Abstract
Bacteria employ a myriad of regulatory mechanisms to adapt to the continuously changing environments that they face. They can, for example, use post-translational modifications, such as Nε-lysine acetylation, to alter enzyme activity. Although a lot of progress has been made, the extent and role of lysine acetylation in many bacterial strains remains uncharted. Here, we applied stable isotope labeling by amino acids in cell culture (SILAC) in combination with the immunoprecipitation of acetylated peptides and LC-MS/MS to measure the first Pseudomonas aeruginosa PAO1 acetylome, revealing 1076 unique acetylation sites in 508 proteins. Next, we assessed interstrain acetylome differences within P. aeruginosa by comparing our PAO1 acetylome with two publicly available PA14 acetylomes, and postulate that the overall acetylation patterns are not driven by strain-specific factors. In addition, the comparison of the P. aeruginosa acetylome to 30 other bacterial acetylomes revealed that a high percentage of transcription related proteins are acetylated in the majority of bacterial species. This conservation could help prioritize the characterization of functional consequences of individual acetylation sites.
Keywords: Pseudomonas aeruginosa, PAO1, PA14, Nε-lysine acetylation, acetylome, interstrain analysis
The acetylome of Pseudomonas aeruginosa strain PAO1 was measured for the first time and compared to available strain PA14 acetylomes, revealing extensive acetylation diversity within this species that does not seem to be driven by strain-specific factors.
Introduction
In continuously changing environments, post-translational modifications (PTMs; i.e. alterations to proteins after their synthesis) provide cells with a powerful and swift adaptation strategy. Indeed, while generally small, PTMs impact protein function, activity, interactions, and/or localization (Macek et al. 2019). Moreover, by modifying the pool of pre-existing proteins, PTMs outpace transcriptional and translational regulation that act through the time- and energy-consuming process of protein synthesis.
A common bacterial PTM is Nε-lysine acetylation, which is the reversible addition of an acetyl-group to the sidechain of lysine, elongating the residue and neutralizing its positive charge (Hentchel and Escalante-Semerena 2015, Macek et al. 2019). This modification results from either nonenzymatic processes or enzymatic reactions carried out by acetyltransferases (Lammers 2021). While this PTM was originally discovered in bacteria in 1992 (Barak et al. 1992), proteome-wide analyses of bacterial acetylation sites were only charted from 2008 onwards (Yu et al. 2008), following key technical developments. Indeed, due to the low stoichiometry of this PTM, thorough enrichment protocols, high accuracy mass spectrometers and software advances proved critically important (Weinert et al. 2017, Virág et al. 2020).
To date, the acetylomes of over 75 different bacterial species have been charted. The list of bacteria with characterized acetylomes entails a wide diversity of species consisting of model organisms including Escherichia coli (Yu et al. 2008, Zhang et al. 2009, 2013a, Castaño-Cerezo et al. 2014, Meyer et al. 2016, Weinert et al. 2017, Christensen et al. 2018), Salmonella enterica (serovar Typhimurium) (Wang et al. 2010), and Bacillus subtilis (Kosono et al. 2015, Carabetta et al. 2016, Nakayasu et al. 2017, Ravikumar et al. 2018, Reverdy et al. 2018), notorious pathogens including Pseudomonas aeruginosa (Ouidir et al. 2015, Gaviard et al. 2018, 2019) and Mycobacterium tuberculosis (Liu et al. 2014, Xie et al. 2015, Birhanu et al. 2017, Yang et al. 2018), as well as bacteria relevant to biotechnology and industry, such as Thermus thermophilus (Okanishi et al. 2013). The percentage of acetylated proteins reported in these different species varies greatly, ranging from 5% in Borrelia burgdorferi (Bontemps-Gallo et al. 2018), 45% in Spiroplasma eriocheiris (Meng et al. 2016) to 80.5% in Mycoplasma genitalium (Chen et al. 2016). Common acetylation targets include enzymes of the central metabolism, in which a high degree of conservation of acetylation sites at catalytic residues has been reported (Wang et al. 2010, Nakayasu et al. 2017), as well as proteins involved in quorum sensing (Sun et al. 2019), chemotaxis (Barak et al. 1992), virulence and antibiotic resistance (Luu and Carabetta 2021).
