Abstract
Full-length VP22 is necessary for efficient spread of herpes simplex virus type 1 (HSV-1) from cell to cell during the course of productive infection. VP22 is a virion phosphoprotein, and its nuclear localization initiates between 5 and 7 h postinfection (hpi) during the course of synchronized infection. The goal of this study was to determine which features of HSV-1 infection function to regulate the translocation of VP22 into the nucleus. We report the following. (i) HSV-1(F)-induced microtubule rearrangement occurred in infected Vero cells by 13 hpi and was characterized by the loss of obvious microtubule organizing centers (MtOCs). Reformed MtOCs were detected at 25 hpi. (ii) VP22 was observed in the cytoplasm of cells prior to microtubule rearrangement and localized in the nucleus following the process. (iii) Stabilization of microtubules by the addition of taxol increased the accumulation of VP22 in the cytoplasm either during infection or in cells expressing VP22 in the absence of other viral proteins. (iv) While VP22 localized to the nuclei of cells treated with the microtubule depolymerizing agent nocodazole, either taxol or nocodazole treatment prevented optimal HSV-1(F) replication in Vero cells. (v) VP22 migration to the nucleus occurred in the presence of phosphonoacetic acid, indicating that viral DNA and true late protein synthesis were not required for its translocation. Based on these results, we conclude that (iv) microtubule reorganization during HSV-1 infection facilitates the nuclear localization of VP22.
The molecular mechanism of herpes simplex virus type 1 (HSV-1) tegument and envelope assembly is poorly understood. Most of the major tegument proteins exhibit nuclear or perinuclear distributions late in infection (1, 8, 10–12, 26, 33, 37, 43), and there is general agreement that primary envelopment occurs as the capsid exits the nucleus (7, 19, 21, 42, 46) (for a detailed review, see reference 17).
Work in our laboratory has focused on the major tegument protein VP22 and its function in virion assembly and virus replication. Full-length VP22 is necessary for efficient spread of the virus from cell to cell, inasmuch as a recombinant virus producing a truncated form of the protein exhibits a decreased plaque size in Vero cells compared to that of wild-type virus (36). In an earlier study, we reported that the nuclear localization of VP22 initiates between 5 and 7 h postinfection (hpi) during the course of synchronized infection (37). This period corresponds to the peak of viral β protein production and DNA synthesis (39). Recently, computer analyses predicted two nuclear localization signals (NLS) in the primary structure of VP22 (22). Although transiently expressed VP22 can be detected in the nuclei of transfected cells (15, 22), these proposed NLS have not yet been characterized in nuclear import assays. VP22 is a 301-amino-acid protein with a predicted size of 32,000 Da (28). Thus, VP22 is below the size exclusion limit (40,000 to 45,000 Da) for passive diffusion through the nuclear pore (13). That VP22 accumulates in the cytoplasm of infected cells prior to 5 hpi suggests its translocation to the nucleus is regulated during infection. One possible means of regulation could be active retention through binding of VP22 to a cytoplasmic structure.
In this study, we sought to determine which features of HSV-1 replication are involved in the redistribution of VP22 during productive infection. Our investigations show the following. Regulated VP22 nuclear localization initiates after 5 hpi with HSV-1 and is independent of viral DNA and true late protein synthesis. We propose that HSV-1 induced microtubule restructuring releases VP22 from the cytoskeleton, allowing its entry into the nucleus. Stabilization of microtubules during infection or in VP22-expressing cells increases VP22 retention in the cytoplasm. Based on these results, we conclude that microtubule reorganization during herpes HSV-1 infection acts to facilitate the nuclear localization of VP22.
MATERIALS AND METHODS
Cells and virus.
African green monkey kidney (Vero) cells were obtained from the American Type Culture Collection and passaged in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal bovine serum. VP22-expressing V49 cells (36) were passaged in DMEM supplemented with 5% fetal bovine serum and 1 mg of G418 per ml (Gibco-BRL). The prototype HSV-1(F) (14) and gE-minus recombinant virus HSV-1(R7032) (31) were provided by Bernard Roizman, University of Chicago. To obtain virus stocks, subconfluent Vero monolayer cells (approximately 3 × 106) were inoculated with virus at a multiplicity of infection (MOI) of 0.01 for 2 h at 37°C in 199V medium (Life Technologies) supplemented with 1% newborn calf serum (NBCS). The inoculum was then removed, fresh DMEM supplemented with 5% NBCS was added, and the cells were incubated at 37°C in 5% CO2. Virus stocks were prepared once the infection reached a cytopathic effect of 100%, the titer was determined on Vero cells, and aliquots were stored at −80°C. All MOIs were derived from the number of plaque-forming units (PFU) of virus on Vero cells.
Synchronized infections.
Synchronous infections are defined as uniform staining in all cells in a microscopic field at a given time postinfection, as determined by indirect immunofluorescence with a single antibody specific for a unique HSV-1 polypeptide (37). Vero cells were seeded the day before infection in either six-well dishes containing 22-mm2 coverslips for indirect immunofluorescence or 25-cm2 flasks for infected-cell extracts. For synchronized infections, cells were incubated on ice on an orbital shaker at 4°C for 15 min prior to the addition of virus. The cells were then inoculated while still on ice and returned to the shaker at 4°C. After allowing the virus to adsorb for 1 h, the cells were rinsed with 4°C phosphate-buffered saline (PBS). Cells were then removed from the ice, 37°C medium was added immediately, and the cells were returned to a 37°C incubator.
Induction of synchronous infection by adsorption of the inoculum at 4°C is routine and has little or no effect on cells in culture. However, it should noted that there is some evidence that the 4°C incubation used to synchronize the infections may have a transient effect on the monolayer cells (37). Cells fixed immediately after the temperature shift to 37°C with formaldehyde and permeablized with acetone have a slightly more diffuse α-tubulin staining pattern than that observed in the cells held at 37°C (37). This finding is consistent with the fact that the dynamics of polymerization and depolymerization of microtubules are energy dependent and are expected to be reduced at the lower (4°C) temperature. This apparent change in the α-tubulin organization was not observable by 4 hpi, and the two sets of cells (4 versus 37°C) were indistinguishable based on α-tubulin staining (37). Thus, the cells recover from the 4°C shock by this time.
Immunological reagents.
RGST49 is a rabbit polyclonal antibody directed against a GST-VP22 fusion protein (4). Affinity-purified RGST49 antibody was generated as described previously (37) and used at a dilution of 1:10 for immunofluorescence and 1:50 for immunoblotting. Hybridoma supernatant containing G49 monoclonal antibody specific for VP22 has been described previously (37). Monoclonal antibody DM 1A, specific for α-tubulin, was obtained from Sigma and was used at a dilution of 1:500 for immunofluorescence. Anti-VP16(1–21) monoclonal antibody was purchased from Transduction Laboratories and used at a dilution of 1:500. R220/5 polyclonal antiserum to VP13/14 (32) (a kind gift from David Meredith) was used at a dilution of 1:500 (4). Fluorescein isothiocyanate-conjugated anti-rabbit immunoglobulin G (IgG) (H+L) and Texas Red-conjugated anti-mouse IgG (H+L) were purchased from Vector Laboratories (Santa Cruz, Calif.) and used at a dilution of 1:100 in 1% bovine serum albumin (BSA). Alexa568-conjugated highly cross-adsorbed anti-rabbit IgG (Molecular Probes A-11036) was used at a dilution of 1:250 in 1% BSA in the experiments using the VP22-expressing V49 cells. Alexa568 is much more sensitive than Texas Red and is the secondary fluor of choice when visualizing VP22 in noninfected cells (A. Blouin, L. E. Pomeranz, and J. A. Blaho, unpublished observations).
Indirect immunofluorescence and microscopy.
Vero and V49 cells were prepared for indirect immunofluorescence as previously described (37). Briefly, cells rinsed twice in PBS were fixed in 2.5% methanol-free formaldehyde (Polysciences, Inc.) for 20 min at room temperature, rinsed twice again in PBS, and permeabilized in 100% acetone at −20°C for 3 to 5 min. Infected cells were incubated for 16 h in 1% BSA (Sigma) supplemented with 10 μg of pooled human Ig (Sigma) per ml. Treatment with this amount of human Ig was shown by us to be sufficient to neutralize Fc binding by the viral gE and gI proteins (37). The primary antibodies used for immunofluorescence studies were diluted as described above and were added for 1 h. Hoechst 33258 dye (Sigma) was added directly to the primary antibody mixture to facilitate the visualization of the nuclei. After extensive rinsing with PBS, the appropriate secondary antibody was added, and this mixture was incubated for 45 min. Finally, the cells were preserved in a 0.1% solution of Mowiol (Sigma) with 2.5% DABCO (Sigma) used as an antibleaching agent under a fresh coverslip and sealed with nail polish. Cells were visualized on an Olympus IX70/IX-FLA inverted fluorescence microscope, and images were acquired with a Sony DKC-5000 digital photo camera linked to a PowerMac G3 and processed through Adobe Photoshop version 4.0. Supplemental data, including color images of several of the figures, may be accessed through our public domain website at http://www.mssm.edu/micro /blaho/webdata/aketaldns.shtml.
Chemical treatments during infection.
