Abstract
This study aimed to develop a machine learning (ML) model for predicting pulmonary embolism (PE) in patients with gastrointestinal cancers, a group at increased risk for PE. We conducted a retrospective, multicenter study analyzing patients who underwent computed tomographic pulmonary angiography (CTPA) between 2010 and 2020. The study utilized demographic and clinical data, including the Wells score and D-dimer levels, to train a random forest ML model. The model’s effectiveness was assessed using the area under the receiver operating curve (AUROC). In total, 446 patients from hospital A and 139 from hospital B were included. The training set consisted of 356 patients from hospital A, with internal validation on 90 and external validation on 139 patients from hospital B. The model achieved an AUROC of 0.736 in hospital A and 0.669 in hospital B. The ML model significantly reduced the number of patients recommended for CTPA compared to the conventional diagnostic strategy (hospital A; 100.0% vs. 91.1%, P < 0.001, hospital B; 100.0% vs. 93.5%, P = 0.003). The results indicate that an ML-based prediction model can reduce unnecessary CTPA procedures in gastrointestinal cancer patients, highlighting its potential to enhance diagnostic efficiency and reduce patient burden.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-75977-y.
Keywords: Pulmonary embolism, Machine learning, Gastrointestinal cancer, Computed tomographic pulmonary angiography, Random forest model
Subject terms: Gastrointestinal cancer, Gastrointestinal cancer, Embolism, Thromboembolism
Introduction
Venous thromboembolism (VTE) occurs in 15-20% of cancer patients1. Among cancer populations, VTE increases the risk of death2–4. Epidemiological studies have reported that the highest risk of VTE is found in intra-abdominal cancers, including gastrointestinal cancers5–7. Among VTEs, pulmonary embolism (PE) is a clinically important disease that requires urgent management. Early detection of PE is crucial because massive PE can lead to cardiac arrest or circulatory collapse, both of which are associated with high mortality rates8.
Many studies have developed various diagnostic strategies for PE. These strategies aim to identify low-risk patients for PE from whom it is safe to withhold imaging studies and anticoagulation therapy9. Assessments of clinical pretest probability (C-PTP), such as the Wells score, are most often used in combination with the D-dimer test. It has been well established that PE can be ruled out in patients with low C-PTP and D-dimer levels under 500ng per milliliter9–11. However, these patients only constitute approximately 30% of outpatients12.
Computed tomography pulmonary angiography (CTPA) is the method of choice for evaluating pulmonary vasculature in patients with suspected PE10. However, CTPA is associated with issues such as radiation exposure, potential side effects from the contrast media, and high cost. Various efforts have been made to reduce unnecessary CTPA in patients with suspected PE. Several studies have aimed to investigate the potential for modifying the cut-off value of the D-dimer test12,13. Nevertheless, these studies did not analyze patients with cancer, which has limited the ability of their proposed strategies to predict VTE in cancer patients.
More recently, there have been studies intending to use artificial intelligence to improve the diagnosis of PE. One study used machine learning (ML) to support clinical decision-making for CTPA in patients with moderate to high C-PTP14. Another study used ML for risk-stratification of deep vein thrombosis (DVT) using Wells score and D-dimer15. However, there have been few studies using ML to aid in the decision-making of CTPA in cancer patients suspected of having PE.
The existing diagnostic methods for PE have limited applicability to cancer patients. This is primarily because cancer patients tend to have elevated D-dimer levels16. PE is relatively common in gastrointestinal cancers5–7. Therefore, our objective in the current work was to improve the diagnostic strategy used for PE in gastrointestinal cancer patients through the use of ML, with the ultimate aim of reducing unnecessary CTPA scans.
Methods
Study design and patients
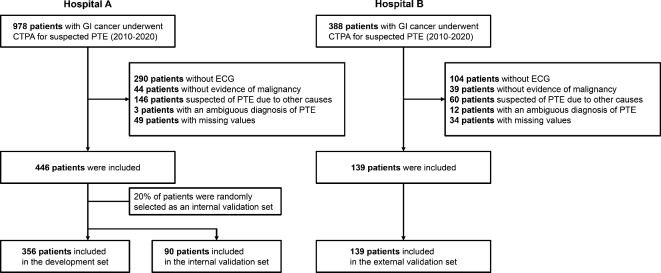
This retrospective, multicenter study investigated patients with gastrointestinal cancer who underwent CTPA between 2010 and 2020 by surveying an electrical medical records database (Fig. 1). Data from Hospital A (Seoul National University Hospital, Seoul, Korea) were used for development and internal validation of the proposed model. Data from Hospital B (Seoul National University Bundang Hospital, Seongnam-si, Korea) were used for external validation of the proposed model. The diagnosis of gastrointestinal cancer was confirmed based on the pathological results. Hepatocellular carcinoma diagnosed based on imaging without pathological confirmation was also included17. Any patients who met the exclusion criteria were not included in the analysis; the exclusion criteria were as follows: suspected PE associated with causes other than cancer or other malignancy, no evidence of malignancy at the time of CTPA, ambiguous diagnosis of PE on CTPA, or missing data. We also checked heart rate through electrocardiography, and we excluded patients who did not have an electrocardiogram at the time of the CTPA. The diagnosis of PE was confirmed by trained experts based on CTPA.
