Abstract
Objectives
Extracting the sample size from randomized controlled trials (RCTs) remains a challenge to developing better search functionalities or automating systematic reviews. Most current approaches rely on the sample size being explicitly mentioned in the abstract. The objective of this study was, therefore, to develop and validate additional approaches.
Materials and Methods
847 RCTs from high-impact medical journals were tagged with 6 different entities that could indicate the sample size. A named entity recognition (NER) model was trained to extract the entities and then deployed on a test set of 150 RCTs. The entities’ performance in predicting the actual number of trial participants who were randomized was assessed and possible combinations of the entities were evaluated to create predictive models. The test set was also used to evaluate the performance of GPT-4o on the same task.
Results
The most accurate model could make predictions for 64.7% of trials in the test set, and the resulting predictions were equal to the ground truth in 93.8%. GPT-4o was able to make a prediction on 94.7% of trials and the resulting predictions were equal to the ground truth in 90.8%.
Discussion
This study presents an NER model that can extract different entities that can be used to predict the sample size from the abstract of an RCT. The entities can be combined in different ways to obtain models with different characteristics.
Conclusion
Training an NER model to predict the sample size from RCTs is feasible. Large language models can deliver similar performance without the need for prior training on the task although at a higher cost due to proprietary technology and/or required computational power.
Keywords: natural language processing, randomized controlled trial, evidence-based medicine, machine learning, transformer, text mining, GPT-4
Introduction
Using natural language processing (NLP) for data mining in biomedical research has been a topic of longstanding interest.1,2 Recently, it has gained new interest due to the emerging capabilities of large language models.3 Furthermore, new technologies that enable more robust data extraction have been developed in recent years.4,5
In clinical research, randomized controlled trials (RCTs) are the gold standard for testing the effectiveness of an intervention.6 Therefore, they are the focus of many meta-research efforts, such as systematic reviews or meta-analyses. Automatically extracting PICO (patient, intervention, control, outcome) characteristics from RCTs using NLP could improve various processes, from screening trials over assessing adherence to reporting standards to ultimately fully automating the process of evidence synthesis.7–9
A key characteristic of an RCT is the sample size, ie, the number of people included in the trial.10 This information is normally already presented in the abstract, which makes it a suitable parameter for data mining based on the abstract. From a practical point of view, the abstract is also usually not locked behind a paywall.
Inclusion in the trial is usually defined as having undergone randomization.5 However, there are different ways to present this information in an RCT. While some trials might explicitly state the number of participants that were randomized, others might just state the number of patients who “were included,” “were analyzed,” or “completed the trial.”
We hypothesized that each of these different phrases carries a different likelihood of the number being presented actually representing the number of patients who were randomized. We, therefore, trained a named entity recognition (NER) model to extract these phrases as different entities and built a prediction model that considers the different performances for predicting the ground truth of how many people underwent randomization. As an additional validation step, we evaluated the performance of a commercial large language model without task-specific pretraining (GPT-4o) on the same task.
Methods
A random sample of 996 RCTs from 7 major journals (British Medical Journal, JAMA, JAMA Oncology, Journal of Clinical Oncology, Lancet, Lancet Oncology, New England Journal of Medicine) published between 2010 and 2022 were labeled. To do so, abstracts were retrieved as a txt file from PubMed and parsed using regular expressions (ie, expressions that match certain patterns in text).
For each trial, the number of people who were randomized was retrieved by looking at the abstract, followed by the full publication if the number could not be determined with certainty from the abstract.