Cell lines or synchronously infected cells were treated with either taxol, nocodazole, or phosphonoacetic acid (PAA) prior to immunofluorescence or immunoblotting experiments. Taxol was obtained from Sigma and was added to cell cultures at a final concentration of 20 μg per ml of medium at the times indicated in the text. It has been reported that this concentration of taxol does not ablate productive HSV-1 replication (3). Nocodazole (Sigma) was used in a similar manner at a final concentration of 0.5 μg per ml. Taxol and nocodazole were added to the synchronized infected cells at 3 hpi to ensure that the cells fully recovered from the temperature shifting before treatment with the drugs. Cells were preincubated for 60 min in the presence of PAA (Sigma) at a final concentration of 300 μg per ml of medium, and the drug was maintained throughout the adsorption and infection stages (5). A PAA concentration of 500 μg/ml was also tested and found to yield identical results, so the lower concentration was chosen for this study (A. Kotsakis and J. A. Blaho, unpublished results).
Infected-whole-cell extracts.
Whole-cell extracts were prepared from approximately 106 cells in a mixture of 140 mM NaCl, 3 mM KCl, 10 mM Na2HPO4, and 1.5 mM KH2PO4 (pH 7.5) (PBS) containing protease inhibitors [10 mM l-1-chlor-3-(4-tosylamido)-7-amino-2-heptanon-hydrochloride (TLCK), 10 mM l-1-chlor-3-(4-tosylamido)-4-phenyl-2-butanone (TPCK), and 100 mM phenylmethylsulfonyl fluoride (PMSF)]. Extracts of the infected cells were pelleted by low-speed centrifugation and resuspension of the pellet in PBS containing 1.0% Triton X-100 plus protease inhibitors. Lysis by sonication was performed with a Bronson Sonifier.
Denaturing gel electrophoresis and immunoblotting.
Protein concentrations of infected cell extracts were determined with a modified Bradford assay (Bio-Rad) according to the manufacturer's specifications. Equal amounts of infected cell protein were separated in sodium dodecyl sulfate (SDS)–15% polyacrylamide gels cross-linked with N,N′-diallyltartardiamide (DATD) (6), electrically transferred to nitrocellulose, and probed for a minimum of 2 h with the appropriate primary antibodies. Horseradish peroxidase-conjugated anti-rabbit or anti-mouse (Amersham) secondary antibodies were diluted 1:1,000 in PBS and incubated with the blots for 1 h. Specific viral bands were detected following development with enhanced chemiluminescence (ECL) reagents (Amersham) and autoradiography at 25°C with X-OMAT film (Kodak, Rochester, N.Y.). Alkaline phosphatase-conjugated goat anti-rabbit and anti-mouse secondary antibodies, used at 1:500 in PBS, were purchased from Southern Biotech (Birmingham, Ala.). Prestained molecular mass markers (Gibco BRL) were included in all acrylamide gels (lanes are not shown in figures).
Analysis of virus yields.
Virus yields in the presence of either PAA, taxol, or nocodazole during synchronized infection were determined as follows. Approximately 106 Vero cells grown to 90% confluence in 25-cm2 flasks were incubated on ice for 2 h with 5.0 PFU of HSV-1(F) per cell, rinsed in PBS, and incubated at 37°C. For PAA, 300 μg of the drug per ml was added 30 min prior to virus adsorption and maintained throughout the infection. Either taxol or nocodazole was added at 3 hpi and maintained until the end of the infection. At 13 hpi, intracellular virus stocks (36) were prepared from each flask, and titers were determined in duplicate on Vero cells.
Computer analysis and imaging.
Analysis of the primary structure of VP22 was performed by using the PSORT II Prediction program (updated 24 November 1999), which is available at the public domain web site http://psort.nibb.ac.jp. An earlier version of this algorithm was used to compare HSV-1 VP22 with its homologue from BHV-1 (22). All immunoblots and autoradiograms were digitized at 600 dots per inch with an AGFA Arcus II scanner linked to a Macintosh G3 PowerPC workstation. Raw digital images, saved as tagged image files with Adobe Photoshop version 5.0, were organized into figures by using Adobe Illustrator version 7.1. Gray scale or color prints of figures were obtained by using a Codonics dye sublimation printer.
RESULTS
HSV-1(F)-induced microtubule reorganization coincides with VP22 localization to the nucleus.
It has been reported that cells transiently expressing VP22 and fixed for indirect immunofluorescence with methanol alone exhibit atypical microtubule morphologies, suggesting that VP22 binds to and rearranges cellular microtubules into “bundles” in the absence of other viral proteins (16, 22). The microtubule network of infected cells undergoes a dramatic rearrangement or fragmentation beginning at approximately 6 hpi and then reorients itself later in infection (at approximately 16 hpi) (3). HSV-1-induced microtubule reorganization is observed in cells infected in the presence of PAA (3), suggesting that true late protein synthesis is not required for the rearrangement. Interestingly, infection-induced microtubule fragmentation occurs at the same time that nuclear localization of VP22 initiates during synchronized infection (37).
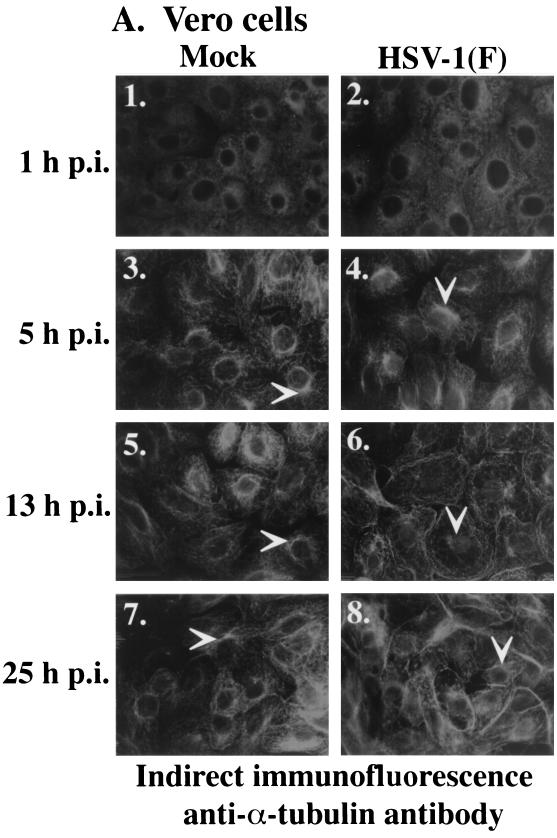
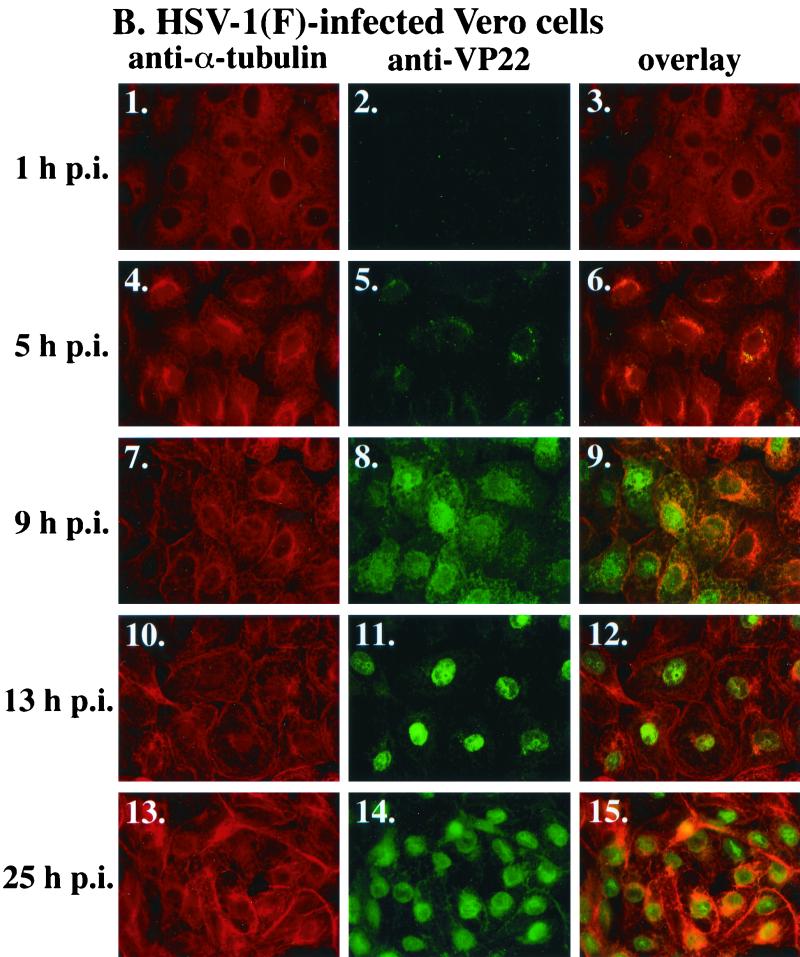
The goal of the following series of experiments was to test the hypothesis that nuclear localization of VP22 is regulated by the reorganization of microtubules. VP22 and cellular microtubules were monitored in Vero cells that were synchronously infected with HSV-1(F). As described previously, this technique involves adsorbing virus at low temperature (4°C) for 1 h and then shifting to a higher temperature by the addition of warm (37°C) medium to allow synchronized entry and infection (37). At 1, 5, 9, 13, and 25 hpi cells were fixed with formaldehyde and permeabilized in acetone (37). To detect VP22 and microtubules by indirect immunofluorescence, cells were stained with anti-VP22 and anti-α-tubulin antibodies, followed by indirect immunofluorescence as described in Materials and Methods. The results of this experiment are shown in Fig. 1. Gray scale images of α-tubulin staining in mock and infected cells are shown in panel A to aid in the visualization of the effect of infection on microtubules. Panel B shows color images of α-tubulin and VP22 staining to emphasize the cytoplasmic and nuclear localizations of VP22. Thus, panels 2, 4, 6, and 8 in Fig. 1A are the same images as panels 1, 4, 10, and 13 in Fig. 1B.
