Abstract
Chronic prostate inflammation in patients with benign prostate hyperplasia (BPH) correlates with the severity of symptoms. How inflammation contributes to prostate enlargement and/or BPH symptoms and the underlying mechanisms remain unclear. In this study, we utilize a unique transgenic mouse model that mimics chronic non-bacterial prostatitis in men and investigate the impact of inflammation on androgen receptor (AR) in basal prostate stem cells (bPSC) and their differentiation in vivo. We find that inflammation significantly enhances AR levels and activity in bPSC. More importantly, we identify interleukin 1 receptor antagonist (IL-1RA) as a crucial regulator of AR in bPSC during inflammation. IL-1RA is one of the top molecules upregulated by inflammation, and inhibiting IL-1RA reverses the enhanced AR activity in organoids derived from inflamed bPSC. Additionally, IL-1RA appears to activate AR by counteracting IL-1α's inhibitory effect. Furthermore, using a lineage tracing model, we observe that inflammation induces bPSC proliferation and differentiation into luminal cells even under castrate conditions, indicating that AR activation driven by inflammation is sufficient to promote bPSC proliferation and differentiation. Taken together, our study uncovers mechanisms through which inflammation modulates AR signaling in bPSC and induces bPSC luminal differentiation that may contribute to prostate hyperplasia.
Subject terms: Stem-cell differentiation, Mechanisms of disease
Inflammation enhances androgen receptor levels and activity in basal prostate stem cells via IL-1RA, promoting their proliferation and differentiation, potentially contributing to benign prostate hyperplasia in mice.
Introduction
The coexistence of inflammation and benign prostate hyperplasia (BPH) is well documented1,2. The majority of BPH patients exhibit chronic inflammation and the extent of inflammation correlates with the severity of symptoms3,4. However, a direct link between inflammation and BPH remains an open question. Inflammation may contribute to BPH through multiple mechanisms2. One important mechanism may be through its modulation of prostate stem cell populations, which have long been implicated in hyperproliferative diseases5,6
Stem cells within the prostate basal epithelium (basal prostate stem cells (bPSC)) can differentiate into both luminal and basal lineages in in vitro organoid cultures and have been shown to contribute to the repair and recovery of the luminal epithelium in various animal models for prostate diseases and injuries. For example, lineage tracing models tracking bPSC have shown that these stem cells contribute to the repair of prostate luminal epithelium in an induced luminal anoikis model7, or to a smaller extent, during the castration-regeneration cycles8,9. We and others have shown that inflammation greatly impacts the behavior of bPSC. In our previous studies, we established a unique mouse model (Prostate Ovalbumin Expressing Transgenic 3 (POET3)) that closely models human chronic non-bacterial prostatitis, the most commonly diagnosed clinical prostatitis10. After adoptive transfer of pre-activated ovalbumin peptide-specific CD8+ T cells isolated from the spleens of OT-1 transgenic mice, this model experiences an influx of inflammatory immune cells into the prostate, including CD8+, CD4+, and Foxp3+/CD4+ T cells as well as myeloid-derived suppressor cells, and an increase in pro-inflammatory cytokine production10, which leads to the development of epithelial and stromal hyperplasia. Along with the induction of inflammation, we observed a significant expansion of bPSC population, which produced larger and more differentiated organoids (resembling type II tubule-like organoids, based on classifications by Kwon et al.11) in a 3D androgen-free culture system12. Inflammation-induced expansion of the bPSC population and enhanced bPSC differentiation capability were also described in other inflammation models, including bacteria-induced acute prostatitis13,14. The mechanisms underlying the impact of inflammation on bPSC and how the bPSC contributes to prostate hyperplasia are still poorly understood.
The androgen receptor (AR) plays a critical role in prostate homeostasis and maintenance. Drugs that target androgen synthesis and signaling remain as important treatments for both benign and malignant prostate diseases. In addition to its strong presence in the epithelium, AR activation in prostate stromal cells releases paracrine growth factors (e.g., NRG, FGF, IGF, EGF), which act on the corresponding receptors on luminal epithelial cells and contribute to epithelial and stromal expansion and the regeneration of luminal epithelium during the castration-regeneration cycle8,9. While strong AR nuclear presence is prevalent in luminal epithelial cells, AR staining is observed in about half of total basal epithelial cells8. Not surprisingly, AR is dispensable in the regression and recovery of basal epithelium during the castration and regeneration process8. However, AR is required for the differentiation of bPSC into luminal epithelial cells during the castration-regeneration cycle, as the number of lineage-traced bPSC in the luminal layer during regeneration was significantly reduced in basal epithelium-specific AR knockout mice8.
In the current study, we determined that both AR expression and activity were markedly stimulated in bPSC isolated from inflamed prostates in our POET3/OT-1 prostatitis model. With both organoid cultures and in vivo lineage tracing models, we have shown that AR signaling is essential for the differentiation of bPSC into luminal epithelial cells during prostatitis and inflammation stimulates AR even under androgen-deprived castrate conditions, which is sufficient to trigger basal to luminal differentiation. Most importantly, to the best of our knowledge, we identified a novel mechanism for AR modulation in bPSC where interleukin-1 receptor antagonist (IL-1RA) expressed by bPSC was shown to enhance nuclear localization of AR, modulate AR-dependent gene expression and function as the driver for bPSC luminal differentiation.
Results
AR activity is elevated in prostate basal stem cells isolated from inflamed POET3 mice
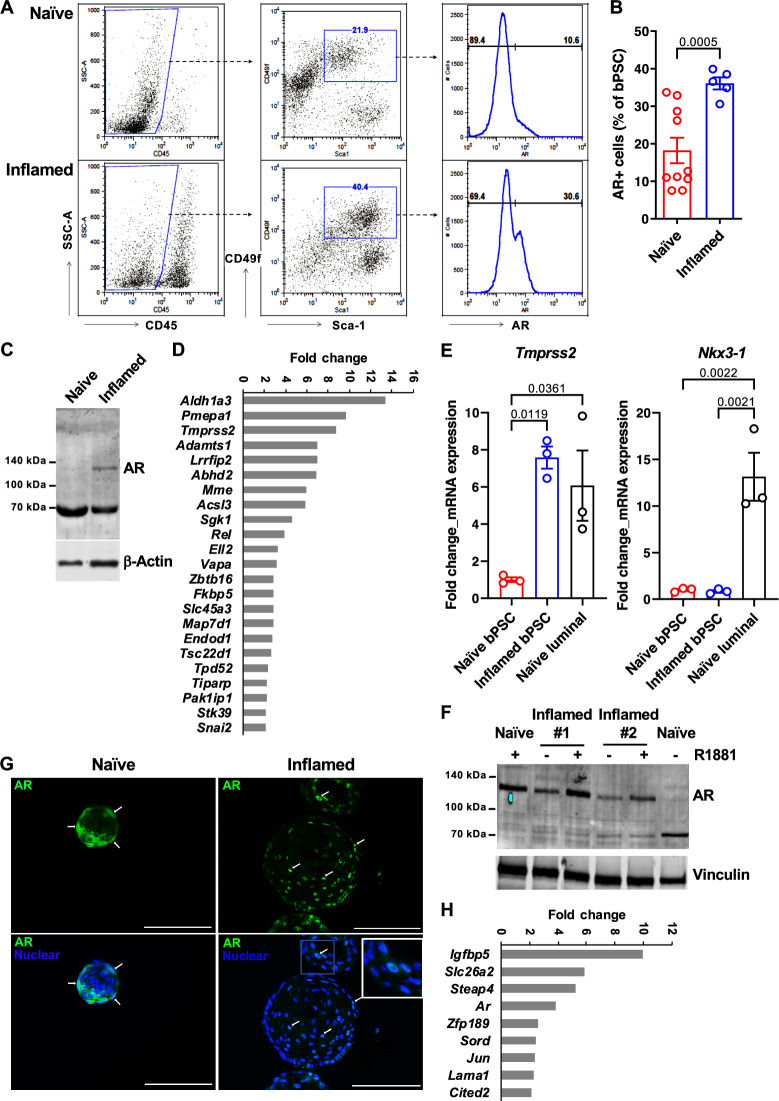
Given the critical function of AR in prostate homeostasis and stem cell differentiation, we assessed the impact of inflammation in our POET3/OT-1 prostatitis model on AR signaling in bPSC population. As shown in Fig. 1A, bPSC were identified via flow cytometry as CD45-/CD31-, Sca-1+, and CD49f+ (middle panels). A significantly higher percentage of the inflamed bPSC was positive for full-length AR identified with an antibody against C-terminal AR (Fig. 1A, right panels, and B). Consistent with the flow cytometry data, immunoblot using an antibody against N-terminal AR detected an AR band at about 120 kDa in inflamed bPSC, while, consistent with previous characterization studies15, the naïve bPSC produced a strong band of degraded AR at about 70 kDa (Fig. 1C). To verify increased AR activity, a quantitative real-time polymerase chain reaction (qRT-PCR) array for AR signaling target genes was used to compare the naïve and inflamed bPSC samples. The resulting heatmap illustrates the differential expression of various AR target genes between naïve and inflamed bPSC (Supplementary Fig. 1A), with twenty-three genes, including Tmprss2, significantly upregulated in inflamed bPSC (Fig. 1D, Supplementary Table 1). The upregulation of Tmprss2 in inflamed bPSC was further verified with qRT-PCR and its level in inflamed bPSC was comparable with the level observed in luminal epithelial cells (Fig. 1E). We further examined other canonical luminal AR target genes via qRT-PCR and found that the canonical target Nkx3-1 remained at similar levels between naïve and inflamed bPSC (Fig. 1E). Therefore, these data demonstrate that both AR levels and activity were increased in inflamed bPSC and AR activation driven by inflammation may trigger a different target gene program.
Fig. 1. AR expression and activity are elevated in bPSC freshly isolated from inflamed prostates and the enhanced AR activity is sustained in inflamed organoids in the absence of androgen.
