Abstract
In vitro experiments have demonstrated intercellular trafficking of the VP22 tegument protein of herpes simplex virus type 1 from infected cells to neighboring cells, which internalize VP22 and transport it to the nucleus. VP22 also can mediate intercellular transport of fusion proteins, providing a strategy for increasing the distribution of therapeutic proteins in gene therapy. Intercellular trafficking of the p53 tumor suppressor protein was demonstrated in vitro using a plasmid expressing full-length p53 fused in-frame to full-length VP22. The p53-VP22 chimeric protein induced apoptosis both in transfected tumor cells and in neighboring cells, resulting in a widespread cytotoxic effect. To evaluate the anti-tumor activity of p53-VP22 in vivo, we constructed recombinant adenoviruses expressing either wild-type p53 (FTCB) or a p53-VP22 fusion protein (FVCB) and compared their effects in p53-resistant tumor cells. In vitro, treatment of tumor cells with FVCB resulted in enhanced p53-specific apoptosis compared to treatment with equivalent doses of FTCB. However, in normal cells there was no difference in the dose-related cytotoxicity of FVCB compared to that of FTCB. In vivo, treatment of established tumors with FVCB was more effective than equivalent doses of FTCB. The dose-response curve to FVCB was flatter than that to FTCB; maximal antitumor responses could be achieved using FVCB at doses 1 log lower than those obtained with FTCB. Increased antitumor efficacy was correlated with increased distribution of p53 protein in FVCB-treated tumors. This study is the first demonstration that VP22 can enhance the in vivo distribution of therapeutic proteins and improve efficacy in gene therapy.
p53 is one of the major cell cycle control genes. The p53 protein is a transcription factor that regulates cell cycle arrest and apoptosis in response to various stimuli (7, 21, 25). p53 is also the most commonly mutated tumor suppressor gene found in human cancers. Gene therapy using adenovirus-delivered p53 has been demonstrated to effectively inhibit growth of human tumor cells with altered p53 function (22, 30, 31). Human tumor lines that express wild-type p53 are more resistant to p53 gene therapy (15, 19), often due to other genetic alterations that enhance the degradation of p53 or impair its ability to trigger an apoptotic response (12, 13, 16). A mechanism to increase delivery of functional p53 protein to these cells could overcome this resistance and allow appropriate apoptosis to occur.
VP22 is a herpes simplex virus type 1 (HSV-1) tegument protein encoded by the UL49 gene (9). This protein has been shown to have the property of intercellular transport and internalization from infected cells to neighboring recipient cells (10). In vitro experiments have shown that VP22 fusion proteins also can undergo intercellular transport while retaining the activity of the fusion partner (6, 10, 24). Intercellular trafficking of the p53 tumor suppressor protein has been demonstrated in vitro using a plasmid expressing full-length p53 fused in-frame to full-length VP22. The p53-VP22 chimeric protein induced apoptosis both in transfected tumor cells and in neighboring cells, resulting in a widespread cytotoxic effect (24).
To evaluate the antitumor activity of p53-VP22 in vivo, we constructed recombinant adenoviruses (Ad) expressing either wild-type p53 (FTCB) or a p53-VP22 fusion protein (FVCB). We focused testing on p53 wild-type tumors to determine if enhanced delivery of p53 protein via VP22-mediated spread would overcome resistance in this tumor cell population. In vitro viral infections were used to compare protein expression levels over time by Western analysis, while dose-dependent apoptosis was measured via annexin V staining. VP22-mediated intercellular spread was confirmed through immunofluorescence analysis of coplating experiments, while safety was assessed in normal human cells using an MTS viability assay. Growth inhibition of in vivo established tumors and intratumoral spread of p53 protein was tested in nude mice.
Our results showed that FVCB expressed higher steady-state levels of p53 protein than FTCB in infected cell populations and that these levels persisted for a longer period of time. We verified that the in vitro spread reported previously using transfection experiments could also be observed in vitro via coplating experiments mixing Ad-infected and noninfected cells. We observed that FVCB infections resulted in significantly higher levels of apoptosis in resistant tumor lines than FTCB infections and that this effect was p53 specific. Despite the high p53 levels and increased apoptosis in p53 wild-type tumor cells, infection of normal human cells resulted in similar low levels of cytopathicity with either virus.
In vivo analysis matched up well with the in vitro findings, demonstrating a greater degree of tumor growth inhibition in an in vivo established tumor with FVCB compared to treatment with the wild-type p53 virus. In addition, we were also able to observe the in vivo spread of p53 protein, correlating this effect with the enhanced tumor killing. Taken together, our results suggest that a recombinant adenovirus expressing a p53-VP22 fusion protein could provide a means of enhancing p53 gene therapy through increased distribution of functional p53 protein.
MATERIALS AND METHODS
Cells.
Recombinant Ad were established and propagated in 293 cells (human embryonal kidney) from Microbix grown in minimum essential medium–10% fetal bovine serum (FBS). Cos-7 (green monkey kidney) cells used in the coplating assay cells were grown in Dulbecco's modified Eagle's medium (DME) supplemented with 10% FBS. The human tumor cell lines SK HEP 1 (liver adenocarcinoma) and RKO (colon carcinoma) were grown in DME–10% FBS, while G55 (glioblastoma) cells were grown in Hams F12/DME supplemented with 10% FBS. Normal human mammary epithelium, lung fibroblasts, and microvascular endothelium were purchased from Clonetics (San Diego, Calif.) and grown in specific growth media as defined by Clonetics.
Viruses.