FIG. 1.
Nuclear localization of VP22 during synchronized infection correlates with HSV-1(F)-induced microtubule fragmentation. Vero cells were synchronously infected with HSV-1(F) or mock infected and fixed for indirect immunofluorescence at the times indicated as described previously (37). Cells were stained with antibodies to α-tubulin (A and B) and VP22 (B) as described in Materials and Methods. (A) Gray scale images of mock-infected and infected cells. White arrows mark perinuclear regions (MtOCs). Panels 2, 4, 6, and 8 in A are the same images as panels 1, 5, 10, and 13 in B. Merged images (overlay) are shown in B, panels 3, 6, 9, 12, and 15. All images were acquired under the same conditions.
As expected, α-tubulin staining was detected in all cells. At 1 hpi, low-temperature-induced microtubule depolymerization (37) was observed in mock (Fig. 1A, panel 1)- and HSV-1(F) (Fig. 1A, panel 2; B, panel 1)-infected cells. Cellular microtubules recovered from the low-temperature-induced depolymerization by 5 hpi, as described previously (37), since mock (Fig. 1A, panel 3)- and HSV-1(F) (Fig. 1A, panel 4; 1B, panel 4)-infected cells displayed the typical microtubule arrangement expected for Vero cells (3). These networks showed characteristic microtubule organizing centers (MtOCs) in the perinuclear regions of the cells (marked by arrows). MtOCs were consistently observed in the mock-infected cells at 13 (Fig. 1A, panel 5) and 25 hpi (Fig. 1A, panel 7). The microtubule structure in HSV-1(F)-infected cells at 5 hpi (Fig. 1A, panel 4) was identical to that observed in the mock-infected cells at 5, 13, and 25 hpi.
Microtubule fragmentation was clearly detected in the synchronously infected Vero cells at 13 hpi (Fig. 1A, panel 6; B, panel 10). The cytoskeletal redistribution due to infection was dramatic, as evidenced by the loss of distinctive MtOCs and an apparent accumulation of disorganized microtubules around the periphery of the cells, as previously reported (3). The arrow in Fig. 1A, panel 6, indentifies a perinuclear region that has no obvious MtOC (compare with Fig. 1A, panel 5). A “haze” in the area of the nucleus was observed in the infected cells (arrow in Fig. 1B, panel 6) and is likely attributed to fragmented microtubules in the cytoplasm either over or under the nucleus, but out of the plane of focus. Microtubule bundles were again observed at 25 hpi in the infected cells (Fig. 1A, panel 8), indicating that reassembly had occurred as previously reported (3). The arrow in Fig. 1A, panel 8, shows an MtOC in an infected cell at 25 hpi. These results represent the first demonstration that HSV-1-induced microtubule reorganization occurs during synchronized infection.
The staining patterns of VP22 in synchronously infected cells were as previously described (37). VP22 was detected in the cytoplasm of infected cells at 5 hpi and localized to a perinuclear region (Fig. 1B, panel 5). This finding is consistent with our earlier results, in which we observed that at early infection times (<5 hpi), VP22 colocalized with the p58 Golgi protein (37). While VP22 does not contain an endoplasmic reticulum (ER) retrieval signal (consensus, KDEL) in its carboxy-terminal end, we have recently detected an ER membrane retention signal at amino acids 2 to 5 in its amino end. Since prediction of retrieval signals in proteins associated with the ER membrane is complex (44), further investigations will determine the significance of these signals in directing the localization of VP22 in the cytoplasm.
At 9 hpi (Fig. 1B, panel 8), most infected cells exhibited the combination of cytoplasmic and nuclear staining we previously defined as diffuse (37). VP22 was predominantly nuclear at 13 (Fig. 1B, panel 11) and 25 (Fig. 1B, panel 14) hpi. These results at the 13- and 25-h time points strongly support our observations that VP22 exists in the cytoplasm early and the nucleus late in infection. Additionally, the fact that the cytoplasm is essentially devoid of VP22 at 13 hpi indicates that viral gE or gI receptor binding to our anti-VP22 antibody is not the basis for the VP22 staining patterns we observe. As expected, no anti-VP22 immune reactivity was detected in mock-infected cells (data not shown).
Comparison of the VP22 and microtubule patterns indicated that the localization of VP22 to the nucleus correlated with HSV-1-induced microtubule fragmentation, as evidenced by merged images of the VP22 and α-tubulin staining (Fig. 1B, panels 3, 6, 9, 12, and 15). Cytoplasmic VP22 (Fig. 1B, panels 6 and 9) was detected prior to infection-induced microtubule reorganization. A perinuclear yellow-orange staining pattern in the infected cells at these time points identified areas of VP22-tubulin colocalization. Upon microtubule rearrangement, VP22 was detected almost exclusively in the nucleus (Fig. 1B, panels 12 and 15). This phenomenon is best illustrated at 13 hpi, when nuclear VP22 stains bright green and the cytoplasm is almost completely red due to tubulin staining only (Fig. 1B, panel 12). No obvious MtOCs were observed in the infected cells at 13 hpi (Fig. 1A, panel 6), indicating that microtubule rearrangement occurred by this time. The 9-h time point likely represents an intermediate stage of the microtubule redistribution and VP22 nuclear translocation (Fig. 1B, panel 9). Based on these findings, we conclude that microtubule reorganization during HSV-1 infection correlates with the nuclear localization of VP22.
Inhibition of microtubule depolymerization by taxol increases the amount of VP22 detected in the cytoplasm during infection.
HSV-1-induced microtubule redistribution is inhibited by treating infected cells with taxol (3). Taxol is a drug that reversibly binds tubulin (35) and stabilizes microtubules into bundles (40), preventing their depolymerization. To test the hypothesis that microtubule reorganization facilitates the nuclear localization of VP22, synchronously infected Vero cells were treated with taxol at 4 hpi fixed at 5, 9, 13, and 25 hpi, and stained for indirect immunofluorescence as described in Materials and Methods. Control cells were mock infected in the presence of taxol and fixed at 25 hpi. The results of this study are shown in Fig. 2.
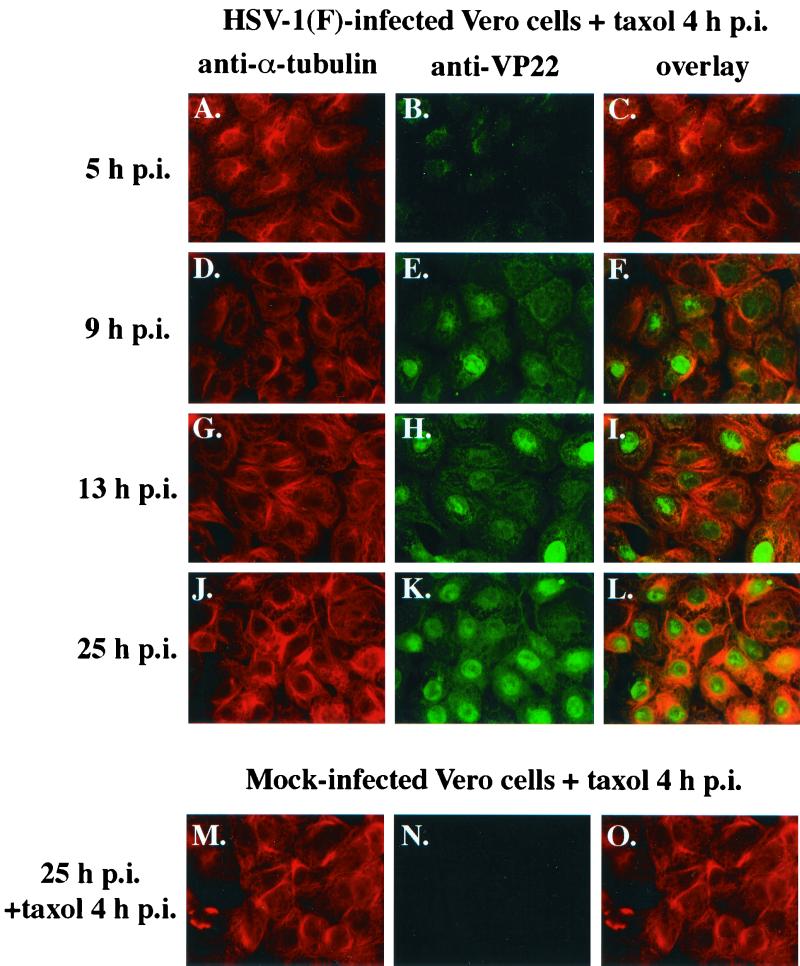
FIG. 2.
Taxol treatment increases the amount of VP22 detected in the cytoplasm of synchronously infected cells. Vero cells were synchronously infected with HSV-1(F) or mock infected, and taxol (20 μg/ml) was added to cells at 4 hpi (3 h post-temperature shift). Cells were stained with antibodies to α-tubulin and VP22 and fixed for indirect immunofluorescence at the times indicated as described in Materials and Methods: α-tubulin staining shown in panels A, D, G, J, and M; VP22 shown in panels B, E, H, K, and N; merged images (overlay) shown in panels C, F, I, L, and O. All images were acquired under the same conditions.
As expected, 1 h of taxol treatment had no effect on infected cell microtubules at 5 hpi (Fig. 2A). In contrast to the results from untreated infected cells shown in Fig. 1, bundles of microtubules were observed in taxol-treated infected cells at 9 (Fig. 2D) and 13 hpi (Fig. 2G). Extensive microtubule bundling was observed in the presence of taxol at 25 hpi (Fig. 2J). In the control, bundles of microtubules were also observed in taxol-treated mock-infected cells at 25 hpi (Fig. 2M). These results indicate that taxol treatment of synchronously infected cells inhibits HSV-1(F)-induced microtubule redistribution. This finding is consistent with previous studies (3).