A Representative flow cytometry dot plots and histograms of intracellular AR in bPSC from naïve and inflamed prostates. B Bar graph showing the percentage of AR+ cells within naïve and inflamed bPSC population as analyzed with flow cytometry (Naïve, n = 10; Inflamed, n = 5). C Immunoblot of full-length AR in naïve and inflamed bPSC lysates. D Fold change of AR target genes significantly upregulated in inflamed bPSC (Naïve or Inflamed, n = 3). E qRT-PCR of conventional AR target genes, Tmprss2 and Nkx3-1, in bPSC. Matched luminal population is included as a control (Naïve or Inflamed, n = 3). F Immunoblot showing increased level of full-length AR in inflamed organoids compared to naïve organoids. Full-length AR levels in both naïve and inflamed organoids were elevated with 1 nM R1881 treatment. G AR immunofluorescence staining in organoids derived from naïve or inflamed bPSC. Representative cells with nuclear-localized AR are indicated with white arrows. AR nuclear localization is also shown in the inset image with higher magnification (Scale bar, 100 μm). H Fold change of AR target genes significantly upregulated in inflamed organoids (Naïve or Inflamed, n = 3). Data presented in this figure are mean ± SEM.
Inflammation-driven AR activity is sustained in bPSC-derived organoids under androgen-deficient conditions and is essential for organoid growth and differentiation
To examine the persistence of altered AR signaling upon removal from the inflammatory environment and circulating androgens, bPSC were isolated from naïve or inflamed prostates and cultured under defined and serum-free organoid culture conditions in the absence of androgen12. AR expression and activity were analyzed in formed organoids. Immunoblot of lysates from naïve or inflamed organoids revealed that full-length AR was elevated in inflamed organoids and its levels were further increased with synthetic AR ligand R1881 (Fig. 1F). R1881 was able to stabilize AR in the naïve organoids, with band shifting from the degraded product at 70 kDa to full-length AR at 120 kDa15 (Fig. 1F). Consistent with the increase in total AR protein level, a significantly higher percentage of cells with overall or nuclear AR staining was observed in organoids derived from inflamed bPSC compared to those from naïve bPSC (Fig. 1G, with quantitation in Fig. 5C, Naïve control and Inflamed control groups), indicating that AR remains active in inflamed organoids even in the absence of androgen and inflammatory cells. Compared to the PCR array data collected from freshly isolated bPSC (Fig. 1D), AR target gene array with the organoids identified a smaller, yet intriguing set of significantly upregulated AR responsive genes in the inflamed organoids (Fig. 1H, Supplementary Fig. 1B, Supplementary Table 2). The modulation of these target genes confirmed the enhanced AR activity in organoids derived from inflamed bPSC. Furthermore, the difference in AR-responsive genes between freshly isolated bPSC and organoids suggests that AR may perform distinct functions in organoid growth and differentiation in androgen-depleted conditions.
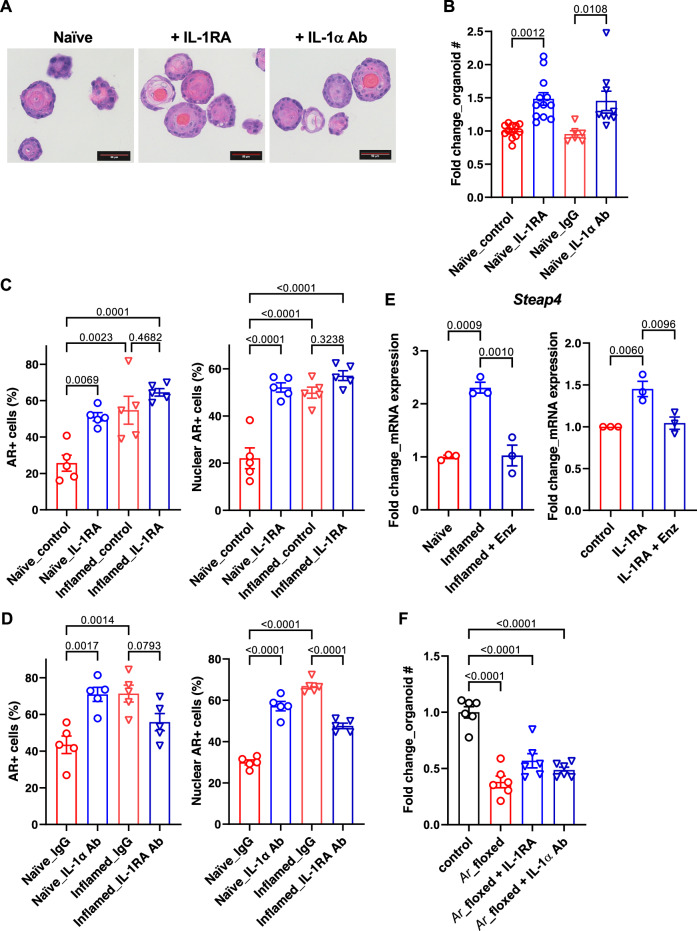
Fig. 5. IL-1RA mediates inflammation-induced differentiation and AR activity in bPSC organoids.
A Representative H&E images of naïve organoids treated with recombinant IL-1RA or neutralizing antibody (Ab) against IL-1α, showing an increase in stratification of treated organoids (Scale bar, 50 μm). B Organoid formation with naïve bPSC was significantly increased with recombinant IL-1RA (50 ng/mL) or IL-1α neutralizing Ab treatment (5 μg/mL) (Naïve_control, n = 12; Naïve_IL-1RA, n = 12; Naïve_IgG, n = 6; Naïve_IL-1α Ab, n = 9). C Quantitation of AR+ (both overall and nuclear) cells in naïve or inflamed organoids treated with IL-1RA (50 ng/mL) (All groups, n = 5). D AR staining (overall and nuclear) was increased in naïve organoids treated with an IL-1α neutralizing Ab (5 μg/mL) and decreased in inflamed organoids treated with an IL-1RA neutralizing Ab (5 μg/mL) (All groups, n = 5). E qRT-PCR showing that the AR target gene Steap4, which was induced in inflamed organoids and suppressed with Enz (left, n = 3), was upregulated with IL-1RA in naïve organoids and Enz treatment abolished the upregulation (right, n = 3). F The effects of IL-1RA or IL-1α neutralizing Ab were lost with naïve organoids derived from Ar_flox/y bPSC which were deprived of AR (All groups, n = 6). Data presented in this figure are mean ± SEM.
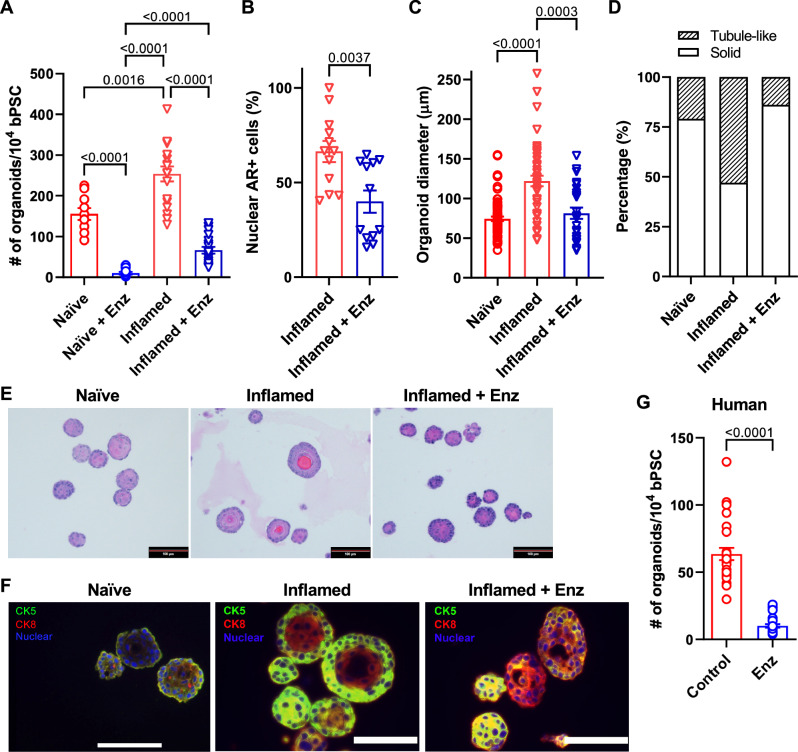
Given the strong expression of full-length AR in the inflamed bPSC and organoids, coupled with induced AR target gene expression and increased bPSC differentiation, we reasoned that the effects inflammation exerted on bPSC may be mediated through AR. To verify the requirement for AR signaling in inflammation-driven organoid proliferation and differentiation, naïve or inflamed bPSC were treated with the clinical AR antagonist, Enzalutamide (Enz), for the duration of organoid culture. AR inhibition resulted in a significant 74% reduction in inflamed organoid formation and 94% reduction in naïve organoid formation (Fig. 2A). These data suggest that the presence of low AR activity in naïve bPSC and organoids is essential for organoid formation, which is consistent with a previous report on the indispensable role of AR during basal to luminal differentiation using conditional AR knockout mouse models8. Of note, inflamed bPSC were able to form a significantly larger number of organoids compared to naïve bPSC in the presence of Enz, suggesting that inflamed bPSC are more resistant to Enz treatment. Consistently, ~40% of cells in Enz-treated inflamed organoids maintained their AR nuclear localization compared to 66% of cells in untreated inflamed organoids (Fig. 2B). In addition to reduced organoid number, Enz-treated inflamed organoids showed a significant reduction in size (Fig. 2C). Histopathological assessment further revealed an altered organoid morphology, with Enz-treated inflamed organoids more closely resembling naïve organoids. Organization and cellular stratification present in the inflamed organoids were reduced with Enz treatment, as reflected by the decreased percentage of tubule-like organoids with hollow lumens (Fig. 2D, E). Immunofluorescence staining verified the enhanced luminal differentiation in inflamed organoids with distinct central Cytokeratin 8 (CK8) (luminal epithelial marker) positive layers and peripheral CK5 (basal epithelial marker) positive layers (Fig. 2F). Enz treatment resulted in more solid organoids composed of CK8 and CK5 double-positive cells (Fig. 2D–F), indicating that Enz blocked the complete differentiation into CK8+, CK5- luminal cells and trapped the stem cells in a transitional state. In addition to treatments with pharmacological AR inhibitors, bPSC were also isolated from Ar_flox/y mice and infected with lentivirus encoding Cre recombinase. The consequent knockout of AR resulted in significantly reduced number of organoids (Fig. 5F, control and Ar_floxed groups). Furthermore, based on a protocol described by Henry et al.16, we enriched PSC from human prostate samples with flow cytometry as CD45-, EpCAM+, CD26-, and CD49f+ cells. Treatment with Enz significantly decreased human organoid formation by 84% (Fig. 2G). Taken together, these results from the inflamed bPSC-derived organoids with manipulated AR established the underlying role of AR in inflammation-driven organoid differentiation and AR may be linked to enhanced basal stem cell proliferation.