Fig. 1.
Study flow chart. GI cancer gastrointestinal cancer, PE pulmonary embolism, CTPA computed tomographic pulmonary angiography, ECG electrocardiography.
This study was approved by the institutional review board (IRB) of the Seoul National University Hospital, Korea (IRB No.2009-146-1159) and the Seoul National University Bundang Hospital, Korea (IRB No.B-2111-721-401). The need for informed consent was waived by the IRB of Seoul National University Hospital and the Seoul National University Bundang Hospital. The study was conducted in accordance with the Declaration of Helsinki.
Data collection and definition
Patient characteristics were retrospectively collected, including age, sex, cancer diagnosis, Wells score and components of the Wells score, and D-dimer. Gastrointestinal cancers were defined as cancers of gastrointestinal tract from esophagus to anus as well as cancers of liver and pancreatobiliary system18. The Wells score was calculated using a method that has previously been used in patients suspected of having PE19. The components of the Wells score included the signs and symptoms of VTE, alternative diagnosis less likely than pulmonary embolism, heart rate > 100 beats per minute, history of VTE, immobilization, malignancy, and hemoptysis. Among them, we did not include malignancy because all considered participants were diagnosed with cancer. History of VTE was defined as a previous history of PE or DVT19. A low C-PTP was defined as a Wells score of 0 to 1.5, a moderate C-PTP was defined as a Wells score from 2.0 to 6.0, and a high C-PTP was defined as a Wells score of 6.5 or higher19. According to the conventional PE diagnostic strategy, PE could be ruled out in patients with low to moderate C-PTP and D-dimer levels of less than 500ng per milliliter. Otherwise, CTPA is required to identify PE10. (Supplementary Fig. 1).
Model development
This study used four machine learning algorithms: logistic regression, XGBoost, LightGBM, and random forest. The interpretability of the results produced by logistic regression is particularly important in the context of clinical decision-making. XGBoost and LightGBM are gradient boosting methods that have exhibited high performance and efficiency20,21. The random forest (RF) model demonstrated satisfactory performance in both regression and classification, particularly in the healthcare field, where deep learning is not accessible due to a lack of data22,23. Four models were trained, and the one showing the best performance was selected for external validation.
Study outcome measures
The primary outcome considered in this study was the area under the receiver operating characteristics curve (AUROC) and the accuracy of the ML model for the diagnosis of PE in patients with gastrointestinal cancer.
As a secondary outcome, we compared the number of CTPA performed for PE in the ML model with the conventional diagnostic strategy. We also investigated the feature importance of the ML model.
Statistical analysis
To compare the baseline characteristics, the Student’s t-test and the Chi-square test were used for continuous and dichotomous variables, respectively. If any subgroups had less than four subjects, the Fisher’s exact test was used instead of a Chi-square test.
We utilized the components of the Wells score and incorporated D-dimer as continuous variables in machine learning training. We evaluated our model using the hold-out test method. We performed a five-fold cross validation on 80% of the subjects to determine the optimal hyperparameters. Subsequently, we evaluated the model using the remaining 20% of the subjects. To evaluate this, we measured the performance of the model using the area under the receiver operating characteristics curve (AUROC), sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). To determine the optimal cutoff, we identified the point at which the sensitivity reached 1.024. Moreover, to evaluate the net benefits of the proposed ML model and the conventional diagnostic strategy, we performed a decision curve analysis (DCA)25. The benefit was defined as accurately identifying PE patients while reducing unnecessary CTPA scans, while the harm was defined as either missing PE cases or performing unnecessary CTPA scans.
P-values lower than 0.05 were considered to indicate statistically significant findings. Statistical calculations were performed using SPSS and scikit-learn’s random forest model. To measure feature importance, the SHAP algorithm was used. Lastly, statistical analysis based on the Delong test was performed to compare prediction ROC curves between models.