If the localization of VP22 to the cell nucleus during infection is influenced by microtubule reorganization, we would predict that VP22 retention in the cytoplasm would be increased when microtubule depolymerization is inhibited. We observed that the subcellular distribution of VP22 was similar in the presence (Fig. 2B and E) and absence (Fig. 1B, panels 5 and 8) of taxol at 5 and 9 hpi. This finding is supported by the results of the overlays where the images are similar with (Fig. 2C and F) and without (Fig. 1B, panels 6 and 9) the drug. Together, these results indicate that prior to 9 hpi, the presence of taxol did not have a dramatic effect on the localization of VP22 in the infected cells.
However, in contrast to the results in Fig. 1 where VP22 is predominantly nuclear after 13 hpi (Fig. 1B, panel 11), taxol-treated cells at 13 hpi exhibited a diffuse VP22 staining pattern (Fig. 2H). Although VP22 staining could be detected in the nuclei of taxol-treated cells at 13 hpi (Fig. 2H), significantly more VP22 was observed in the cytoplasm of these cells than that found in the nontreated cells at 13 hpi (compare Fig. 2H with Fig. 1B, panel 11). The increased retention of VP22 in the cytoplasm in the taxol-treated cells compared to untreated cells was also observed at 25 hpi (compare Fig. 2K with Fig. 1B, panel 14). Importantly, no VP22 staining was detected in mock-infected cells (Fig. 2N), indicating that the anti-VP22 reactivity observed in infected cells did not result from the high α-tubulin concentration in bundled microtubules. The retention of cytoplasmic VP22 in cells synchronously infected in the presence of taxol was even more striking in the overlays of the late time points of infection; the orange-yellow cytoplasmic staining indicates that VP22 localizes with microtubules in these cells (compare Fig. 2I and L with Fig. 1B, panels 12 and 15, respectively). As expected, no orange-yellow staining was observed in overlays of control mock-infected cells (Fig. 2O).
The results presented in Fig. 1 and 2 support the hypothesis that microtubule-associated VP22 localizes to the nucleus when microtubules fragment during HSV-1 infection. This model is based on two observations. First, the localization of VP22 to the nucleus correlates temporally with HSV-1(F)-induced microtubule disassembly. Second, the addition of an agent that acts to inhibit microtubule depolymerization leads to an increase in the amount of VP22 detected in the cytoplasm of infected cells.
Cytoplasmic VP22 observed in VP22-expressing V49 cells treated with taxol.
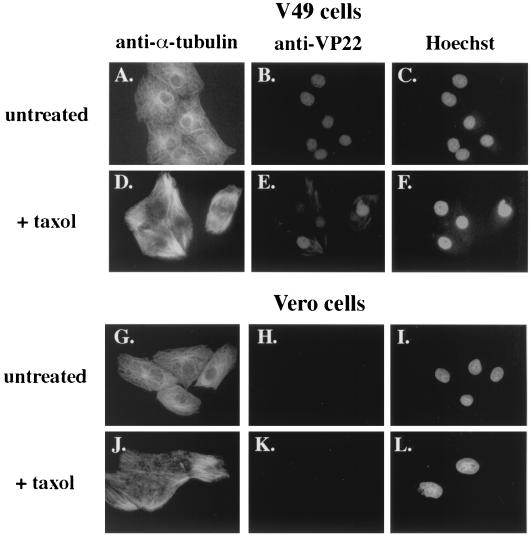
Since the exact mechanism by which HSV-1 rearranges microtubules is unknown, the role of specific viral proteins in the process is not clear. It is conceivable that specific viral gene products may act on microtubules in a yet undetermined manner and that VP22 simply associates with these viral polypeptides. To investigate whether the microtubule-associated mechanism of VP22 localization to the nucleus requires the presence of other viral proteins, we took advantage of the VP22-expressing Vero (V49) cells we generated for an earlier study (36). Vero cells that do not express VP22 or V49 cells were left untreated or treated with taxol for 20 h prior to fixing and staining cells for indirect immunofluorescence with anti-VP22 and anti-α-tubulin antibodies as described in Materials and Methods. As an additional control for cell morphology, DNA was stained with Hoechst 33258 dye as described previously (2).
The results of this experiment (Fig. 3) showed that the microtubule structure of V49 cells (Fig. 3A) was indistinguishable from that observed in Vero cells (Fig. 3G). Taxol-induced microtubule bundling was observed in both V49 (Fig. 3D) and Vero (Fig. 3J) cells. As described previously (36), VP22 was detected in untreated V49 cells (Fig. 3B), and this staining was exclusively nuclear in all cells as evidenced by the Hoechst stain (Fig. 3C). In contrast, taxol treatment of V49 cells resulted in the appearance of VP22 in the cytoplasm (Fig. 3E). Under the taxol treatment conditions, VP22 localized to areas where α-tubulin staining was most intense (compare Fig. 3D and E). As expected, VP22 staining was not detected in non-VP22-expressing Vero cells (Fig. 3H and K). Based on these results, we conclude that the ability of VP22 to be retained in the cytoplasm of taxol-treated cells, which we first observed during viral infection (Fig. 2), does not require the presence of additional viral gene products. It is currently not clear which, if any, cellular factors besides α-tubulin might be involved in this VP22-cytoplasmic retention process.
FIG. 3.
Increased detection of cytoplasmic VP22 in taxol-treated V49 cells. VP22-expressing V49 cells (A to F) and Vero cells (G to L), which do not express VP22, were left untreated or treated (+ taxol) with 20 μg of taxol per ml for 20 h. Cells were fixed and stained for indirect immunofluorescence with anti-α-tubulin (A, D, G, and J) and anti-VP22 (B, E, H, and K) antibodies. DNA was stained with Hoechst dye (C, F, I, and L).
It should be noted that a significant amount of VP22 was still detected in the nuclei of the taxol-treated cells. It is conceivable that the VP22 retained in the cytoplasm of the taxol-treated cells represents VP22 that was newly synthesized after the addition of the taxol. Thus, the VP22 was probably present in the nuclei prior to the addition of the drug. We have no evidence that VP22 shuttles between the nucleus and the cytoplasm, so the possibility that nuclear VP22 migrates to the cytoplasm and binds microtubules during taxol treatment seems unlikely.
Nuclear VP22 observed in VP22-expressing V49 cells treated with nocodazole.
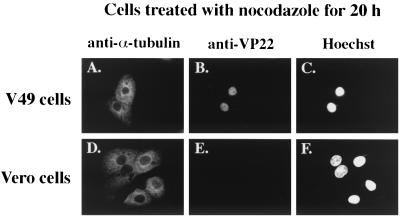
The previous results (Fig. 3) indicated that in the presence of taxol, VP22 could be detected in both the nucleus and the cytoplasm. This finding raised the possibility that VP22 might actually migrate from the nucleus to the cytoplasm. The goal of this experiment was to determine whether VP22 localized to the cytoplasm in cells in which the microtubules have been completely depolymerized. Vero or V49 cells were treated with nocodazole for 20 h prior to fixing and staining cells with Hoechst 33258 dye or anti-VP22 and anti-α-tubulin antibodies for indirect immunofluorescence as described in Materials and Methods.
The data (Fig. 4) indicate that nocodazole treatment of both Vero and V49 cells results in complete depolymerization of microtubules (Fig. 4A and D). No obvious MtOCs could be detected in these cells, and no filamentous microtubule structures were observed. However, the nuclear structures of the cells were retained as evidenced by DNA staining (Fig. 4C and F). VP22 was detected exclusively in the nuclei of the V49 cells treated with nocodazole (Fig. 4B). Taken together with the results in Fig. 3, we conclude that stabilized microtubules and not ones that have been depolymerized act to retain VP22 in the cytoplasm of cells. This theory is consistent with our earlier results (Fig. 1) showing that nuclear translocation of VP22 during infection coincides with HSV-1-induced microtubule rearrangement.
FIG. 4.
Detection of nuclear VP22 in nocodazole-treated V49 cells. VP22-expressing V49 cells (A to C) and Vero cells (D to F), which do not express VP22, were treated with 0.5 μg of nocodazole per ml for 20 h. Cells were fixed and stained for indirect immunofluorescence with anti-α-tubulin (A and D) and anti-VP22 (B and E) antibodies. DNA was stained with Hoechst dye (C and F).
Virus yields in the presence of taxol, nocodazole, and PAA.
The previous experiments (Fig. 2 and 3) indicated that the addition of taxol to cells results in the retention of a detectable amount of VP22 in the cytoplasm. In contrast, no VP22 was detected in the cytoplasm when the microtubule depolymerizing agent nocodazole was added to the cells (Fig. 4). In order for us to understand the significance of VP22 retention in the cytoplasm in HSV-1 replication, it was necessary to test the effect that these drugs have on productive viral infection. It has been previously reported that taxol does not ablate HSV-1 infection (3), and our results showing VP22 expression during infection in the presence of taxol at 25 hpi (Fig. 2L) support this finding. Vero cells were synchronously infected with HSV-1(F), and at 3 hpi either taxol or nocodazole was added to the medium. The concentrations of the drugs were identical to those used in Fig. 2 to 4. Control infected cells (untreated) were also mock treated at 3 hpi. At 13 hpi, virus stocks were made and titers were determined on Vero cells in duplicate as described in Materials and Methods. As an additional control, cells were also pretreated with the viral DNA polymerase inhibitor PAA 1 h prior to infection, and the PAA was maintained in the medium throughout the infection to 13 hpi.