Fig. 2. Inhibition of AR activity reduces the differentiation of inflamed bPSC in organoid cultures.
A Number of organoids formed with naïve, inflamed bPSC with or without 10 μM Enz treatment (Naïve groups, n = 10; Inflamed groups, n = 18). B Quantitation of nuclear AR staining in inflamed organoids treated with Enz (n = 12). C Diameter of organoids and (D) percentage of organoids with tubule-like structures in naïve, inflamed organoids or inflamed organoids treated with Enz (Naïve, n = 74; Inflamed, n = 47; Inflamed+Enz, n = 25). E Representative H&E images of naïve, inflamed organoids or inflamed organoids treated with Enz (Scale bar, 100 μm). F CK5 (green) and CK8 (red) staining showing lack of organization in Enz-treated inflamed organoids (Scale bar, 25 μm). G Enz significantly reduces the number of organoids formed with human PSC (control or Enz, n = 27). Data presented in this figure are mean ± SEM.
IL-1RA is upregulated in bPSC from inflamed prostates and identified as an AR target gene
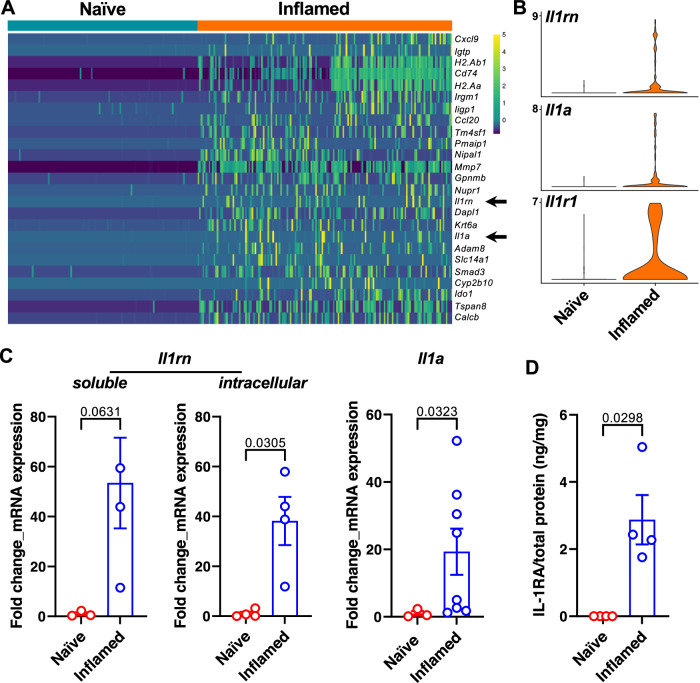
We next investigated the mechanisms that underlie the inflammation-driven changes in the basal stem cells, including the enhanced AR activity and differentiation, by conducting a single-cell RNA sequencing (scRNA-seq) analysis comparing freshly sorted naïve and inflamed bPSC. Along with a multitude of intriguing gene expression changes (Supplementary Data 1), IL-1 receptor antagonist (IL-1RA, gene name Il1rn) and IL-1α (gene name Il1a) were identified among the top differentially expressed genes between naïve and inflamed bPSC (Fig. 3A, arrows, and Fig. 3B), whereas IL-1β was not detectable in either naïve or inflamed bPSC. IL-1 receptor 1 (IL-1R1, gene name Il1r1) was also identified as significantly upregulated in inflamed bPSC by scRNA-seq analysis (Fig. 3B). The upregulation of Il1rn (both the soluble and intracellular isoforms) and Il1a was also confirmed with qRT-PCR (Fig. 3C). Consistent with its transcript expression, IL-1RA protein could be detected using ELISA in the lysates from inflamed bPSC but was undetectable in naïve bPSC lysates (Fig. 3D). IL-1α protein was not detectable by ELISA in the lysates of either naïve or inflamed bPSC, possibly because most of IL-1α was released by the bPSC.
Fig. 3. IL-1RA is upregulated in inflamed bPSC.
A Il1rn and Il1a (arrows) are among the most upregulated genes in the inflamed bPSC compared to naïve bPSC as analyzed with scRNA-seq. B Violin plots of Il1rn, Il1a and Il1r1 expression in naïve and inflamed bPSC. C qRT-PCR of Il1rn isoforms and Il1a in naïve and inflamed bPSC (Naïve, n = 3–4; Inflamed, n = 3–8). D Upregulation of IL-1RA protein is confirmed using ELISA with lysates collected from naïve or inflamed bPSC (Naïve or Inflamed, n = 4). Data presented in this figure are mean ± SEM.
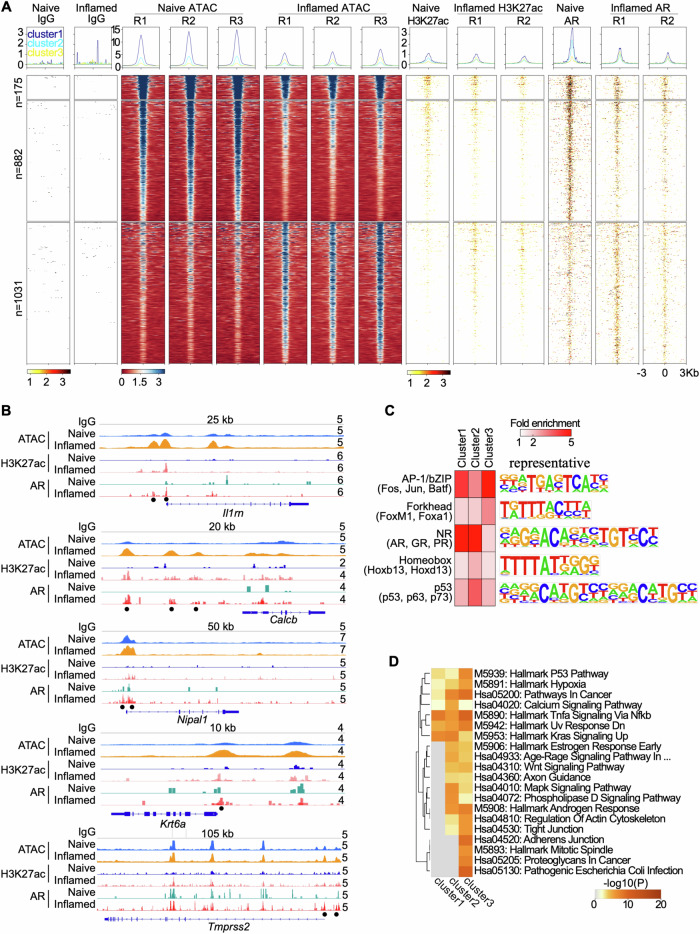
To better understand how inflammation triggers transcriptional changes across the genome, more specifically, AR-related transcriptional changes, we performed a set of genome-wide assays on inflamed and naïve bPSC, including Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) to measure chromatin accessibility and Cleavage Under Targets and Tagmentation (CUT&Tag) to measure H3K27ac histone modification and AR binding. Overall, ATAC-seq revealed 6502 loci with changes in chromatin accessibility when comparing inflamed to naïve bPSC, with nearly half of these loci (3188) more accessible in inflamed bPSC (Supplementary Data 2). Of the loci with chromatin changes, approximately one-third had AR binding, the majority of which were co-localized with regions marked by H3K27ac. AR-specific binding mirrored H3k27ac modification, which marks transcriptional activity, in inflamed or naïve bPSC (Fig. 4A).
Fig. 4. Genome-wide profiling of chromatin accessibility and AR binding.
A Histograms (above) and heatmaps (below) showing signal intensity of IgG, ATAC, H3K27ac, and AR peaks across naïve and inflamed bPSC, with biological replicates indicated as R1, R2, and R3. Differential peaks between naïve and inflamed bPSC with AR binding are displayed in three clusters. B Genome browser tracks at the Il1rn, Calcb, Nipal1, Krt6a, and Tmprss2 loci showing IgG, ATAC, H3K27ac, and AR signal in naïve and inflamed bPSC. Black dots represent peaks induced in inflamed bPSC. Track heights are indicated on the right corner for each track. C Enriched transcription factor (TF) DNA motifs at peak loci in each cluster. Shown are TF families on the left with representative TF members in parenthesis and a representative TF logo on the right. D Enrichment of KEGG and Hallmark pathways in genes adjacent to peaks from each cluster.
Clustering of loci with altered accessibility identified three groups: cluster 1 and 2 showed reduced chromatin accessibility and AR binding, while cluster 3 showed enhanced chromatin accessibility and AR binding in inflamed cells (Fig. 4A). Notably, some of the top induced genes in inflamed bPSC, including Il1rn, Calcb, Nipal1, Krt6a, and Tmprss2 (Figs. 3A and 1D), had increased chromatin accessibility, H3K27ac modification, as well as AR binding in their promoter and/or enhancer regions (Fig. 4B). These data indicate that AR directly regulates these genes and underscore the critical role of AR in inflammation-induced changes in bPSC.
To identify other regulatory factors potentially contributing to bPSC inflammation, we next examined the transcription factor (TF) binding motifs enriched in the three clusters and found that cluster 3 was enriched with Forkhead and AP-1 TF family motifs (Fig. 4C), although the AR binding motif was not enriched. FOXA1, a member of the Forkhead TF family, acts as a pioneering factor for AR by remodeling compact chromatin, opening up previously inaccessible regions, and facilitating AR recruitment to its target genes17–19. The AP-1 TF complex serves as a coregulator of AR by binding to it and modulating its transcriptional activity20–23. The presence of FOXA1 and AP-1 may enable AR to bind to less favorable sites and initiate a transcription program that includes non-traditional targets.
Finally, to further understand signaling pathways associated with changes in inflamed bPSC, we performed pathway analysis on the genes adjacent to loci identified in the three clusters (Fig. 4D). Consistent with enhanced AR binding in cluster 3, Androgen Response pathway was found to be enriched in this cluster. Pathways reflecting inflammation status (Pathogenic Escherichia Coli Infection) or involved in cell movements (Regulation of Actin Cytoskeleton, Tight Junction, and Adherens Junction) and proliferation (P53, Mitotic Spindle) were also enriched in cluster 3. Importantly, Wnt Signaling and Hypoxia pathways, which are implicated in stem cell maintenance and differentiation24–27, were also enriched in cluster 3. Collectively these data may provide further insights into how inflammation regulates prostate stem cell behavior and differentiation.