Results
Baseline characteristics of the study population
In hospital A, 708 patients diagnosed with gastrointestinal cancer underwent CTPA; of these, 262 were excluded because of the exclusion criteria, thus leaving a total of 446 patients. In hospital B, 388 patients diagnosed with gastrointestinal cancer underwent CTPA; 249 of these were excluded (Fig. 1). In the training set, 103 (28.9%) patients were diagnosed with PE. Table 1 presents a comparison of the baseline characteristics according to PE in the development dataset. Pancreatic cancer was the most common type of cancer in patients with PE (33.0%). Patients with PE had higher C-PTP (40.8% vs. 23.3%, P < 0.001). More patients were suspected to have DVT among the patients with PE (48.5% vs. 26.9%, P < 0.001). History of VTE (2.4% vs. 14.6%, P < 0.001) was more common in patients with PE. There were no significant differences between the two groups in the other variables.
Table 1.
Baseline characteristics of patients according to the presence of pulmonary embolism.
| All patients | Patients without PE | Patients with PE | P value | |
|---|---|---|---|---|
| (n = 356) | (n = 253) | (n = 103) | ||
| Age (mean) | 66.5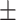 11.1 11.1 |
66.8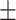 10.9 10.9 |
65.5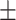 11.6 11.6 |
0.318 |
| Male (%) | 243 (68.3) | 177 (70.0) | 66 (64.1) | 0.339 |
| Cancer (%) | < 0.001 | |||
| Gastric cancer | 70 (19.7) | 58 (22.9) | 12 (11.7) | |
| Colon cancer | 72 (20.2) | 49 (19.4) | 23 (22.3) | |
| Hepatocellular carcinoma | 81 (22.8) | 65 (25.7) | 16 (15.5) | |
| Pancreatic cancer | 71 (19.9) | 37 (14.6) | 34 (33.0) | |
| Cholangiocarcinoma | 40 (11.2) | 26 (10.3) | 14 (13.6) | |
| Others | 22 (6.2) | 18 (7.1) | 4 (3.9) | |
| Components of Wells score (%) | ||||
| Signs and symptoms of DVT | 118 (33.1) | 68 (26.9) | 50 (48.5) | < 0.001 |
| Alternative diagnosis less likely than PE | 168 (47.2) | 114 (45.1) | 54 (52.4) | 0.252 |
| Heart rate > 100/min | 160 (44.9) | 113 (44.7) | 47 (45.6) | 0.961 |
| Immobilization | 136 (38.2) | 91 (36.0) | 45 (43.7) | 0.215 |
| History of VTE | 21 (5.9) | 6 (2.4) | 15 (14.6) | < 0.001 |
| Hemoptysis | 3 (0.8) | 1 (0.4) | 2 (1.9) | 0.202 |
| Wells score (%) | 0.001 | |||
| Low to intermediate | 225 (71.6) | 194 (76.7) | 61 (59.2) | |
| High | 101 (28.4) | 59 (23.3) | 42 (40.8) | |
D-dimer  500 ng/mL (%) 500 ng/mL (%) |
348 (97.8) | 246 (97.2) | 102 (99.0) | 0.447 |
DVT deep vein thrombosis, PE pulmonary embolism, VTE venous embolism.
Demographics were evaluated on the internal validation set and on the external validation set (Table 2). The patients who were treated at hospital B were older than the patients who were treated at hospital A (hospital A; 65.6 ± 12.6 vs. hospital B; 69.4 ± 10.9, P = 0.021). There were also differences in the types of cancer among hospitals. The patients who were treated at hospital B had higher incidences of pancreatic cancer (20.0% vs. 29.5%) and cholangiocarcinoma (6.7% vs. 12.2%), while they exhibited lower prevalence of colon cancer (22.2% vs. 16.5%) and hepatocellular carcinoma (27.8% vs. 12.2%). The patients who were treated at hospital A had higher C-PTP than hospital B (35.6% vs. 15.8%, P < 0.001). There were more patients who were suspected to have DVT (36.7% vs. 13.7%, P < 0.001) and who were reported to have a history of VTE (13.3% vs. 1.4%, P < 0.001) among the patients who were treated at hospital A.
Table 2.