In addition, 6 different entities were tagged in each abstract, independent of whether the information was presented using words or integers. If the number of people who were randomized was explicitly stated (eg, using the words “randomly,” “randomized,” etc.), this was tagged as “RANDOMIZED_TOTAL.” If the number of people who were analyzed was presented, this was tagged as “ANALYSIS_TOTAL.” If the number of people who completed the trial or a certain follow-up period was presented, this was tagged as “COMPLETION_TOTAL.” If the number of people who were part of the trial without being more specific was presented, this was tagged as “GENERAL_TOTAL.” If the number of people who were assigned to an arm of the trial was presented, this was tagged as “ARM.” Lastly, if the number of patients who were assigned to an arm was presented in the context of how many patients experienced an event, this was tagged as “ARM_EVENT.” As a hypothetical example, in the sentence “50 of 200 people in the intervention arm and 20 of 203 people in the control arm experienced treatment-related toxicity,” 200 and 203 would be tagged as ARM_EVENT. If the abstract did not contain the aforementioned entities, the manuscript was added to the dataset without any tags.
Note that while the dataset and code use the all-caps names for the entities, we will use normal names due to readability for the rest of the paper.
Annotation was carried out independently by 2 physician annotators and conflicts were resolved by discussing the differences afterwards.
150 annotated examples were randomly assigned to an unseen test set. The remaining 846 examples were used to train and validate an NER model using a random 85:15 split into training and validation. The transformer model RoBERTa-base was trained using Adam as the optimizer.11,12 The detailed configuration file with all parameters used for training and validation is available from the code repository at https://github.com/windisch-paul/sample_size_extraction.
In addition to training an NER model, we also built a system of regular expressions and conditional statements for cleaning the entities. This includes a function to turn numbers written as words (eg, due to being at the beginning of a sentence) into integers and code to remove the “n=” from an entity that was extracted as “n = 934,” as well as other unwanted characters, commas, or spaces. The system was developed iteratively on the training set and is presented in its entirety in the analysis.ipynb file in the repository.
After training the model, inference was done on the unseen test set. For each entity, we assessed its agreement with the ground truth. If the same entity was extracted several times from the same trial, we used 2 different approaches: In the case of entities that are supposed to indicate the total number of people in the trial, such as randomized total, analysis total, completion total, or general total, we used the maximum number of each respective entity. In the case of entities where each entity only presents a part of the people in the trial, such as arm and arm event, we summed up all instances of the same entity.
After assessing the performance of the individual entities, we combined them into different models for a final prediction of the ground truth (ie, how many people underwent randomization) using different conditions. The different models were supposed to have different strengths and weaknesses to allow for different use cases. For the first model, the ordered model, we simply ordered the entities according to their performance and built a model that returns the best-performing entity that is present in a publication. If no entity is identified, no prediction is made. For the second model, the accurate entities model, we only chose the 3 best-performing entities and instructed the model to refrain from making a prediction if none of these entities is found. For the third model, the conditional mode, we only allowed the model to make a prediction if either the best-performing entity was found or the second and third best-performing entity were both found and within 10% agreement of each other.
As an additional validation step, Generative Pretrained Transformer 4 Omni (GPT-4o, OpenAI, San Francisco, United States) was evaluated on the test set. We used 2 different system prompts.
The regular prompt allowed GPT-4o to infer the number of people who underwent randomization based on other numbers in the abstract if it wasn’t explicitly mentioned (“You will be provided with the abstract of a randomized controlled clinical trial. Your task will be to extract the number of people who underwent randomization. If this number is not explicitly mentioned, you may use other numerical information [eg, the number of total participants or adding up the number of patients in each arm] to infer that number. Please return only the number as a single integer. If no information is available, please return null.”).
The strict prompt allowed GPT-4o to only return a number if the number of people who underwent randomization was explicitly mentioned (“You will be provided with the abstract of a randomized controlled clinical trial. Your task will be to extract the number of people who underwent randomization. Please return only the number as a single integer. If this number is not explicitly mentioned, please return null.”). The user prompt was the abstract of the respective publication. The temperature was set to 0.2 as per the API documentation that mentions this value as an example to make the model’s output “more focused and deterministic.”
Training, validation, and testing were performed in Python (version 3.11.5) using, among others, the pandas (version 2.1.0), spacy (version 3.7.4), spacy-transformers (1.2.5), and openai (version 1.40.3) packages.