IL-1RA regulates AR activity and differentiation in bPSC-derived organoids
Given the impact of IL-1 on AR expression in prostate cancer cells and its impact on prostate cancer cell proliferation28–31, we reasoned that IL-1RA or IL-1α may modulate AR in inflamed bPSC and influence organoid growth and differentiation. We first initiated studies to evaluate the impact of IL-1RA on differentiation and AR activity in naïve organoids. Treatment of naïve bPSC cultured for organoid development with IL-1RA recombinant protein produced organoids with distinct cellular stratification, more closely resembling those from inflamed bPSC (Fig. 5A). IL-1RA treatment also induced a significant increase in the number of formed organoids (Fig. 5B). Naïve organoids treated with an IL-1α neutralizing antibody (Ab), which blocks the binding of IL-1α to its receptor, IL-1R1, and mimics the function of IL-1RA, showed more differentiated structure and increased number as well, resembling the results from IL-1RA treatment (Fig. 5A, B). Consistent with the inhibitory function of IL-1α in organoid formation, treatment with IL-1α significantly reduced the number of organoids formed with both naïve and inflamed bPSC (Supplementary Fig. 2). These data suggest that IL-1RA promotes bPSC organoid growth and differentiation by antagonizing the inhibitory effects of IL-1α.
We next assessed the impact of IL-1RA and IL-1α on AR in bPSC-derived organoids. AR levels and nuclear localization in organoids were analyzed with immunofluorescence. The number of cells with positive AR staining, and more importantly, the number of cells with AR nuclear localization were significantly increased in naïve organoids treated with IL-1RA to the level comparable with inflamed organoids (Fig. 5C). Addition of IL-1RA to organoids derived from inflamed bPSC that expressed a higher level of IL-1RA did not have any significant effects on AR (Fig. 5C). The effects of IL-1RA on AR levels in naïve organoids were mirrored by those induced by an IL-1α neutralizing Ab (Fig. 5D), suggesting that IL-1RA exerts its action on AR by blocking IL-1α. Steap4 was identified as one of the top upregulated genes in inflamed organoids (Fig. 1H). Enz treatment suppressed its upregulation in inflamed organoids (Fig. 5E, left panel), confirming that it is an AR target gene and its expression correlates with AR activity. Consistent with IL-1RA’s effects on AR nuclear localization, Steap4 was found to be significantly induced in naïve organoids with IL-1RA treatment and the upregulation was abolished in the presence of Enz (Fig. 5E, right panel). In contrast, an antibody that neutralized IL-1RA activity was observed to inhibit both total AR level and AR nuclear localization in inflamed organoids, demonstrating that IL-1RA is the driver of AR activation during inflammation (Fig. 5D). Data supporting the indispensable role of AR downstream of IL-1RA were also obtained with genetic deletion of AR in the organoids. When bPSC were isolated from Ar_flox/y transgenic mice and AR was removed from the organoids with Cre recombinase lentivirus infection, IL-1RA or IL-1α neutralizing Ab could no longer stimulate growth in AR knockout organoids (Fig. 5F). These data demonstrate that IL-1RA, which is upregulated in bPSC during inflammation, is responsible for activated AR in this stem cell population and enhanced organoid growth and differentiation.
Inflammation in a prostatitis mouse model promotes the basal to luminal differentiation
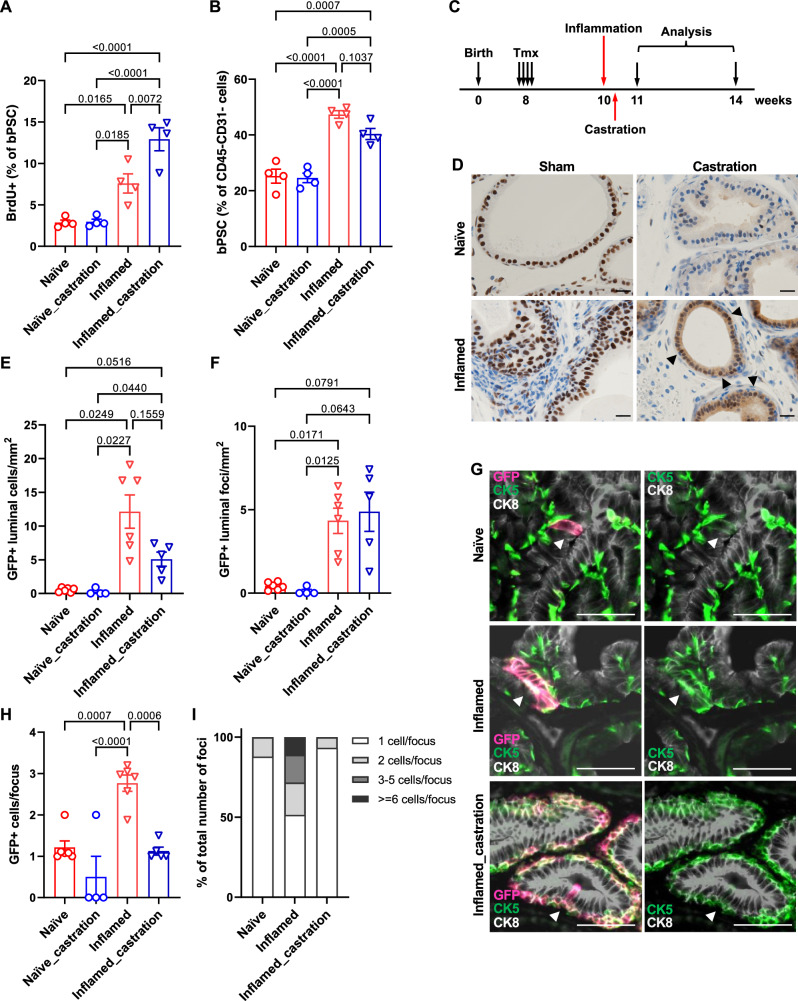
We have previously reported in our POET3/OT-1 prostatitis model that prostate inflammation expanded the size of the bPSC population in vivo12. To investigate whether inflammation increases the number of bPSC by stimulating their proliferation, POET3 mice were injected with pre-activated OT-1 T cells, and BrdU incorporation was analyzed 7 days later at the peak of inflammation. The percentage of proliferating BrdU+ cells in the bPSC population was found to be significantly increased in inflamed prostates compared to naïve prostates (Fig. 6A).
Fig. 6. Inflammation mimicking non-bacterial prostatitis promotes the proliferation of basal stem cells and the basal to luminal differentiation in vivo.
A Quantitation of BrdU+ bPSC in naïve and inflamed prostates, with or without castration (All groups, n = 4). B Percentage of bPSC in naïve and inflamed prostates, with or without castration (All groups, n = 4). C Experimental scheme showing time points of inflammation, castration and analysis. D AR staining shows maintenance of AR nuclear localization (arrow heads) in castrated inflamed prostates (Scale bar, 20 μm). E, F Quantitation of GFP+ cells or foci within the luminal layer (Naïve, n = 6; Naïve_castration, n = 4; Inflamed, n = 6; Inflamed_castration, n = 5). G Representative immunofluorescent images from naïve and inflamed prostates with or without castration showing the presence of GFP + CK5+ cells within the CK8+ luminal layer (arrowheads) (Scale bar, 50 μm). H, I Size of GFP+ foci and percentage of GFP+ foci of different sizes within the luminal layer (Naïve, n = 6; Naïve_castration, n = 4; Inflamed, n = 6; Inflamed_castration, n = 5). Data presented in this figure are mean ± SEM.
In addition to having a larger population size, we have also shown previously that bPSC isolated from inflamed POET3 mice exhibited enhanced growth and differentiation in organoid cultures, as reflected by increased organoid number, larger organoid size, and more distinct differentiation of the CK8+ luminal layer and CK5+ basal layer within the organoids12. To follow the basal to luminal differentiation upon inflammation in vivo and verify the results from the in vitro organoid cultures, we established a lineage tracing model, by crossing the POET3 mice with the mTmG;Krt5-creERT2 mice in which the Tomato protein expressed in basal epithelial cells switched to green fluorescent protein (GFP) upon the tamoxifen (Tmx)-induced Krt5 promoter-directed expression of Cre recombinase32. Lineage tracing was initiated in the resulting transgenic colony (POET3het;mTmGhet;Krt5-creERT2het) with intraperitoneal injections of Tmx, then activated OT-1 T cells were injected intravenously 2 weeks later to induce inflammation in the prostates. Animals were euthanized a month after inflammation initiation for both histological and flow cytometry analysis (Fig. 6C). Tmx injection induced GFP expression in ~90% of prostate basal epithelial cells as demonstrated by immunofluorescent staining with both anti-GFP and -CK5 antibodies (Supplementary Fig. 3A). Induction of GFP in basal epithelial cells was also confirmed with flow cytometry (Supplementary Fig. 3B). To assess the extent of basal to luminal differentiation after inflammation, prostate sections were stained with antibodies against GFP, CK8 and CK5. GFP+ luminal cells were counted in anterior lobes. We focused on anterior lobes because of their relative larger area, even under castrate conditions as described later. Total number of GFP+ cells found within the CK8+ luminal layer (GFP + CK8+) per mm2 of acinar area was dramatically increased in inflamed prostates (Fig. 6E). Increased number of GFP+ luminal cells in inflamed prostates was not only due to increased basal to luminal differentiation, as reflected by the number of GFP+ foci in the luminal layer (Fig. 6F), but also due to increased proliferation of GFP+ cells after they differentiated into luminal cells, as reflected by the number of cells within each GFP+ focus (Fig. 6G, H). Most (88%) GFP+ luminal cells found in naïve prostates were singles and the rest of the GFP+ luminal foci contained at most two cells (Fig. 6G, I), whereas GFP+ luminal cells found in inflamed prostates clustered in groups averaging 2.8 cells/cluster (Fig. 6G, H). 17% and 11% of GFP+ luminal foci in inflamed prostates were found to contain 3–5 cells and over 6 cells, respectively (Fig. 6I). Increased number of luminal GFP+ cells (identified as EpCAM+/CD49flow/GFP+) in inflamed prostates were also confirmed with flow cytometry (Supplementary Fig. 3C). Of note, nearly all of the GFP+ luminal cells expressed both luminal and basal markers (positive for both CK8 and CK5) (Fig. 6G) in both naïve and inflamed prostates, suggesting luminal cells derived from basal stem cells retained basal features and were of a transitional phenotype, at least at the 1-month time point. Taken together, data collected from our lineage tracing model, together with those from the BrdU incorporation experiment and organoid cultures, demonstrate that inflammation promotes bPSC proliferation, their basal to luminal differentiation, and proliferation of the luminal epithelial cells.