Baseline characteristics for internal and external validation datasets.
| All patients | Hospital A | Hospital B | P value | |
|---|---|---|---|---|
| (n = 229) | (n = 90) | (n = 139) | ||
| Age (mean) | 67.9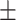 11.7 11.7 |
65.6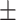 12.6 12.6 |
69.4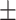 10.9 10.9 |
0.021 |
| Male (%) | 143 (62.4) | 63 (70.0) | 80 (57.6) | 0.078 |
| PE (%) | 72 (31.4) | 31 (34.4) | 41 (29.5) | 0.521 |
| Cancer (%) | 0.018 | |||
| Gastric cancer | 48 (21.0) | 18 (20.0) | 30 (21.6) | |
| Colon cancer | 43 (18.8) | 20 (22.2) | 23 (16.5) | |
| Hepatocellular carcinoma | 42 (18.3) | 25 (27.8) | 17.(12.2) | |
| Pancreatic cancer | 59 (25.8) | 18 (20.0) | 41 (29.5) | |
| Cholangiocarcinoma | 23 (10.0) | 6 (6.7) | 17 (12.2) | |
| Others | 14 (6.1) | 3 (3.3) | 11 (7.9) | |
| Components of wells score (%) | ||||
| Signs and symptoms of DVT | 52 (22.7) | 33 (36.7) | 19 (13.7) | < 0.001 |
| Alternative diagnosis less likely than PE | 99 (43.2) | 46 (51.1) | 53 (38.1) | 0.072 |
| Heart rate > 100/min | 120 (52.4) | 47 (52.2) | 73 (52.5) | 1.000 |
| Immobilization | 79 (34.5) | 35 (38.9) | 44 (31.7) | 0.326 |
| History of VTE | 14 (6.1) | 12 (13.3) | 2 (1.4) | 0.001 |
| Hemoptysis | 2 (0.9) | 2 (2.2) | 0 (0) | 0.153 |
| Wells score (%) | 0.001 | |||
| Low to intermediate | 175 (76.4) | 58 (64.4) | 117 (84.2) | |
| High | 54 (23.6) | 32 (35.6) | 22 (15.8) | |
D-dimer 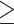 500 ng/mL (%) 500 ng/mL (%) |
228 (99.6) | 89 (98.9) | 139 (100.0) | 0.393 |
DVT deep vein thrombosis, PE pulmonary embolism, VTE venous embolism.
Primary study outcomes
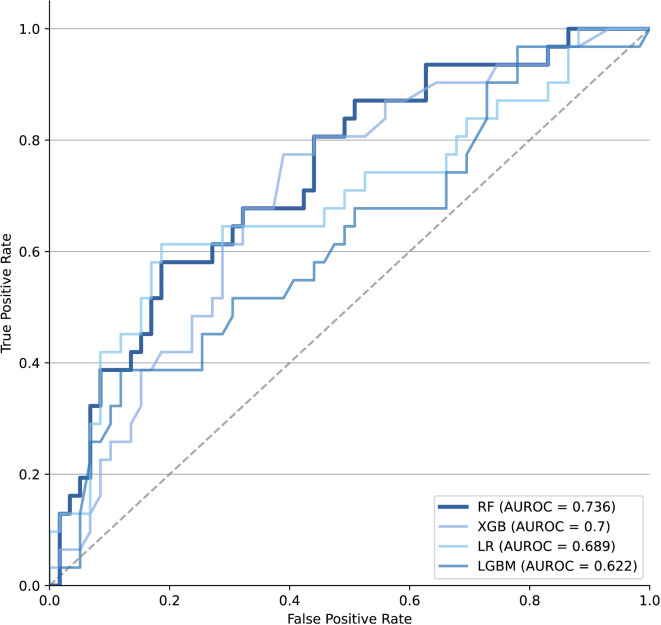
We have trained the data using several machine learning models. Among them, we employed the random forest model with the highest AUROC (Supplementary Table 1). The performance of the ML model is summarized in Table 3; Fig. 2. The AUROC and accuracy of the ML model during internal validation were 0.736 and 0.433, respectively. In the external validation, the AUROC and accuracy of the ML model were 0.669 and 0.345, respectively. We validated this model at an operating point with high sensitivity (1.000) for use as a screening tool, and the results of external validation showed a sensitivity of 0.976.
Table 3.
Performance of machine learning model for predicting PE.
| Features | Hospital A | Hospital B |
|---|---|---|
| WC + D-dimer | WC + D-dimer | |
| AUROC | 0.736 (0.624–0.838) | 0.669 (0.557–0.767) |
| Accuracy | 0.433 (0.333–0.533) | 0.345 (0.226–0.424) |
| Sensitivity | 1.000 (1.000–1.000) | 0.976 (0.921-1.000) |
| NPV | 1.000 (1.000–1.000) | 0.889 (0.625-1.000) |
| Specificity | 0.136 (0.053–0.231) | 0.082 (0.030–0.136) |
| PPV | 0.378 (0.275–0.482) | 0.308 (0.231–0.388) |
PE pulmonary embolism, WC components of Wells score, AUROC area under receiver operating curve, NPV negative predictive value, PPV positive predictive value.