Results
Annotators disagreed on the number of randomized participants in 28 (2.81%) of trials. The mean difference across all trials was 1.9% of the actual number of randomized participants. In trials where there was a difference between the annotators, the mean difference was 66.4% and the median difference was 21.6%. Annotation disagreements were mostly related to transposed digits or picking an incorrect sample size, eg, when multiple sample sizes (such as the number of participants who were randomized, the number of participants who completed the trial, or the number of participants who were analyzed) were mentioned.
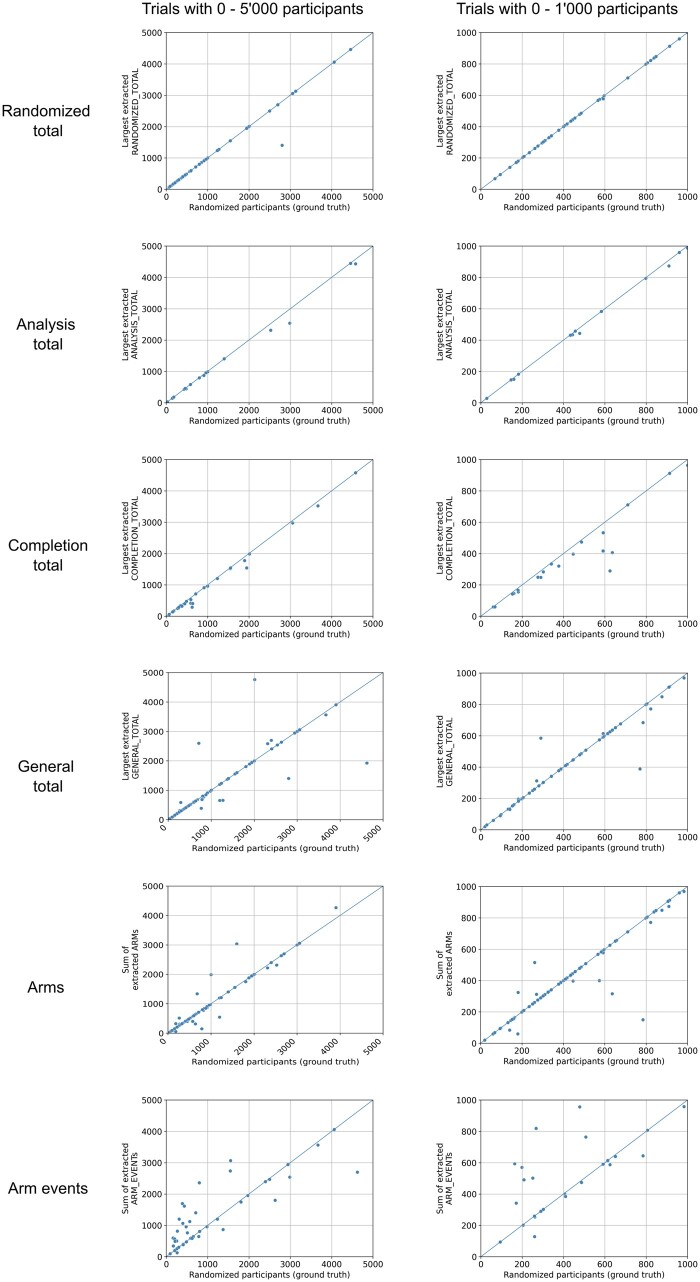
The distribution of different entities in the training and test set and performance of the NER model when extracting the entities from the test set is presented in Table 1. The performance of the different entities at predicting the ground truth is presented in Table 2. The arms entity was most frequently identified (in 74.0% of trials in the test set), followed by the general total (65.3%) and the randomized total (42.0%). The best performance in terms of predicting the ground truth was demonstrated by the randomized total, the arms, and the general total, which all demonstrated a median absolute percentage error of 0.0% and mean absolute percentage errors of 2.4%, 11.1%, and 19.0%, respectively. Scatterplots depicting the agreement between the extracted entities and the ground truth are presented in Figure 1.