The results were as follows. The infected cells that were mock treated yielded a titer of 1.0 × 108 PFU/ml. The control PAA-addition infection yielded a titer of 1.0 × 105 PFU/ml, consistent with a significant reduction in virus production due to the inhibition of the viral DNA polymerase. The taxol- and nocodazole-treated infected cells yielded titers of 8.4 × 106 and 8.8 × 106 PFU/ml, respectively. Thus, the amount of virus produced in the presence of these drugs was almost 3 logs higher than that produced in the presence of PAA. By this definition, neither taxol nor nocodazole was a potent inhibitor of HSV-1 replication. However, the virus yields in the presence of taxol and nocodazole were both more than a log lower than that with the untreated control. One of the surprising features of these results is that the titers with taxol and nocodazole were essentially the same. These results suggest that either stabilized (by taxol) or depolymerized (by nocodazole) microtubules can reduce optimal viral replication in Vero cells.
Partitioning of VP22 in nuclear and cytoplasmic fractions in cells infected in the presence of taxol and nocodazole.
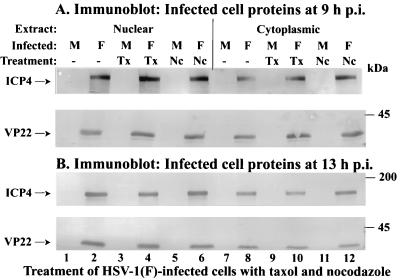
The previous virus yield experiments indicated that the presence of taxol or nocodazole reduces optimal HSV-1 replication. The goal of this series of experiments was to determine the effect of these drugs on the accumulation of VP22 in infected-cell fractions. Vero cells were synchronously mock or HSV-1(F) infected in the absence or presence of taxol or nocodazole. At 9 and 13 hpi nuclear and cytoplasmic extracts were prepared, and polypeptides were separated in denaturing gels, transferred to nitrocellulose, and probed with antibodies to VP22 and ICP4 as described in Materials and Methods. The subcellular extracts were probed with antibody to the immediate-early ICP4 protein to control for the effects of the drugs.
The results at 9 hpi (Fig. 5A) show that slightly more VP22 was present in the infected cytoplasmic extract than in the nuclear fraction in the presence of taxol (compare lane 10 with lane 4). Approximately equal amounts of VP22 were observed in each extract in the untreated (compare lanes 2 and 8) and nocodazole-treated (compare lanes 6 and 12) cells. At 13 hpi (Fig. 5B), more VP22 was detected in the nucleus than in the cytoplasm for all treatments. However, slightly less VP22 appeared in the taxol-treated nuclear extract (lane 4) than in the untreated (lane 1) and nocodazole-treated (lane 6) extracts. These findings are consistent with our earlier immunofluorescence results presented in Fig. 2 showing increased cytoplasmic VP22 in infected cells in the presence of taxol at late infection times. As expected, more ICP4 was observed in the nucleus than in the cytoplasm at 9 and 13 hpi. Based on these findings, we conclude that taxol treatment of infected cells can lead to retention of VP22 in the cytoplasm of cells.
FIG. 5.
Partitioning of VP22 in nuclear and cytoplasmic fractions in cells infected in the presence of taxol and nocodazole. Vero cells were synchronously mock (M) or HSV-1(F) infected in the absence or presence of taxol (Tx) or nocodazole (Nc). At 9 (A) and 13 (B) hpi, nuclear and cytoplasmic fractions (Extract) were prepared, and polypeptides were separated in denaturing gels, transferred to nitrocellulose, and probed with antibodies to VP22 and ICP4 as described in Materials and Methods. Locations of the migrations of molecular mass markers are indicated in the right margin.
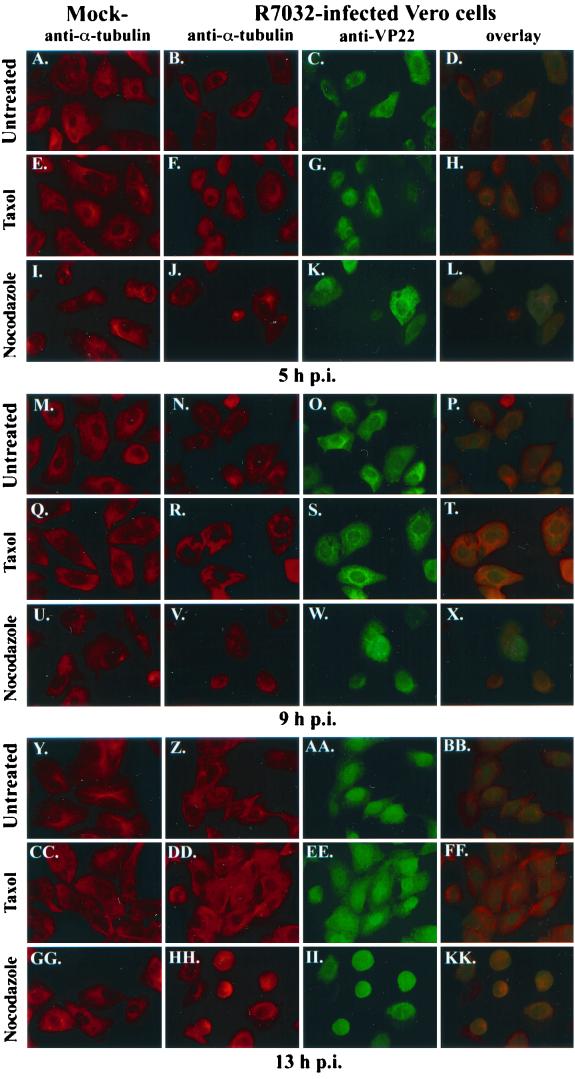
Indirect immunofluorescence of VP22 during HSV-1(R7032) infection in the presence of taxol and nocodazole.
Up to this point, our study has focused on the behavior of VP22 during wild-type HSV-1(F) infection. Our next goal was to confirm our findings by using the related HSV-1(R7032) virus, which contains a deletion of the gene encoding gE (31) and is therefore unable to form a gE-gI complex. Vero cells were synchronously mock- or HSV-1(R7032)-infected in the absence or presence of taxol or nocodazole prior to fixing the cells at 5, 9, and 13 hpi for immunofluorescence with antitubulin and anti-VP22 antibodies as described in Materials and Methods. The results of this study (Fig. 6) were as follows.
FIG. 6.
Indirect immunofluorescence of VP22 during HSV-1(R7032) infection in the presence of taxol and nocodazole. Vero cells were synchronously mock or HSV-1(R7032) infected without (Untreated) or with taxol and nocodazole, fixed at 5 (A to L), 9 (M to X), and 13 (Y to KK) hpi for indirect immunofluorescence at the times indicated, and stained with antibodies to α-tubulin and VP22 as described in Materials and Methods. Merged images (overlay) are shown in panels D, H, L, P, T, X, BB, FF, and KK. All images were acquired under the same conditions.
As expected, the addition of taxol led to the formation of tightly bundled microtubules (Fig. 6Q and CC), and nocodazole (Fig. 6U and GG) generated diffuse structures in mock-infected cells at 9 and 13 hpi. Consistent with the results in Fig. 2, minimal effects of the drugs on the mock-infected cells were observed at 5 hpi (Fig. 6E and I), and obvious MtOCs were detected in the untreated mock-infected cells at all time points (Fig. 6A, M, and Y). These results indicate that our drug treatment strategy is reproducible and consistent.
As we observed in Fig. 1, discrete MtOCs were detected in the untreated HSV-1(R7032)-infected cells at 5 hpi (Fig. 6B). These structures were lost at 9 and 13 hpi (Fig. 6N and Z). This finding differs from what we saw with HSV-1(F) where the loss of MtOCs was seen at 13 hpi (Fig. 1A, panel 6; Fig. 1B, panel 10). Although unexpected based on our data with the wild-type (parental) virus, our findings with HSV-1(R7032) are identical to those of Avitabile and colleagues (3), who also analyzed HSV-1(R7032). The basis for this difference between the two viruses is unknown and likely represents a function dependent on gE.
Consistent with our original findings (37) and those in Fig. 1, VP22 localized in the cytoplasm to a perinuclear region at 5 hpi in the untreated cells (Fig. 6C). This finding is important, since it confirms our previous argument that the cytoplasmic localization of VP22 is real and not related to binding of VP22 antibody by the gE-gI complex. VP22 is observed in the nucleus at 9 hpi (Fig. 6O), and it predominates there at 13 hpi (Fig. 6AA) in the untreated cells. The translocation of VP22 to the nucleus in the untreated cells was readily observed in the overlay images (compare Fig. 6D, P, and BB). Thus, while the kinetics of the translocation of VP22 to the nucleus during HSV-1(R7032) infection differs from that of HSV-1(F), the general pattern is the same in that it exists in the cytoplasm early and in the nucleus late.