Fig. 2.
AUROC of conventional model and machine learning model. AUROC area under the receiver operating characteristics curve, RF random forest, LR logistic regression, XGB xgboost, LGBM light gradient boosting model.
The factor analysis revealed that the variables “less likely than PE” and “hemoptysis” were only present in the identical factors. Consequently, when the RF model was analyzed without hemoptysis, the AUROC remained consistent (Supplemental Table 2 and Supplemental Fig. 2).
Secondary study outcomes
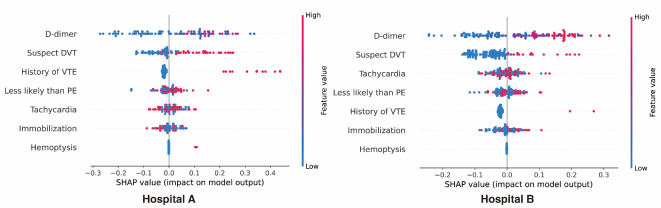
Figure 3 illustrates the feature importance of the ML model. According to the SHAP analysis of the ML model, high levels of D-dimer as well as positive signs and symptoms of DVT have the greatest impacts on the prediction of PE in both hospitals. Clinically, D-dimer, which has been a pivotal biomarker in stratifying patients for CTPA, was also found to be the most important variable in the ML model. We also analyzed the variables important for predicting PE in the traditional logistic model and Mean Decrease Gini, and the results were consistent with the SHAP analysis. (Supplemental Table 3 and Supplemental Fig. 3).
Fig. 3.
Feature importance after machine learning. HR heart rate, VTE venous embolism, DVT deep vein thrombosis, PE pulmonary embolism.
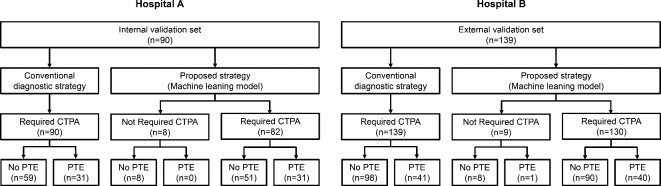
In accordance with the conventional diagnostic strategy, all patients from both hospitals were required to undergo a CTPA for a PE to be diagnosed. On the other hand, the number of patients classified as requiring CTPA during the diagnosis of PE was significantly reduced in both hospitals after the implementation of the ML model (hospital A; 100.0% vs. 91.1%, P < 0.001, hospital B; 100.0% vs. 93.5%, P = 0.003) (Fig. 4).
Fig. 4.
Comparison of patients who needed CT pulmonary angiography in different hospitals between conventional diagnostic strategy and machine learning model. PE pulmonary embolism, CTPA computed tomographic pulmonary angiography.
The DCA indicates that our ML model maintains a higher net benefit across various thresholds, while the conventional diagnostic strategy’s net benefit declines rapidly and becomes negative beyond a threshold of 0.3; these findings demonstrate the superior clinical utility of the ML model (Supplementary Fig. 4).
Discussion
In gastrointestinal cancer patients, PE is one of the most critical complications that necessitates early diagnosis and treatment. However, the diagnostic strategy that is currently used for PE has exhibited unsatisfactory performance in ruling out PE, which has led to unnecessary CTPA scans in cancer patients. The current study showed that the ML model enhances the performance of ruling out PE and reduces unnecessary CTPA scans.
Several studies have reported that the ML model can be used to enhance the diagnostic strategies used for PE. One study demonstrated that a neural network model utilizing raw structured electronic medical record data can be used to predict the risk of PE14. They developed a predictive model that was specifically tailored to moderate to high C-PTP patients. Willian et al. showed that a ML model can outperform existing risk assessment scoring in excluding DVT15. Humberto et al. reported that a ML model outperformed traditional risk scores such as Wells score combined with D-dimer, the revised Geneva score, and the pulmonary embolism rule-out criteria score (PERC)26. This study also showed that ML models could improve the diagnosis of PE compared to conventional diagnostic strategies, even though it was based on patients with gastrointestinal cancer. We selected the RF model because it showed the best performance among the machine learning models. Compared to deep learning, RF models can be used on a smaller number of patients, which is advantageous for diseases with relatively low incidence, such as PE22,23. The RF model can also track which features were mainly considered by the model in the process of making decisions. This is a form of explainable artificial intelligence, which can solve the model reliability problem that is being faced by most deep learning algorithms.
Among the patients who were diagnosed with cancer in South Korea in 2018, gastric cancer and colorectal cancer were the most common, with gastrointestinal cancer accounting for 36% of all cancer cases27. Based on that study, the ML model proposed herein has the potential to reduce unnecessary CTPA scans in thousands of gastrointestinal cancer patients per year in South Korea. Considering that a CTPA scan costs approximately 400 US dollars in South Korea, this could result in significant cost savings in healthcare expenses. It could also decrease the medical burden by reducing complications associated with contrast media, such as nephropathy and anaphylaxis. The DCA also showed that the ML model offers a greater net benefit than the traditional diagnostic strategy. However, the small number of variables considered limits the interpretability of the results from DCA, and this warrants further study.
In this study, pancreatic cancer was the most common type of cancer in patients with PE. Previous studies have reported that pancreatic cancer carries the highest risk of VTE among all cancer types7,28. Signs and symptoms of DVT, along with history of VTE, are factors that have previously been indicated to be associated with PE10,19,29. While immobilization is a strong risk factor for PE19, there was no significant difference in immobilization according to PE in this study. This could be attributed to the fact that the present study focused on cancer patients. Cancer itself is a risk factor for PE19, and it can also influence the performance status of patients.
In the analysis of SHAP values, high D-dimer levels as well as positive signs and symptoms of DVT were found to be the most important variables in predicting PE. This was observed in both groups, i.e., those in hospital A and hospital B. The link between DVT and PE is well known. A PE occurs when part of the DVT clot breaks off and travels along the blood vessels to the lungs30. D-dimer is also known to be a very important consideration affecting the diagnosis of PE. D-dimer was of even greater importance in the current study because it focused on cancer patients16. Many prior studies have attempted to achieve improved diagnostic rates by altering the cut-off value of D-dimer. One study enhanced the diagnostic rate by adjusting the D-dimer cutoff value based on age31. Another study ruled out more patients suspected of having PE by raising the D-dimer cut-off value to 1000 ng per milliliter in the low probability group based on their Wells scores12. Based on these results, modifying the cut-off value of the D-dimer test has the potential to enhance the diagnostic rate for PE, particularly in cancer patients who exhibit elevated D-dimer levels. We did not use a previously established D-dimer cut-off value in this study, as we instead let the ML model adjust the cut-off value based on other variables, which may have contributed to the reduction in the number of CTPA scans required by the ML model.
This study has several limitations. First, as a retrospective study, it may potentially involve unexpected bias. Since our analysis focused on patients who had undergone CTPA scans, there could be selective bias inherent in the sample. Second, the AUROC values of the ML model are relatively lower than those seen in other studies14,31. This discrepancy is attributed to the fact that the subjects of this study were cancer patients, who typically exhibit higher D-dimer levels than non-cancer patients16. We used ML to address these D-dimer-related limitations. However, as most artificial intelligence—including ML—operates in the form of a black-box model, the precise cutoff value for D-dimer within the ML model remains unknown. Nevertheless, we underscored the significance of D-dimer using feature importance, which is an explainable artificial intelligence technique. Third, the study’s sample size is relatively small. PE is a relatively rare disease with an annual incidence of approximately 0.1%32. Nevertheless, despite using smaller sample sizes, previous studies have successfully developed ML models with robust performance33,34. Fourth, we compared the ML model with the Wells score combined with D-dimer. However, aside from the Wells score, other assessment tools such as the revised Geneva score and PERC are also used for PE risk assessment35. Further research is warranted to make comparisons between these scoring systems with ML model. Fifth, we only used ML models in this study. Various AI models have recently been introduced, and many studies have reported using them36–40. Since we only used variables that are used in existing PE diagnostic methods, we chose the ML model, taking into account the data structure. However, further performance improvements can be achieved by using a deep learning model with medical imaging data and monitoring data such as electrocardiograms. We plan to conduct future studies involving different variables and models.
Despite these limitations, this study has several strengths. To our knowledge, this study is the first to develop a ML model to predict PE in gastrointestinal cancer patients. We applied ML to a simplified model that has already been demonstrated in prior work, which makes it more easily accessible in the emergent department. Further, since the weight of each variable in the Wells score and the cut-off value of D-dimer are adjusted in the ML model, the limitations of the existing diagnostic system can be overcome. Therefore, a large prospective study in the future would be warranted to verify these results.
Conclusion
We developed an ML-based model to predict PE in patients with gastrointestinal cancer. D-dimer was found to be the most important variable in the feature importance analysis. This machine learning model has the potential to both enhance diagnostic strategies for pulmonary embolism (PE) and reduce the number of unnecessary computed tomography pulmonary angiography (CTPA) procedures in the diagnostic process for gastrointestinal cancer patients. It is necessary to verify these results in a prospective cohort study in the future. We plan to improve the performance of the model by including additional variables and considering a larger cohort in the future.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Author contributions
J.S.K., S.H.L. and S-B.L. devised and designed the study. D.K., M.W.L., N.P., J.H.C. and E.S.J. collected data and analyzed data. J.S.K., D.K. and K.K. wrote the manuscript. K.K., E.S.J., I.R.C., W.H.P, J.K.L., J.K.R. and Y-T.K. edited the manuscript. S.H.L. takes full responsibility for the study. All authors have read and approved the manuscript.
Funding
This study was supported by Seoul National University Hospital Research Fund (Grant Number 0420212090).
Data availability
The anonymized datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Joo Seong Kim, Doyun Kwon, Kyungdo Kim, Sang Hyub Lee and Seung-Bo Lee.
Contributor Information
Sang Hyub Lee, Email: gidoctor@snu.ac.kr.
Seung-Bo Lee, Email: koreateam23@gmail.com.
References
- 1.Blom, J. W. et al. Incidence of venous thrombosis in a large cohort of 66,329 cancer patients: Results of a record linkage study. J. Thromb. Haemost. 4(3), 529–535 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Gross, C. P., Galusha, D. H. & Krumholz, H. M. The impact of venous thromboembolism on risk of death or hemorrhage in older cancer patients. J. Gen. Intern. Med. 22(3), 321–326 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sørensen, H. T., Mellemkjaer, L., Olsen, J. H. & Baron, J. A. Prognosis of cancers associated with venous thromboembolism. N Engl. J. Med. 343(25), 1846–1850 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Kim, J. S. et al. Clinical significance of venous thromboembolism in patients with Advanced Cholangiocarcinoma. Gut Liver. 18(1), 165–173 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohashi, Y. et al. Venous thromboembolism in cancer patients: Report of baseline data from the multicentre, prospective Cancer-VTE Registry. Jpn J. Clin. Oncol. 50(11), 1246–1253 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulder, F. I. et al. Venous thromboembolism in cancer patients: A population-based cohort study. Blood. 137(14), 1959–1969 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Cohen, A. T., Katholing, A., Rietbrock, S., Bamber, L. & Martinez, C. Epidemiology of first and recurrent venous thromboembolism in patients with active cancer. A population-based cohort study. Thromb. Haemost. 117(1), 57–65 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Mukai, M. & Oka, T. Mechanism and management of cancer-associated thrombosis. J. Cardiol. 72(2), 89–93 (2018). [DOI] [PubMed] [Google Scholar]
- 9.van Es, N. et al. Wells Rule and d-Dimer Testing to Rule out Pulmonary Embolism: A systematic review and individual-Patient Data Meta-analysis. Ann. Intern. Med. 165(4), 253–261 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Konstantinides, S. V. et al. 2019 ESC guidelines for the diagnosis and management of acute pulmonary embolism developed in collaboration with the European Respiratory Society (ERS): The Task Force for the diagnosis and management of acute pulmonary embolism of the European Society of Cardiology (ESC). Eur. Respir. J. 54(3). (2019). [DOI] [PubMed]
- 11.Hendriksen, J. M. et al. Diagnostic prediction models for suspected pulmonary embolism: Systematic review and independent external validation in primary care. Bmj. 351, h4438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kearon, C. et al. Diagnosis of pulmonary embolism with d-Dimer adjusted to clinical probability. N Engl. J. Med. 381(22), 2125–2134 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Takach Lapner, S., Julian, J. A., Linkins, L. A., Bates, S. & Kearon, C. Comparison of clinical probability-adjusted D-dimer and age-adjusted D-dimer interpretation to exclude venous thromboembolism. Thromb. Haemost. 117(10), 1937–1943 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Banerjee, I. et al. Development and performance of the pulmonary embolism result Forecast Model (PERFORM) for computed tomography clinical decision support. JAMA Netw. Open. 2(8), e198719 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willan, J., Katz, H. & Keeling, D. The use of artificial neural network analysis can improve the risk-stratification of patients presenting with suspected deep vein thrombosis. Br. J. Haematol. 185(2), 289–296 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Crawford, F. et al. D-dimer test for excluding the diagnosis of pulmonary embolism. Cochrane Database Syst. Rev. 2016(8), Cd010864 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruix, J. & Sherman, M. Management of hepatocellular carcinoma: An update. Hepatology. 53(3), 1020–1022 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, K. et al. Oral health and gastrointestinal cancer: A nationwide cohort study. J. Clin. Periodontol. 47(7), 796–808 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Di Nisio, M., van Es, N. & Büller, H. R. Deep vein thrombosis and pulmonary embolism. Lancet. 388(10063), 3060–3073 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Zheng, J. et al. Clinical Data based XGBoost Algorithm for infection risk prediction of patients with decompensated cirrhosis: A 10-year (2012–2021) Multicenter Retrospective Case-control study. BMC Gastroenterol. 23(1), 310 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rufo, D. D., Debelee, T. G., Ibenthal, A. & Negera, W. G. Diagnosis of diabetes mellitus using gradient boosting machine (LightGBM). Diagnostics. 11(9), 1714 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pourghasemi, H. R. et al. Spatial modeling, risk mapping, change detection, and outbreak trend analysis of coronavirus (COVID-19) in Iran (days between February 19 and June 14, 2020). Int. J. Infect. Dis. 98, 90–108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar, M., Kumar, R. & Kaur, P. A healthcare monitoring system using random forest and internet of things (IoT). Multimedia Tools Appl. ;78. (2019).