Given that inflammation modulates AR level and activity in bPSC, we next examined whether inflammation, by boosting AR signaling independent of androgen, is able to promote bPSC expansion and their differentiation into luminal cells in vivo under castrate conditions. Castration was performed 2 days after the initiation of inflammation so that the inflammation was accompanied with reducing levels of androgen. BrdU incorporation was analyzed 7 days after the initiation of inflammation and the presence of GFP+ luminal cells was analyzed 1 month later (Fig. 6C). We first examined AR intensity and nuclear localization with immunohistochemistry to confirm diminished AR activity after castration. AR nuclear staining in the epithelial cells was found to be more intense in inflamed prostates 1 week after the initiation of inflammation compared to naïve prostates. A faint, diffuse cytoplasmic AR staining pattern with little nuclear localization was observed in naïve prostates at merely 5 days after castration, indicating a sharp reduction in circulating androgen, whereas the nuclear AR staining persisted in inflamed prostates in castrated mice (Fig. 6D). Stronger cytoplasmic AR staining was also observed in castrated inflamed prostates.
Inflammation-induced expansion of bPSC persisted with castration (Fig. 6B) and bPSC proliferation as shown by BrdU incorporation remained significantly increased in inflamed prostates even after castration (Fig. 6A), consistent with our previous findings that bPSC population size in inflamed prostates remained unaffected with castration conducted 2 weeks before inflammation12. Interestingly, castration further increased the percentage of proliferating bPSC in inflamed prostates, suggesting a synergistic effect of inflammation and androgen deprivation on bPSC proliferation (Fig. 6A). Flow cytometry analysis of CD45+ leukocytes infiltrated into the prostates confirmed that robust inflammation was induced under the castrate condition (Supplementary Fig. 4). In our lineage tracing model, neither total number of GFP+ luminal cells or number of GFP+ foci was significantly altered with castration in inflamed prostates (Fig. 6E, F). However, the size of GFP+ foci within the luminal layer in inflamed prostates was significantly reduced with castration (Fig. 6H), with 93% of GFP+ luminal foci containing only one cell, similar to naïve prostates (Fig. 6G, I). These data demonstrate that the reduced level of androgen during inflammation does not affect the proliferation of bPSC, nor does it affect their basal to luminal differentiation. However, normal levels of androgen are essential for the proliferation of luminal cells during inflammation, as the transdifferentiated GFP+ cells stopped growing into multi-cell clusters with castration.
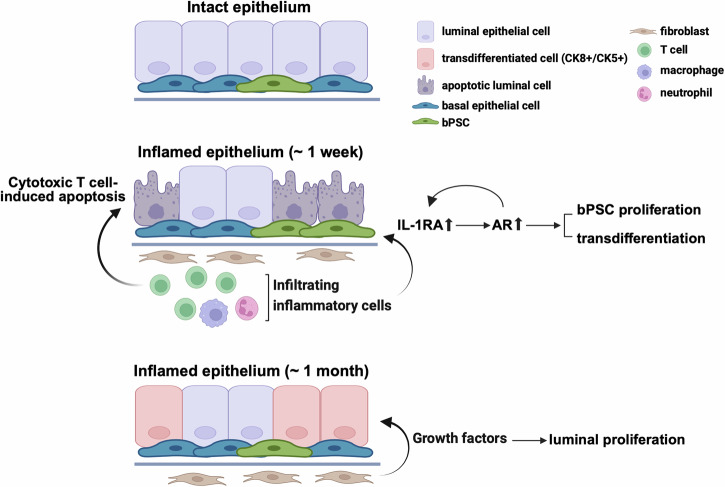
In summary, we have demonstrated in a prostatitis model that AR activity is stimulated in basal stem cells by inflammation and is essential for the luminal differentiation of these stem cells. IL-1RA, which is strongly upregulated during inflammation, was identified as the driver for AR activation independently of androgen in these stem cells and consequently responsible for inflammation-induced differentiation (Fig. 7). Il1rn was also identified as one of the AR target genes (Fig. 4B), suggesting a positive feedback loop between AR and IL-1RA in bPSC during inflammation. Given the identical results from IL-1RA or IL-1α neutralizing Ab treatments, IL-1RA may trigger the downstream pathway that leads to AR activation by antagonizing IL-1α. Furthermore, we have demonstrated that inflammation, by stimulating AR signaling in an androgen-independent fashion, promotes the proliferation and differentiation of bPSC, even under castrate conditions.
Fig. 7. Diagram of enhanced AR signaling, bPSC proliferation, and differentiation that are mediated through IL-1RA during prostate inflammation that mimics non-bacterial prostatitis.
(Created in BioRender. Yang, J. (2023) BioRender.com/b35v181).
Discussion
Although strong correlations have been reported between prostate inflammation and BPH risk and progression1–4, how inflammation contributes to the etiology of BPH remains poorly understood. Data reported herein identify a mechanism, in which inflammation drives the proliferation and differentiation of the stem cell population within the basal epithelial layer, by inducing IL-1RA, which results in the enhancement of AR activation (Fig. 7). In addition, the CUT&Tag and ATAC-seq data suggest that IL-1RA is an AR-dependent gene in bPSC, which is the first demonstration of the potential AR regulation of IL-1RA (Fig. 4B). The proposed mechanism was supported by data from both in vitro organoid cultures and an in vivo lineage tracing model. Using organoid cultures for mechanistic studies, we have demonstrated that IL-1RA is not only sufficient to induce AR activation independently of androgen and promote differentiation in naïve bPSC-derived organoids, but also an essential mediator in inflammation-driven AR activation, as inhibition of IL-1RA abrogated AR activation in inflamed bPSC-derived organoids. In our lineage tracing model, inflammation greatly promoted the basal to luminal differentiation even under castrate conditions by stimulating AR in bPSC independently of androgen. Furthermore, we have demonstrated that by eliminating AR through both pharmacological inhibition and genetic deletion, differentiation of bPSC into the luminal lineage was significantly inhibited in organoid cultures, which corroborates a previous report on the indispensable role of AR in the basal to luminal differentiation8 and further suggests that inflammation is an important contributor to epithelial hyperplasia through the bPSC to luminal differentiation process. Further studies are necessary to better understand the biological implications of basal to luminal differentiation relative to homeostatic maintenance via luminal progenitors9,33.
In our inflammation model, while castration did not impact inflammation-induced basal to luminal differentiation, it inhibited the proliferation of differentiated luminal cells. Luminal GFP+ cells differentiated from the basal layer remained as single cells, in contrast to the multi-cell luminal GFP+ clusters observed in non-castrate inflamed prostates. These data suggest that inflammation may not affect AR activity in fibroblasts where AR activation induces fibroblast secretion of growth factors such as NRG, IGF1, or FGF10. These growth factors bind to their corresponding receptors on luminal epithelial cells and are critical for luminal proliferation and replenishment during the prostate castration and regeneration cycle9 and in non-cell autonomous prostate growth in sub-renal capsule simulated prostate growth34. Under castrate conditions, inflammation is not sufficient to sustain AR in fibroblasts and provide paracrine signals for luminal proliferation. Notably, the lineage tracing data in castrate and intact mice show that inflammation-induced luminal cell hyperplasia requires two signals, one from the inflammatory process and the second from androgen (Fig. 6D–I), suggesting a possible link to glandular hyperplasia in BPH patients.
IL-1RA exists as both a secreted form (sIL-1RA) and multiple intracellular non-secreted forms35,36. The soluble extracellular sIL-1RA acts as an antagonist for IL-1R1 and competes with IL-1α or IL-1β for IL-1R1 binding. The intracellular IL-1RAs have been shown to inhibit IL-1R1-mediated target gene transcription but without affecting surface IL-1R1 engagement37,38. Although IL-1RA (either the precursor of the soluble isoform or intracellular isoforms) could be easily detected in the lysates of inflamed bPSC (Fig. 3D), consistent with the dramatic upregulation of Il1rn transcripts with inflammation as revealed by our scRNA-seq analysis and qRT-PCR, it is more likely that the extracellular soluble form mediates the effects of inflammation. First of all, enhanced differentiation and AR activation observed in inflamed organoids could be recapitulated in naïve organoids with the addition of recombinant IL-1RA in the culture media. Conversely, the aforementioned phenotypes of inflamed organoids could be reversed with the addition of a neutralizing antibody against IL-1RA. Secondly, the effects of recombinant IL-1RA resemble those from an IL-1α neutralizing antibody, suggesting that sIL-1RA functions by antagonizing IL-1α extracellularly and relieving IL-1α’s suppression of AR activity29–31. IL-1R1 is likely to be the link connecting IL-1RA with the subsequent intracellular events as our previous findings have demonstrated with a bacteria-induced prostate inflammation model that inflammation-induced expansion of basal stem cell population was significantly reduced in IL-1R1 knockout mice14. In vivo infiltrating leukocytes such as myeloid cells in the inflamed prostates might also be the source for sIL-1RA39,40. The expression pattern of IL-1RA in inflamed prostates, how IL-1RA from different sources contributes to stem cell AR activation and differentiation, and other inflammatory factors41–43 that may play critical functions in AR stabilization during prostate hyperplasia are currently under investigation.
In contrast to the presence of AR in luminal prostate cells and some mature basal cells, it has been reported by Xin et al. that only degraded AR was present in freshly isolated naïve bPSC and in organoid cultures derived from them15. Our data have shown the presence of full-length AR in the lysates from both inflamed bPSC and organoids, suggesting that inflammation may promote the stabilization of AR (Fig. 1C, F). The active involvement of AR in modulating genes in inflamed bPSC was further demonstrated in CUT&Tag and ATAC-seq studies where AR modulation of gene expression was implicated in cluster 3, which showed increased chromatin accessibility, H3K27ac modification, as well as AR binding in their promoter and/or enhancer regions (Fig. 4). Inflammatory signals such as cytokines and growth factors have been observed in the prostate cancer setting to stabilize AR independent of androgen through phosphorylation events44–48. It is possible that IL-1RA may bind to IL-1R, modulating NFκB pathway members31, altering the phosphorylation status of AR, and ultimately leading to AR stabilization. Understanding the mechanisms connecting inflammation, IL-1RA and AR will potentially benefit the treatment of not only the benign prostate diseases such as BPH but also malignant castration-resistant prostate cancer.