As expected from our findings in Fig. 2, taxol treatment had little effect on the translocation of VP22 at 5 hpi (Fig. 6G). However, nocodazole treatment resulted in more VP22 being detected in the nucleus than with no treatment or in the taxol-treated cells (compare Fig. 6K with C and G). In addition, we found that the cell morphologies of the nocodazole-treated, infected cells differed from those of the other cells in that they were more rounded and had lost their classic fibroblast appearance. Consistent with Fig. 2, taxol treatment led to more cytoplasmic VP22 than in the untreated controls at 9 and 13 hpi (compare Fig. 6S with O and EE with AA). This phenomenon was also observed in the overlays where more yellow staining in the cytoplasm was seen in the taxol-treated cells (compare Fig. 6T with P and FF with BB). Although it appeared that more nuclear VP22 was present in the nocodazole-treated cells at 9 (Fig. 6W) and 13 (Fig. 6II) hpi compared to the other treatments, most of the cells appeared to be rounded up, and many had become detached from the coverslips. Thus, it seems that nocodazole treatment may act to increase the viral cytopathic effect as assessed by cell morphology. It is conceivable that a consequence of this effect of nocodazole is the reduction in virus yield that we observed above in our virus growth experiments in the presence of this drug.
Based on these results, we conclude the following. (i) Since HSV-1(R7032) does not produce viral Fc receptor, VP22 localization in the cytoplasm at 5 hpi detected by our indirect immunofluorescence techniques has been validated. (ii) Wild-type (parental) HSV-1(F) and the gE-deletion mutant HSV-1(R7032) are both able to induce microtubule reorganization during productive infection, but the times postinfection at which the processes are observed differ. (iii) Modulation of microtubule structure by the addition of either taxol or nocodazole influences the translocation of VP22 into the nucleus. Finally, (iv) these observations support our hypothesis that microtubule reorganization during HSV-1 infection facilitates VP22's nuclear translocation.
VP22 detected in the nuclear fraction following HSV-1(F) infection in the presence of PAA.
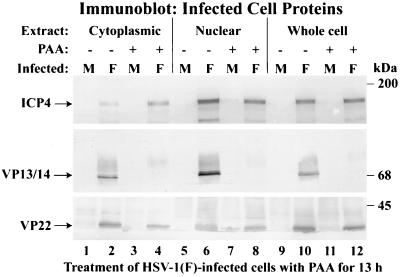
The previous findings support the model that the reorganization of microtubules late in infection facilitates the nuclear translocation of VP22. At least two important issues remain unknown. (i) Does VP22 retention in the cytoplasm during infection require additional viral proteins? (ii) What viral proteins participate in the reorganization of the microtubules? As an initial investigation to address these issues, we tested whether VP22 localization to the nucleus was dependent on HSV-1 true late (γ2) protein synthesis. Vero cells were synchronously mock or HSV-1(F) infected in the absence or presence of PAA (5) as described in Materials and Methods. Cells were pretreated with PAA for 1 h prior to the addition of virus, and the drug was maintained throughout the remainder of the infection. At 13 hpi, whole-cell extracts and nuclear and cytoplasmic fractions were prepared as described previously (37). Equal amounts of protein were separated in a denaturing gel, transferred to nitrocellulose, and probed with monoclonal antibody G49 specific for VP22 (37). As a control, the blot was also probed with antibodies specific for immediate-early ICP4 and VP13/14, a true late (γ2) tegument protein (29). The results (Fig. 7) of this study were as follows.
FIG. 7.
VP22 partitioning in the nuclear fractions of HSV-1(F)-infected cells does not require viral DNA or true late protein synthesis. Vero cells were synchronously mock (M) or HSV-1(F) infected in the absence (−) or presence (+) of PAA. At 13 hpi, whole-cell, nuclear, and cytoplasmic fractions (Extract) were prepared, and polypeptides were separated in denaturing gels, transferred to nitrocellulose, and probed with antibodies to VP22, VP13/14, and ICP4 as described in Materials and Methods. Locations of the migrations of molecular mass markers are indicated in the right margin.
As expected, VP22, VP13/14, and ICP4 were detected in the nuclear fraction of cells infected in the absence of PAA (Fig. 7, lane 6). VP22 was also detected in the infected-cell nuclear fraction in the presence of PAA (Fig. 7, lane 8). However, the amount of VP22 observed following PAA treatment was slightly reduced. A similar reduction in the amount of VP22 was observed in whole extracts prepared from cells infected in the presence of PAA (Fig. 7, lane 12). The decrease in the amount of VP22 synthesized during infection in the presence of PAA was expected (9) and is typical of the γ1 group of viral genes, whose expression is not strictly dependent on viral DNA replication (41). No VP13/14 was detected in PAA-treated whole-cell (Fig. 7, lane 12), nuclear (lane 8), and cytoplasmic (lane 4) extracts, confirming that VP13/14 was produced as a true late protein. This result confirms that the PAA-dependent inhibition of viral DNA synthesis was successful. Minor amounts of immune reactivity with the anti-VP13/14 antibody were observed on the blot above the VP13/14 region (at approximately 70 to 80 kDa) in the PAA-containing infected samples (Fig. 7, lanes 4, 8, and 12). This immunostain was a cross-reaction of the antibody with unspecified viral protein and was observed previously with infected-cell extracts by using a VP13/14-null virus (4). As expected, more nuclear than cytoplasmic ICP4 was detected in the presence and absence of PAA. No immune reactivity was observed with mock-infected cell proteins. Based on these results, we conclude that the amount of PAA and the incubation conditions used were appropriate to differentiate between the γ1 and γ2 gene classes. The detection of VP22 in the nuclear fraction of PAA-treated cells suggests that both viral DNA and true late protein synthesis are not required for the nuclear localization of VP22.
VP22 localization in nuclei of HSV-1(R7032)-infected cells does not require viral DNA or true late protein synthesis.
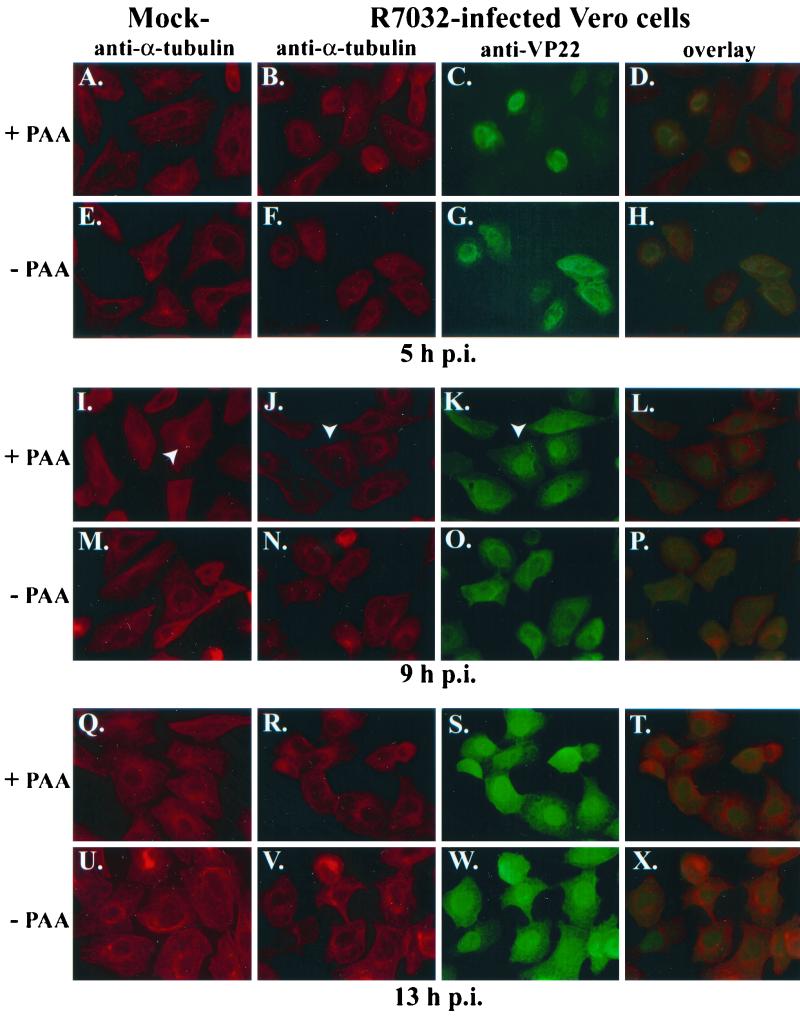
To confirm the results of our subcellular fractionation experiments which showed VP22 in infected-cell nuclear extracts in the presence of PAA, we performed indirect immunofluorescence studies with HSV-1(R7032). Vero cells were synchronously mock or HSV-1(R7032) infected in the presence or absence of PAA prior to fixing the cells at 5, 9, and 13 hpi for immunofluorescence with antitubulin and anti-VP22 antibodies as described in Materials and Methods.
The results (Fig. 8) of this experiment showed that VP22 localized to perinuclear regions in the presence (Fig. 8C and D) and absence (Fig. 8G and H) of PAA. These observations corroborate our findings shown in Fig. 1 with HSV-1(F). At 9 and 13 hpi, obvious nuclear VP22 was detected with (Fig. 8K and S) and without (Fig. 8O and W) PAA treatment. Comparison of the overlays either with (Fig. 8L and T) or without (Fig. 8P and X) PAA indicates that essentially identical amounts of VP22 were present in the nuclei. Consistent with our earlier findings (Fig. 1 and 6), obvious MtOCs were observed with mock-infected cells at 9 hpi (arrow in Fig. 8I), while none were detected in the infected cells (arrow in Fig. 8J), and VP22 was nuclear (arrow in Fig. 8K). As expected, no VP22 was detected in mock-infected cells (data not shown). Based on the results presented in Fig. 7 and 8, we conclude that HSV-1 DNA synthesis and true late protein synthesis are not required for the nuclear translocation of VP22.
FIG. 8.