The recombinant Ad expressing either wild-type p53 (A/C/N/53, designated FTCB here), empty control virus (ACN, designated ZZCB here), and the control reporter green fluorescent protein gene (GFCB) have been described previously (5, 29, 30). The recombinant adenovirus expressing the p53-VP22 fusion protein under the control of the cytomegalovirus (CMV) promoter was constructed as follows. VP22 was isolated from HSV-1 DNA (a kind gift from Chiang; Syntro) by PCR using primers which added on 5′ XbaI and 3′ BamHI restriction sites at the 5′ and 3′ ends of the VP22 coding sequence, respectively. The human p53 cDNA was isolated from the plasmid pA/C/N/53 described previously (30) via PCR using primers which removed the p53 stop codon and added on XbaI restriction sites at each end. This allowed an in-frame fusion between the 3′ end of the p53 coding sequence and the 5′ initiation codon of VP22. After PCR amplification, the fragments were isolated by gel purification, digested with XbaI plus BamHI or XbaI alone, respectively, and repurified from agarose gels. The plasmid pTG5623b (supplied by Transgene, Strasbourg, France) was digested with XbaI plus BamHI followed by calf intestinal alkaline phosphatase (CIP) treatment and ethanol precipitation. The VP22 fragment was first cloned into the XbaI, BamHI orientation, followed by insertion of the XbaI-p53 fragment into the XbaI-digested 5623-VP22 plasmid using standard cloning procedures (26). The resulting plasmid was characterized by restriction enzyme analysis and sequencing. This plasmid, designated p5623p53VP22, was then digested with XmnI and PmeI and transformed with ClaI-digested, CIP-treated plasmid 4213 (supplied by Transgene) into the Escherichia coli strain BJ 5183 cells. p4213 encodes for the entire Ad type 5 sequence, except for an E1 deletion of approximately 2.9 kb and an E3 deletion of approximately 1.9 kb. Homologous recombination in E. coli between the two DNA fragments was as described previously (4). The resulting plasmid encodes the entire Ad type 5 sequence, except for an E1 deletion where the Ad sequence is replaced by an expression cassette encoding the in-frame p53-VP22 fusion protein driven by the human CMV promoter/enhancer and the E3 deletion described above. This plasmid was linearized by PacI digestion and used to transfect 293 cells to produce viable, replication-deficient Ad-expressing p53-VP22 fusion protein. This virus was then designated FVCB. Identity of the correct sequence of the expression cassette was verified by sequencing. Recombinant Ad were isolated and purified by column chromatography as described previously (18, 27). All virus infections were carried out using purified virus dosed by concentration of virus particles. This was as recommended by the Center for Biologics Evaluation and Research and the Food and Drug Administration as published previously (3). Purified viruses were also characterized for their infectious units per milliliter as described previously (23). The characterization of both virus particle numbers per milliliter and infectious units per milliliter allow a ratio to be determined relating the physical characteristics of the virus to its biological property of infection. For FTCB and FVCB, this virus particle to infectious unit per milliliter ratio was 65:1 and 67:1, respectively.
Protein expression.
SK HEP 1 cells were seeded in 6-well plates at a density of 100,000 cells per well. The next day, the cells were infected with the indicated viruses at a concentration of 3 × 108 virus particles/ml (P/ml) for 1 h, washed once in phosphate-buffered saline (PBS), and returned to 37°C incubation in fresh medium. The cells were then harvested at 24, 48, or 72 h postinfection. Equal amounts of protein, as determined by Bradford assay (Bio-Rad cat. #500-0006), were loaded onto an 8% gel and transferred onto an Immobilon-P filter. The filter was probed first with mouse monoclonal p53 antibody (Ab) (Novocastra NCL-p53-1801) and then washed and re-probed with mouse monoclonal β-actin Ab (Sigma cat. #A-5441).
Human mammary epithelial cells were seeded in 6-well plates at a density of 900,000 cells per well. The next day the cells were infected with the indicated viruses at a concentration of 109 P/ml for 1 h, washed once in PBS, and returned to 37°C incubation in fresh medium. The cells were then harvested at 24 h postinfection. Equal amounts of protein were loaded onto an 8% gel and transferred onto an Immobilon-P filter. The filter was probed simultaneously with both mouse monoclonal p53 Ab (Novocastra NCL-p53-1801) and mouse monoclonal β-actin Ab (Sigma cat. #A-5441). To determine p21 activity, SK HEP 1 cells were seeded in a 6-well plate at a density of 120,000 cells per well and infected as before with 109-P/ml concentrations of indicated virus. The cells were harvested at 24, 48, and 72 h postinfection, and equal amounts of protein were loaded onto a 4 to 20% gel. The protein was transferred onto an Immobilon-P filter and probed with mouse monoclonal p21(187) Ab (Santa Cruz Biotechnology cat. #SC-817).
In vitro spread.
Cos cells were seeded in 6-well plates at a density of 250,000 cells/well and allowed to adhere overnight. The next day the cells were infected with a 5 × 108-P/ml concentration of either FTCB or FVCB and incubated overnight. The Cos cells were then trypsinized and coplated with uninfected SK HEP1 cells at a ratio of 1:50 (COS:SK HEP1), and 50,000 cells were seeded onto an 8-chamber slide. After a 24-h incubation, the cells were washed twice with PBS, fixed with ice-cold methanol for 10 min, and washed again with PBS. The cells were then blocked with 10% FBS, followed by the addition of primary Abs; mouse anti-simian virus 40 large T-antigen Ab (PharMingen 14111A) and rabbit polyclonal p53 Ab (Novocastra NCL-p53-CM1). After 30 min at room temperature, the cells were washed with PBS and the secondary Abs were added (rabbit anti-mouse rhodamine-tetramethyl rhodamine isothiocyanate [TRITC] [Jackson ImmunoResearch #315-025-045] and donkey anti-rabbit-aminocoumarin [AMCA] [Jackson ImmunoResearch #711-156-152]). After a 20-min incubation at room temperature, the cells were washed with PBS and fluorescent signals were observed. Images were captured using a Hammamatsu 3CCD analog camera and controller.
Apoptosis.
G55 and RKO cells were seeded in 6-well plates at a density of 50,000 cells/well while SK HEP 1 cells were seeded at a density of 70,000 cells /well and allowed to adhere. The following day the cells were pulsed for 1 h at the indicated virus doses and then were washed with PBS. Fresh medium was added and the cells were harvested 48 or 72 h postinfection. The media, PBS wash, and trypsinized cells were pooled and spun at 1,000 rpm on a clinical centrifuge (Beckman TJ-6). The cell pellet was resuspended and incubated in 50 μl (1:50 dilution) of annexin-V Fluos (Boehringer Mannheim cat. #1828681) followed by the addition of 400 μl of annexin binding buffer (10mM HEPES [pH 7.4], 140 mM NaCl, 5 mM CaCl2). The cells were then analyzed by flow cytometric analysis for apoptosis using the FL-1 channel to compare subpopulation frequencies.
Caspase 9 activation.