However, it has come to light that interstrain differences might also contribute to some strain-dependent phenotypic differences. For example, Fang et al. (2022) showed that the increased acetylation of pyruvate kinase contributed to the antibiotic resistance of several resistant strains. Similarly, large acetylome differences were found between an antibiotic resistant and sensitive Actinobacter baumannii strain by Kentache et al. (2016) and Liao et al. (2017). In contrast, Birhanu et al. (2017) found extensive overlap between the acetylome of Lineage 7 and four M. tuberculosis strains, although admittedly, this publication also included O-acetylation. Nonetheless, due to the small number of studies and their contrasting results, the significance of these interstrain differences remains elusive.
Pseudomonas aeruginosa is a Gram-negative opportunistic human pathogen, characterized by its metabolic versatility and extensive genome plasticity. It is particularly difficult to eradicate, due to the presence of multiple drug resistance mechanisms, and presents a major health hazard for infected patients (Qin et al. 2022). The most commonly studied P. aeruginosa strains are PAO1 and PA14. The latter is a virulent strain isolated from human patients and is known to possess two additional pathogenicity islands (Mikkelsen et al. 2011, Grace et al. 2022). The acetylome of PA14 has previously been charted (Ouidir et al. 2015, Gaviard et al. 2018, 2019), and revealed that many of the proteins involved in translation (Gaviard et al. 2018) and carbon metabolism (Ouidir et al. 2015, Gaviard et al. 2018) were acetylated. The strains PAO1 and PA14 each represent one of the two largest groups in the five-group population structure of P. aeruginosa, which represent the majority of all sequenced isolates to date (Freschi et al. 2019). The acetylome of the laboratory strain PAO1 has not yet been studied and the interstrain acetylation differences within P. aeruginosa remain unclear. In addition, it is unclear if bacterial acetylation is widely conserved between different species or species-specific acetylation pattern exist.
To address this knowledge gap, we here present the first acetylome of P. aeruginosa strain PAO1, containing 1076 unique acetylation sites in 508 proteins. We compare the interstrain acetylome differences between PAO1 and PA14 obtained by Ouidir et al. (2015) and Gaviard et al. (2018), and postulate that the overall acetylation patterns are not driven by strain-specific factors. Additionally, the comparison of the P. aeruginosa acetylome to 30 other bacterial acetylomes reveals that high acetylation levels for transcription related proteins are shared across bacterial species. This conserved acetylome can help prioritize the characterization of functional consequences of individual acetylation sites.
Materials and methods
Acetylomics
To analyse the acetylome of P. aeruginosa PAO1, a SILAC labeling strategy was used together with immunoprecipitation and mass spectrometry. This labeling strategy was used to quantify the relative acetylome changes upon overexpression of an unspecified gene. However, in this study, we only focused on the nonoverexpression condition.
PAO1 condition 1 cells (no overexpression) and PAO1 condition 2 cells were grown overnight at 37°C in MMP medium (30 mM Na2HPO4, 14 mM KH2PO4, 1 mM MgSO4, 20 mM NH4SO4, 20 mM glucose, and 20 µM FeSO4) supplemented with 0.025% of ‘light’ Lysine (Lys0) and 0.025% of the ‘heavy’ Lysine (Lys4), respectively. From these overnight cultures, precultures were inoculated in the same medium, grown until an OD600 = 0.5 and used to inoculate two flasks containing 500 ml of MMP medium supplemented with 0.025% Lys0 and 0.025% Lys4, respectively. At an OD600 = 0.25, 1 mM IPTG was added to the flask containing Lys4. After 45 min of induction the cells were pelleted by centrifugation (15 min, 4600 rpm, 4°C). Lysis of this pellet was achieved using the Y-PERTM Yeast Protein Extraction Reagent protocol (Thermo Scientific). Briefly, lysozyme was added to a final concentration of 50 μg/ml and 1800 μl Y-PER Reagent (Thermo Scientific) was added per 500 mg of pellet. The mixture was agitated for 20 min at 37°C, sonicated (30 s, 40%) and pelleted by centrifugation at 14 000 × g for 30 min.
Proteins were then extracted from these cells using a chloroform/methanol protein precipitation as described before (Soufi and Macek 2014). The protein precipitates were stored at −80°C. Proteins were resolved in digestion buffer (6 M urea, 2 M thiourea, 10 mM Tris, and pH 8.0) and mixed (1:1 ratio according to protein amounts). 14 mg of the mixture was digested in solution with trypsin as described previously (Borchert et al. 2010).