Indirect immunofluorescence of VP22 during HSV-1(R7032) infection in the presence of PAA. Vero cells were synchronously mock or HSV-1(R7032) infected with (+) or without (−) PAA, fixed for indirect immunofluorescence at the times indicated, and stained with antibodies to α-tubulin and VP22 as described in Materials and Methods. White arrowheads mark cells referred to in the text. Merged images (overlay) are shown in panels D, H, L, P, T, and X. All images were acquired under the same conditions.
DISCUSSION
The goal of this study was to identify factors that might regulate the nuclear localization of VP22 during HSV-1 infection. The subcellular localization of VP22 was compared with the dynamic architecture of cytoplasmic microtubules during synchronized HSV-1 infections. The significant features of our studies may be summarized as follows.
(i) HSV-1(F)-induced microtubule rearrangement occurred in infected Vero cells by 13 hpi and was characterized by the loss of obvious MtOCs.
Reformation of MtOCs was detected by 25 hpi. In contrast, reorganization was clearly observed in HSV-1(R7032)-infected cells at 9 hpi (Fig. 6 and 8) The latter result is consistent with the original findings of Avitable and colleagues (3), who also investigated HSV-1(R7032)-infected cells. It is unclear why the microtubule rearrangement process seems to be “delayed” with the wild-type (and parental) HSV-1(F) virus, but this may reflect a previously unrealized function of the viral gE protein. During productive wild-type HSV-1 infection, gE associates with gI in a complex (24) at the trans-Golgi network (30). It is conceivable that the gE-gI complex might represent a viral component that acts in, and perhaps regulates, the microtubule rearrangement process. Additional biochemical and cell biological experiments must be performed to assess the validity of this intriguing hypothesis.
(ii) VP22 was observed in the cytoplasm of cells prior to microtubule rearrangement and localized in the nucleus following the process.
The previously reported association of VP22 with cellular microtubules (16) led us to hypothesize that microtubule fragmentation during HSV-1 infection (3) is a key step in the mechanism that regulates the nuclear localization of VP22. This model is supported by our data which indicate that the nuclear localization of VP22 during synchronized infection (37) correlates with HSV-1-induced microtubule reorganization (Fig. 1, 6, and 8).
Currently, the mechanism by which VP22 is actually imported into the nucleus remains unknown. A reductionist model suggests that VP22 is simply small enough to passively diffuse through the nuclear pore and is actively retained in the cytoplasm prior to 5 hpi via its association with cytoskeleton components or factors. While the VP22 homologue from bovine herpesvirus 1 (BVP22) does not contain any regions consistent with NLS (22), computational analyses of the HSV-1 VP22 primary structure (Materials and Methods) indicates the following. VP22 does not contain any regions that are consistent with a biphasic basic NLS. The prototype for this motif is nucleoplasmin (KRPAATKKAGQAKKKK, where underlined residues form the bipartite structure), which has two blocks of 2 or more basic amino acids separated by 10 amino acids (38). It appears that a spacer region of 9 to 11 amino acids is absolutely required for this motif (A. Radu, Mount Sinai School of Medicine, personal communication).
The best-defined NLS is the simple basic motif, which is exemplified by the simian virus 40 large T antigen (PKKKRKV) (25). This NLS is generally a short sequence that contains a cluster of basic amino acids, which is often preceded by either a proline or an acidic amino acid. While proteins that have a cumulative sum of K and R residues that is higher than 20% are considered to have a high possibility of being nuclear by the PSORT II algorithm (23), VP22 has a 15% basic amino acid content. To identify possible simple basic NLS motifs, PSORT II searches for two separate patterns (23). The first is termed pat4 and is either a four-residue pattern composed of four basic amino acids or a pattern composed of three basic amino acids and possessing either an H or P residue. The second is termed pat7 and is a pattern starting with a P followed within three residues by a basic segment containing three K/R residues out of four. VP22 contains one pat7 motif starting at amino acid 82 (PRTRRPV) and one pat4 motif starting at amino acid 295 (RPRR).
Neither of these motifs has been used to generate an artificial import substrate, as was done with the T antigen NLS (20), and caution is advised (Radu, personal communication) in basing nuclear import predictions solely on sequence analysis, since many well-defined import mediators do not recognize the classic types of NLS (27). Nevertheless, full-length VP22 is capable of recruiting fused jellyfish green fluorescent protein to the nuclei of cells during transfection and transient expression experiments (18). An important remaining question is the following. Even if VP22 could passively diffuse or be actively transported into the nucleus, what is the mechanism that retains it in the cytoplasm? In an earlier study, we originally reported that prior to 5 hpi, VP22 localizes in the cytoplasm to a perinuclear region, and additional investigations led us to conclude that it colocalized with a marker for the Golgi apparatus (37). While VP22 does not possess a cleavable signal sequence or a KDEL ER retrieval signal (44), our computational analysis (Materials and Methods) of its primary structure also identified an ER membrane retention motif in its amino terminus (TSRR starting at amino acid 2). Studies are currently under way to determine the significance of this observation in the productive replication of HSV-1.
(iii) Stabilization of microtubules by the addition of taxol increased the accumulation of VP22 in the cytoplasm either during infection or in cells expressing VP22 in the absence of other viral proteins.
These findings are consistent with those of studies which indicate that both transiently (15) and stably expressed (36) VP22 associates with cellular chromatin during mitosis. While the retention of microtubule-associated VP22 can occur in the absence of other HSV-1 proteins, these results do not exclude the possibility that other viral proteins are involved in regulating VP22's subcellular distribution during infection.
(iv) While VP22 localized to the nuclei of cells treated with the microtubule-depolymerizing agent nocodazole, either taxol or nocodazole treatment prevented optimal HSV-1(F) replication in Vero cells.
Although our results indicate that HSV-1(F) produced large amounts of infectious particles in the presence of either taxol or nocodazole, the yields were less than that obtained without the drugs. The interpretation of the taxol results that we favor is that the stabilization of microtubules by the drug leads to a reduction in the nuclear import of VP22 and thus limits its participation in virion assembly. At this time, we do not know whether additional virion components are also affected by taxol. However, the fact that an increase of VP22 in the cytoplasm correlates with reduced virus yield is consistent with our earlier finding that VP22 is required for efficient cell-to-cell spreading (36). The interpretation of the nocodazole results is complicated by the fact that the drug affects the cytopathic effect of the virus (Fig. 6). Thus, while it appears that more VP22 enters nuclei in the presence of the drug compared to in untreated cells, a consequence of the lack of a defined microtubule network in the cytoplasm may be a reduction in virion egress, which, in turn, could explain the drop in virus yield. These results are somewhat different from those of Avitabile and colleagues (3), who reported that the drugs did not significantly affect the release of virus. This conclusion was based on measurements of free virus released into the medium of infected cells. Our differences may reflect the fact that our data were obtained under conditions in which the infections were synchronized and our numbers considered cell-associated virus due to its dependence on VP22. Thus, in our experimental system, the dynamic state of the cytoskeleton appears to be a determinant of the efficiency of infectious virion production.
(v) VP22 migration to the nucleus occurred in the presence of PAA, indicating that viral DNA and true late protein synthesis were not required for its translocation.
This conclusion is based on the fact that we did not detect any representative VP13/14 true late protein in our studies in the presence of PAA, while VP22 was found in cell nuclei. Since microtubule reorganization was also observed during HSV-1 infection in the presence of PAA, true late viral proteins do appear to be required for this process as well. Together, these observations support our earlier findings that the nuclear translocation of VP22 correlates with the restructuring of microtubules.
Based on these results, we conclude that microtubule reorganization during HSV-1 infection facilitates the nuclear localization of VP22. Regulated VP22 nuclear localization initiates after 5 hpi with HSV-1 and is independent of viral DNA and true late protein synthesis. HSV-1-induced microtubule reorganization releases VP22 from the cytoskeleton, allowing its entry into the nucleus. Thus, stabilizing microtubules during infection or in VP22-expressing cells increases VP22 retention in the cytoplasm. During HSV-1 infection, microtubule interaction may present a means by which VP22 avoids nuclear localization during the early phase of the replication cycle. Experiments designed to determine whether the nuclear localization of other tegument proteins is similarly regulated are currently under way. Selective cytoplasmic retention of major tegument proteins may represent a novel mechanism for regulating infectious virion assembly.
ACKNOWLEDGMENTS
We thank (i) Aurelian Radu (Mount Sinai School of Medicine) for important discussions regarding nuclear localization motifs and the reliability of motif prediction algorithms, (ii) Jamie Yedowitz (Mount Sinai School of Medicine) for technical assistance in gathering data with our fluorescence microscope, and (iii) Bernard Roizman (University of Chicago) for providing low-passage isolates of HSV-1 (R7032), which was originally generated by Richard Longnecker, Penelope Mavromara-Nazos, and Amy Sears in his laboratory (31), and its parental strain HSV-1(F).
These studies were supported in part by grants from the United States Public Health Service (AI38873) and the American Cancer Society (JFRA 634). L.E.P. was supported in part by a United States Public Health Service Institutional Research Training Award (GM08553). A.B. was supported in part by an American Society for Microbiology Undergraduate Research Fellowship. J.A.B. thanks the Lucille P. Markey Charitable Trust and the National Foundation for Infectious Diseases for their support.