Table 1.
Distribution of different entities in the training and test set and performance of the NER model when extracting the entities from the test set.
| Training set—n (%) | Test set—n (%) | Precision | Recall | F1-Score | |
|---|---|---|---|---|---|
| Randomized total | 440 (52.0%) | 64 (42.7%) | 0.98 | 0.97 | 0.98 |
| Analysis total | 144 (17.0%) | 26 (17.3%) | 0.95 | 0.69 | 0.80 |
| Completion total | 92 (10.9%) | 26 (17.3%) | 0.87 | 1.00 | 0.93 |
| General total | 489 (57.8%) | 95 (63.3%) | 0.89 | 0.92 | 0.90 |
| Arms | 620 (73.3%) | 109 (72.7%) | 0.96 | 0.98 | 0.97 |
| Arm events | 307 (36.3%) | 55 (36.7%) | 0.92 | 0.98 | 0.95 |
Abbreviation: NER, named entity recognition.
Table 2.
Performance of different entities at predicting the ground truth.
| Extracted from (%) | Mean absolute percentage error (%) | Median absolute percentage error (%) | Extracted entity within 10% from ground truth (%) | Extracted entity within 1% from ground truth (%) | Extracted entity equal to ground truth (%) | |
|---|---|---|---|---|---|---|
| Randomized total | 42.0 | 2.4 | 0.0 | 96.8 | 95.2 | 95.2 |
| Analysis total | 12.7 | 3.1 | 1.3 | 94.7 | 47.4 | 26.3 |
| Completion total | 20.0 | 9.5 | 6.2 | 70.0 | 20.0 | 10.0 |
| General total | 65.3 | 19.0 | 0.0 | 85.7 | 74.5 | 70.4 |
| Arms | 74.0 | 11.1 | 0.0 | 82.9 | 71.2 | 66.7 |
| Arm events | 39.3 | 61.7 | 7.0 | 50.8 | 27.1 | 20.3 |
The “Extracted from” column indicates the percentage of trials in which the respective entity was found. The remaining columns indicate the accuracy of the respective entity in predicting the ground truth, ie, how many people were randomized.
Figure 1.
Scatterplots of different extracted entities. Each dot represents a trial with the ground truth, ie, the number of participants who were randomized being its x-coordinate. The y-coordinate is the respective entity. For the randomized total, analysis total, completion total, and general total entities, the largest respective number was used in case multiple numbers were extracted from the same trial. In case of the arm and arm event entities, all extracted