- 24.Kline, J. A. et al. Prospective multicenter evaluation of the pulmonary embolism rule-out criteria. J. Thromb. Haemost. 6(5), 772–780 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Siddiqui, M. M. et al. Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. Jama. 313(4), 390–397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Villacorta, H. et al. Machine learning with D-dimer in the risk stratification for pulmonary embolism: A derivation and internal validation study. Eur. Heart J. Acute Cardiovasc. Care. 11(1), 13–19 (2022). [DOI] [PubMed] [Google Scholar]
- 27.Hong, S. et al. Cancer statistics in Korea: Incidence, mortality, survival, and prevalence in 2018. Cancer Res. Treat. 53(2), 301–315 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frere, C. et al. Incidence of venous thromboembolism in patients with newly diagnosed pancreatic Cancer and factors Associated with outcomes. Gastroenterology. 158(5), 1346–58e4 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Wells, P. S. et al. Derivation of a simple clinical model to categorize patients probability of pulmonary embolism: Increasing the models utility with the SimpliRED D-dimer. Thromb. Haemost. 83(3), 416–420 (2000). [PubMed] [Google Scholar]
- 30.Jiménez, D. et al. Prognostic significance of deep vein thrombosis in patients presenting with acute symptomatic pulmonary embolism. Am. J. Respir Crit. Care Med. 181(9), 983–991 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Nagel, S. N., Steffen, I. G., Schwartz, S., Hamm, B. & Elgeti, T. Age-dependent diagnostic accuracy of clinical scoring systems and D-dimer levels in the diagnosis of pulmonary embolism with computed tomography pulmonary angiography (CTPA). Eur. Radiol. 29(9), 4563–4571 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Keller, K. et al. Trends in thrombolytic treatment and outcomes of acute pulmonary embolism in Germany. Eur. Heart J. 41(4), 522–529 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Lu, Z. et al. Prediction of immune checkpoint inhibition with immune oncology-related gene expression in gastrointestinal cancer using a machine learning classifier. J. Immunother Cancer ;8(2). (2020). [DOI] [PMC free article] [PubMed]
- 34.Yokoyama, S. et al. Predicted prognosis of patients with pancreatic Cancer by machine learning. Clin. Cancer Res. 26(10), 2411–2421 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Freund, Y., Cohen-Aubart, F. & Bloom, B. Acute Pulmonary Embolism: A review. Jama. 328(13), 1336–1345 (2022). [DOI] [PubMed] [Google Scholar]
- 36.Shi, B. et al. Prediction of recurrent spontaneous abortion using evolutionary machine learning with joint self-adaptive sime mould algorithm. Comput. Biol. Med. 148, 105885 (2022). [DOI] [PubMed] [Google Scholar]
- 37.Li, Y. et al. Epileptic seizure detection in EEG signals using sparse multiscale radial basis function networks and the Fisher vector approach. Knowl. Based Syst. 164, 96–106 (2019). [Google Scholar]
- 38.Chen, Y. et al. LDANet: Automatic lung parenchyma segmentation from CT images. Comput. Biol. Med. 155, 106659 (2023). [DOI] [PubMed] [Google Scholar]
- 39.Zhou, T. et al. Deep learning methods for medical image fusion: A review. Comput. Biol. Med. 160, 106959 (2023). [DOI] [PubMed] [Google Scholar]
- 40.Shi, B. et al. Evolutionary warning system for COVID-19 severity: Colony predation algorithm enhanced extreme learning machine. Comput. Biol. Med. 136, 104698 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The anonymized datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.