SK HEP 1 cells were treated with either FTCB, FVCB, or control virus ZZCB at a 6 × 108 P/ml concentration. After 48 h, 106 cells per assay were lysed in 50 μl of cell lysis buffer (Clontech, Inc.). To the cell lysates, 20 mM of substrate (Enzyme Systems) for caspase 9 (Ac-LEHD-AFC) in reaction buffer (Clontech, Inc.) was incubated at 37° for 1 h. Arbitrary fluorescence units were determined on a Cytofluor fluorescence multiwell plate reader (PerSeptive Biosystems) on a 400-nm excitation filter and 505-nm emission filter.
MTS assay in normal cells.
Primary normal human mammary epithelial cells, normal human lung fibroblasts, and normal human microvascular endothelial cells were obtained (Clonetics) and cultured in specific growth medium (Clonetics). The cells were seeded at 5 × 103 cells per well in 96-well plates (Nunc) and incubated until they were 50 to 75% confluent. Adenoviral constructs were serially diluted and added to the cells. The plates were allowed to incubate at 37°C in 7% CO2. After 1 h, the viruses were removed and the cells were fed with 200 μl of growth medium and reincubated at 37°C. After a 3-day incubation period, 40 μl of MTS (Cell Titer 96 Aqueous nonradioactive cell proliferation assay; Promega) was added to the wells and the plates were incubated for an additional 4 h at 37°C. Absorbance was read using a SpectraMax 250 plate reader (Molecular Devices) at 490 nm. The values of the treated wells were calculated as a percent of untreated control.
In vivo tumor growth inhibition.
SK HEP 1 cells were inoculated into the flanks of female nude mice (5 × 106 cells/200 μl, subcutaneous injection). Ten days after tumor cell inoculation, animals were randomized by tumor size (approximately 70 mm3) and a cycle of five intratumoral injections were given beginning on day 0. Injections were given either daily or every other day as indicated in a 100-μl volume per injection. The concentration of virus used was 5 × 109, 1 × 109, or 5 × 108 P/injection of FTCB or FVCB. A high dose of 1 × 1010 of the control virus GFCB was used in the dose-response experiment, while a 5 × 109 dose was used in the time course/in vivo spread experiment. Five animals were used per dose per group of FTCB or FVCB, while four animals received each GFCB dose. An additional five animals received PBS buffer control injections. Tumor measurements were taken weekly over a 7-week period following the initial virus injections.
In vivo p53 protein spread.
SK HEP 1 cells were inoculated into nude mice as before, and two animals per group were infected with either FTCB, FVCB, or control virus GFCB. Each animal was given a daily injection of 5 × 109 P/injection for 5 days and was sacrificed 48 h after the last virus injection. Their tumors were harvested and divided into two sections for immunohistochemistry for p53 protein expression and for DNA extraction for PCR quantification of viral hexon DNA. p53 immunohistochemistry was performed on formalin-fixed paraffin-embedded tissue. Tissues were sectioned at 5 μm onto Superfrost Plus slides (Fisher Scientific, Pittsburg, Pa.). Slides were deparaffined in Propar clearant (Anatech Ltd., Battle Creek, Mich.) and rehydrated through decreasing alcohol steps to water. Antigen retrieval was performed in a microwave using Zymed citrate buffer, pH 6.0 (San Francisco, Calif.), in deionized water. Slides were cycled on low power for 1 min and then on no power for 2 min for a total of 5 cycles, and then they were cooled in the buffer in an ice bath for 15 min. Slides were then rinsed for 15 min in three changes of PBS. Tissues were blocked with Zymed CAS Block for 30 min in a humid chamber, and then the block was aspirated off. The primary Ab used was the anti-p53 CM1 rabbit polyclonal Ab from Novocastra Laboratories Ltd. and was applied at a 1:1,000 dilution in PBS and incubated in the humid chamber for 60 min. Slides were washed for 15 min in three changes of PBS. The Texas Red-conjugated goat anti-rabbit secondary Ab from Molecular Probes (Eugene, Oreg.) was applied at a dilution of 1:200 in 1% goat serum in PBS and incubated for 30 min in a humid chamber in the dark. Slides were washed for 15 min in three changes of PBS and then mounted with Prolong Antifade (Molecular Probes) containing a 1:1,000 dilution of 4′, 6′-diamidino-2-phenylindole (DAPI) nuclear counterstain (Molecular Probes). Fluorescent signal was visualized using a Nikon Optiphot-2 upright microscope with a mercury lamp, fitted with Nikon single band filter blocks B-2A (Long Pass filter) for Texas Red and DAPI (Band Pass filter). Images were captured using a Hammamatsu 3CCD analog camera and controller.
In vivo hexon DNA PCR.
The tumor tissue excised for PCR was snap frozen in liquid nitrogen. DNA was extracted using the Qiagen Mini Kit (Qiagen) per the manufacturer's protocol. Quantification of Ad hexon was performed using real-time quantitative PCR (26, 27) utilizing the 5′ nuclease activity of Taq polymerase to detect PCR products. Real-time quantitative PCR was performed using the Taqman PCR Core Reagent (PE Applied BioSystems). Hexon PCR was performed in a total volume of 50 μl containing 1.0 μl of total DNA extracted from 1.0 × 1011 to 1.0 × 1012 total viral particles, 200 μM concentrations of (each) dATP, dGTP, or dCTP and 400 μM of dUTP, 1.25 U of AmpliTag Gold DNA Polymerase, 6 U of AmpErase UNG, 300 nM concentrations of each primer, a 300 nM concentration of probe, 1% Tween, and 0.05% gelatin. The following thermal cycler conditions were optimized for the above genes: 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 62°C. Viral DNA was extracted from gradient purified replication-competent Ad type 5 (Advanced Biotechnologies Inc.) as described previously. Viral DNA was then quantified using a PicoGreen dsDNA Quantitation Kit (Molecular Probes) per the manufacturer's protocol. The gene copy number or particle number of hexon was estimated using Avogadro's number. Serial dilutions of viral DNA were used in the PCR assays as standards for quantitation of hexon. The sequence of the oligonucleotides used for hexon detection in the assay above are the following: hexon 5′ primer, 5′-ACTATATGGACAACGTCAACCCATT-3′; hexon 3′ primer, 5′-ACCTTCTGAGGCACCTGGATGT-3′; and probe, FTM-ACCACCGCAATGCTGGCCTGC-TAMRA.