Lysine-acetylated peptides were enriched by immunoprecipitation using anti-AcK antibodies as described before by Choudhary et al. (2017), with slight modifications. Peptides were desalted using solid phase extraction and dissolved in IP buffer (50 mM MOPS pH 7.2, 10 mM sodium phosphate and 50 mM sodium chloride). Peptides were incubated overnight at 4°C with agarose conjugated antiacetyllysine antibody (ImmuneChem) on a rotation wheel. The immunoprecipitates were washed four times with IP buffer and twice with water. Lysine-acetylated peptides were eluted three times with 0.1% TFA in water. Pooled peptides were purified using stage tips (Rappsilber et al. 2007) and analysed on an EasyLC nano-HPLC (Thermo Scientific) coupled to an LTQ Orbitrap Elite mass spectrometer (Thermo Scientific) as described previously (Franz-Wachtel et al. 2012). The peptide mixture was injected onto the column in HPLC solvent A (0.5% acetic acid) at a flow rate of 500 nl/min and subsequently eluted with a 227 min segmented gradient of 5%–33% to 50%–90% HPLC solvent B (80% acetonitrile in 0.5% acetic acid). During peptide elution the flow rate was kept constant at 200 nl/min. The mass spectrometer was operated in the positive ion mode. Full scan was acquired in the mass range from m/z 300–2000 at a resolution of 120 000 followed by HCD fragmentation of the 15 most intense precursor ions. High-resolution HCD MS/MS spectra were acquired with a resolution of 15 000. The target values for the MS scan and MS/MS fragmentation were 106 and 40 000 charges, respectively. Precursor ions were excluded from sequencing for 60 s after MS/MS.
Acquired MS spectra were processed with the MaxQuant software package version 1.2.2.9 (Cox and Mann 2008) with integrated Andromeda search engine (Cox et al. 2011). Database searches were performed against a target decoy (Elias and Gygi 2007) P. aeruginosa PAO1 database obtained from Uniprot (on 23 June 2014) (The UniProt Consortium 2023), containing 16 883 protein entries and 248 commonly observed contaminants. In database search, full tryptic specificity was required and up to two missed cleavages were allowed. Carbamidomethylation of cysteine was set as fixed modification; protein N-terminal acetylation, oxidation of methionine, and acetylation of lysine were set as variable modifications. Initial precursor mass tolerance was set to 6 ppm at the precursor ion and 20 ppm at the fragment ion level. False discovery rates were set to 1% at peptide, acetylation site, and protein group level. Subsequently, the acetylation sites identified in condition 1 (intensity light >0) were further filtered based on the site localization probability (>0.75) and PEP (<0.05), similar to Birhanu et al. (2017).
Bioinformatics and biostatistics analyses
Enrichment analysis
Once the data was obtained, an enrichment analysis for GO and KEGG terms (Ashburner et al. 2000, Kanehisa and Goto 2000, Kanehisa et al. 2017, 2021, The Gene Ontology Consortium et al. 2021) was performed using the functional annotation tool of the DAVID Bioinformatics database v.2021 (Huang et al. 2009, Sherman et al. 2022). This tool uses a modified version of the Fisher’s Exact test, called the EASE score, to identify enriched terms. The internal list of all P. aeruginosa PAO1 proteins was used as the background. The effect of multiple testing was corrected by using a false discovery rate threshold of 0.05.
Functional acetylation sites
First, a list of potentially functional lysine acetylation sites in bacteria was collated. This was accomplished by searching PubMed using the query ‘[N-ε-lysine acetylation OR (lysine AND acetylat*)] AND (bacter*)’ on PubMed on 08 March 2023. Subsequently, the obtained research papers were screened for information concerning the effect of acetylation, the method used to check the effect of lysine acetylation and the conditions under which it was tested. Sites for which acetylation influenced the behavior of the protein and for which the effect of acetylation of the specific site was tested using mutational experiments, either via blocking or mimicking the acetylation, or a genetic code expansion strategy were included in our list.
Next, the collected sites were mapped to their respective sites in PAO1 homologs using the BLAST+ software v2.15.0+. Specifically, the protein BLAST algorithm—with a word size of 5, a gap open cost of 11, and a gap extend cost of 1 (default values of the online BLAST tool)—was used to find homologs of the proteins containing functional acetylation site (retrieved from UniProt on 9 December 2023) in the PAO1 proteome (UP000002438, retrieved from UniProt on 23 December 2021). Proteins were only deemed homologs if they had an E-value smaller than 10−5, a query coverage larger or equal to 70% and a percentage identity larger or equal to 35%. Afterwards, the BLAST alignments were used to find the lysine residues in the PAO1 homologs that correspond with the functional lysine residues in the composed list. A misalignment of one amino acid was allowed between the lysine in the original and homologues protein.