numbers from the same entity for a trial were summed up. Plots on the left show trials with 0-5000 participants, while plots on the right show only trials with 0-1000 participants to allow for a better assessment of the performance in the range that most trials fall into. The diagonal lines indicate perfect predictions.
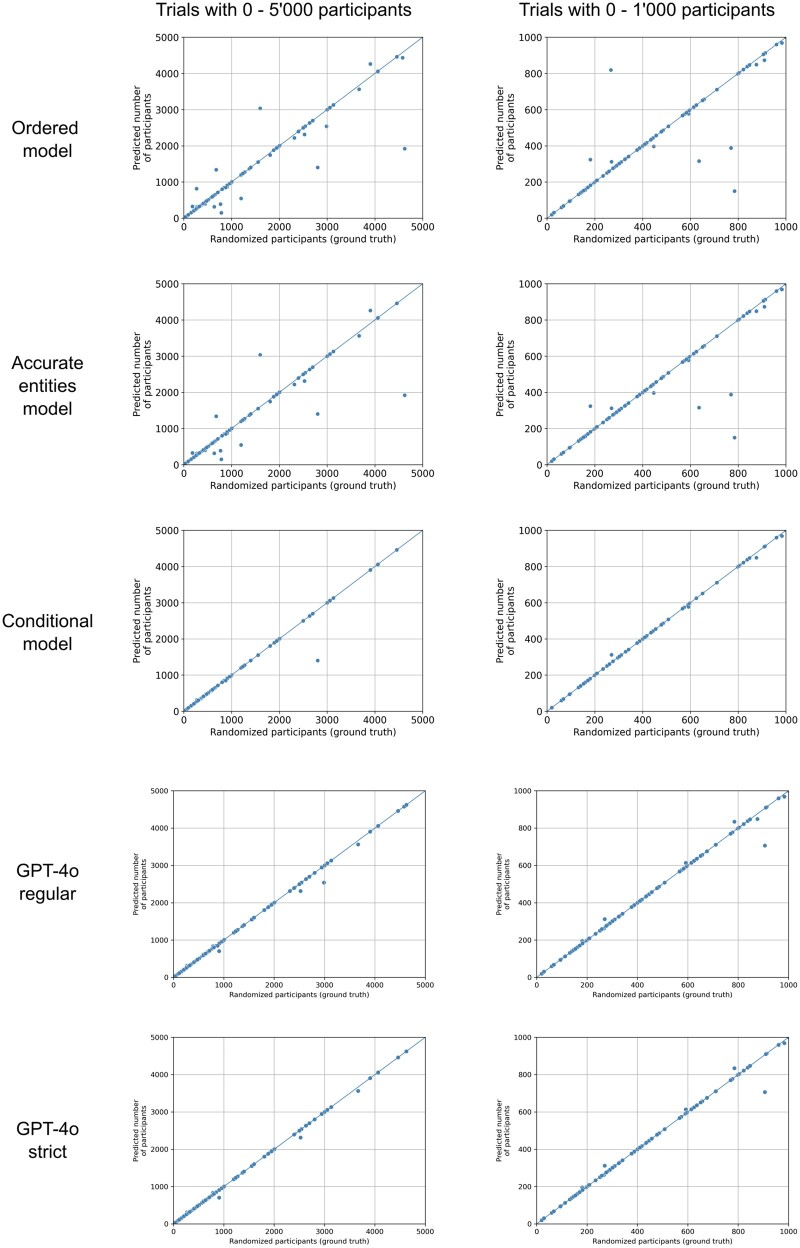
The performance of the 3 models is depicted in Table 3 and Figure 2. While the conditional model exhibited the best performance and made predictions that were equal to or within 1% of the ground truth in 93.8% and within 10% in 96.9%, it could only make predictions on 64.7% of all the trials in the test set due to the strict conditions regarding entities that need to be present for making predictions.
Table 3.
Performance of different models.
| Predicted in (%) | Mean absolute percentage error (%) | Median absolute percentage error (%) | Prediction within 10% of ground truth (%) | Prediction within 1% of ground truth (%) | Prediction equal to ground truth (%) | |
|---|---|---|---|---|---|---|
| Ordered model | 98.0 | 9.2 | 0.0 | 87.1 | 78.2 | 76.9 |
| Accurate entities model | 96.0 | 7.8 | 0.0 | 88.2 | 79.9 | 78.5 |
| Conditional model | 64.7 | 1.7 | 0.0 | 96.9 | 93.8 | 93.8 |
| GPT-4o regular prompt | 99.3 | 1.5 | 0.0 | 96.6 | 90.6 | 88.6 |
| GPT-4o strict prompt | 94.7 | 0.6 | 0.0 | 98.6 | 92.3 | 90.8 |
The “Predicted in” column indicates the percentage of trials for which a prediction could be made. The remaining columns indicate the accuracy of the respective model in predicting the ground truth, ie, how many people were randomized.
Figure 2.
Scatterplots of different models. Each dot represents a trial with the ground truth, ie, the number of participants who were randomized being its x-coordinate. The y-coordinate is the prediction by the model. Plots on the left show trials with 0-5000 participants, while plots on the right show only trials with 0-1000 participants to allow for a better assessment of the performance in the range that most trials fall into. The diagonal lines indicate perfect predictions.
On the other end of the spectrum, the ordered model was able to make a prediction on 98.0% of trials in the test set. However, these predictions were less accurate, with 76.9 being equal to the ground truth, 78.2% being within 1%, and 87.1% within 10%.
The accurate entities model represents a compromise. It was able to make a prediction on 96.0% of trials, with 78.5% of its predictions being equal to the ground truth, 79.9% being within 1%, and 88.2% within 10%.
GPT-4o with the regular prompt made a prediction for almost all trials in the test set (99.3%). Its predictions were equal to the ground truth in 88.6% of cases, with 90.6% being within 1% and 96.6% within 10%. GPT-4o with the strict prompt made a prediction for 94.7% of trials. Its predictions were equal to the ground truth in 90.8% of trials, with 92.3% being within 1%, and 98.6% within 10%. The cost for classifying the 100 abstracts in the test set was USD 0.38.
Discussion
This study presents an NER model that can extract 6 different entities that can be used to predict the sample size from the abstract of an RCT. The entities can be combined in different ways to obtain models with different characteristics.
The fact that the randomized total entity demonstrated the best performance is unsurprising, considering that the number of people who were randomized was the ground truth and that sentences that explicitly state how many people were randomized should be very indicative of this. The completion and analysis total entities showed larger discrepancies. In the case of the completion total, the scatterplot suggests that this number is often smaller than the ground truth, which makes sense as not every patient who is randomized completes a trial. The same discrepancy can be seen with the analysis total, as not every trial reports its results by the intention-to-treat (which should be equal to the number of people who were randomized) but rather conducts a per-protocol analysis for which the number of included patients is often smaller. For the general total entity, there were also outliers where the extracted entity was larger than the ground truth. This happened, for example, when a trial stated that “X patients were included,” while the number X was actually the number of patients who were screened. In the case of the arm and arm event entities, the arm entity performs better and arm events produce more outliers where the entity was actually larger than the ground truth. This can happen due to multiple arm events being present. As an example, in a 2-arm trial, the results might first report the results for the primary endpoint, then the rate of grade 2 toxicities and the rate of grade 3 toxicities. If, in each of those sentences, the number of patients in the arm is reported, all of those entities are extracted, and the sum of those entities is obviously larger than the actual number of patients in the trial. A possibility to improve this could be to train a model to predict how many arms a trial has and to only add up as many arm entities as there are arms in the trial.
The 3 models that leverage the entities to make a prediction that were presented have different strengths and weaknesses that can support different use cases. In addition, combinations of the models (or other models based on the extracted entities) could be used to allow for explainable prediction workflows, where every prediction that is made is associated with the model that made the prediction so that the person reviewing the prediction knows which entities the prediction is based on and how certain a prediction is based on the characteristics of the respective model. These workflows could then be used, eg, to create a database of DOIs and sample sizes so that future projects can rely on a simple lookup instead of always having to run a computationally expensive inference.
The conditional model creates predictions that are highly accurate, but these predictions can only be made for around two-thirds of publications. A model like this could be used in a workflow where a human annotator manually annotates the other publications. The ordered and the accurate entities models could be used in a workflow where either a human annotator is not feasible, but accuracy is not as critical or where a human annotator manually annotates all trials and the tool pre-completes the annotations so that the annotator can accomplish his tasks with fewer clicks and, in turn, faster.13 However, it should be noted that GPT-4o, while performing slightly worse than the conditional model in terms of the number of its predictions equal to the ground truth, substantially outperformed both the ordered and accurate entities model. Therefore, it is likely the better option if cost is not a concern, since the cost for GPT-4o was USD 0.38 per 100 abstracts while the task-specific models could perform the inference on the authors’ local machine. While there are open source LLMs that could be deployed locally which might offer comparable performance to OpenAI’s proprietary model, computational power will remain a consideration that favors task-specific models. RoBERTa-base has 125 million parameters while the exact size of GPT-4o has not been published. However, the open source Llama 3.1 405B which can compete with GPT-4o on several benchmarks has 405 billion parameters thus requiring considerably more resources.14
Comparing our models’ performances to previously published research is difficult. Lin and colleagues, as well as Kiritchenko et al, and Marshall et al and, in the publication proposing Trialstreamer report precision and recall and not continuous measures of performance compared to the ground truth.5,10,15 This makes sense for them, as their approach is to extract the sample size if it is explicitly mentioned in the paper. If the correct number is extracted, it is a true positive. If no sample size is present and the model does not extract a number, it is a true negative. Our approach is different in that we also predict the sample size even if it cannot be extracted directly, eg, by extracting the number of patients in each arm and adding them up to allow for retrieving a sample size for more publications. Also, for some use cases, it might be acceptable to just get a rough estimate of how many patients were randomized, eg, by using the number of patients who were analyzed or who completed the trial. However, the previous research results already supported the feasibility of this task in general, with a recall and precision of 0.79 and 0.88 presented by Marshall et al and similar results presented by Lin and colleagues.