RESULTS
Expression of p53-VP22 fusion protein.
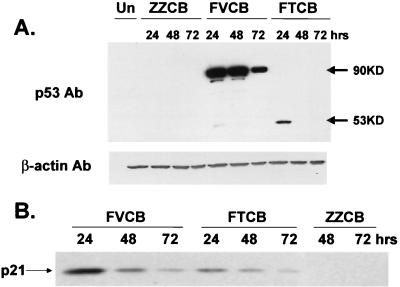
In the p53 wild-type tumor line SK HEP 1, added wild-type p53 protein undergoes rapid degradation and loss. Although p53 protein expression was evident from both constructs at 24 h postinfection (Fig. 1A), it was lost below detection levels by 48 h postinfection for FTCB. In contrast, protein expression levels were easily detectable through 72 h at the same dose of FVCB and were at initially higher levels than those of FTCB as well (Fig. 1A). This would be expected from an accumulation of p53 within a greater number of cells due to its spread by VP22. The shift in protein size for FVCB to a 90-kDa band represents intact VP22 (38 kDa) and p53 (53 kDa) fusion proteins. A β-actin probe was used as a control to ensure equal loading between samples.
FIG. 1.
Protein expression and transcriptional activation from wild-type p53 construct FTCB and p53-VP22 fusion construct FVCB. (A) SK HEP 1 cells were infected with 3 × 108-P/ml concentrations of either FTCB, FVCB, or control virus ZZCB. Lysates were harvested at 24, 48, or 72 h postinfection as indicated. Equal amounts of total protein were loaded per well, and p53 protein was detected by Western analysis. A secondary probing with Ab versus β-actin was used to verify equal loading between wells. (B) SK HEP 1 cells were infected with 109-P/ml concentrations of virus as indicated, and cell lysates were harvested at 24, 48, or 72 h postinfection. Equal amounts of total protein were loaded per lane, and p21 protein levels were detected by Western analysis using a monoclonal Ab against p21.
Induction of p21 expression by both FTCB and FVCB.
One of the hallmarks of p53 function is the ability to induce p21 expression (8, 20). We therefore tested the ability of both FTCB and FVCB to induce p21 expression following viral infection. SK HEP 1 cells were infected with FTCB, FVCB, or empty control virus ZZCB at concentrations of 109 P/ml for 1 h, followed by cell harvest at 24, 48, or 72 h postinfection. Western analysis showed that both FTCB and FVCB were able to induce p21 expression in a similar fashion over the time tested, while the control virus did not (Fig. 1B). These results indicate that the p53 expressed from the VP22 fusion construct FVCB was active and retained the ability to induce transcription despite being part of a fusion protein.
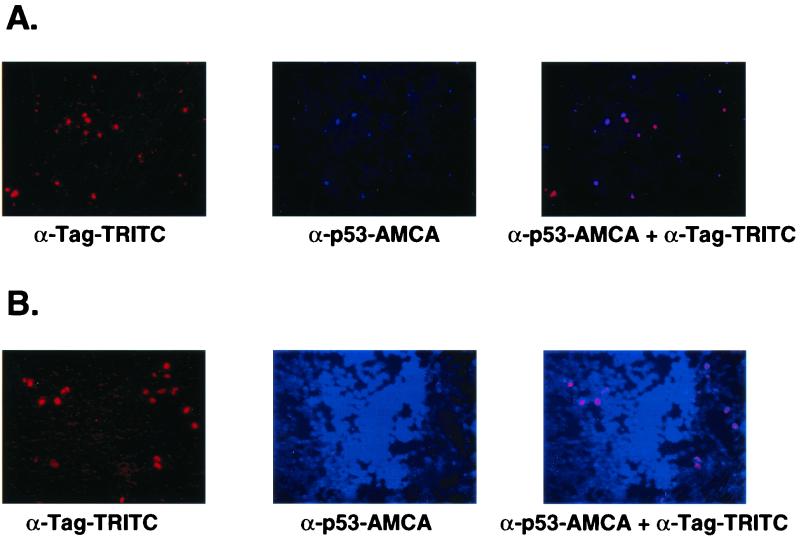
In vitro spread of p53 by VP22.
In vitro spread of p53-VP22 protein expressed from the FVCB recombinant Ad was demonstrated by mixing infected Cos cells with an excess of noninfected SK HEP 1 cells. The Cos cells could be identified by using fluorescent-tagged antibody against large T antigen and thus could be distinguished from the SK HEP 1 cells. When FTCB was used to infect Cos cells followed by coplating with noninfected SK HEP 1 cells, p53 protein and T antigen were colocalized to the Cos cells (Fig. 2A; red signal identifies T antigen, blue signal identifies p53). When FVCB was used to infect the Cos cells prior to being mixed with SK HEP 1 cells, the p53-VP22 fusion protein was clearly seen to have spread beyond the T antigen-identified Cos cells and into the surrounding SK HEP 1 cells (Fig. 2B). This intercellular spread of protein expressed from a recombinant Ad is similar to that reported by Elliot and O'Hare (10) and Phelan et al. (24) in coplating experiments using transfected VP22 fusion plasmids. Similar results were seen when mixing FTCB- versus FVCB-infected Cos cells with an excess of noninfected PC-3 or HeLa cells (data not shown).
FIG. 2.
In vitro spread of p53 by VP22. (A) Cos cells were first infected with 5 × 108-P/ml concentrations of FTCB virus, and then were replated the next day with a 50-fold excess of uninfected SK HEP 1 cells. Twenty-four hours later, cells were analyzed by immunohistochemistry for Cos cell identification (red-TRITC anti-T-antigen [Tag] detection) and p53 protein expression (blue-AMCA anti-p53 detection). (B) The conditions and treatments were the same as above except that FVCB virus was used to infect the Cos cells.
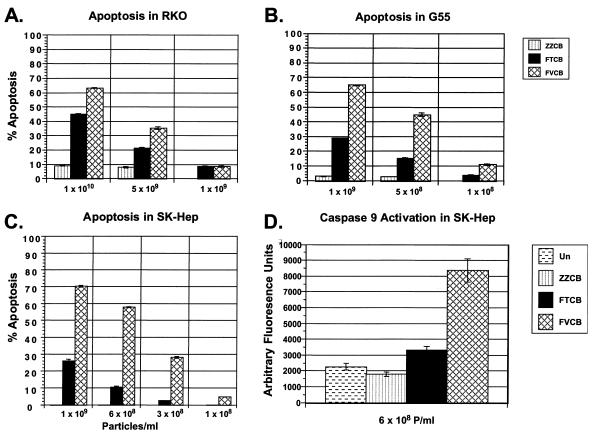
Apoptosis in p53 wild-type tumor cells.