Taken together, our studies have identified IL-1RA as an AR modulator in the inflamed prostate. IL-1RA is both sufficient and necessary for the stimulation of AR expression and activity in bPSC and may be critical in inflammation-induced bPSC expansion and differentiation and prostate hyperplasia.
Methods
Animal studies
Male mice aged 8–12 weeks were utilized for all studies. All animals were housed and maintained under pathogen-free conditions with 12 h-light/dark cycles. All procedures were performed in accordance with protocols approved by Purdue University Animal Care and Use Committee (PACUC). We have complied with all relevant ethical regulations for animal use. Mice were euthanized prior to harvest by CO2 asphyxiation followed by secondary cervical dislocation.
POET3 mice (C57BL/6 background) were generated as previously described10. Mice were maintained in homozygous colonies. Inflammation was induced by adoptive transfer of 5 × 106 pre-activated OT-1 cells (described below) via retro-orbital injection. Inflammation was allowed to develop for 7 days before prostate harvest for bPSC analysis and organoid culture. For inflammation/castration studies, mice were inflamed by adoptive transfer 2 days before surgical orchiectomy. BrdU labeling and analysis were performed according to instructions provided with APC BrdU Flow Kit (Becton Dickinson). Mice were injected with BrdU intraperitoneally 2 h before euthanasia.
OT-1 mice were purchased from Jackson Laboratories (Strain #:003831). As previously described10,12, spleens from OT-1 mice were ground between frosted slides in RPMI-1640 medium (Gibco) with 10% fetal bovine serum (FBS) (Corning). The resulting cell slurry was filtered through a 70 μm cell strainer before treatment with Ammonium-Chloride-Potassium (ACK) lysis buffer to remove red blood cells. Remaining splenocytes were re-suspended in RPMI-1640 medium and plated at 1 × 106/well in 24-well plates with 1:1000 beta-mercaptoethanol (Gibco) and 0.2 μg/mL SIINFEKL peptide (Ova peptide 257-264, AnaSpec). Cells were activated for 48 h before purification using Ficoll (Cytiva) according to manufacturer’s protocol.
For lineage tracing studies, POET3 mice were crossed with mTmG mice (Strain #:007676, Jackson) and Krt5-CreERT2 mice (Strain #:029155, Jackson) to generate POET3het;mTmGhet;Krt5-creERT2het mice that were used in the studies. To initiate lineage tracing, mice were injected intraperitoneally with tamoxifen (Tmx) (Sigma-Aldrich) dissolved in corn oil at the dosage of 3 mg/40 g (body weight) for four consecutive days. Inflammation was induced via injection of pre-activated OT-1 T cells 2 weeks after Tmx injection and mice were euthanized for analysis 1 month after inflammation.
Ar_flox/y mouse strain was a generous gift from Dr. Robert E. Braun at The Jackson Laboratories. AR deletion was achieved with Cre-GFP lentivirus (SignaGen Laboratories) infection. The protocol of lentiviral infection was adopted and modified from ref. 49. Briefly, 105 bPSC isolated from Ar_flox/y mice were mixed with 100 μL lentivirus and 4 µg/mL polybrene and centrifuged at 1800 rpm for 90 min. Cells were washed once with DMEM media (Gibco) and re-suspended for organoid culture as described below.
Orchiectomy: Mice were anesthetized using isoflurane gas. After thorough cleaning of the surgical area, a small incision was made at the base of the scrotum. Another smaller incision was made in the inner membrane surrounding the testicle, which was pushed out by gentle pressure on the abdomen. Once extracted, the connective tissue and blood vessels were cauterized with heated forceps before cutting. This was repeated for the second testicle. The incisions were sutured shut. Flunixin and Bupivacaine were administered as analgesics.
Isolation of bPSC population
This process was a modification of a well-established protocol12,50. Briefly, minced prostate tissues were digested in 1 mg/mL collagenase (Sigma-Aldrich) in RPMI-1640 (Gibco) media containing 10% FBS (Corning) with shaking at 37 °C for 2 h, followed by trypsinization. Dissociated cells were passed through 20G needles and 40 µm cell strainers to eliminate aggregates, followed by removal of red blood cells by ACK buffer. To enrich murine bPSC, isolated cells were stained with Zombie Violet Live/Dead Fixable Viability Dye (Biolegend) in PBS, followed by incubation with 1:100 diluted fluorescence-conjugated specific antibodies (Biolegend): CD45-FITC (#103108), CD31-FITC (#102506), Sca-1-APC (#122512) and CD49f-PE (#313612). Human prostate samples from unidentified patients were acquired from Indiana University School of Medicine Tissue Repository under IRB-approved protocols. Isolated human prostate cells were stained with Zombie UV Fixable Viability Dye (Biolegend), CD45-FITC (#304006), EpCAM-PE (#324206), CD26-APC (#302710) and CD49f-BV421 (#313624) as described in detail by Strand et al.51. Once stained, fluorescence-activated cell sorting was performed on the BD FACSAria under sterile conditions.
Organoid culture
Organoid culture protocol was adapted from ref. 50. Briefly, sorted bPSC were counted and resuspended in a 1:2 mixture of Prostate Epithelial Growth Medium (PrEGM, Lonza) and Matrigel (growth factor reduced, Corning). The mixture was deposited in a ring around the edge of a refrigerated 12-well plate at 104 cells/120 μL/well and allowed to solidify for 15 min at 37 °C before addition of pre-warmed PrEGM with or without treatments. Growth medium with treatments was changed every 2–3 days before counting and harvesting on day 7. The following treatments were used with vehicle controls: Enzalutamide (Selleckchem), R1881 (Sigma-Aldrich), recombinant mouse IL-1RA (R&D Systems), recombinant mouse IL-1α (R&D), IL1-RA neutralizing antibody (#AF-480-NA, R&D) and IL-1α neutralizing antibody (#AF-400-NA, R&D). Intact organoids were released from Matrigel with Dispase (Gibco), fixed in 10% neutral buffered formalin followed by 70% ethanol, and embedded in HistoGel (ThermoFisher Scientific).
AR target gene PCR array
Total RNA was isolated from freshly sorted bPSC or organoids using TRIzol Reagent (Invitrogen) according to manufacturer’s protocol. cDNA was synthesized using M-MuLV Reverse Transcriptase (New England Biolabs). Global AR target gene array analysis was conducted with Qiagen RT2 Profiler PCR Array-Mouse Androgen Receptor Signaling Targets (#PAMM-142ZF) and Qiagen online data analyzer. Differentially expressed genes in both naïve and inflamed groups (threshold cycle <35) were further subjected to statistical analysis, where the significance was determined by α = 0.05.
Quantitative real-time polymerase chain reaction
qRT-PCR was conducted using PerfeCTa qPCR FastMix II (QuantaBio) together with 6-FAM/ZEN/IBFQ primer/probe sets (PrimeTime qPCR Assays, IDT) and VIC-labeled TaqMan Ribosomal RNA Control Reagents (Applied Biosystems) on Roche LightCycler 96 according to manufacturers’ protocols. Triplicate samples from three independent mice were analyzed for each group. Relative gene expression was calculated using the formula 2^-[Ct(target gene)-Ct(18S)], where Ct refers to the cycle threshold.
Western blot
Protein was isolated from freshly sorted bPSC or organoids using TRIzol Reagent (Invitrogen) according to manufacturer’s protocol and 30-50 μg protein lysates were used. AR protein was detected using rabbit anti-AR (N-20) antibody (#sc-816, Santa Cruz Biotechnology) at 1:200 dilution followed by IRDye 680RD goat anti-rabbit IgG antibody (Li-Cor) at 1:10,000 dilution. The blot was then incubated with mouse anti-β-Actin or mouse anti-Vinculin antibody (Sigma-Aldrich) followed by IRDye 800CW goat anti-mouse antibody (Li-Cor). Blots were imaged using Odyssey CLx Infrared Imaging system (Li-Cor).
ELISA
Protein was isolated from freshly sorted bPSC using TRIzol Reagent (Invitrogen). IL-1RA and IL-1α in bPSC lysates were detected using Mouse IL-1ra/IL-1F3 Quantikine ELISA Kit and Mouse IL-1 alpha/IL-1F1 Quantikine ELISA Kit (R&D), respectively, according to manufacturer’s instructions.
Flow cytometry analysis
Protocols were adopted from our previous study12. Prostate tissues were processed as described above for bPSC isolation and isolated cells were stained with the following fluorescence conjugated antibodies (Biolegend): CD45-PerCP (#103130), Sca-1-APC (#122512) and CD49f-PE (#313612) before fixation with 10% neutral buffered formalin. To detect AR protein, fixed samples were stained with unconjugated rabbit anti-AR (#ab52615, Abcam, 1:50) followed by secondary stain with Alexa Fluor 488 (Invitrogen, 1:500). Prostate cells isolated from lineage tracing mice were stained with the following fluorescence conjugated antibodies (Biolegend): EpCAM-PE/Cyanine7 (#18215) and CD49f-BV421(#313623). Flow cytometry analyses were performed using BD LSRFortessa.