REFERENCES
- 1.Albright A G, Jenkins F J. The herpes simplex virus UL37 protein is phosphorylated in infected cells. J Virol. 1993;67:4842–4847. doi: 10.1128/jvi.67.8.4842-4847.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubert M, Blaho J A. The herpes simplex virus type 1 regulatory protein ICP27 is required for the prevention of apoptosis in infected human cells. J Virol. 1999;73:2803–2813. doi: 10.1128/jvi.73.4.2803-2813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avitabile E, Di Gaeta S, Torrisi M R, Ward P L, Roizman B, Campadelli-Flume G. Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J Virol. 1995;69:7472–7482. doi: 10.1128/jvi.69.12.7472-7482.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaho J A, Mitchell C, Roizman B. An amino acid sequence shared by the herpes simplex virus 1 alpha regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylylation of the UL21, UL31, UL47, and UL49 gene products. J Biol Chem. 1994;269:17401–17410. [PubMed] [Google Scholar]
- 5.Blaho J A, Mitchell C, Roizman B. Guanylylation and adenylylation of the α regulatory proteins of herpes simplex virus require a viral β or γ function. J Virol. 1993;67:3891–900. doi: 10.1128/jvi.67.7.3891-3900.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaho J A, Roizman B. Analyses of HSV proteins for posttranslational modifications and enzyme functions. Methods Mol Med. 1998;10:237–256. doi: 10.1385/0-89603-347-3:237. [DOI] [PubMed] [Google Scholar]
- 7.Browne H, Bell S, Minson T, Wilson D W. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J Virol. 1996;70:4311–4316. doi: 10.1128/jvi.70.7.4311-4316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang Y E, Roizman B. The product of the UL31 gene of herpes simplex virus 1 is a nuclear phosphoprotein which partitions with the nuclear matrix. J Virol. 1993;67:6348–6356. doi: 10.1128/jvi.67.11.6348-6356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conley A J, Knipe D M, Jones P C, Roizman B. Molecular genetics of herpes simplex virus. VII. Characterization of a temperature-sensitive mutant produced by in vitro mutagenesis and defective in DNA synthesis and accumulation of γ polypeptides. J Virol. 1981;37:191–206. doi: 10.1128/jvi.37.1.191-206.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunningham C, Davison A J, MacLean A R, Taus N S, Baines J D. Herpes simplex virus type 1 gene UL14: phenotype of a null mutant and identification of the encoded protein. J Virol. 2000;74:33–41. doi: 10.1128/jvi.74.1.33-41.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daikoku T, Ikenoya K, Yamada H, Goshima F, Nishiyama Y. Identification and characterization of the herpes simplex virus type 1 UL51 gene product. J Gen Virol. 1998;79:3027–3031. doi: 10.1099/0022-1317-79-12-3027. [DOI] [PubMed] [Google Scholar]
- 12.Daikoku T, Shibata S, Goshima F, Oshima S, Tsurumi T, Yamada H, Yamashita Y, Nishiyama Y. Purification and characterization of the protein kinase encoded by the UL13 gene of herpes simplex virus type 2. Virology. 1997;235:82–93. doi: 10.1006/viro.1997.8653. [DOI] [PubMed] [Google Scholar]
- 13.Davis L I. The nuclear pore complex. Annu Rev Biochem. 1995;64:865–896. doi: 10.1146/annurev.bi.64.070195.004245. [DOI] [PubMed] [Google Scholar]
- 14.Ejercito P M, Kieff E D, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 15.Elliott G, O'Hare P. Cytoplasm-to-nucleus translocation of a herpesvirus tegument protein during cell division. J Virol. 2000;74:2131–2141. doi: 10.1128/jvi.74.5.2131-2141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott G, O'Hare P. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J Virol. 1998;72:6448–6455. doi: 10.1128/jvi.72.8.6448-6455.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enquist L W, Husak P J, Banfield B W, Smith G A. Infection and spread of alphaherpesviruses in the nervous system. Adv Virus Res. 1998;51:237–347. doi: 10.1016/s0065-3527(08)60787-3. [DOI] [PubMed] [Google Scholar]
- 18.Fang B, Xu B, Kock P, Roth J A. Intercellular trafficking of VP22-GFP fusion proteins is not observed in cultured mammalian cells. Gene Ther. 1998;5:1420–1424. doi: 10.1038/sj.gt.3300741. [DOI] [PubMed] [Google Scholar]
- 19.Gershon A A, Sherman D L, Zhu Z, Gabel C A, Ambron R T, Gershon M D. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J Virol. 1994;68:6372–6390. doi: 10.1128/jvi.68.10.6372-6390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldfarb D S, Gariepy J, Schoolnik G, Kornberg R D. Synthetic peptides as nuclear localization signals. Nature. 1986;322:641–644. doi: 10.1038/322641a0. [DOI] [PubMed] [Google Scholar]
- 21.Granzow H, Weiland F, Jöns A, Klupp B G, Karger A, Mettenleiter T C. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harms J S, Ren X, Oliveira S C, Splitter G A. Distinctions between bovine herpesvirus 1 and herpes simplex virus type 1 VP22 tegument protein subcellular associations. J Virol. 2000;74:3301–3312. doi: 10.1128/jvi.74.7.3301-3312.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horton P, Nakai K. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc Int Conf Intell Syst Mol Biol. 1997;5:147–152. [PubMed] [Google Scholar]
- 24.Johnson D C, Frame M C, Ligas M W, Cross A M, Stow N D. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J Virol. 1988;62:1347–1354. doi: 10.1128/jvi.62.4.1347-1354.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalderon D, Roberts B L, Richardson W D, Smith A E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 26.MacLean C A, Rixon F J, Marsden H S. The products of gene US11 of herpes simplex virus type 1 are DNA-binding and localize to the nucleoli of infected cells. J Gen Virol. 1987;68:1921–1937. doi: 10.1099/0022-1317-68-7-1921. [DOI] [PubMed] [Google Scholar]
- 27.Mattaj I W, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 28.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 29.McLean G, Rixon F, Langeland N, Haarr L, Marsden H. Identification and characterization of the virion protein products of herpes simplex virus type 1 gene UL47. J Gen Virol. 1990;71:2953–2960. doi: 10.1099/0022-1317-71-12-2953. [DOI] [PubMed] [Google Scholar]
- 30.McMillan T N, Johnson D C. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J Virol. 2001;75:1928–1940. doi: 10.1128/JVI.75.4.1928-1940.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meignier B, Longnecker R, Mavromara-Nazos P, Sears A E, Roizman B. Virulence of and establishment of latency by genetically engineered deletion mutants of herpes simplex virus 1. Virology. 1988;162:251–254. doi: 10.1016/0042-6822(88)90417-5. [DOI] [PubMed] [Google Scholar]
- 32.Meredith D M, Lindsay J A, Halliburton I W, Whittaker G R. Post-translational modification of the tegument proteins (VP13 and VP14) of herpes simplex virus type 1 by glycosylation and phosphorylation. J Gen Virol. 1991;72:2771–2775. doi: 10.1099/0022-1317-72-11-2771. [DOI] [PubMed] [Google Scholar]
- 33.Morrison E E, Stevenson A J, Wang Y F, Meredith D M. Differences in the intracellular localization and fate of herpes simplex virus tegument proteins early in the infection of Vero cells. J Gen Virol. 1998;79:2517–2528. doi: 10.1099/0022-1317-79-10-2517. [DOI] [PubMed] [Google Scholar]
- 34.Mossman K L, Sherburne R, Lavery C, Duncan J, Smiley J R. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J Virol. 2000;74:6287–6299. doi: 10.1128/jvi.74.14.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parness J, Horwitz S B. Taxol binds to polymerized tubulin in vitro. J Cell Biol. 1981;91:479–487. doi: 10.1083/jcb.91.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pomeranz L E, Blaho J A. Assembly of infectious herpes simplex virus type 1 virions in the absence of full-length VP22. J Virol. 2000;74:10041–10054. doi: 10.1128/jvi.74.21.10041-10054.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pomeranz L E, Blaho J A. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J Virol. 1999;73:6769–6781. doi: 10.1128/jvi.73.8.6769-6781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robbins J, Dilworth S M, Laskey R A, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 39.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, editor. Fundamental virology. Philadelphia, Pa: Lippencott-Raven Publishers; 1996. pp. 1043–1107. [Google Scholar]
- 40.Schiff P B, Fant J, Horwitz S B. Promotion of microtubule assembly in vitro by taxol. Nature. 1979;277:665–667. doi: 10.1038/277665a0. [DOI] [PubMed] [Google Scholar]
- 41.Silver S, Roizman B. γ2-Thymidine kinase chimeras are identically transcribed but regulated as γ2 genes in herpes simplex virus genomes and as β genes in cell genomes. Mol Cell Biol. 1985;5:518–528. doi: 10.1128/mcb.5.3.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stackpole C W. Herpes-type virus of the frog renal adenocarcinoma. I. Virus development in tumor transplants maintained at low temperature. J Virol. 1969;4:75–93. doi: 10.1128/jvi.4.1.75-93.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taus N S, Salmon B, Baines J D. The herpes simplex virus 1 UL 17 gene is required for localization of capsids and major and minor capsid proteins to intranuclear sites where viral DNA is cleaved and packaged. Virology. 1998;252:115–125. doi: 10.1006/viro.1998.9439. [DOI] [PubMed] [Google Scholar]
- 44.Teasdale R D, Jackson M R. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the golgi apparatus. Annu Rev Cell Dev Biol. 1996;12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- 45.Weinheimer S P, Boyd B A, Durham S K, Resnick J L, O'Boyle D R., II Deletion of the VP16 open reading frame of herpes simplex virus type 1. J Virol. 1992;66:258–269. doi: 10.1128/jvi.66.1.258-269.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whiteley A, Bruun B, Minson T, Browne H. Effects of targeting herpes simplex virus type 1 gD to the endoplasmic reticulum and trans-Golgi network. J Virol. 1999;73:9515–9520. doi: 10.1128/jvi.73.11.9515-9520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]