Our study is limited by the fact that we only used trials from 7 journals for training and testing. While these are journals that publish many practice-changing RCTs, we can’t assess the model’s ability to generalize to trials from other journals, especially those that use unstructured abstracts. The strengths of this study include the use of a dedicated unseen test set and the high degree of reproducibility as all code and annotated data are shared in a public repository. The NER model returns entities that can be tailored by other researchers to make predictions based on the characteristics of the task that needs to be completed.
As an outlook, one could try to link trial publications to their respective entry on clinicaltrials.gov and use the information there if the sample size cannot be inferred from the abstract. As a general observation, more strict enforcement of guidelines such as CONSORT could greatly improve the performance of text-mining efforts in evidence-based medicine.16,17 To enable readers to judge the model’s performance, a sample-size filter based on the model presented herein can be tested at https://www.scantrials.com/.
In conclusion, training an NER model to predict the sample size from RCTs is feasible, not only if the sample size is explicitly mentioned but also if the sample size can be calculated, eg, by adding up the number of patients in each arm. Being able to extract the sample size automatically could support various meta-research efforts. Large language models can deliver similar performance without the need for prior training on the task although at a higher cost due to proprietary technology and/or required computational power.
Contributor Information
Paul Windisch, Department of Radiation Oncology, Cantonal Hospital Winterthur, 8400 Winterthur, Switzerland.
Fabio Dennstädt, Department of Radiation Oncology, Inselspital, Bern University Hospital, University of Bern, 3012 Bern, Switzerland.
Carole Koechli, Department of Radiation Oncology, Cantonal Hospital Winterthur, 8400 Winterthur, Switzerland.
Robert Förster, Department of Radiation Oncology, Cantonal Hospital Winterthur, 8400 Winterthur, Switzerland; Department of Radiation Oncology, Inselspital, Bern University Hospital, University of Bern, 3012 Bern, Switzerland.
Christina Schröder, Department of Radiation Oncology, Cantonal Hospital Winterthur, 8400 Winterthur, Switzerland.
Daniel M Aebersold, Department of Radiation Oncology, Inselspital, Bern University Hospital, University of Bern, 3012 Bern, Switzerland.
Daniel R Zwahlen, Department of Radiation Oncology, Cantonal Hospital Winterthur, 8400 Winterthur, Switzerland.
Author contributions
Paul Windisch, Fabio Dennstädt (Conceptualization); Paul Windisch, Daniel R. Zwahlen (Methodology); Paul Windisch, Daniel R. Zwahlen (Formal analysis); Paul Windisch (Data curation); Paul Windisch (Writing—original draft preparation); Fabio Dennstädt, Carole Koechli, Robert Förster, Christina Schröder, Daniel M. Aebersold, Daniel R. Zwahlen (Writing—review and editing); Daniel R. Zwahlen (Supervision); Daniel M. Aebersold, Daniel R. Zwahlen (Project administration); All authors read and approved the final manuscript.
Funding
No funding was received for this project.
Conflicts of interest
P.W. has a patent application titled “Method for detection of neurological abnormalities” outside of the submitted work. The remaining authors declare no conflict of interest.
Data availability
All data and code used to obtain this study’s results have been uploaded to https://github.com/windisch-paul/sample_size_extraction.
References
- 1. Wallace BC, Small K, Brodley CE, et al. Deploying an interactive machine learning system in an evidence-based practice center: abstrackr. In: Proceedings of the 2nd ACM SIGHIT International Health Informatics Symposium. Association for Computing Machinery; 2012.
- 2. Vaswani A, Shazeer NM, Parmar N, et al. Attention is all you need. Adv Neural Inf Process Syst. 2017;30:5998-6008. [Google Scholar]
- 3. Dennstädt F, Zink J, Putora PM, et al. Title and abstract screening for literature reviews using large language models: an exploratory study in the biomedical domain. Syst Rev. 2024;13:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beltagy I, Lo K, Cohan A. SciBERT: a pretrained language model for scientific text. arXiv [csCL]. 2019. Accessed October 1, 2024. https://arxiv.org/abs/1903.10676
- 5. Marshall IJ, Nye B, Kuiper J, et al. Trialstreamer: a living, automatically updated database of clinical trial reports. J Am Med Inform Assoc. 2020;27:1903-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jones DS, Podolsky SH.. The history and fate of the gold standard. Lancet. 2015;385:1502-1503. [DOI] [PubMed] [Google Scholar]
- 7. Kilicoglu H, Rosemblat G, Hoang L, et al. Toward assessing clinical trial publications for reporting transparency. J Biomed Inform. 2021;116:103717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schmidt L, Sinyor M, Webb RT, et al. A narrative review of recent tools and innovations toward automating living systematic reviews and evidence syntheses. Z Evid Fortbild Qual Gesundhwes. 2023;181:65-75. [DOI] [PubMed] [Google Scholar]
- 9. Hoang L, Guan Y, Kilicoglu H.. Methodological information extraction from randomized controlled trial publications: a pilot study. AMIA Annu Symp Proc. 2022;2022:542-551. [PMC free article] [PubMed] [Google Scholar]
- 10. Lin F, Liu H, Moon P, et al. A sample size extractor for RCT reports. MEDINFO 2021: One World, One Health—Global Partnership for Digital Innovation. IOS Press; 2022:617-621. [DOI] [PubMed] [Google Scholar]
- 11. Liu Y, Ott M, Goyal N, et al. RoBERTa: a robustly optimized BERT pretraining approach. arXiv [csCL]. 2019. Accessed October 1, 2024. https://arxiv.org/abs/1907.11692 [Google Scholar]
- 12. Kingma DP, Ba J. Adam: a method for stochastic optimization. arXiv [csLG]. 2014. Accessed October 1, 2024. https://arxiv.org/abs/1412.6980
- 13. Reidsma D, Hofs DHW, Jovanovic N. Designing focused and efficient annotation tools. In: Measuring Behaviour, 5th International Conference on Methods and Techniques in Behavioral Research. Noldus Information Technology; 2005:149-152.
- 14.Introducing Llama 3.1: our most capable models to date. Meta AI; 2024. Accessed October 1, 2024. https://ai.meta.com/blog/meta-llama-3-1/
- 15. Kiritchenko S, de Bruijn B, Carini S, et al. ExaCT: automatic extraction of clinical trial characteristics from journal publications. BMC Med Inform Decis Mak. 2010;10:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hopewell S, Clarke M, Moher D, et al. ; CONSORT Group. CONSORT for reporting randomised trials in journal and conference abstracts. Lancet. 2008;371:281-283. [DOI] [PubMed] [Google Scholar]
- 17. Begg C, Cho M, Eastwood S, et al. Improving the quality of reporting of randomized controlled trials. The CONSORT statement. JAMA. 1996;276:637-639. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and code used to obtain this study’s results have been uploaded to https://github.com/windisch-paul/sample_size_extraction.