Histology
Formalin-fixed samples were paraffin-embedded and sectioned at 4 μm onto charged microscope slides. Tissues were deparaffinized through multiple changes of Xylene and 100% Ethanol. Immunohistochemistry: Anti-AR antibody (#ab133273, Abcam) was applied at 1:500 dilution for 30 min and the secondary antibody ImmPRESS HRP Goat anti-Rabbit (#MP-7451-50, Novus Biologicals) was also applied for 30 min. Following this, the DAB chromogen was applied for 5 min. Slides were double rinsed with Tris buffer between all steps. Slides were counterstained with hematoxylin before covered with resinous mounting media. Immunofluorescence: The primary GFP antibody (#A-11122, Invitrogen) was applied at a 1:250 dilution for 30 min followed by incubation with a secondary Goat anti-Rabbit antibody conjugated with Alexa Fluor 647 (Invitrogen). Slides were blocked twice again with 2.5% normal rabbit serum followed by AffiniPure Fab Goat anti-Rabbit (#111-007-003, Jackson ImmunoResearch Laboratories) at 20 μg/mL. Primary antibodies anti-CK5 (1:500) (#905501, Biolegend) and anti-CK8 (1:1000) (#904804, Biolegend) were applied for 60 min. The secondary antibodies Goat anti-Rabbit conjugated with Dylight 488 and Goat anti-Mouse conjugated with Alexa Fluor 555 (Invitrogen) were applied for 30 min. Slides received a DAPI counterstain at 1 μg/mL for 10 min and were covered with ProLong Gold Antifade Mountant (Invitrogen). Slide digitization was conducted on the Leica Aperio VERSA 8 Whole Slide Scanner in the appropriate brightfield and fluorescent settings. Digital images were uploaded to eSlide Manager for analysis. For AR immunofluorescent staining, sections were incubated with the indicated primary antibodies overnight at 4 °C: rat anti-AR (#MA1-150, Invitrogen, 4 μg/mL) or rabbit anti-AR (#PA5-16363, Invitrogen, 2 μg/mL). Species-specific Alexa 488 and Alexa 594-conjugated secondary antibodies (Invitrogen) were applied for 1 h at room temperature at a dilution of 1:200. Nuclei were stained by incubation with Hoechst 33258 nuclear stain (Sigma-Aldrich) at a concentration of 1 μg/mL. All specimens were visualized using immunofluorescence intensity with the Leica 6000 epifluorescence/confocal microscope.
Single-cell RNA sequencing
Data were collected at the Purdue Cell Cytometry Facility using the C1 Fluidigm instrument with SMARTer chemistry (Clontech) to generate cDNA from captured single cells. The Purdue Genomics Facility prepared libraries using a Nextera kit (Illumina). Single-end 1 × 50 bp reads were sequenced using the HiSeq2500 on rapid run mode. FastX-Toolkit v. 0.0.13.2 (Gordon, A., FastX-Toolkit. 2009) quality trimmer was used to further trim reads based on quality score and FASTX-Toolkit quality chart was used to make read per-base quality plots. A trimscore and a trim length of 30 were used. Tophat2 was used to align reads to the Mus musculus GRCm38.p6 reference genome52,53. Tophat2 was run with defaults except that the number of mismatches allowed was 1. The htseq-count script in HTSeq v.0.6.154 and Biopython v.2.7.3 were used to generate a counts matrix in “intersection-nonempty” mode, with feature set to “exon” and attribute parameter set to “gene_id”. R version R-3.4.1 and the package Seurat v. 1.4 were used to cluster cells into subpopulations55 using the first 4 principal components, a resolution of 0.5, and k.param = 4. The FindAllMarkers() function within Seurat was used to identify cluster markers using the zero-inflated “bimod” model and a false discovery rate (FDR) of 0.01. FindAllMarkers() was likewise used to identify differentially expressed genes between inflamed and naïve cells, controlling the FDR at 5%. Ingenuity Pathway Analysis (IPA, Qiagen) was used to perform pathway analyses and enrichment analyses.
ATAC-seq sample preparation and analyses
Methods for sample preparation were adapted from ref. 56. Briefly, nuclei from freshly sorted naïve or inflamed bPSC were prepared using ice cold lysis buffer (10 mM Tris-Cl, pH 7.4, 10 mM NaCl, 3 mM MgCl2 and 0.1% IGEPAL CA-630) before immediate transposition of DNA in 25 μL 2x TD buffer, 2.5 μL Transposase (Illumina) and 22.5 μL of nuclease free H2O for 30 min at 37 °C. Samples were then purified using Mini Elute Kit (Qiagen). PCR amplification and library prep were performed using 10 μL transposed DNA, 9.7 μL nuclease-free H2O, 0.3 μL 100x SYBR Green I (Invitrogen), 25 μL NEBNext High-Fidelity 2x PCR Master Mix (New England Biolabs), and 2.5 μL each of individual 25 μM dual unique Nextera PCR primers in a 50 μL reaction. ATAC-seq libraries were sequenced using HiSeq 2500.
The quality control for ATAC-seq data was performed using FastQC v0.11.6. Raw sequencing reads were aligned to mm10 using BWA mem v0.7.17-r118857 with “-M” with other parameters kept as default. Duplicate reads were removed using PicardTools v2.12.2 (http://broadinstitute.github.io/picard/). Properly mapped pairs were extracted by SAMtools58. Reads were offset by using the publicly available script ATAC_BAM_shifter_gappedAlign.pl and those with TLEN < 30 bp were removed. Required tag directories were generated by makeTagDirectory from HOMER v4.859. Differential chromatin accessible regions/peaks between naïve and inflamed bPSC were obtained using getDifferentialPeaksReplicates.pl from HOMER v4.859 with “-f 1.5 -edgeR -balanced”. pos2bed.pl was used to generate merged peak files from differential peaks for each sample. makeUCSCfile was used to generate bedGraph files from tag directories. bedGraphToBigWig60 was then used to generate bigwig files from bedGraph files with the mm10 chromosome size file.
CUT&Tag sample preparation and analysis
CUT&Tag was performed per the published protocol61 (https://www.protocols.io/view/bench-top-cut-amp-tag-kqdg34qdpl25/v3) on freshly sorted naïve or inflamed bPSC. In brief, nuclei were isolated from the bPSC and lightly fixed with paraformaldehyde for 2 min at room temperature. Fixed nuclei for each target (~80 K for AR and ~10 K for H3K27ac and IgG) were incubated with primary antibodies (dilution 1:50) for AR (#ab133273, Abcam), H3K27ac (#ab4729, Abcam), and IgG (#31235, Invitrogen) at 4 °C on a nutating mixer overnight. After washing, nuclei were incubated with secondary antibody (#ABIN101961, Antibodies-online) (dilution 1:100) at room temperature for 1 h while nutating. Preloaded pA-Tn5 (#C01070001, Diagenode) was tethered to antibodies for 1 h followed by tagmentation at 37 °C for 1 h and DNA isolation. PCR was done with the isolated DNA samples (23 μL DNA, 0.5 μL H2O, 1.5 μL primer from 24 UDI for Tagmented libraries Set I, Diagenode, 25 μL NEBNext Ultra II Q5 Master Mix, New England Biolabs) for 13 cycles. PCR products were cleaned with 1.3× Mag-Bind TotalPure NGS beads (Omega). TAPE was run using Agilent 4200 TapeStation with D1000 reagent to confirm the success of CUT&Tag for the positive (H3K27ac) and negative (IgG) control. Barcoded libraries were mixed to get an equal representation of each sample followed by 1x bead clean-up to remove any remaining primers. Libraries were sequenced using NextSeq 2000.
CUT&Tag reads were aligned to the mm10 genome build using Bowtie2 (v2.5.1)62 with the “--local --very-sensitive --no-mixed --no-discordant --phred33 -I 10 -X 700” parameters. The resulting BAM files were sorted with SAMtools (v1.17)58 and duplicate reads were marked using Picard MarkDuplicates (v3.1.1) (http://broadinstitute.github.io/picard/). BigWig files were generated from the aligned reads using DeepTools (v3.5.1)63 with the “--normalizeUsing CPM --minMappingQuality 30” parameters. The differential peaks from ATAC-seq were grouped into three clusters using the computeMatrix and plotHeatmap functions from Deeptools. These peak clusters were further analyzed by subclustering CUT&Tag AR samples to identify ATAC-seq differential peaks that contain AR binding. findMotifsGenome.pl from HOMER v4.1159 was used to find motifs enriched in the peaks of different clusters and annotatePeaks.pl was used to annotate the peaks and identify adjacent genes. Metascape web tool64 was used to assess enrichment in KEGG and Hallmark pathways.
Statistics and reproducibility
Statistical analyses were performed using GraphPad Prism (v.9.5.1). Two-tailed unpaired Student’s t test or Welch’s t test was performed for comparisons between two groups. Unpaired ordinary one-way analysis of variance (ANOVA) followed by multiple comparisons was performed for comparisons among three or more groups. P values less than 0.05 are considered as significant. Data are presented as means ± SEM (standard error of mean) of at least three independent experiments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank Dr. Jill Hutchcroft at Purdue Flow Cytometry & Cell Separation Facility for cell sorting; Histology Research Laboratory at Purdue College of Veterinary Medicine, a core facility of the NIH-funded Indiana Clinical and Translational Science Institute, for histology processing, immunohistochemistry and immunofluorescence staining. This project was supported by NIH R01DK084454 and R01DK126478 to TLR, R35GM138283 to MK, the Purdue Institute for Cancer Research P30CA023168, and the Walther Cancer Foundation. This project was also funded with support from the Indiana Clinical and Translational Sciences Institute, which is funded in part by Award Number UM1TR004402 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors also gratefully acknowledge the SIRG Graduate Research Assistantships Award to BY. This work is dedicated to the loving memory of our dear friend and colleague, Dr. Hsing-Hui Wang. We will forever cherish the memories of the time we shared in and out of the lab.
Author contributions
T.L.R., T.J.J., J.Y., and P.O.C designed the study and interpreted the data. J.Y., P.O.C, and T.L.R. wrote the manuscript. P.O.C., J.Y., H-H.W., M.M.B., G.D.A., G.M.C., L.W., and S.A. carried out the experiments. E.G., N.A.L., R.W.D., and F.Z. analyzed the scRNA-seq data. S.M.J., S.S.S, B.Y., and M.K. prepared CUT&Tag library and analyzed the ATAC-seq and CUT&Tag data. Ar_flox/y mice were provided by R.E.B. and lineage-tracing mice were generated by J.Y. and P.O.C. L.C. provided histological analyses and S.A.C. participated in study design and data interpretation.
Peer review
Peer review information
Communications Biology thanks Zeyad Nassar and the other, anonymous, reviewer for their contribution to the peer review of this work. Primary Handling Editors: Jesmond Dalli and Dario Ummarino.
Data availability
The scRNA-seq, ATAC-seq, and CUT&Tag data acquired in this study have been deposited in the Gene Expression Omnibus database under the accession codes GSE271146, GSE269582, and GSE269548, respectively. Uncropped Western blot images are included as Supplementary Fig. 5. The source data underlying the figures in this paper are provided in Supplementary Data 3. All other data are available from the corresponding authors upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Deceased: Dr. Hsing-Hui Wang.
These authors contributed equally: Paula O. Cooper, Jiang Yang, Hsing-Hui Wang.
These authors jointly supervised this work: Jiang Yang, Travis J. Jerde, Timothy L. Ratliff.
Contributor Information
Jiang Yang, Email: yang1491@purdue.edu.
Travis J. Jerde, Email: tjjerde@iu.edu
Timothy L. Ratliff, Email: tlratliff@purdue.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07071-y.
References
- 1.Gandaglia, G. et al. The role of prostatic inflammation in the development and progression of benign and malignant diseases. Curr. Opin. Urol.27, 99–106 (2017). [DOI] [PubMed] [Google Scholar]
- 2.De Nunzio, C., Presicce, F. & Tubaro, A. Inflammatory mediators in the development and progression of benign prostatic hyperplasia. Nat. Rev. Urol.13, 613–626 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Nickel, J. C. et al. The relationship between prostate inflammation and lower urinary tract symptoms: examination of baseline data from the REDUCE trial. Eur. Urol.54, 1379–1384 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robert, G. et al. Inflammation in benign prostatic hyperplasia: a 282 patients’ immunohistochemical analysis. Prostate69, 1774–1780 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawson, D. A. et al. Basal epithelial stem cells are efficient targets for prostate cancer initiation. Proc. Natl. Acad. Sci. USA107, 2610–2615 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crowell, P. D. et al. Expansion of luminal progenitor cells in the aging mouse and human prostate. Cell Rep.28, 1499–1510.e1496 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toivanen, R., Mohan, A. & Shen, M. M. Basal progenitors contribute to repair of the prostate epithelium following induced luminal anoikis. Stem Cell Rep.6, 660–667 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie, Q. et al. Dissecting cell-type-specific roles of androgen receptor in prostate homeostasis and regeneration through lineage tracing. Nat. Commun.8, 14284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karthaus, W. R. et al. Regenerative potential of prostate luminal cells revealed by single-cell analysis. Science368, 497–505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haverkamp, J. M. et al. An inducible model of abacterial prostatitis induces antigen specific inflammatory and proliferative changes in the murine prostate. Prostate71, 1139–1150 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon, O. J., Zhang, L. & Xin, L. Stem cell antigen-1 identifies a distinct androgen-independent murine prostatic luminal cell lineage with bipotent potential. Stem Cells34, 191–202 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang, H. H. et al. Characterization of autoimmune inflammation induced prostate stem cell expansion. Prostate75, 1620–1631 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon, O. J., Zhang, L., Ittmann, M. M. & Xin, L. Prostatic inflammation enhances basal-to-luminal differentiation and accelerates initiation of prostate cancer with a basal cell origin. Proc. Natl. Acad. Sci. USA111, E592–E600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, L. et al. Expansion of prostate epithelial progenitor cells after inflammation of the mouse prostate. Am. J. Physiol. Ren. Physiol.308, F1421–F1430 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xin, L., Lukacs, R. U., Lawson, D. A., Cheng, D. & Witte, O. N. Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem Cells25, 2760–2769 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Henry, G. H. et al. A cellular anatomy of the normal adult human prostate and prostatic urethra. Cell Rep.25, 3530–3542.e3535 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, N. et al. The role of hepatocyte nuclear factor-3 alpha (Forkhead Box A1) and androgen receptor in transcriptional regulation of prostatic genes. Mol. Endocrinol.17, 1484–1507 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Lupien, M. et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell132, 958–970 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jozwik, K. M. & Carroll, J. S. Pioneer factors in hormone-dependent cancers. Nat. Rev. Cancer12, 381–385 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Sato, N. et al. Androgenic induction of prostate-specific antigen gene is repressed by protein-protein interaction between the androgen receptor and AP-1/c-Jun in the human prostate cancer cell line LNCaP. J. Biol. Chem.272, 17485–17494 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Lin-Tsai, O. et al. Surgical intervention for symptomatic benign prostatic hyperplasia is correlated with expression of the AP-1 transcription factor network. Prostate74, 669–679 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thiel, G., Welck, J., Wissenbach, U. & Rossler, O. G. Dihydrotestosterone activates AP-1 in LNCaP prostate cancer cells. Int. J. Biochem. Cell Biol.110, 9–20 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Tang, F. et al. Chromatin profiles classify castration-resistant prostate cancers suggesting therapeutic targets. Science376, eabe1505 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei, X. et al. Spatially restricted stromal wnt signaling restrains prostate epithelial progenitor growth through direct and indirect mechanisms. Cell Stem Cell24, 753–768.e756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bisson, I. & Prowse, D. M. WNT signaling regulates self-renewal and differentiation of prostate cancer cells with stem cell characteristics. Cell Res.19, 683–697 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Mohyeldin, A., Garzon-Muvdi, T. & Quinones-Hinojosa, A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell7, 150–161 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Mazumdar, J. et al. O2 regulates stem cells through Wnt/beta-catenin signalling. Nat. Cell Biol.12, 1007–1013 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Culig, Z. et al. Interleukin 1beta mediates the modulatory effects of monocytes on LNCaP human prostate cancer cells. Br. J. Cancer78, 1004–1011 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staverosky, J. A., Zhu, X. H., Ha, S. & Logan, S. K. Anti-androgen resistance in prostate cancer cells chronically induced by interleukin-1beta. Am. J. Clin. Exp. Urol.1, 53–65 (2013). [PMC free article] [PubMed] [Google Scholar]
- 30.Chang, M. A. et al. IL-1beta induces p62/SQSTM1 and represses androgen receptor expression in prostate cancer cells. J. Cell Biochem.115, 2188–2197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas-Jardin, S. E. et al. RELA is sufficient to mediate interleukin-1 repression of androgen receptor expression and activity in an LNCaP disease progression model. Prostate80, 133–145 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis45, 593–605 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Crowley, L. & Shen, M. M. Heterogeneity and complexity of the prostate epithelium: new findings from single-cell RNA sequencing studies. Cancer Lett.525, 108–114 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, B. et al. Non-cell-autonomous regulation of prostate epithelial homeostasis by androgen receptor. Mol. Cell63, 976–989 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gabay, C., Porter, B., Fantuzzi, G. & Arend, W. P. Mouse IL-1 receptor antagonist isoforms: complementary DNA cloning and protein expression of intracellular isoform and tissue distribution of secreted and intracellular IL-1 receptor antagonist in vivo. J. Immunol.159, 5905–5913 (1997). [PubMed] [Google Scholar]
- 36.Martin, P. et al. Intracellular IL-1 receptor antagonist isoform 1 released from keratinocytes upon cell death acts as an inhibitor for the alarmin IL-1alpha. J. Immunol.204, 967–979 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Watson, J. M. et al. The intracellular IL-1 receptor antagonist alters IL-1-inducible gene expression without blocking exogenous signaling by IL-1 beta. J. Immunol.155, 4467–4475 (1995). [PubMed] [Google Scholar]
- 38.Vecile, E. et al. Intracellular function of interleukin-1 receptor antagonist in ischemic cardiomyocytes. PLoS ONE8, e53265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giavridis, T. et al. CAR T cell-induced cytokine release syndrome is mediated by macrophages and abated by IL-1 blockade. Nat. Med.24, 731–738 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gander-Bui, H. T. T. et al. Targeted removal of macrophage-secreted interleukin-1 receptor antagonist protects against lethal Candida albicans sepsis. Immunity56, 1743–1760 e1749 (2023). [DOI] [PubMed] [Google Scholar]
- 41.Lee, S. O., Lou, W., Hou, M., Onate, S. A. & Gao, A. C. Interleukin-4 enhances prostate-specific antigen expression by activation of the androgen receptor and Akt pathway. Oncogene22, 7981–7988 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Hobisch, A. et al. Interleukin-6 regulates prostate-specific protein expression in prostate carcinoma cells by activation of the androgen receptor. Cancer Res.58, 4640–4645 (1998). [PubMed] [Google Scholar]
- 43.Culig, Z. et al. Androgen receptor activation in prostatic tumor cell lines by insulin-like growth factor-I, keratinocyte growth factor, and epidermal growth factor. Cancer Res.54, 5474–5478 (1994). [PubMed] [Google Scholar]
- 44.Koryakina, Y., Ta, H. Q. & Gioeli, D. Androgen receptor phosphorylation: biological context and functional consequences. Endocr. Relat. Cancer21, T131–T145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willder, J. M. et al. Androgen receptor phosphorylation at serine 515 by Cdk1 predicts biochemical relapse in prostate cancer patients. Br. J. Cancer108, 139–148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ponguta, L. A., Gregory, C. W., French, F. S. & Wilson, E. M. Site-specific androgen receptor serine phosphorylation linked to epidermal growth factor-dependent growth of castration-recurrent prostate cancer. J. Biol. Chem.283, 20989–21001 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo, Z. et al. Regulation of androgen receptor activity by tyrosine phosphorylation. Cancer Cell10, 309–319 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Gioeli, D. et al. Androgen receptor phosphorylation. Regulation and identification of the phosphorylation sites. J. Biol. Chem.277, 29304–29314 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Shahi, P., Seethammagari, M. R., Valdez, J. M., Xin, L. & Spencer, D. M. Wnt and Notch pathways have interrelated opposing roles on prostate progenitor cell proliferation and differentiation. Stem Cells29, 678–688 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lukacs, R. U., Goldstein, A. S., Lawson, D. A., Cheng, D. & Witte, O. N. Isolation, cultivation and characterization of adult murine prostate stem cells. Nat. Protoc.5, 702–713 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strand, D. W., Aaron, L., Henry, G., Franco, O. E. & Hayward, S. W. Isolation and analysis of discreet human prostate cellular populations. Differentiation91, 139–151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol.14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol.33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kent, W. J., Zweig, A. S., Barber, G., Hinrichs, A. S. & Karolchik, D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics26, 2204–2207 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun.10, 1930 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res.44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun.10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The scRNA-seq, ATAC-seq, and CUT&Tag data acquired in this study have been deposited in the Gene Expression Omnibus database under the accession codes GSE271146, GSE269582, and GSE269548, respectively. Uncropped Western blot images are included as Supplementary Fig. 5. The source data underlying the figures in this paper are provided in Supplementary Data 3. All other data are available from the corresponding authors upon reasonable request.