Bacterial acetylome datasets
To compare our results to others, a list of acetylome datasets was compiled by searching the PubMed and ProteomeXchange database (5 May 2022). The query terms used in this search can be found in Supplementary Table S13. Only bacterial datasets that provided information on acetylated positions and for which a proteome was available [either defined in the publication or available on UniProt (The UniProt Consortium 2023)] were retained.
Subsequently, the protein identifiers and the position of the acetylated amino acids were extracted. The acetylation sites were then processed by mapping the identifiers to UniProt identifiers (The UniProt Consortium 2023) (e.g. by searching for the protein ID that belongs with the gene ID provided in the paper in the UniProt proteome that was used in the same paper) and converting peptide positions to protein positions, depending on the dataset and the proteome that the creators of the dataset used. In the end, a list of 30 datasets containing a total of 52 796 acetylation sites was obtained (Kim et al. 2013, Lee et al. 2013, Okanishi et al. 2013, Castaño-Cerezo et al. 2014, Pan et al. 2014, Kosono et al. 2015, Ouidir et al. 2015, Xie et al. 2015, Carabetta et al. 2016, Chen et al. 2016, 2017, Kentache et al. 2016, Birhanu et al. 2017, Weinert et al. 2017, Gaviard et al. 2018, Jers et al. 2018, Liu et al. 2018, Ravikumar et al. 2018, Türkowsky et al. 2018, Xu et al. 2018, 2020, Yang et al. 2018, 2021, Sun et al. 2019, Wang et al. 2019, Marakasova et al. 2020, Novak et al. 2020, Lei et al. 2021).
Comparison of the acetylomes of P. aeruginosa strain PAO1 and PA14
Two P. aeruginosa strain PA14 acetylome datasets were acquired using the previously described process (Ouidir et al. 2015, Gaviard et al. 2018). These were subsequently compared to our PAO1 acetylome. To start, the site per protein distribution of these datasets was analysed.
Next, the overlap between the acetylated sites was assessed. To enable this, the acetylation sites in PA14 proteins were mapped to their corresponding sites in PAO1 orthologs. These orthologs were identified using OrthoFinder v2.5.4 (Emms and Kelly 2019). Next, the acetylation positions in the PAO1 orthologs were determined by aligning the PA14 proteins to their PAO1 counterparts using the pairwise alignment algorithm implemented in the Bio.Align package of biopython v1.78 (Cock et al. 2009). To be more precise, this algorithm was used in its ‘global’ mode with the BLOSUM62 matrix as the substitution matrix. Additionally, a gap opening score of 10 and extension score of 0.5 was used during the alignment process, similar to Chaudhuri et al. (2015). For the remaining parameters (e.g. the mismatch score) the default values were used. A lysine in a PAO1 ortholog was then considered a match with the acetylated lysine in the PA14 protein, if it the lysine was either directly aligned with the PA14 lysine or flanked the amino acid aligned with the PA14 lysine.
Finally, the pathway coverage by acetylation—calculated as the fraction of the genes in a pathway for which its protein was acetylated—was compared between the three datasets for each KEGG pathway. The pathway information needed for this analysis—meaning the genes of P. aeruginosa strain PAO1 (pae) and strain PA14 (pau), and the pathways to which they belong—was directly retrieved from the KEGG database (Kanehisa and Goto 2000, Kanehisa et al. 2017, 2021) (release 104: 2022/10). The fraction could be calculated after the pathway information was linked to the UniProt IDs. This linking was accomplished using the UniProt mapping tool, as it allowed us to find the KEGG gene id for each UniProt protein ID (The UniProt Consortium 2023).
Comparison of the bacterial acetylomes
Similar to the PAO1–PA14 comparison, the pathway coverage was analysed for and compared between each actylomics dataset. However, in contrast to the previous analysis, we looked at pathway coverage from a ‘pathway group’ level. Specifically, the groups defined in the KEGG PATHWAY Database for the pathways involved in metabolism, genetic information processing, environmental information processing and cellular processing (e.g. carbohydrate metabolism) (Kanehisa and Goto 2000, