of allelic diversity may be less important in generating a detectable CTL response to the single, unchanging set of peptides artificially assembled in a vaccine than in retarding escape by evolving virus from immune containment (17, 32). While apparent irrelevance of class I homozygosity for vaccine-induced CTL does not ensure its irrelevance in the context of a live-virus challenge, confirmation that class I homozygosity is not overtly disadvantageous may simplify vaccine development and application.
We have uncovered noteworthy similarities as well as differences between class I HLA-mediated responses to natural HIV-1 and to certain immunogenic HIV-1 components incorporated into the ALVAC-HIV canarypox vaccine. Future clinical trials should clarify whether these similarities and differences are generalizable to other HIV-1 vaccine formulations. Beyond HIV-1 vaccines, dissection of the molecular interactions of B∗27, B∗57, and others that might be associated with favorable response to both replicating virus and stable antigenic peptides could have implications for our understanding of HLA determinants of vaccine-induced CTL response in general.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI 45209 to M.J.M. and by grant AI 41951 (in part) to R.A.K. from the National Institute for Allergy and Infectious Diseases.
REFERENCES
- 1.Allen T M, O'Connor D H, Jing P, Dzuris J L, Mothe B R, Vogel T U, Dunphy E, Liebl M E, Emerson C, Wilson N, Kunstman K J, Wang X, Allison D B, Hughes A L, Desrosiers R C, Altman J D, Wolinsky S M, Sette A, Watkins D I. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi: 10.1038/35030124. [DOI] [PubMed] [Google Scholar]
- 2.Barouch D H, Santra S, Schmitz J E, Kuroda M J, Fu T-M, Wagner W, Bilska M, Craiu A, Zheng X X, Krivulka G R, Beaudry K, Lifton M A, Nickerson C E, Trigona W L, Punt K, Freed D C, Guan L M, Dubey S, Casimiro D, Simon A, Davies M-E, Chastain M, Strom T B, Gelman R S, Montefiori D C, Lewis M G, Emini E A, Shiver J W A, Letvin N L. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science. 2000;290:486–492. doi: 10.1126/science.290.5491.486. [DOI] [PubMed] [Google Scholar]
- 3.Belshe R B, Gorse G J, Mulligan M J, Evans T G, Keefer M C, Excler J L, Duliege A M, Tartaglia J, Cox W I, McNamara J, Hwang K L, Bradney A, Montefiori D, Weinhold K J. Induction of immune responses to HIV-1 by canarypox virus (ALVAC) HIV-1 and gp120 SF-2 recombinant vaccines in uninfected volunteers. NIAID AIDS Vaccine Evaluation Group. AIDS. 1998;12:2407–2415. doi: 10.1097/00002030-199818000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Berzofsky J A, Ahlers J D, Derby M A, Pendleton C D, Arichi T, Belyakov I M. Approaches to improve engineered vaccines for human immunodeficiency virus and other viruses that cause chronic infections. Immunol Rev. 1999;170:151–172. doi: 10.1111/j.1600-065x.1999.tb01336.x. [DOI] [PubMed] [Google Scholar]
- 5.Beyrer C, Artenstein A W, Rugpao S, Stephens H, VanCott T C, Robb M L, Rinkaew M, Birx D L, Khamboonruang C, Zimmerman P A, Nelson K E, Natpratan C Chiang Mai HEPS Working Group. Epidemiologic and biologic characterization of a cohort of human immunodeficiency virus type 1 highly exposed, persistently seronegative female sex workers in northern Thailand. J Infect Dis. 1999;179:59–67. doi: 10.1086/314556. [DOI] [PubMed] [Google Scholar]
- 6.Bodmer J G, Marsh S G E, Albert E D, Bodmer W F, Bontrop R E, Dupont R E, Erlich H A, Hansen J A, Mach B, Mayr W R, Parham P, Petersdorf E W, Sasazuki T, Schreuder G M T, Strominger J L, Svejgaard A, Terasaki P I. Nomenclature for factors of the HLA system, 1998. Tissue Antigens. 1999;53:407–446. doi: 10.1034/j.1399-0039.1999.530421.x. [DOI] [PubMed] [Google Scholar]
- 7.Borrow P, Lweicki H, Wei X, Horwitz M, Peffer N, Meyers H, Nelson J A, Gairin J E, Hahn B H, Oldstone M B A, Shaw G H. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 8.Carrington M, Nelson G W, Martin M P, Kissner T, Vlahov D, Goedert J J, Kaslow R, Buchbinder S, Hoots K, O'Brien S J. HLA and HIV-1: heterozygosity advantage and B∗35-Cw∗04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 9.Cereb N, Maye P, Lee S, Kong Y, Yang S Y. Locus-specific amplification of HLA class I genes from genomic DNA: locus-specific sequences in the first and third introns of HLA-A, and -C alleles. Tissue Antigens. 1995;45:1–11. doi: 10.1111/j.1399-0039.1995.tb02408.x. [DOI] [PubMed] [Google Scholar]
- 10.Clements-Mann M L, Weinhold K, Matthews T J, Graham B S, Gorse G J, Keefer M C, McElrath M J, Hsieh R H, Mestecky J, Zolla-Pazner S, Mascola J, Schwartz D, Siliciano R, Corey L, Wright P F, Belshe R, Dolin R, Jackson S, Xu S, Fast P, Walker M C, Stablein D, Excler J L, Tartaglia J, Paoletti E. Immune responses to human immunodeficiency virus (HIV) type 1 induced by canarypox expressing HIV-1MN gp120, HIV-1SF2 recombinant gp120, or both vaccines in seronegative adults. NIAID AIDS Vaccine Evaluation Group. J Infect Dis. 1998;177:1230–1246. doi: 10.1086/515288. [DOI] [PubMed] [Google Scholar]
- 11.Costello C, Tang J, Rivers C, Karita E, Meizen-Derr J, Allen S, Kaslow R A. HLA-B∗5703 independently associated with slower HIV-1 disease progression in Rwandan women. AIDS. 1999;13:1990–1991. doi: 10.1097/00002030-199910010-00031. [DOI] [PubMed] [Google Scholar]
- 12.Evans D T, O'Connor D H, Jing P, Dzuris J L, Sidney J, de Silva J, Allen T M, Horton H, Venham J E, Rudersdorf R A, Vogel T, Pauza C D, Bontrop R E, DeMars R, Sette A, Hughes A L, Watkins D I. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat Med. 1999;11:1270–1276. doi: 10.1038/15224. [DOI] [PubMed] [Google Scholar]
- 13.Evans T G, Keefer M C, Weinhold K J, Wolff M, Montefiori G, Gorse G J, Graham B S, McElrath M J, Clements-Mann M L, Mulligan M J, Fast P, Walker M C, Excler J L, Duliege A M, Tartaglia J A. A canarypox vaccine expressing multiple human immunodeficiency virus type 1 genes alone or with rgp120 elicits broad and durable CD8+ cytotoxic T lymphocyte responses in seronegative volunteers. J Infect Dis. 1999;180:290–298. doi: 10.1086/314895. [DOI] [PubMed] [Google Scholar]
- 14.Ferrari G, Humphery W, McElrath M J, Excler J L, Duliege A M, Clements M L, Corey L C, Bolognesi D P, Weinhold K J. Clade B-based vaccines elicit cross-clade T lymphocyte reactivities in uninfected volunteers. Proc Natl Acad Sci USA. 1997;94:1396–1401. doi: 10.1073/pnas.94.4.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Flores-Villaneuva P O, Yunis E J, Delgado J C, Vittinghoff E, Buchbinder S, Leung J Y, Uglialoro A M, Clavijo O P, Rosenberg E S, Kalams S A, Braun J D, Boswell S L, Walker B D, Goldfeld A E. Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc Natl Acad Sci USA. 2001;98:5140–5145. doi: 10.1073/pnas.071548198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14b.Gao X, Nelson G W, Karacki P, Martin M P, Phair J, Kaslow R, Goedert J J, Buchbinder S, Hoots K, Vlahov D, O'Brien S, Carrington M. Effect of a single amino acid change in MHC class I molecules on the rate of progression of AIDS. N Engl J Med. 2001;344:1668–1675. doi: 10.1056/NEJM200105313442203. [DOI] [PubMed] [Google Scholar]
- 15.Geczy A F, Kuipers H, Coolen M, Ashton L J, Kennedy C, Ng G, Dodd R, Wallace R, Le T, Raynes-Greenow C H, Dyer W B, Learmont J C, Sullivan J S. HLA and other host factors in transfusion-acquired HIV-1 infection. Hum Immunol. 2000;61:172–176. doi: 10.1016/s0198-8859(99)00142-1. [DOI] [PubMed] [Google Scholar]
- 16.Gorse G J, Patel G B, Newman F K, Mandava M, Belshe R B. Recombinant gp160 vaccination schedule and MHC HLA type as factors influencing cellular responses to HIV-1 envelope glycoprotein. NIAID AIDS Vaccine Clinical Trials Network. Vaccine. 1995;13:1170–1179. doi: 10.1016/0264-410x(95)00020-2. [DOI] [PubMed] [Google Scholar]
- 17.Goulder P J, Phillips R E, Cobert R A, McAdam S, Ogg G, Nowak M A, Glangrande P, Luzzi G, Morgan B, Edwards A, McMichael A J, Rowland-Jones S. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 18.Goulder P J R, Brander C, Annamalai K, Mngqundaniso N, Govender U, Tang Y, He S, Hartmen K E, O'Callaghan C A, Ogg G S, Altfeld M A, Rosenberg E S, Cao H, Kalams S A, Hammond M, Bunce M, Pelton S I, Burchett S A, Mclntosh K, Coovadia H M, Walker B D. Differential narrow focusing of immunodominant human immunodeficiency virus Gag-specific cytotoxic T-lymphocyte responses in infected African and Caucasoid adults and children. J Virol. 2000;74:5679–5690. doi: 10.1128/jvi.74.12.5679-5690.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goulder P J R, Bunce M, Krausa P, Mclntyre K, Crowley S, Morgan B, Edwards A, Giangrande P, Phillips R E, McMichael A. Novel, cross-restricted, conserved and immunodominant epitopes. I. Slow progressors in HIV infection. AIDS Res Hum Retrovirus. 1996;12:1691–1698. doi: 10.1089/aid.1996.12.1691. [DOI] [PubMed] [Google Scholar]
- 20.Goulder P J R, Edwards A, Phillips R E, McMichael A J. Identification of a novel HLA-B∗3501-restricted cytotoxic T lymphocyte epitope using overlapping peptides. AIDS. 1997;11:930–932. [PubMed] [Google Scholar]
- 21.Hayney M S, Poland G A, Dimanlig P, Schaid D J, Jacobson R M, Lipsky J J. Polymorphisms of the TAP2 gene may influence antibody response to live measles vaccine virus. Vaccine. 1997;15:3–6. doi: 10.1016/s0264-410x(96)00133-8. [DOI] [PubMed] [Google Scholar]
- 22.Hendel H, Caillat-Zucman S, Lebuanec H, Carrington M, O'Brien S, Andrieu J M, Schachter F, Zagury D, Rappaport J, Winkler C, Nelson G W, Zagury J F. New class I and II HLA alleles strongly associated with opposite patterns of progression to AIDS. J Immunol. 1999;162:6942–6946. [PubMed] [Google Scholar]
- 23.Itescu S, Mathur-Wagh U, Skovron M L, Brancato L J, Marmor M, Zeleniuch-Jacquotte A, Winchester R. HLA-B35 is associated with accelerated progression to AIDS. J Acquir Immune Defic Syndr. 1992;5:37–45. [PubMed] [Google Scholar]
- 24.Jin X, Bauer D E, Tuttleton S E, Lewin S, Gettie A, Blanchard J, Irwin C E, Safrit J T, Mittler J, Weinberger L, Kostrikis L G, Zhang L, Perelson A S, Ho D D. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson R P, Glickman R L, Yang J Q, Kaur A, Dion J T, Mulligan M M, Desrosiers R C. Induction of vigorous cytotoxic T lymphocyte responses by live attenuated simian immunodeficiency virus. J Virol. 1997;71:7711–7718. doi: 10.1128/jvi.71.10.7711-7718.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaslow R A, Carrington M, Apple R, Park L, Munoz A, Saah A J, Goedert J J, Winkler C, O'Brien S L, Rinaldo C, Detels R, Blattner W, Phair J, Erlich H, Mann D L. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 27.Kaslow R A, Duquesnoy R, VanRaden M, Kingsley L, Marrari M, Su S, Saah A J, Detels R, Phair J P, Rinaldo C R., Jr Combinations of A1, Cw7, B8, DR3 HLA antigens associated with rapid decline of T-helper lymphocytes in HIV-1 infected homosexual men: a report from the Multicenter AIDS Cohort Study. Lancet. 1990;335:927–930. doi: 10.1016/0140-6736(90)90995-h. [DOI] [PubMed] [Google Scholar]
- 28.Kaul R, Plummer F A, Kimani J, Dong T, Kiama P, Rostron T, Njagi E, MacDonald K S, Bwayo J J, McMichael A J, Rowland-Jones S L. HIV-1-specific mucosal CD8(+) lymphocyte responses in the cervix of HIV-1-resistant prostitutes in Nairobi. J Immunol. 2000;164:1602–1611. doi: 10.4049/jimmunol.164.3.1602. [DOI] [PubMed] [Google Scholar]
- 29.Keet I P M, Klein M R, Just J J, Kaslow R A. The role of host genetics in the natural history of HIV-1 infection: the needles in the haystack. AIDS. 1996;10(Suppl. A):S59–S67. doi: 10.1097/00002030-199601001-00009. [DOI] [PubMed] [Google Scholar]
- 30.Keet I P M, Tang J, Klein M R, LeBlanc S, Enger C, Rivers C, Apple R J, Mann D, Goedert J J, Miedema F, Kaslow R A. Consistent associations of HLA class I and class II and transporter gene products with progression of human immunodeficiency virus-1 infection in homosexual men. J Infect Dis. 1999;180:299–309. doi: 10.1086/314862. [DOI] [PubMed] [Google Scholar]
- 31.Kelleher A D, Long C, Holmes E C, Allen R L, Wilson J, Conlon C, Workman C, Shaunak S, Olson K, Goulder P, Brander C, Ogg G, Sullivan J S, Dyer W, Jones I I, McMichael A J, Rowland-Jones S, Phillips R E. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J Exp Med. 2001;193:375–386. doi: 10.1084/jem.193.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khanna R, Burrows S R, Burrows J M. The role of cytotoxic T lymphocytes in the evolution of genetically stable viruses. Trends Microbiol. 1997;5:64–69. doi: 10.1016/S0966-842X(96)10081-0. [DOI] [PubMed] [Google Scholar]
- 33.Klein M R, van Baalen C, Holwerda A M, Kerkhof Garde S R, Bende R J, Keep I P, Eeftinck-Schattenkerk J K, Osterhaus A D, Schuitemaker H, Miedema F. Kinetics of gag-specific cytotoxic T lymphocyte responses during the clinical cohorts of HIV-infection: a longitudinal analysis of rapid progressors and long-term asymptomatics. J Exp Med. 1995;181:1365–1372. doi: 10.1084/jem.181.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koup R A, Safrit J T, Cao Y, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kruskall M S, Alper C A, Awdeh Z, Yunis E J, Marcus-Bagley D. The immune response to hepatitis B vaccine in humans: inheritance patterns in families. J Exp Med. 1992;175:495–502. doi: 10.1084/jem.175.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacDonald K S, Fowke K R, Kimani J, Dunand V A, Nagelkerke N J D, Ball T B, Oyugi J, Njagi E, Gaur L K, Brunham R C, Wade J, Luscher M A, Hrausa P, Rowland-Jones S, Ngugi E, Bwayo J J, Plummer F A. Influence of HLA supertypes on susceptibility and resistance to human immunodeficiency virus type 1 infection. J Infect Dis. 2000;181:1581–1589. doi: 10.1086/315472. [DOI] [PubMed] [Google Scholar]
- 37.Magierowska M, Theodorou I, Debre P, Sanson F, Autran B, Riviere Y, Charron D, Costagliola D. Combined genotypes of CCR5, CCR2, SDF1, and HLA genes can predict the long-term nonprogressor status in human immunodeficiency virus-1-infected individuals. Blood. 1999;93:936–941. [PubMed] [Google Scholar]
- 38.Marsh S G E, Parham P, Barber L D. The HLA facts book. San Diego, Calif: Academic Press; 2000. [Google Scholar]
- 39.McElrath M J, Siliciano R F, Weinhold K J. HIV type 1 vaccine-induced cytotoxic T cell responses in phase I clinical trials: detection, characterization, and quantitation. AIDS Res Hum Retrovirus. 1997;13:211–216. doi: 10.1089/aid.1997.13.211. [DOI] [PubMed] [Google Scholar]
- 40.McNeil A J, Yap P L, Gore S M, Brettle R P, McColl M, Wyld R, Davidson S, Weightman R, Richardson A M, Robertson J R. Association of HLA types A1–B8-DR3 and B27 with rapid and slow progression of HIV disease. Q J Med. 1996;89:177–185. doi: 10.1093/qjmed/89.3.177. [DOI] [PubMed] [Google Scholar]
- 41.Metzner K J, Jin X, Lee F V, Gettie A, Bauer D E, DiMascio M, Perelson A S, Marx P A, Ho D D, Kostrikis L G, Connor R I. T-cell depletion on virus replication in rhesus macaques immunized with a live, attenuated siminan immunodeficiency virus vaccine. J Exp Med. 2000;191:1921–1932. doi: 10.1084/jem.191.11.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Migueles S A, Sabbaghian M S, Shupert W L, Bettinotti M P, Marincola F M, Martino L, Hallahan C W, Selig S M, Schwartz D, Sullivan J, Connors M. HLA B∗5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci USA. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulligan M J, Webber J. Human trials of HIV-1 vaccines. AIDS. 1999;13(Suppl. A):S105–S112. [PubMed] [Google Scholar]
- 44.Nelson G W, Goedert J J, Kaslow R A, Mann D L. Frequency of HLA allele-specific peptide motifs in HIV-1 proteins correlates with the allele's association with relative rates of disease progression after HIV-1 infection. Proc Natl Acad Sci USA. 1997;94:9802–9807. doi: 10.1073/pnas.94.18.9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinto L A, Sullivan J, Berzofsky J A, Clerici M, Kessler H A, Landay A L, Shearer G M. Env-specific cytotoxic T lymphocyte responses in HIV seronegative health care workers occupationally exposed to HIV contaminated body fluids. J Clin Investig. 1995;96:867–876. doi: 10.1172/JCI118133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rinaldo C, Huang X L, Fan Z F, Ding M, Beltz L, Logar A, Panicali D, Mazzara G, Liebmann J, Cottrill M. High levels of anti-human immunodeficiency virus type 1 (HIV-1) memory cytotoxic T-lymphocyte activity and low viral load are associated with lack of disease in HIV-1-infected long-term nonprogressors. J Virol. 1995;69:5838–5842. doi: 10.1128/jvi.69.9.5838-5842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohowsky-Kochan C, Skurnick J, Molinaro D, Louria D. HLA antigens associated with susceptibility/resistance to HIV-1 infection. Hum Immunol. 1998;59:802–815. doi: 10.1016/s0198-8859(98)00086-x. [DOI] [PubMed] [Google Scholar]
- 48.Rowland-Jones S L, Dong T, Dorrell L, Ogg G, Hansasuta P, Krausa P, Kimani J, Sabally S, Ariyoshi K, Oyugi J, MacDonald K S, Bwayo J, Whittle H, Plummer F A, McMichael A J. Broadly cross-reactive HIV-specific cytotoxic T-lymphocytes in highly-exposed persistently seronegative donors. Immunol Lett. 1999;66:9–14. doi: 10.1016/s0165-2478(98)00179-5. [DOI] [PubMed] [Google Scholar]
- 49.Rowland-Jones S L, Dong T, Fowke K R, Kimani J, Krausa P, Newell H, Blanchard T, Ariyoshi K, Oyugi J, Ngugi E, Bwayo J, MacDonald K S, McMichael A J, Plummer F A. Cytotoxic T cell responses to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J Clin Invest. 1998;102:1758–1765. doi: 10.1172/JCI4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmitz J E, Kuroda M J, Veazey R S, Seth A, Taylor W M, Nickerson C E, Lifton M A, Dailey P J, Forman M A, Racz P, Tenner-Racz K, Letvin N L. Simian immunodeficiency virus (SIV)-specific CTL are present in large numbers in livers of SIV-infected rhesus monkeys. J Immunol. 2000;164:6015–6019. doi: 10.4049/jimmunol.164.11.6015. [DOI] [PubMed] [Google Scholar]
- 51.Seth A, Ourmanov I, Schmitz J E, Kuroda M J, Lifton M A, Nickerson C E, Wyatt L, Carroll M, Moss B, Venzon D, Letvin N L, Hirsch V M. Immunization with a modified vaccinia virus expressing simian immunodeficiency virus (SIV) Gag-Pol primers for an anemnestic Gag-specific cytotoxic T-lymphocyte response and is associated with reduction of viremia after SIV challenge. J Virol. 2000;74:2502–2509. doi: 10.1128/jvi.74.6.2502-2509.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang J, Costello C, Keet I P M, Rivers C, LeBlanc S, Karita E, Allen S, Kaslow R A. HLA class I homozygosity accelerates disease progression in human immunodeficiency virus type 1 infection. AIDS Res Hum Retrovirus. 1999;15:317–324. doi: 10.1089/088922299311277. [DOI] [PubMed] [Google Scholar]
- 53.Turner S, Ellexson M E, Hickman H, Sidebottom D A, Fernandez-Vina M, Confer D L, Hildebrand W H. Sequence-based typing provides a new look at HLA-C diversity. J Immunol. 1998;161:1406–1413. [PubMed] [Google Scholar]
- 54.Wilson J D K, Ogg G S, Allen R L, Davis C, Shaunak S, Downie J, Dyer W, Workman C, Sullivan J S, McMichael A J, Rowland-Jones S L. Direct visualization of HIV-1-specific cytotoxic T lymphocytes during primary infection. AIDS. 2000;14:225–233. doi: 10.1097/00002030-200002180-00003. [DOI] [PubMed] [Google Scholar]