Abstract
Zta has a dual role in the Epstein-Barr virus (EBV) lytic cycle, acting as a key regulator of EBV lytic gene expression and also being essential for lytic viral DNA replication. Zta's replication function is mediated in part through interactions with the core viral replication proteins. We now show interaction between Zta and the helicase (BBLF4) and map the binding region to within amino acids (aa) 22 to 86 of the Zta activation domain. In immunofluorescence assays, green fluorescent protein (GFP)-tagged BBLF4 localized to the cytoplasm of transfected cells. Cotransfection of Zta resulted in translocation of BBLF4-GFP into the nucleus indicating interaction between these two proteins. However, Zta with a deletion of aa 24 to 86 was unable to mediate nuclear translocation of BBLF4-GFP. Results obtained with Zta variants carrying deletions across the aa 24 to 86 region indicated more than one contact site for BBLF4 within this domain, and this was reinforced by the behavior of the four-point mutant Zta (m22/26,74/75), which was severely impaired for BBLF4 interaction. Binding of BBLF4 to Zta was confirmed using GST affinity assays. In both cotransfection-replication assays and replication assays performed in EBV-positive P3HR1 cells, the Zta (m22/26,74/75) mutant was replication defective. In Zta-transfected D98-HR1 cells, replication compartments could be detected by immunofluorescence staining using anti-BMRF1 monoclonal antibody. Cells transfected with Zta variants that were defective for helicase binding still formed replication compartments, but Zta was excluded from these compartments. These experiments reveal a role for the Zta-helicase interaction in targeting Zta to sites of viral DNA replication.
The Epstein-Barr virus (EBV) lytic regulatory protein Zta (BZLF1, ZEBRA, Z) plays a key role in both regulation of EBV lytic gene expression and in lytic viral DNA replication. Zta is related to the Fos/CEBP family of cellular bZIP transcription factors but contains a unique coiled-coil dimerization domain that lacks the standard heptad leucine repeat (13, 19, 21). As a consequence, Zta forms homodimers and does not heterodimerize with the cellular Jun, Fos, and CREB bZIP proteins. Zta binds to both AP-1 sites and related sequences called Zta response elements (ZREs) (19, 38, 57). Both DNA binding and Zta activity are modulated by phosphorylation (7, 31). Zta activates transcription through stabilization of a TFIIA-TFIIID complex (17, 37, 40) and by recruiting the CREB-binding protein CBP (1, 14, 62). CBP and the related p300 possess intrinsic histone acetylase activity. They stimulate transcription through acetylation of histones, which leads to chromatin remodeling, through the acetylation of nonhistone transcription factors and by serving as a bridging factor between transcription factors and polymerase II (6, 33, 46, 48).
Two cis-acting regions have been defined as essential components of the EBV lytic origin of replication, oriLyt (25, 54). One of these comprises the promoter for the BHLF1 open reading frame which contains four ZREs and the second region, which is located 530 bp distally and does not contain ZREs, is required for oriLyt replication but not for transcriptional regulation of BHLF1 (8, 47). Mutation of all four of the ZREs in the promoter proximal region abolished oriLyt replication, indicating the necessity for Zta binding to this region for origin function (53). In a complementary approach, examination of the trans-acting EBV proteins needed for replication of an oriLyt containing plasmid in a Challberg cotransfection assay in EBV-negative cells also revealed an absolute requirement for Zta. In this assay, oriLyt replication was dependent on the presence of Zta plus six core replication proteins, the DNA polymerase (BALF5), polymerase processivity factor (BMRF1), single-stranded DNA-binding protein (BALF2), helicase (BBLF4), primase (BSLF1), and the primase-associated factor (BBLF2/3) (20, 52).
Zta is likely to contribute to oriLyt replication through a variety of mechanisms. Zta induces growth arrest, in part through posttranscriptional p53 stabilization and upregulation of the cyclin-dependent kinase inhibitors p21 and p27 (11, 49). Unlike the smaller DNA viruses, herpesviruses encode many of the enzymes necessary for DNA replication, and it may be advantageous for these viruses to replicate their genomes at a time when they are not competing with cellular DNA replication. The Zta activation domain is not required for Zta-induced growth arrest (12). Recent work has revealed that incoming viral genomes of adenovirus, simian virus 40 (SV40), herpes simplex virus (HSV), human cytomegalovirus (HCMV), and Kaposi's sarcoma-associated herpesvirus (KSHV) accumulate and replicate at promyelocytic leukemia protein (PML)-associated nuclear bodies called PODs (PML oncogenic domains) or ND10 (nuclear domain 10) (4, 10, 28, 42, 56, 59). In HSV- and HCMV-infected cells the POD structure is disrupted prior to lytic DNA replication (3, 32, 43), but replication compartments were detected in KSHV-infected cells that were decorated on the periphery with PODs (59). Induction of lytic replication in EBV-positive D98-HR1 cells revealed a complex process in which the POD protein Sp100 was dispersed prior to the onset of DNA replication, but PML dispersion and loss of POD structure was delayed until after the detection of replicating viral DNA (9). Zta appears to be the EBV protein that mediates late-stage dispersion of PODs, and the N terminus of the Zta activation domain is required for this function (2). The ability of Zta to disperse PML may be related to competition between Zta and PML for SUMO-1 modification (2).
Zta may also contribute to origin function through its transcriptional activity. Transcription factors frequently bind to auxiliary sequences flanking the minimal origin region where they stimulate replication efficiency (15, 24, 45) by contributing to the relief of nucleosomal repression (27, 35, 44). Initial deletion analyses of the Zta transcriptional activation domain found concurrent reductions in both the transcription and the replication functions of Zta (5, 53). However, there were indications that the Zta transactivation domain might have a role in oriLyt replication beyond that made by its contribution to transcriptional activation. An oriLyt plasmid in which the ZRE sites were converted to binding sites for either human papillomavirus E2 or yeast Gal4 could not be efficiently replicated by these proteins but was replicated by Zta fusion proteins targeted to the altered binding sites (53). Further, an activation domain deletion, Zta(Δ11–25) that had no negative effects on transactivation function resulted in a Zta protein that was replication deficient in cotransfection replication assays (51).
In herpesvirus-infected cells, lytic viral DNA replication takes place in globular or kidney-shaped nuclear subdomains that stain for viral replication proteins (3, 41, 52, 58, 59, 66). Replication compartment formation has not been extensively examined in EBV-infected cells but Zta and BMRF1 were shown to colocalize in replication compartments formed after induction of the lytic cycle in EBV-infected Akata cells (55). In the present study we examined the interaction between the EBV helicase BBLF4 and Zta. The BBLF4 interaction site mapped to the amino acid (aa) 22 to 86 segment of the Zta transactivation domain adjacent to the interaction site for the BSLF1-BBLF2/3 primase subcomplex, further illustrating the complexity of contacts made by the core replication proteins. Zta mutants that were defective for helicase interaction failed to efficiently associate with replication compartments in Zta-transfected D98-HR1 cells, providing evidence for an additional role for the helicase in the intranuclear targeting of Zta.
MATERIALS AND METHODS
Plasmid constructions.
A 750-bp BglII fragment containing the green fluorescent protein (GFP) coding sequences was obtained by introducing a 10-mer BglII linker into the NheI site of pEGFP-C1 (Clontech, Inc.) and cleaving with BglII. BBLF4-GFP was constructed by ligating the 750 bp fragment into BclI-cleaved pRTS28 (23). The Zta inserts from the plasmids pPL228 (39), pZQ239, and pZBSΔ39–45 (obtained from P. Lieberman) were transferred as EcoRI or BamHI fragments, respectively, into the appropriate sites of the SG5 vector (Stratagene) to generate Zta Δ24–86, Zta Δ3–39, and Zta Δ39–45 eukaryotic expression plasmids. Zta Δ61–81 was constructed by PCR amplification of Zta aa 1 to 60 using the primers 5′-CAGTGGATCCTAATGATGGACCCAAACTCG and 5′-GCTAGCGGCCGCTGGCCTTGTGGCAGA, and Zta aa 82 to 245 was constructed using the primers 5′-GCTAGCGGCCGCCTCCTGAGAATGCTTAT and 5′-GCTAGGATCCTTACTTGTCATCGTCGTCC. The NotI-cleaved PCR products were ligated, and the Zta aa 1 to 60 plus 82-to-245 fragment was purified and ligated into the BamHI site of the SG5 vector. A pPL230 (51) EcoRI fragment was ligated into pGH416 to create the glutathione S-transferase (GST)–Zta Δ94–140 fusion protein expression plasmid. GST-Zta(wt)(pDH237), GST-Zta(26–133)(pDH279), and GST-Zta(1–133)(pDH245) have been described elsewhere (23), as have the oriLyt-chloramphenicol acetyltransferase (CAT) reporter, pDH123, and the expression plasmid for Zta(wt), pRTS21, and Zta(Δ2–25). (51). The Zta(m22/24,74/75) and GST-Zta(22/24,74/75) plasmids were a gift from Paul M. Lieberman (40).
Immunofluorescence assays.
Vero cells were seeded at 8 × 104 cells per well in two-well slide chambers. Cells were transfected with a maximum of 3 μg of DNA by the calcium phosphate procedure. After transfection, cells were incubated in Dulbecco modified Eagle medium plus 10% fetal bovine serum for 16 h at 35°C in 3% CO2, followed by a medium change and a further 24-h incubation. When appropriate, bromodeoxyuridine (BrdU) was added to the culture medium at a final concentration of 10 μM for 45 min before fixation. Cells were washed in phosphate-buffered saline (PBS; 0.144 g of KH2PO4, 9.0 g of NaCl, and 0.795 g of Na2HPO4 · 7H2O per liter), fixed with 1% paraformaldehyde in PBS for 10 min at room temperature, and permeabilized for 20 min on ice in 0.2% Triton X-100 in PBS. For BrdU staining, pulse-labeled cells were incubated with 4 N HCl for 10 min at room temperature in order to expose incorporated BrdU residues and then washed in PBS three times for 5 min each time before permeabilization. Cells were incubated with primary antibody for 60 min at 37°C and with secondary antibody at 37°C for 30 min. The antibodies used were anti-BMRF1 monoclonal antibody (1:200; ABI Advanced Biotechnologies, Inc., Columbia, Md.), anti-BZLF1 polyclonal antibody (1:800; a gift of Marie Hardwick, Johns Hopkins School of Hygiene and Public Health), and sheep anti-BrdU antibody (1:200; Fitzerald Industries International, Inc., Concord, Mass.), fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G (IgG; 1:200; Cappel Organon Teknika, Durham, N.C.), FITC-conjugated donkey anti-rabbit IgG (1:200), rhodamine-conjugated donkey anti-mouse immunoglobulin (1:200) and anti-rabbit immunoglobulin (1:200), and rabbit anti-sheep IgG (1:200; Chemicon, Temecula, Calif.).
CAT assay.
Hela cells were plated in six-well cluster dishes at 2 × 105 cells per well 16 h before transfection, with a medium change 4 h before transfection. Cells were transfected by calcium phosphate precipitation with the oriLyt-CAT reporter (pDH123, 1 μg) and Zta or Zta variants. Vector SG5 DNA was used to equalize the amount of DNA in each transfection to 5 μg. Each experiment was repeated at least two times. CAT assays were performed as previously described (26). Total protein was measured using the BCA Protein Assay Reagent (Pierce, Rockford, Ill.), and equal amounts of protein were analyzed for CAT activity.
GST-protein affinity assay.
GST and GST-Zta fusion proteins were induced by growth in medium containing 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 30°C. Pelleted bacteria were resuspended in binding buffer (50 mM Tris-HCl [pH 7.9], 100 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, 2 mM dithiothreitol [DTT], 0.2% Nonidet P-40) and sonicated. Cell debris was removed by centrifugation at 10,000 × g for 10 min. The supernatant was incubated with glutathione agarose beads (Sigma, St. Louis, Mo.) at 4°C overnight, followed by three washes in binding buffer. The amount of protein bound to the beads was determined by Coomassie brilliant blue staining of protein separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Equal amounts of each GST protein were used in the affinity assays.
293T cells in 100-mm dishes were transfected with a maximum of 15 μg per dish of DNA, and cells were harvested 40 h after transfection. Cells were lysed in 2.2 ml of lysis buffer (50 mM Tris-HCl [pH 7.4], 100 mM NaCl, 0.5 mM MgCl2, 1 mM EDTA, 2 mM DTT, and 0.2% Nonidet P-40). Cell extract was incubated with the GST fusion protein-glutathione agarose beads overnight at 4°C, after which the complex was washed five times in binding buffer. The complex was dissociated from the beads by boiling for 5 min in 2× SDS-PAGE loading buffer (2% SDS, 10% glycerol, 100 mM DTT, 60 mM Tris [pH 6.8], 0.02% bromophenol blue), and the proteins were separated by SDS-PAGE on a 10% gel. Proteins were transferred to a nitrocellulose membrane (Bio-Rad, Hercules, Calif.), and BBLF4-GFP and GFP proteins were detected by incubation with an anti-GFP monoclonal antibody (1:5,000; Clontech), followed by visualization using enhanced chemiluminescence (Amersham Life Science, Buckinghamshire, England).
DNA replication assays.
The DNA transfection-replication assay was performed using a modification of the previously described protocol (51, 52). Briefly, 1.5 × 106 Vero cells per 100-mm dish were transfected with 10 μg of pEF52(ori-Lyt) DNA, 1.6 μg of Zta expression plasmid, and 0.8 μg of expression plasmids for each of the six core replication proteins, as well as Mta and Rta. At 80 h after the posttransfection medium change, the cell monolayer was washed twice with PBS and scraped into 4 ml of 40 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 150 mM NaCl. The cells were then pelleted and lysed in 2 ml of lysis buffer (10 mM Tris-HCl [pH 8.0], 10 mM EDTA, 2% SDS, 100 μg of proteinase K per ml). After overnight incubation at 37°C, the samples were diluted to 4 ml with Tris-EDTA (pH 8.0); extracted with phenol, phenol-chloroform, and chloroform; and ethanol precipitated after the addition of sodium acetate (pH 5.2) to a final concentration of 0.3 M. The DNA pellets were resuspended in 450 μl of distilled H2O, treated with 100 μg of RNase A per ml, ethanol precipitated, and resuspended in 300 μl of H2O. Then, 10 μg of extracted cellular DNA was digested overnight with 30 U of DpnI at 37°C. Replicated DNA was detected by PCR amplification using oligonucleotide primers specific for pBR322 DNA. The primers 5′-GAAGCCAGTTACCTTCGG and 5′-GCAGGACCACTTCTGCG amplified a 510-bp fragment containing seven DpnI restriction sites, while the primers 5′-CTGTGGAACACCTACATCTG and 5′-AGATGTCTGCCTGTTCATCC amplified a 330-bp control fragment that lacked DpnI sites. The products were visualized by electrophoresis on a 1.2% agarose gel containing 500 ng of ethidium bromide per ml.
In the induction-replication assay, EBV-infected P3HR-1 cells were transfected with Zta or Zta variants by electroporation (300 V; capacitance, 950 μF; DNA, 2 μg). Total cellular DNA was extracted as described above, and an equal amount of the DNA as determined by spectrophotometry and ethidium bromide fluorescence quantitation was digested with BamHI. The cellular DNA was resolved by electrophoresis on a 1.0% agarose gel at 80 V for 6 h. After treatment of the gel at 20°C for 45 min in 1.5 mol of NaCl and 0.5 mol of NaOH per liter and incubation in 1.5 mol of NaCl and 1 mol of Tris-HCl (pH 7.4) per liter for 15 min, the agarose gel was then transferred to a nitrocellulose membrane in the presence of 10× SSC (1.5 M NaCl plus 0.15 M sodium citrate) for 16 to 20 h. After the membrane was dried completely at 20°C, the DNA was irreversibly cross-linked by UV radiation (Stratalinker; Stratagene). For hybridization, the membrane was incubated for 3 h at 65°C in 15 ml of buffer consisting of 6× SSPE (900 mM NaCl, 60 mM Na2HPO4, 6 mM Na2EDTA), 5× Denhardt solution, 0.5% SDS, and 91 μg of calf thymus DNA (Sigma Chemical Co., St. Louis, Mo.) per ml. Approximately 25 ng of a gel-purified EBV genomic EcoRI-Dhet fragment was radiolabeled with [α-32P]dCTP by random priming to a specific activity of 108 cpm/μg (Boehringer Mannheim-Roche Kit). The membrane was then incubated at 65°C with 106 cpm of denatured radiolabeled EcoRI-Dhet probe DNA per ml in fresh hybridization buffer. Following hybridization for 16 h, the membrane was washed in 2× SSC–0.5% SDS at room temperature for 5 min, in 2× SSC–0.1% SDS for 5 min, and in 0.1× SSC–0.5% SDS at 37°C for 30 min and then again at 65°C for 45 min. The membrane was exposed to Kodak XAR5 film for 12 h at −80°C using an intensifying screen.
RESULTS
Zta interaction with the EBV helicase, BBLF4.
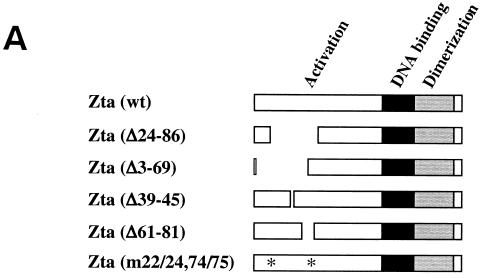
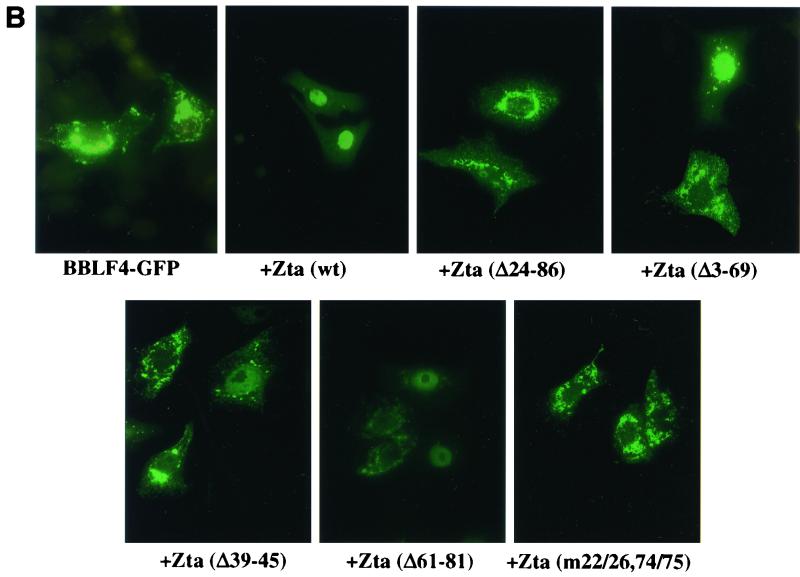
The EBV helicase, BBLF4, has previously been shown to localize to the cytoplasm of transfected cells (23). Cotransfection with Zta was found to result in nuclear translocation of BBLF4, suggesting an interaction between these two proteins (23). A BBLF4-GFP expression vector was generated to further characterize this interaction. BBLF4-GFP was transfected into Vero cells alone or with the series of Zta expression plasmids shown in Fig. 1A and the intracellular localization of BBLF4-GFP was determined by fluorescence microscopy (Fig. 1B). In transfected cells, BBLF4-GFP showed a cytoplasmic distribution. This localization was converted to nuclear when BBLF4-GFP was cotransfected with wild-type Zta. However, Zta(Δ24–86) was ineffective at mediating nuclear localization of BBLF4-GFP. Deletions were created across the aa 24 to 86 region in an attempt to further define the sequences required for BBLF4 binding. Deletion of either the front portion of this domain [Zta(Δ3–69)] or the back portion [Zta(Δ61–81)] had a similar effect. In each case, cotransfection with BBLF4-GFP led to the detection of nuclear BBLF4-GFP, although the efficiency of nuclear translocation was reduced, and cytoplasmic BBLF4-GFP was also detected in a significant proportion of the cells (Fig. 1B; Table 1). Changing the spacing between the boundaries of the domain by introducing a small 6-aa deletion in the central region [Zta(Δ39–45)] also had the effect of reducing the effectiveness of the interaction as measured by BBLF4-GFP nuclear localization (Fig. 1B; Table 1). These results suggested that BBLF4 might make contacts within each half of the aa 24 to 86 region of Zta. In support of this conclusion, a mutant [Zta(m22/26,74/75)] that carried a pair of mutations at each margin of the aa 24 to 86 region was severely impaired for nuclear translocation of BBLF4-GFP (Fig. 1B) (Table 1).
FIG. 1.
Effect of cotransfected Zta on the intracellular localization of BBLF4-GFP. (A) Diagrammatic representation of the Zta plasmids used in the cotransfection assays. (B) Representative photomicrographs of Vero cells transfected with BBLF4-GFP either alone or in the presence of the indicated wt or mutant Zta plasmids. The intracellular localization of BBLF4-GFP was determined by fluorescence microscopy.
TABLE 1.
Effect of Zta deletions and mutations on intracellular localization of BBLF4
| Zta deletion or mutation | % Localizationa of BBLF4
|
|
|---|---|---|
| Nucleusb | Cytoplasmc | |
| Controld | 0 | 100 |
| Zta(wt) | 81 | 19 |
| Zta(Δ24–86) | 3 | 97 |
| Zta(Δ3–69) | 38 | 62 |
| Zta(Δ39–45) | 55 | 45 |
| Zta(Δ61–81) | 46 | 54 |
| Zta(m22/26,74/75) | 10 | 90 |
Percent GFP-positive cells visualized after excitation at 490 nm. Data are from four separate experiments in which 100 to 600 positive cells were counted.
Nuclear or nuclear is greater than cytoplasmic staining.
Cytoplasmic or cytoplasmic is greater than nuclear staining.
BBLF4-GFP alone.
Mapping the interaction domain for BBLF4-GFP using GST affinity assays.
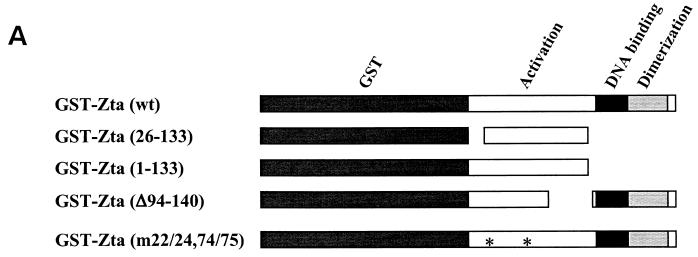
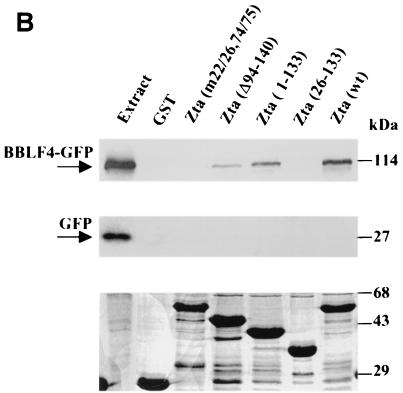
To provide additional evidence that Zta binds to BBLF4 and that the aa 22 to 86 region of Zta was required for the interaction. GST affinity assays were performed using the GST-Zta constructions illustrated in Fig. 2A. Extracts from 293T cells transfected with either BBLF4-GFP or a GFP control vector were incubated with glutathione beads containing equal amounts of the different GST-Zta proteins. The relative amounts of GST-Zta proteins used were determined by Coomassie brilliant blue staining of the SDS-PAGE-separated proteins (Fig. 2B, lower panel). Western blot analysis to detect protein binding to the GST-Zta proteins was performed using anti-GFP antibody and a chemiluminescence visualization procedure. Wild-type GST-Zta bound BBLF4-GFP, as did the GST-Zta activation domain construction, Zta(1–133) (Fig. 2B, upper panel). Zta carrying an internal deletion of the carboxy-terminal region of the activation domain, GST-Zta(Δ94–140), also bound BBLF4-GFP, although with slightly reduced affinity (Fig. 2B, upper panel). Neither GST-Zta(26–133) nor Zta(m22/26,74/75) bound BBLF4-GFP (Fig. 2B, upper panel), again emphasizing the importance of sequences in the aa 22 to 26 and aa 74 to 86 regions of Zta for BBLF4 binding. There was no binding of BBLF4-GFP to GST alone (Fig. 2B, upper panel), nor did the control GFP protein bind to any of the GST constructions (Fig. 2B, middle panel).
FIG. 2.
Binding of BBLF4-GFP to GST-Zta constructions. (A) Diagrammatic representation of the GST-Zta constructions used. (B, upper panel) GST affinity assay in which a BBLF4-GFP transfected 293T cell extract was incubated with equal amounts of GST or the indicated GST-Zta proteins bound to glutathione beads. Bound protein was detected by Western blot analysis using anti-GFP antibody and a chemiluminescence visualization procedure. A total of 5 μl of extract was directly loaded in the extract lane. (Middle panel) GST affinity assay performed as described above using an extract of 293T cells transfected with control vector expressing GFP. (Lower panel) SDS-polyacrylamide gel stained with Coomassie brilliant blue showing the GST and GST-Zta proteins used in the binding assays presented in the upper and middle panels.
Zta(m22/26,74/75) is defective for DNA replication.
To examine the functional consequences of loss of BBLF4 binding, Zta(m22/26,74/75) was tested for its ability to support lytic EBV DNA replication. We first used a PCR-based cotransfection-replication assay to compare the replication function of wild-type Zta with that of the Zta mutant Zta(m22/26,74/75). Vero cells were transfected with an oriLyt containing plasmid, along with expression plasmids for the six core EBV replication proteins, Rta and Mta plus either wild-type Zta, Zta(m22/26,74/75) or vector DNA. At 80 h posttransfection the cells were harvested, the DNA was extracted, and the concentration was determined by absorbance at 260 nm. Equal amounts of DNA from each transfection were either digested overnight with DpnI or were left untreated. Segments of the oriLyt plasmid DNA backbone were then amplified in a PCR reaction using two different sets of primers. The first set of primers amplified a 510-bp segment of the oriLyt plasmid backbone. This 510-bp region contains seven DpnI sites. The bacterially grown oriLyt plasmid DNA that is transfected into the cells is methylated and will be cleaved by DpnI, whereas DNA that has been replicated in the transfected mammalian cells will no longer carry the bacterially imposed methylation pattern and will not be cleaved. Thus, after DpnI digestion of the DNA, the 510-bp fragment should be readily amplifiable only from cells in which Zta has been able to support oriLyt DNA replication. On the other hand, in the absence of DpnI digestion the 510-bp fragment should be equally amplifiable from each of the DNA samples. A second set of PCR primers was used to control for any nonspecific effects of DpnI digestion. This primer pair amplified a 330-bp segment of the oriLyt plasmid backbone that does not contain any DpnI sites. In this case, the DpnI-digested DNA samples should be equally amplifiable regardless of whether or not the oriLyt plasmid had been replicated.
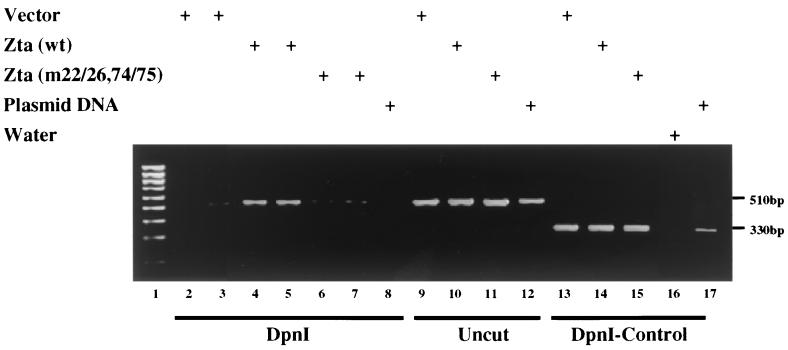
An example of the PCR cotransfection-replication assay is shown in Fig. 3. When DpnI-digested DNA was used as the template for the PCR amplification, the 510-bp fragment was readily amplified from Vero cells that had been transfected with wild-type Zta (Fig. 3, lanes 4 and 5) but was inefficiently amplified from Vero cells that had been transfected with either vector DNA (Fig. 3, lanes 2 and 3) or Zta(m22/26,74/75) (Fig. 3, lanes 6 and 7). In the absence of DpnI digestion (Fig. 3, lanes 9 to 11), the 510-bp PCR fragment was amplified equally from each of the three transfected DNA samples, indicating that the lack of amplification seen in lanes 6 and 7 was linked to DpnI cleavage of the template and not to any inherent inability to amplify the 510-bp PCR fragment from the DNA of Zta(m22/26,74/75)-transfected cells. Similarly, the 310-bp control PCR fragment that lacked DpnI cleavage sites was equally amplified from each of the three DpnI-digested DNA samples (Fig. 3, lanes 13 to 15).
FIG. 3.
Zta(m22/26,74/75) is unable to replicate an oriLyt plasmid. Ethidium bromide-stained gel of electrophoretically separated PCR-amplified fragments of the non-EBV backbone sequences of the oriLyt plasmid pEF52. Vero cells were transfected with pEF52, expression plasmids for the six core replication genes, Mta and Rta plus either control vector DNA or expression plasmids for wild-type Zta or Zta(m22/26,74/75). DNA was isolated from the transfected cells, and segments of the oriLyt plasmid backbone were amplified by PCR either before or after digestion of the DNA with the methylation-sensitive restriction enzyme DpnI. The test primers amplify a 510-bp fragment that contains seven DpnI sites, while the control primers amplify a 330-bp fragment that does not contain any DpnI sites. After DpnI digestion of the transfected cell DNA, the 510-bp fragment should only be amplifiable if the input DNA has been replicated in the transfected cells. Lane 1, marker DNA ladder; lanes 2 to 8, DpnI-digested DNA amplified with the test primers; lanes 9 to 12, undigested DNA amplified with the test primers; lanes 13 to 17, DpnI-digested DNA amplified with the control primers. Plasmid DNA, the PCR reaction was performed directly on the input plasmid DNA; water, no DNA added.
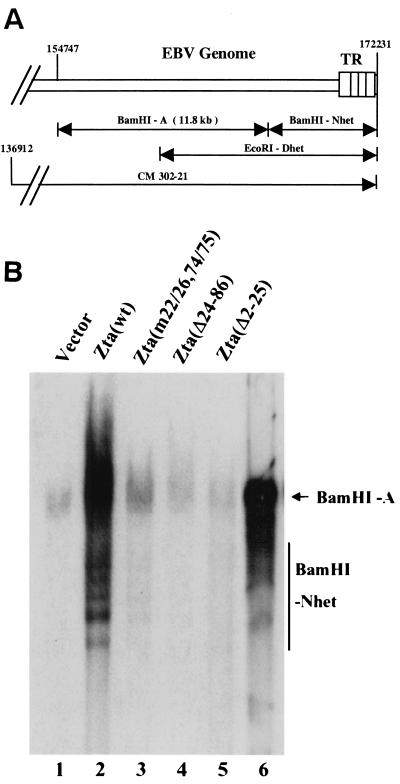
The results of the cotransfection-replication assay suggested that the Zta(m22/26,74/75) mutant that was impaired for binding to BBLF4 was also impaired for oriLyt DNA replication. We next evaluated the ability of Zta(m22/26,74/75) and Zta(Δ24–86) to replicate the endogenous EBV genomes in P3HR1 cells. Transfection of wild-type Zta into P3HR1 cells leads to induction of the lytic cycle. The ability to evaluate Zta mutants in a setting in which endogenous EBV genomes are present derives from the observation that exogenous Zta activates the full pattern of lytic gene expression with the key exception of the expression of endogenous Zta (30). The presence of low-affinity Zta binding sites overlapping the Zta mRNA start site (36) may be responsible, since binding of Zta to these sites in situations of Zta overexpression would result in transcriptional occlusion. The amplification of linear viral DNA in Zta-transfected P3HR1 cells can be assessed by Southern blotting. The linear EBV genome is bounded by 500-bp terminal repeats. The numbers of repeats at each end vary and hence restriction fragments such as BamHI-Nhet and EcoRI-Dhet that encompass the termini display size heterogeneity (Fig. 4A). The induction of DNA replication of the endogenous viral genomes in P3HR1 was examined by isolating DNA from transfected cells, digesting the DNA with BamHI, and probing Southern blots of the separated DNA fragments with 32P-labeled EcoRI-Dhet to determine the relative amounts of the BamHI-A and BamHI-Nhet fragments. The BamHI-A fragment serves as a marker for the relative number of genomes in the cell, while the BamHI-Nhet fragment is diagnostic for linear EBV genomes.
FIG. 4.
Zta(m22/26,74/75) and Zta(Δ24–86) are unable to replicate endogenous P3HR1 genomes. (A) Schematic representation of the righthand end of the EBV genome showing the relative locations of the terminal EcoRI and BamHI restriction fragments. TR, terminal repeats. (B) Southern blot of DNA isolated from P3HR1 cells electroporated with the indicated plasmids. After cleavage with BamHI, the electrophoretically separated DNA fragments were transferred to a nitrocellulose membrane and probed with a 32P-labeled EcoRI-Dhet probe. As indicated by the increase in the amount of the BamHI-Nhet fragment, linear EBV genomes were efficiently amplified only in wild-type Zta-transfected cells. Lanes 1 to 5, DNA isolated from electroporated P3HR1 cells; lane 6, cosmid 302–21 DNA digested with BamHI.
The concentration of the DNA isolated from transfected cells was measured by the absorbance at 260 nm and checked by gel electrophoresis and ethidium bromide staining for DNA quality. Equal amounts of each sample were then subjected to restriction enzyme digestion and analyzed by Southern blotting. The DNA isolated after transfection of wild-type Zta into P3HR1 cells showed greatly increased levels of both the BamHI-A and BamHI-Nhet fragments compared to the levels seen in the control vector transfected cells (Fig. 4B). Zta(Δ2–25) had previously been found to be replication defective (51) and, as expected, there was no increase in BamHI-Nhet DNA in P3HR1 cells transfected with Zta(Δ2–25) (Fig. 4B). There was only a marginal increase in BamHI-Nhet DNA in cells transfected with Zta(m22/26,74/75) (Fig. 4B). Zta(Δ24–86) was also unable to support replication of the endogenous genomes in P3HR1 cells (Fig. 4B). Thus, the two mutants that showed loss of BBLF4 (helicase) binding were both replication defective in this assay.
Zta(m22/26,74/75) and Zta(Δ24–86) retain transactivation activity.
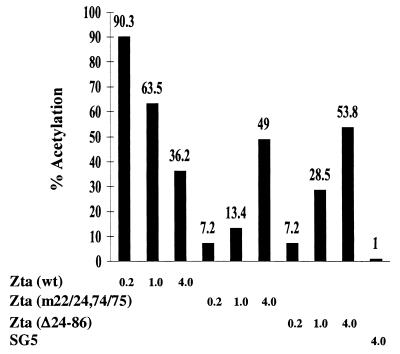
Replication assays performed in P3HR1 or D98-HR1 cells are dependent on the expression of the core EBV replication proteins from the endogenous genomes. The Zta(m22/26,74/75) and Zta(Δ24–86) mutations lie within the transcriptional activation domain of Zta. The activation domain has been shown to have a modular construction, and deletion of any one segment reduces, but does not eliminate, transcriptional activity (16, 37). The behavior of the Zta(m22/24,74/75) and Zta(Δ24–86) mutants compared to wild-type Zta is illustrated in the reporter assay in Fig. 5. At low concentrations of transfected effector DNA, the Zta(m22/24,74/75) and Zta (Δ24–86) mutants were severely impaired in their ability to activate expression from an oriLytp-CAT reporter relative to wild-type Zta. However, the deficiency could be partially compensated for by increasing the dose of input DNA. This was not the case for wild-type Zta, which gave maximal transactivation responses at low levels of input plasmid DNA. The wild-type and Zta(m22/24,74/75) and Zta(Δ24–86) mutants express comparable levels of Zta protein in transfected cells as measured by Western blotting.
FIG. 5.
CAT reporter assay comparing transactivation activity of wild-type Zta and the Zta(m22/24,74/75) and Zta(Δ24–86) mutants. HeLa cells were transfected with an oriLytp-CAT reporter (1 μg) and increasing amounts of expression vectors for wild-type or mutant Zta. Cells were harvested for determination of CAT activity 40 h after transfection.
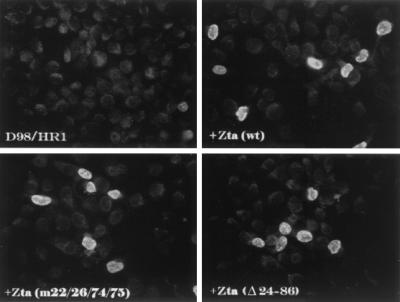
Since Zta-induced replication in P3HR1 or D98-HR1 cells requires the induction of EBV replication gene expression from endogenous genomes rather than transfected DNA, the effect of the Zta(m22/26,74/75) and Zta(Δ24–86) mutations on the induction of one of the core replication proteins, the BMRF1 polymerase processivity factor, was also examined by immunofluorescence analyses. Untransfected D98-HR1 rarely express BMRF1. Transfection of wild-type Zta resulted in the induction of BMRF1 in approximately 7% of the cells. In double-staining experiments, all BMRF1-positive cells were also Zta positive (data not shown). D98-HR1 transfected with either Zta(m22/26,74/75) or Zta(Δ24–86) showed induction of BMRF1 expression that was only marginally reduced from that seen with wild-type Zta (Fig. 6). Thus, the inability of Zta(m22/26,74/75) and Zta(Δ24–86) to support replication of the endogenous genomes in D98-HR1 cells is likely to be a true replication deficit and not due solely to transcriptional impairment.
FIG. 6.
Zta(m22/26,74/75) and Zta(Δ24–86) activate expression of BMRF1 from endogenous EBV genomes. Immunofluorescence assay performed on D98-HR1 cells before and after transfection with the indicated Zta plasmids. Cells were stained with anti-BMRF1 (polymerase processivity factor) monoclonal antibody and FITC-conjugated secondary antibody.
Zta(m22/26,74/75) and Zta(Δ24–86) fail to target efficiently to replication compartments.
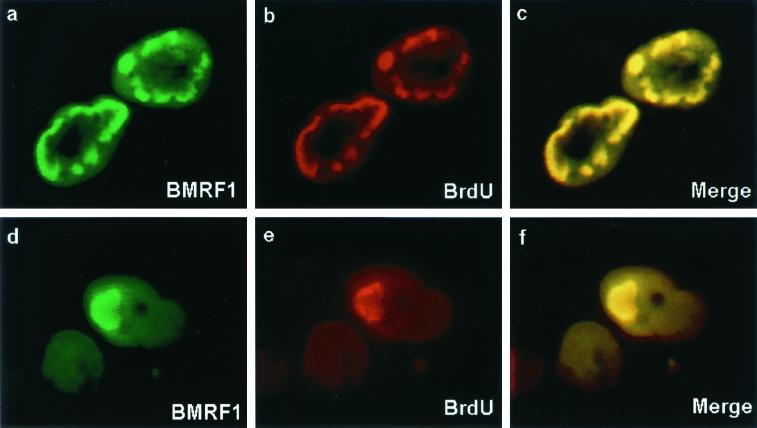
We wished to address the requirements for DNA replication compartment formation in transfected D98-HR1 cells. Replication compartments are visible in immunofluorescence assays as large intranuclear bodies. Replication compartments were detected in D98-HR1 cells transfected with wild-type Zta by staining for BMRF1 (polymerase processivity factor) (Fig. 7). These structures were shown to represent functional replication compartments by incubating the cells with BrdU and double staining for sites of BrdU incorporation with anti-BrdU antibody. The BrdU staining colocalized with BMRF1 in the intranuclear bodies (Fig. 7).
FIG. 7.
Demonstration of replication compartment formation in D98-HR1 cells. Immunofluorescence assay performed on D98-HR1 cells that were transfected with wild-type Zta and incubated with BrdU 45 min prior to fixation. Cells were double stained for the polymerase processivity factor BMRF1 (a and d) using anti-BMRF1 monoclonal antibody and FITC-conjugated anti-mouse immunoglobulin secondary antibody and for BrdU (b and e) using anti-BrdU sheep antibody and rhodamine-conjugated anti-sheep immunoglobulin as the secondary antibody. Merge, overlays of the FITC and rhodamine images (c and f). Replication bodies are visible that double stain for BrdU incorporation and the presence of viral replication proteins, as exemplified by BMRF1.
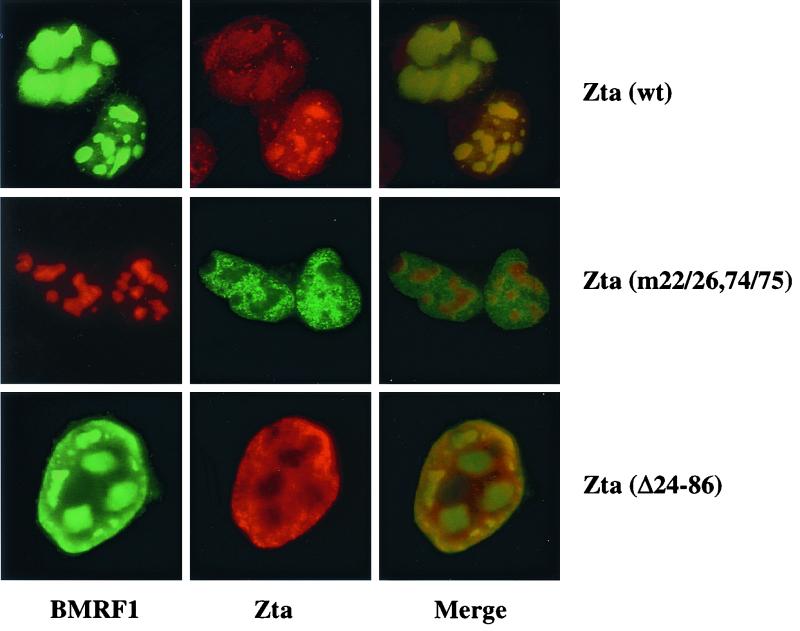
Replication compartments were also detected in D98-HR1 cells transfected with Zta(m22/26,74/75) or Zta(Δ24–86) and stained with anti-BMRF1 antibody (Fig. 8). This reinforces the belief that there is not a profound deficit in induction of early replication proteins in Zta(m22/26,74/75)- and Zta(Δ24–86)-transfected cells. However, double staining of these cells for Zta revealed a distinct difference in Zta distribution (Fig. 8). Whereas wild-type Zta was localized predominantly within the replication compartments, Zta(m22/26,74/75) and Zta(Δ24–86) were excluded from the replication compartments and were instead distributed throughout the remainder of the nucleoplasm. These experiments suggest that BBLF4 binding is necessary for Zta to be efficiently incorporated into replication compartments. The replication deficit of the Zta(m22/26,74/75) and Zta(Δ24–86) mutants is therefore linked to the inability of these Zta mutants to physically associate with sites of viral DNA replication.
FIG. 8.
Zta(m22/26,74/75) and Zta(Δ24–86) fail to associate with replication compartments. An immunofluorescence assay was performed on D98-HR1 cells transfected with expression vectors for wild-type or mutant Zta. Cells were stained for BMRF1 using anti-BMRF1 monoclonal antibody and for Zta using rabbit anti-Zta antiserum. Wild-type Zta colocalized with the replication compartments, whereas the two Zta mutants were excluded from these bodies. Merge, overlays of the FITC and rhodamine images.
DISCUSSION
A central role for Zta in lytic EBV DNA replication lies with the recruitment and assembly of a replication complex on oriLyt. The interactions that have been described between Zta and the core replication proteins and between individual viral replication proteins indicate that complex formation relies on multiple contact points among the participating proteins. The helicase (BBLF4), primase (BSLF1), and primase-associated factor (BBLF2/3) form a tripartite complex, as do the equivalent proteins in HSV (18). In DNA-transfected cells in which changes in intracellular localization of the proteins were used as a measure of interaction, BSLF1 was shown to interact with BBLF2/3, and evidence for a tripartite complex was also presented (23). These interactions were confirmed in immunoprecipitation assays using baculovirus-expressed proteins, with additional evidence provided for separate contacts by both BSLF1 and BBLF2/3 with BBLF4 (60). We had previously mapped the site of interaction of the primase subcomplex (BSLF1-BBLF2/3) with Zta to the N terminus of the Zta activation between Zta aa 11 and 25 (23). We now show that BBLF4 interacts with the adjacent region of Zta between aa 22 and 86. The helicase-primase complex has also been found to interact with polymerase (BALF5) (22). Polymerase interacts with the polymerase processivity factor (BMRF1) (29, 34), which in turn interacts with the bZIP domain of Zta (65). The single-stranded DNA-binding protein BALF2 has also been found to contact the BALF5-BMRF1 proteins (61). Thus, direct contacts on Zta are made by two of the proteins of the helicase-primase complex (BBLF2/3 and BBLF4) and by BMRF1, and the other core replication proteins appear to be tethered by interactions to one or more of these three proteins.
The possibility of additional tethering of the replication complex to oriLyt by cellular transcription factors was raised by the observation that cotransfected BMRF1 activated transcription from BHLF1 oriLyt promoter-reporter constructions and that the sequences required for this activation mapped to the essential promoter distal replication domain (63, 64). A yeast one-hybrid screen identified Sp1 and ZBP-89 as cellular transcription factors that bound to the essential promoter distal domain of oriLyt, and these proteins proved to also interact with BMRF1 and BALF5 (7). Thus, multiple protein-protein interactions between individual members of the virally encoded replication complex and between this complex and the DNA-bound Zta and cellular Spl and ZBP-89 transcription factors contribute to the origin tethering of the replication complex.
The mapping of the helicase interaction region to aa 22 to 86 within the Zta activation domain raises questions about the relationship between Zta's transcription and replication functions. The Zta(m22/26,74/75) mutant that lost interaction with BBLF4 also shows some impairment in transactivation function. Zta(m22/26,74/75) was significantly impaired for activation of the BHLF1(oriLyt) promoter in in vitro transcription assays but showed no deficit on an artificial promoter containing seven upstream ZREs (40). In transfection assays the Zta(m22/26,74/75) mutant was less effective in activating the BHLF1 promoter, with the deficit being partially compensated for by increasing the amount of transfected Zta(m22/26,74/75) (40) (Fig. 5). Analysis of the underlying nature of the deficit in Zta(m22/26,74/75) function revealed that this mutant had a reduced ability to stimulate the formation of a stable TFIID-TFILA complex (40) and also was impaired in its ability to recruit the coactivator CBP to Zta (14). The loss of interaction with CBP correlated with a reduced ability to detect Zta-mediated acetylation of histones. The Zta transactivation domain appears to have a modular structure, in that the loss of individual segments can be compensated for by increasing the number of Zta binding sites on the promoter or increasing the copy number of the other domains (16). It seems likely that the contacts made by the coactivators, basal transcription factors, and TAFs (37) are just as complex as those made by the replication proteins. However, the apparent overlap of the binding region for the helicase with that for CBP raises the possibility that individual DNA-bound Zta dimers may be capable of interacting with either a transcriptional complex or a replication complex but not both simultaneously. If such were the case, this might explain the requirement for multiple Zta binding sites within the origin. oriLyt of the EBV-related baboon virus herpesvirus papio is essentially identical in structure to EBV oriLyt and also contains multiple Zta binding sites (50).
Zta(m22/26,74/75) was defective for oriLyt DNA replication in a cotransfection replication assay and was similarly unable to induce replication of the endogenous genomes in P3HR1 cells. However, structures resembling replication compartments were detected in D98-HR1 cells transfected with this Zta mutant. We observed that Zta(m22/26,74/75) was excluded from the replication compartments and concluded that this inability to localize to the sites of DNA replication was a contributing factor in the DNA replication deficit displayed by Zta(m22/26, 74/75). This Zta mutant is also deficient for helicase interaction, and the implication is that the helicase targets Zta to the replication compartments. The formation of replication compartment-like structures in D98-HR1 cells transfected with Zta(m22/26,74/75) also implies that replication compartment formation in EBV does not require the presence of Zta or an active origin of replication. In cells transfected with the HSV replication machinery, replication compartment formation was found to be independent of the origin binding protein UL9 or an origin containing plasmid, and a similar observation was made for KSHV, where transfection of plasmids encoding the six core replication proteins was sufficient to allow the formation of replication compartments (41, 58, 59, 66). In contrast, the formation of CMV replication compartments in transfected cells required the presence of an oriLyt plasmid and both ancillary and core replication proteins (52).
Zta(Δ24–86), which is also defective for helicase interaction, was unable to induce replication of the endogenous genomes in P3HR1 cells and was also not associated with replication compartments in D98-HR1 cells. Previously, Zta(Δ25–86) had been found to replicate an oriLyt-containing plasmid in a cotransfection-replication assay (51). A differential ability to replicate a transfected oriLyt plasmid but not endogenous EBV genomes was also observed with a Zta aa 200 mutation in studies of Zta-BMRF1 interaction (65). The reason for the discrepancy between the two assays is not known but, given the functions mediated by the aa 24 to 86 region of Zta, it is possible to speculate. First, BBLF4 forms a tripartite complex with BSLF1 and BBLF2/3. These latter proteins bind to the aa 11 to 25 segment of Zta and can bind to Zta(Δ24–86). In the presence of high concentrations of the proteins in transfected cells, it seems likely that BBLF4 could be tethered to Zta indirectly through the helicase subcomplex and hence permit Zta to be targeted to replication foci in the normal manner. A second difference between the assays is the presence of oriLyt on a transfected plasmid versus the endogenous and presumably more extensively nucleosome-associated oriLyt present within the episomal EBV genomes. This region of Zta is also involved in the recruitment of CBP, which has intrinsic histone acetylase activity. Ineffective histone acetylation and clearance from the DNA is likely to be a greater handicap for replication of the endogenous genomes than for transfected DNA. The complex nature of the interactions involved in Zta's transcriptional and replication functions has made it difficult to tease apart the contributions of individual domains of the Zta protein. This is exemplified by the aa 22 to 86 domain, where protein-protein interactions mediating replication complex formation, replication compartment targeting, and nucleosome clearance all contribute to oriLyt DNA replication.
ACKNOWLEDGMENTS
We thank Paul Lieberman for his Zta (m22/26,74/75), pZQ239, and pZBSΔ39–45 plasmids; Marie Hardwick for anti-Zta antiserum; Mabel Chiu for technical support; and Feng Chang for assistance with manuscript preparation.
This work was funded by Public Health Service grant CA30356 to S.D.H. F.Y.W. was partially supported by the Anti-Cancer Drug Development Training Program (T32 CA09243).
REFERENCES
- 1.Adamson A L, Kenney S. The Epstein-Barr virus BZLF1 protein interacts physically and functionally with the histone acetylase CREB-binding protein. J Virol. 1999;73:6551–6558. doi: 10.1128/jvi.73.8.6551-6558.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamson A L, Kenney S. Epstein-Barr Virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J Virol. 2001;75:2388–2399. doi: 10.1128/JVI.75.5.2388-2399.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn J H, Hayward G S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn J H, Jang W J, Hayward G S. The human cytomegalovirus IE2 and UL112–113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10) J Virol. 1999;73:10458–10471. doi: 10.1128/jvi.73.12.10458-10471.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Askovic S, Baumann R. Activation domain requirements for disruption of Epstein-Barr virus latency by ZEBRA. J Virol. 1997;71:6547–6554. doi: 10.1128/jvi.71.9.6547-6554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 7.Baumann M, Feederle R, Kremmer E, Hammerschmidt W. Cellular transcription factors recruit viral replication proteins to activate the Epstein-Barr virus origin of lytic DNA replication, oriLyt. EMBO J. 1999;18:6095–6105. doi: 10.1093/emboj/18.21.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumann M, Mischak H, Dammeier S, Kolch W, Gires O, Pich D, Zeidler R, Delecluse H-J, Hammerschmidt W. Activation of the Epstein-Barr virus transcription factor BZLF1 by 12-O-tetradecanoylphorbol-13-acetate-induced phosphorylation. J Virol. 1998;72:8105–8114. doi: 10.1128/jvi.72.10.8105-8114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell P, Lieberman P M, Maul G G. Lytic but not latent replication of Epstein-Barr virus is associated with PML and induces sequential release of nuclear domain 10 proteins. J Virol. 2000;74:11800–11810. doi: 10.1128/jvi.74.24.11800-11810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burkham J, Coen D M, Weller S K. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J Virol. 1998;72:10100–10107. doi: 10.1128/jvi.72.12.10100-10107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cayrol C, Flemington E. G0/G1 growth arrest mediated by a region encompassing the basic leucine zipper (bZIP) domain of the Epstein-Barr virus transactivator Zta. J Biol Chem. 1996;271:31799–31802. doi: 10.1074/jbc.271.50.31799. [DOI] [PubMed] [Google Scholar]
- 12.Cayrol C, Flemington E K. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 1996;15:2748–2759. [PMC free article] [PubMed] [Google Scholar]
- 13.Chang Y-N, Dong D L-Y, Hayward G S, Hayward S D. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J Virol. 1990;64:3358–3369. doi: 10.1128/jvi.64.7.3358-3369.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C J, Deng Z, Kim A Y, Blobel G A, Lieberman P M. Stimulation of CREB binding protein nuclecosomal histone acetyltransferase activity by a class of transcriptional activators. Mol Cell Biol. 2001;21:476–487. doi: 10.1128/MCB.21.2.476-487.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng L, Kelly T J. Transcriptional activator nuclear factor I stimulates the replication of SV40 minichromosomes in vivo and in vitro. Cell. 1989;59:541–551. doi: 10.1016/0092-8674(89)90037-8. [DOI] [PubMed] [Google Scholar]
- 16.Chi T, Carey M. The ZEBRA activation domain: modular organization and mechanism of action. Mol Cell Biol. 1993;13:7045–7055. doi: 10.1128/mcb.13.11.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chi T, Lieberman P, Ellwood K, Carey M. A general mechanism for transcriptional synergy by eukaryotic activators. Nature. 1995;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- 18.Crute J J, Tsurumi T, Zhu L, Weller S K, Olivo P D, Challberg M D, Mocarski E S, Lehman I R. Herpes simplex virus 1 helicase-primase: a complex of three herpes-encoded gene products. Proc Natl Acad Sci USA. 1989;86:2186–2189. doi: 10.1073/pnas.86.7.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farrell P, Rowe D, Rooney C, Kouzarides J. EBV BZLF-1 transactivator specifically binds to consensus AP-1 sites and is related to c-fos. EMBO J. 1989;8:127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fixman E D, Hayward G S, Hayward S D. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J Virol. 1995;69:2998–3006. doi: 10.1128/jvi.69.5.2998-3006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flemington E, Speck S H. Evidence for coiled-coil dimer formation by an Epstein-Barr virus transactivator that lacks a heptad repeat of leucine residues. Proc Natl Acad Sci USA. 1990;87:9459–9463. doi: 10.1073/pnas.87.23.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujii K, Yokoyama N, Kiyono T, Kuzushima K, Homma M, Nishiyama Y, Fujita M, Tsurumi T. The Epstein-Barr virus pol catalytic subunit physically interacts with the BBLF4-BSLF1-BBLF2/3 complex. J Virol. 2000;74:2550–2557. doi: 10.1128/jvi.74.6.2550-2557.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Z, Krithivas A, Finan J E, Semmes O J, Zhou S, Wang Y, Hayward S D. The EBV lytic transactivator Zta interacts with the helicase-primase replication complex. J Virol. 1998;72:8559–8567. doi: 10.1128/jvi.72.11.8559-8567.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Z S, Gutierrez C, Heine U, Sogo J M, Depamphilis M L. Origin auxiliary sequences can facilitate initiation of simian virus 40 DNA replication in vitro as they do in vivo. Mol Cell Biol. 1989;9:3593–3602. doi: 10.1128/mcb.9.9.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 26.Hsieh J J-D, Henkel T, Salmon P, Robey E, Peterson M G, Hayward S D. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Y F, Hao Z L, Li R. Chromatin remodeling and activation of chromosomal DNA replication by an acidic transcriptional activation domain from BRCA1. Genes Dev. 1999;13:637–642. doi: 10.1101/gad.13.6.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiehl A, Dorsky D I. Cooperation of EBV DNA polymerase and EA-D(BMFR1) in vitro, and colocalization in nuclei of infected cells. Virology. 1991;184:330–340. doi: 10.1016/0042-6822(91)90849-7. [DOI] [PubMed] [Google Scholar]
- 30.Kolman J L, Taylor N, Gradoville L, Countryman J, Miller G. Comparing transcriptional activation and autostimulation by ZEBRA and ZEBRA/c-Fos chimeras. J Virol. 1996;70:1493–1504. doi: 10.1128/jvi.70.3.1493-1504.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolman J L, Taylor N, Marshak D R, Miller G. Serine-173 of the Epstein-Barr virus ZEBRA protein is required for DNA binding and is a target for casein kinase II phosphorylation. Proc Natl Acad Sci USA. 1993;90:10115–10119. doi: 10.1073/pnas.90.21.10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korioth F, Maul G G, Plachter B, Stamminger T, Frey J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res. 1996;229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- 33.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 34.Li J-S, Zhou B-S, Dutschman G E, Grill S P, Tan R-S, Cheng Y-C. Association of Epstein-Barr virus early antigen diffuse component and virus-specified DNA polymerase activity. J Virol. 1987;61:2947–2949. doi: 10.1128/jvi.61.9.2947-2949.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li R, Botchan M R. Acidic transcription factors alleviate nucleosome-mediated repression of DNA replication of bovine papillomavirus type 1. Proc Natl Acad Sci USA. 1994;91:7051–7055. doi: 10.1073/pnas.91.15.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lieberman P M, Berk A J. In vitro transcriptional activation, dimerization, and DNA-binding specificity of the Epstein-Barr virus Zta protein. J Virol. 1990;64:2560–2568. doi: 10.1128/jvi.64.6.2560-2568.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lieberman P M, Berk A J. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA-promoter DNA complex formation. Genes Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- 38.Lieberman P M, Hardwick J M, Sample J, Hayward G S, Hayward S D. The Zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J Virol. 1990;64:1143–1155. doi: 10.1128/jvi.64.3.1143-1155.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lieberman P M, O'Hare P, Hayward G S, Hayward S D. Promiscuous trans-activation of gene expression by an Epstein-Barr virus-encoded early nuclear protein. J Virol. 1986;60:140–148. doi: 10.1128/jvi.60.1.140-148.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lieberman P M, Ozer J, Gursel D B. Requirement for transcription factor IIA (TDFIIA)-TFIID recruitment by an activator depends on promoter structure and template competition. Mol Cell Biol. 1997;17:6624–6632. doi: 10.1128/mcb.17.11.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lukonis C J, Burkham J, Weller S K. Herpes simplex virus type 1 prereplicative sites are a heterogeneous population: only a subset are likely to be precursors to replication compartments. J Virol. 1997;71:4771–4781. doi: 10.1128/jvi.71.6.4771-4781.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maul G G. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays. 1998;20:660–667. doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 43.Maul G G, Guldner H H, Spivack J G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 44.Muller K, Mermod N. The histone-interacting domain of nuclear factor I activates simian virus 40 DNA replication in vivo. J Biol Chem. 2000;275:1645–1650. doi: 10.1074/jbc.275.3.1645. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen-Huynh A T, Schaffer P A. Cellular transcription factors enhance herpes simplex virus type 1 oriS-dependent DNA replication. J Virol. 1998;72:3635–3645. doi: 10.1128/jvi.72.5.3635-3645.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogryzko V V, Shiltz R L, Russanova V, Howard B H, Nakatini Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 47.Portes-Sentis S, Sergeant A, Gruffat H. A particular DNA structure is required for the function of a cis-acting component of the Epstein-Barr virus OriLyt origin of replication. Nucleic Acids Res. 1997;25:1347–1354. doi: 10.1093/nar/25.7.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 49.Rodrigiuez A, Armstrong M, Dwyer D, Flemington E. Genetic dissection of cell growth arrest functions mediated by the Epstein-Barr virus lytic gene product, Zta. J Virol. 1999;73:9029–9038. doi: 10.1128/jvi.73.11.9029-9038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryon J J, Fixman E D, Houchens C, Zong J, Lieberman P M, Chang Y-N, Hayward G S, Hayward S D. The lytic origin of herpesvirus papio is highly homologous to Epstein-Barr virus ori-Lyt: evolutionary conservation of transcriptional activation and replication signals. J Virol. 1993;67:4006–4016. doi: 10.1128/jvi.67.7.4006-4016.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sarisky R T, Gao Z, Lieberman P M, Fixman E D, Hayward G S, Hayward S D. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J Virol. 1996;70:8340–8347. doi: 10.1128/jvi.70.12.8340-8347.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarisky R T, Hayward G S. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J Virol. 1996;70:7398–7413. doi: 10.1128/jvi.70.11.7398-7413.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schepers A, Pich D, Hammerschmidt W. Activation of orilyt, the lytic origin of DNA replication of Epstein-Barr virus, by BZLF1. Virology. 1996;220:367–376. doi: 10.1006/viro.1996.0325. [DOI] [PubMed] [Google Scholar]
- 54.Schepers A, Pich D, Mankertz J, Hammerschmidt W. cis-acting elements in the lytic origin of DNA replication of Epstein-Barr virus. J Virol. 1993;67:4237–4245. doi: 10.1128/jvi.67.7.4237-4245.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takagi S, Takad K, Sairenji T. Formation of intranuclear replication compartments of Epstein-Barr virus with redistribution of BZLF1 and BMRF1 gene products. Virology. 1991;185:309–315. doi: 10.1016/0042-6822(91)90778-a. [DOI] [PubMed] [Google Scholar]
- 56.Tang Q, Bell P, Tegtmeyer P, Maul G G. Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J Virol. 2000;74:9694–9700. doi: 10.1128/jvi.74.20.9694-9700.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor N, Flemington E, Kolman J L, Baumann R P, Speck S H, Miller G. ZEBRA and a Fos-GCN4 chimeric protein differ in their DNA-binding specificities for sites in the Epstein-Barr virus BZLF1 promoter. J Virol. 1991;65:4033–4041. doi: 10.1128/jvi.65.8.4033-4041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uprichard S L, Knipe D M. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology. 1997;229:113–125. doi: 10.1006/viro.1996.8430. [DOI] [PubMed] [Google Scholar]
- 59.Wu F Y, Ahn J H, Alcendor D J, Jang W J, Xiao J, Hayward S D, Hayward G S. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J Virol. 2001;75:1487–1506. doi: 10.1128/JVI.75.3.1487-1506.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yokoyama N, Fujii K, Hirata M, Tamai K, Kiyono T, Kuzushima K, Nishiyama Y, Fujita M, Tsurumi T. Assembly of the Epstein-Barr virus BBLF4, BSLF1 and BBLF2/3 proteins and their interactive properties. J Gen Virol. 1999;80:2879–2887. doi: 10.1099/0022-1317-80-11-2879. [DOI] [PubMed] [Google Scholar]
- 61.Zeng Y, Middeldorp J, Madjar J J, Ooka T. A major DNA binding protein encoded by BALF2 open reading frame of Epstein-Barr virus (EBV) forms a complex with other EBV DNA-binding proteins: DNase, EA-D, and DNA polymerase. Virology. 1997;239:285–295. doi: 10.1006/viro.1997.8891. [DOI] [PubMed] [Google Scholar]
- 62.Zerby D, Chen C-J, Poon E, Shiekhattar R, Lieberman P M. The amino-terminal C/H1 domain of CREB binding protein mediates Zta transcriptional activation of latent Epstein-Barr virus. Mol Cell Biol. 1999;19:1617–1626. doi: 10.1128/mcb.19.3.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Q, Holley-Guthrie E, Dorsky D, Kenney S. Identification of transactivator and nuclear localization domains in the Epstein-Barr virus DNA polymerase accessory protein, BMRF1. J Gen Virol. 1999;80:69–74. doi: 10.1099/0022-1317-80-1-69. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Q, Holley-Guthrie E, Ge J Q, Dorsky D, Kenney S. The Epstein-Barr virus (EBV) DNA polymerase accessory protein, BMRF1, activates the essential downstream component of the EBV oriLyt. Virology. 1997;230:22–34. doi: 10.1006/viro.1997.8470. [DOI] [PubMed] [Google Scholar]
- 65.Zhang Q, Hong Y, Dorsky D, Holley-Guthrie E, Zalani S, Elshiekh N A, Kiehl A, Le T, Kenney S. Functional and physical interactions between the Epstein-Barr virus (EBV) proteins BZLF1 and BMRF1: effects on EBV transcription and lytic replication. J Virol. 1996;70:5131–5142. doi: 10.1128/jvi.70.8.5131-5142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhong L, Hayward G S. Assembly of complete, functionally active herpes simplex virus DNA replication compartments and recruitment of associated viral and cellular proteins in transient cotransfection assays. J Virol. 1997;71:3146–3160. doi: 10.1128/jvi.71.4.3146-3160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]