Ad-delivered p53 has been demonstrated to be an effective inhibitor of growth of human tumor cells with altered p53 status (30, 31). However, human tumor lines expressing endogenous wild-type p53 are often resistant to exogenous p53 treatment (15, 19). Although these lines contain endogenous p53, other alterations can result which impair the ability of p53 to trigger an appropriate apoptotic response and render them less sensitive to exogenous p53 treatment. In Fig. 3A through C, we have compared the abilities of FTCB and FVCB to induce apoptosis in a variety of different tumor cell types which are p53 wild type. Although FTCB was able to trigger apoptosis over levels seen with control virus, FVCB was able to reach greater levels of apoptosis than FTCB at multiple doses. Treatment with FVCB at 109 P/ml induced high levels of apoptosis: 65% of G55 cells, 70% of SK HEP 1 cells, and 63% of RKO cells. In contrast, treatment with FTCB produced levels of only 29, 26, and 45% in the same cell types. These findings were consistent across the different tumor types (colorectal, glioblastoma, and hepatocellular carinoma), indicating that this increased efficacy should be applicable to the vast majority of p53 wild-type tumors. In p53-altered tumor cell lines which are sensitive to p53 treatment, FVCB and FTCB effects were both effective at similar levels (data not shown).
FIG. 3.
Apoptosis and caspase 9 activation. (A through C) Different p53 wild-type human tumor cell types as indicated that were infected with either control virus ZZCB (striped column), FTCB (solid column), or FVCB (hatched column) at the doses indicated (virus particles per milliliter). Harvest points were either at 48 h postinfection (A) or at 72 h (B and C). Uninfected (Un) cells had undetectable levels of apoptosis under these conditions. Results plotted are from triplicate experiments ± standard deviations. (D) SK HEP 1 cells infected with 6 × 108-P/ml concentrations of either control virus ZZCB (striped column), FTCB (solid column), or FVCB (hatched column). After 48 h, cells were harvested and analyzed for caspase 9 activation. Results are plotted in arbitrary fluorescence units versus uninfected (dashed column) or virus treatment. Results are plotted from the average from four experiments ± standard deviations.
p53-specific apoptosis through caspase 9 activation.
Caspase 9 is reported to be an essential and p53-specific mediator of apoptosis (28). To ensure that the increase in apoptosis seen in Fig. 3A through C was p53 specific, we looked for specific activation of caspase 9 from FTCB- and FVCB-infected tumor cells. Using p53-resistant SK HEP 1 cells, an increase in caspase 9 activity was seen for FTCB relative to that of uninfected or ZZCB control virus-treated cells. The increase in caspase 9 activity seen in the FVCB-treated cells was also significantly greater than that seen with FTCB (Fig. 3D). This increase correlated with the increase in apoptosis measured by annexin V staining of FVCB over FTCB in the same cells, indicating p53-specific induced apoptosis.
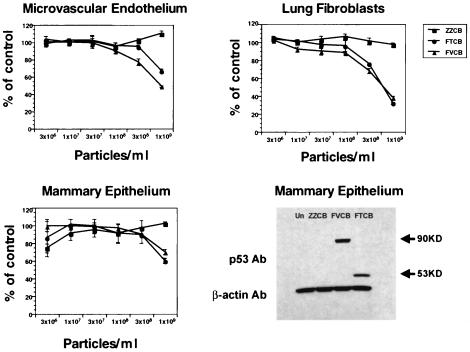
Safety in normal cells.
Because we were seeing an increase in apoptosis in p53 wild-type tumor cells with FVCB, it was necessary to verify that this did not lead to an increase in cytotoxicity in normal, p53 wild-type cells. We cultured primary normal human epithlium, fibroblasts, and endothelial cells and infected them with increasing concentrations of either the wild-type p53 virus FTCB or the p53-VP22 fusion construct FVCB or control ZZCB virus. An MTS-based assay was used to assess cell viability 3 days after virus infection to determine if there was any increase in cytotoxicity between wild-type p53 and the p53-VP22 fusion protein. Figure 4 shows that there was no increase in cytotoxicity in cells infected with FVCB relative to those infected with FTCB. As a control, we infected additional human mammary epithelial cells with the highest dose of viruses used in the MTS experiments. Western analysis verified transduction and p53 protein expression in these cells, with greater expression evident from the FVCB-infected cells, as was demonstrated earlier in tumor cells (Fig. 1). Therefore, despite increased protein expression and apoptosis in p53 wild-type human tumor cells infected with FVCB, the increased killing by FVCB appears specific for tumor cells.
FIG. 4.
Viability of normal human cells treated with virus. Normal human mammary epithelial cells, lung fibroblasts, or microvascular endothelial cells were cultured and infected with increasing concentrations of control virus ZZCB (squares), FTCB (circles), or FVCB (triangles) as indicated and assayed 3 days postinfection for cell viability via an MTS assay. Cell viability is plotted as the percent of untreated control cells ± standard deviations versus increasing concentrations of virus. Additional human mammary epithelial cells were plated and infected with 109-P/ml concentrations of ZZCB, FTCB, or FVCB virus and were harvested 24 h postinfection. Equal amounts of total protein were loaded per well, and p53 protein was detected by Western analysis. A secondary Ab versus β-actin was used to verify equal loading between wells.
In vivo tumor growth inhibition.
To determine whether or not the increase in apoptosis observed in vitro with FVCB would also be seen in vivo, a subcutaneous model for tumor growth inhibition was set up. SK HEP 1 cells were used to establish tumors that would subsequently be treated with virus or controls. Established tumors were injected either daily or every other day for 5 total doses of either FTCB, FVCB, or control GFCB virus. When injected daily, FVCB results in a significant increase in tumor growth inhibition over that of FTCB (P = .0002) over the course of the experiment (Fig. 5A). In a dose-response experiment (Fig. 5B), both FTCB and FVCB were able to repress tumor growth at the high dose. However, as the virus dose decreased, FTCB began to lose its ability to inhibit tumor growth, and by the lowest dose tested, FTCB treatment was as ineffective as control virus or buffer. In contrast, FVCB treatment was still able to retain its tumor growth inhibition, even at the lowest dose tested. The degree of tumor growth inhibition by FVCB at the lowest dose tested (5 × 108 P/injection) was, in fact, similar to that seen with FTCB at a 1 log higher dose (5 × 109 P/injection), indicating an approximate 10-fold increase in efficacy with FVCB. Results from a high dose of control virus (GFCB, 1010 P/injection) were similar to those from vPBS treatment, indicating that virus alone did not repress this tumors growth and that the inhibition was p53 specific.
FIG. 5.
In vivo efficacy and dose response. (A) SK HEP 1 cells were inoculated into the flanks of nude mice and allowed to establish for 10 days. A series of 5 daily intratumoral injections were then given at a dose of 5 × 109 P of either control virus GFCB (square), FTCB (circle), or FVCB (triangle) per dose. Tumor volumes were measured weekly. Results shown plot the tumor volumes versus days postinjection for each dosing group over the time course of the experiment. (B) SK HEP 1 cells were inoculated into the flanks of nude mice and allowed to establish for 10 days. A series of 5 intratumoral injections were then given every other day at the indicated doses of either FTCB (circle) or FVCB (triangle). Control virus GFCB (square) was administered under the same schedule at a dose of 1010 P/dose. Control injections of vPBS (cross) were also administered to one group, and all tumor volumes were measured weekly. Results shown plot the tumor volumes at the end of the experiment at day 49 postinfection for FTCB and FVCB at each dose and for the GFCB- and vPBS-treated animals. s.e.m., standard error of the mean.
In vivo spread of p53 by VP22.
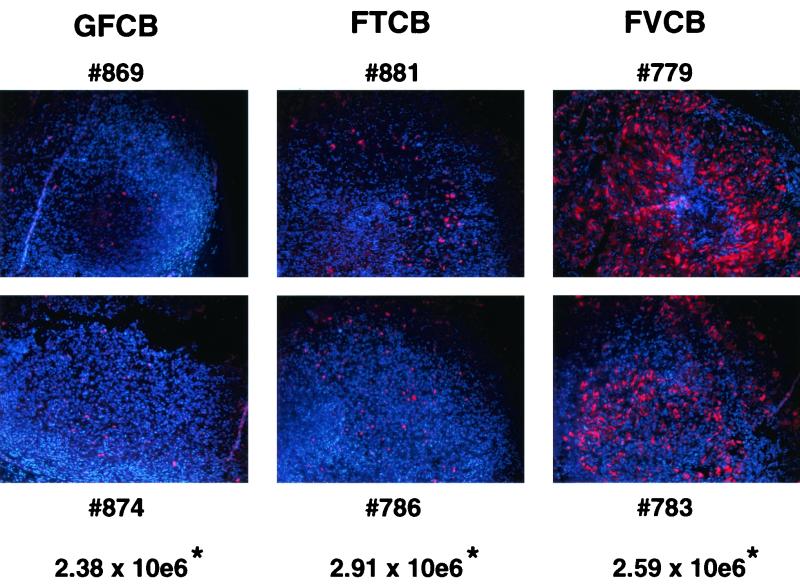
While in vitro spread mediated by VP22 has been demonstrated both here and elsewhere (2, 10), direct evidence for in vivo spread by a VP22 fusion protein has been lacking. Furthermore, while there has been some evidence for spread and increased effects with VP22 fusion protein treatment in vitro (6, 24), an in vivo treatment of an established tumor with a VP22 construct has not been reported. SK HEP 1 tumors from animals treated with 5 daily doses of virus as in experiments depicted in Fig. 5A were excised 48 h post-viral infection and were divided in half. One half was snap frozen in liquid nitrogen for DNA extraction while the other half was fixed in formalin for immunohistochemistry to determine in vivo p53 expression. In Fig. 6 the results are shown for two animals per group treated with FTCB, FVCB, or control virus GFCB. p53 expression is indicated in red against a blue DAPI counterstain. A slight increase in p53-expressing cells can be seen with FTCB treatment over that with control, GFCB-treated tumors. In the case of the FVCB-treated tumors, a striking increase in p53 expression was seen in both tumors. Quantitative PCR for copies of viral hexon DNA performed on the other half of the tumors indicated that similar amounts of virus were present for each treatment group. Therefore, the increase in p53 expression was not due to increased amounts of virus but instead reflects the in vivo counterpart to the in vitro spread demonstrated earlier. This in vivo spread of the p53-VP22 fusion protein correlates with the increased in vivo efficacy seen with FVCB treatment.
FIG. 6.
In vivo spread of p53 protein via VP22. SK HEP 1 cells were inoculated into the flanks of nude mice and allowed to establish for 10 days. A series of 5 daily intratumoral injections were then given at 5 × 109 of either control virus GFCB, FTCB, or FVCB per injection (n = 2 mice/group) as indicated. Tumors were excised 48 h after the last virus injection and processed for immunohistochemistry for p53 protein detection and for DNA extraction for PCR quantification of Ad hexon gene copies. Tumor sections from all six animals (identified by numbers) are shown along with the average copy number of hexon genomes for both animals per group per milligram of tissue (∗).
DISCUSSION
A present limitation of gene therapy is the ability to deliver sufficient amounts of active proteins to target cells. While secreted proteins can overcome this limitation to some degree, this is particularly a problem for nonsecreted proteins, such as p53. In these cases, the potentially therapeutic protein is only active in those cells in which it is initially delivered. When coupled with a cellular environment that limits the activity of the delivered gene product, such as MDM2 amplification associated with enhanced degradation of p53, potential efficacy can be further reduced. It has been suggested that VP22 fusion proteins could provide an approach to this problem. By increasing distribution through intercellular transport, more cells could potentially reach the levels of therapeutic proteins necessary for efficacy, leading to an overall enhancement of biological effects in the target population. While in vitro experiments have been promising, improved distribution and efficacy of VP22 fusion proteins has not been demonstrated in vivo. In the present study, recombinant Ad expressing either wild-type p53 (FTCB) or a p53-VP22 fusion protein (FVCB) were constructed and their activities were compared both in vitro and in vivo.
We observed that in vitro steady-state levels of the p53-VP22 protein from FVCB were elevated and persisted longer than those of wild-type p53 protein from FTCB. Both proteins were transcriptionally active. Ribonuclease protection assays from infected SK HEP 1 cells showed equivalent message for p53 transgene from both CMV promoter-driven constructs (data not shown). We verified earlier reports of in vitro spread by VP22 fusion proteins (10, 24), this time using our recombinant Ad in coplating experiments. We have recently described a recombinant Ad, p53 d13–19, that expresses a variant p53 protein containing a deletion in the MDM2 binding site (1). The p53 protein expressed by this virus is resistant to MDM2-mediated degradation and accumulates to higher levels than the wild-type p53 expressed by FTCB. Although the levels of p53 protein expressed by p53 d13–19 were similar to those produced by FVCB, there was no evidence for spread in coplating experiments as described in Fig. 2 (data not shown). Our results show that the activities of both VP22 and p53 are intact in FVCB and that differences in protein levels do occur which may result in differences in efficacy.
To evaluate whether or not higher protein expression levels correlated with increased apoptotic activity, several different tumor cell types were infected with either FTCB or FVCB. We observed that FVCB infection resulted in enhanced levels of apoptosis over a range of doses relative to that of FTCB. Increases of two- to fivefold higher apoptosis with FVCB were observed under several conditions in tumor cells normally resistant to p53 addition. As before, higher levels of protein expression were observed with FVCB relative to those with FTCB at the doses used (data not shown). In addition, the apoptosis we observed was p53 specific, as demonstrated by activation of the caspase 9 pathway (28). Control experiments using recombinant Ad expressing either a green fluorescent protein-VP22 fusion protein or just green fluorescent protein alone did not cause apoptosis in infected cells, while FVCB given at a log lower dose did, indicating that VP22 was not causing the toxicity (data not shown). Despite the increased expression of the p53 fusion protein and accompanying apoptosis in p53 wild-type tumor cells, safety did not appear to be compromised, as we did not observe any increased cytopathicity in normal, p53 wild-type human cells over that observed with FTCB.
By using a recombinant Ad as a delivery vehicle for the p53-VP22 fusion protein, we were able to extend our observations to the in vivo situation. We conducted in vivo tumor growth inhibition studies and monitored for in vivo spread of p53 protein. Treatment of established tumors with specific doses of virus clearly showed a loss of efficacy for wild-type p53 as the FTCB dose was decreased. At the lowest dose used, FTCB activity was similar to that of buffer control. In contrast, FVCB retained its efficacy, even as its dose was reduced 10-fold. The similar degree of tumor growth inhibition by FVCB when given at a log lower dose than that of FTCB suggests that lower doses may be used clinically as well, potentially improving both safety and cost of treatment.
This study was also the first demonstration that the intercellular trafficking by VP22 seen in vitro could also be observed in vivo. There have been some differences in the ability to detect spread in vitro (2, 11), and there has been a clear lack of direct in vivo observation of intracellular spread thus far. In our findings, sections from tumors treated with either FTCB, FVCB, or control virus, followed by immunohistochemistry for p53 expression, clearly showed in vivo spread of p53 protein with FVCB beyond that seen with wild-type FTCB. PCR analysis was used to rule out viral replication or an increased number of virus-infected cells as the cause, since equal levels of viral DNA were present in each tumor sample despite showing the different levels of p53 protein.
By utilizing the intercellular trafficking ability of HSV-VP22 coupled to the high-efficiency delivery of recombinant Ad, we were able to augment delivery of functional p53 to tumors normally resistant to exogenous p53. Since both infected and uninfected neighboring tumor cells were recipient cells for the fusion protein, functional p53 was able to reach effective levels in more cells, triggering apoptosis and destroying more tumor cells than with comparable infections using an Ad expressing wild-type, nonspreading p53. In addition, the confirmation of in vivo spread mediated by HSV-VP22 should also be important for other fusion constructs, providing a strategy for enhancing the efficacy of gene therapy with other nonsecreted proteins.
ACKNOWLEDGMENTS
We acknowledge Canji Cell Core for providing cells, Process Sciences for growth and purification of virus, and the Animal Facility for the care and treatment of the animals. We also thank Murali Ramachandra, Loretta Nielsen, and Peter O'Hare for scientific discussions.
REFERENCES
- 1.Atencio, I. A., J. B. Avanzini, D. Johnson, S. Neuteboom, M. T. Vaillancourt, L. L. Nielsen, G. Hajian, S. Sutjipto, B. J. Sugarman, J. Philopena, D. L. McAllister, J. C. Beltran, M. Nodelman, M. Ramachandra, and K. N. Wills. Enhanced apoptotic activity of a p53 variant in tumors resistant to wild-type p53 treatment. Mol. Ther., in press. [DOI] [PubMed]
- 2.Brewls N, Phelan A, Webb J, Drew J, Elliot G, O'Hare P. Evaluation of VP22 spread in tissue culture. J Virol. 2000;74:1051–1056. doi: 10.1128/jvi.74.2.1051-1056.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Center for Biologics Evaluation and Research, Food and Drug Administration. Guidance for human somatic cell therapy and gene therapy. Hum Gene Ther. 1998;9:1513–1524. doi: 10.1089/hum.1998.9.10-1513. [DOI] [PubMed] [Google Scholar]
- 4.Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, Mehtali M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J Virol. 1996;70:4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheney I W, Johnson D, Vaillancourt M, Avanzini J, Morimoto A, Demers B, Wills K, Shabram P, Bolen J B, Tavtigian S V, Bookstein R. Suppression of tumorigenicity of glioblastoma cells by adenovirus-mediated MMAC1/PTEN gene transfer. Cancer Res. 1998;58:2331–2334. [PubMed] [Google Scholar]
- 6.Dilber M S, Phelan A, Aints A, Mohamed A J, Elliot G, Smith C I E, O'Hare P. Intercellular delivery of thymidine kinase prodrug activating enzyme by the herpes simplex virus protein, VP22. Gene Ther. 1999;6:12–21. doi: 10.1038/sj.gt.3300838. [DOI] [PubMed] [Google Scholar]
- 7.El-Deiry W S. Regulation of p53 downstream genes. Semin Cancer Biol. 1998;8:345–357. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 8.El-Deiry W S, Harper J W, O'Connor P M, Velculescu V E, Canman C E, Jackman J, Pietenpol J A, Burrell M, Hill D E, Wang Y, Wiman K G, Mercer W E, Kastan M B, Kohn K W, Elledge S J, Kintzler K W, Vogelstein B. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 9.Elliot G D, Meredith D M. The herpes simplex virus type 1 tegument protein VP22 is encoded by the gene UL49. J Gen Virol. 1992;73:723–726. doi: 10.1099/0022-1317-73-3-723. [DOI] [PubMed] [Google Scholar]
- 10.Elliot G, O'Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 11.Fang B, Xu B, Koch P, Roth J A. Intercellular trafficking of VP22-GFP fusion proteins is not observed in cultured mammalian cells. Gene Ther. 1998;5:1420–1424. doi: 10.1038/sj.gt.3300741. [DOI] [PubMed] [Google Scholar]
- 12.Florenes V A, Maelandsmo G M, Forus A, Andreassen A, Myklebost O, Fodstad O. MDM2 gene amplification and transcript levels in human sarcomas: relationship to TP53 gene status. J Natl Cancer Inst. 1994;86:1297–1302. doi: 10.1093/jnci/86.17.1297. [DOI] [PubMed] [Google Scholar]
- 13.Fulci G, Labuhn M, Maier D, Lachat Y, Hausmann O, Hegi M E, Janzer R C, Merio A, Van Meir E G. P53 gene mutation and ink-4a-arf deletion appear to be two mutually exclusive events in human glioblastoma. Oncogene. 2000;19:3816–3822. doi: 10.1038/sj.onc.1203700. [DOI] [PubMed] [Google Scholar]
- 14.Gibson UEM, Heid C A, Williams P M. A novel method for real-time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 15.Harris M P, Sutjipto S, Wills K N, Hancock W, Cornell D, Johnson D E, Gregory R J, Shepard H M, Maneval D C. Adenovirus-mediated p53 gene transfer inhibits growth of human tumor cells expressing mutant p53 protein. Cancer Gene Ther. 1996;3:121–130. [PubMed] [Google Scholar]
- 16.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 17.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′-3′exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huyghe B G, Liu X, Sutjipto S, Sugarman B J, Horn M T, Shepard H M, Scandella C J, Shabram P W. Purification of type-5 recombinant adenovirus encoding human p53 by column chromatography. Hum Gene Ther. 1995;6:1403–1416. doi: 10.1089/hum.1995.6.11-1403. [DOI] [PubMed] [Google Scholar]
- 19.Kock H, Harris M P, Anderson S, Machemer T, Hancock W, Sutjipto S, Wills K N, Gregory R J, Maneval D C, Westphal M, Shepard H M. Adenovirus-mediated p53 gene transfer suppresses growth of human glioblastoma cells in vitro and in vivo. Int J Cancer. 1996;67:808–815. doi: 10.1002/(SICI)1097-0215(19960917)67:6<808::AID-IJC9>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 20.Kuwano K, Hagimoto N, Nomoto Y, Kawasaki M, Kunitake R, Fujita M, Miyazaki H, Hara N. P53 and p21 (Waf1/Cip1) mRNA expression associated with DNA damage and repair in acute immune complex alveolitis in mice. Lab Investig. 1997;76:161–169. [PubMed] [Google Scholar]
- 21.Lane D P, Lu X, Hupp T, Hall P A. The role of the p53 protein in the apoptotic response. Philos Trans R Soc Lond B Sci. 1994;345:277–280. doi: 10.1098/rstb.1994.0106. [DOI] [PubMed] [Google Scholar]
- 22.Neilsen L L, Maneval D C. P53 tumor suppressor gene therapy for cancer. Cancer Gene Ther. 1998;5:52–63. [PubMed] [Google Scholar]
- 23.Nyberg-Hofman C, Shabram P, Li W, Giroux D, Aguilar-Cordova E. Sensitivity and reproducibility in adenoviral infectious titer determination. Nat Med. 1997;3:808–811. doi: 10.1038/nm0797-808. [DOI] [PubMed] [Google Scholar]
- 24.Phelan A, Elliot G, O'Hare P. Intercellular delivery of functional p53 by the herpesvirus protein VP22. Nat Biotechnol. 1998;16:440–443. doi: 10.1038/nbt0598-440. [DOI] [PubMed] [Google Scholar]
- 25.Prives C, Hall P A. The p53 pathway. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Shabram P, Giroux D, Goudreau A M, Gregory R J, Horn M T, Huyghe B G, Liu X, Nunnally M H, Sugarman B J, Sutjipto S. Analytical anion-exchange HPLC of recombinant type-5 adenoviral particles. Hum Gene Ther. 1997;8:453–465. doi: 10.1089/hum.1997.8.4-453. [DOI] [PubMed] [Google Scholar]
- 28.Soengas M S, Alarcon R M, Yoshida H, Giaccia A J, Hakem R, Mak T W, Lowe S W. Apaf-1 and caspase-9 in p53-dependent apoptosis and tumor inhibition. Science. 1999;284:156–159. doi: 10.1126/science.284.5411.156. [DOI] [PubMed] [Google Scholar]
- 29.Wills K N, Huang W M, Harris M P, Machemer T, Maneval D C, Gregory R J. Gene therapy for hepatocellular carcinoma: chemosensitivity conferred by adenovirus mediated transfer of the HSV-1 thymidine kinase gene. Cancer Gene Ther. 1995;2:191–197. [PubMed] [Google Scholar]
- 30.Wills K N, Maneval D C, Menzel P, Harris M P, Sutjipto S, Vaillancourt M T, Huang W M, Johnson D E, Anderson S C, Wen S F, Bookstein R, Shepard H M, Gregory R J. Development and characterization of recombinant adenoviruses encoding human p53 for gene therapy of cancer. Hum Gene Ther. 1994;5:1079–1088. doi: 10.1089/hum.1994.5.9-1079. [DOI] [PubMed] [Google Scholar]
- 31.Zhang W W, Fang X, Mazur W, French B A, Georges R N, Roth J A. High-efficiency gene transfer and high-level expression of wild-type p53 in human lung cancer cells mediated by recombinant adenovirus. Cancer Gene Ther. 1994;1:5–13. [PubMed] [Google Scholar]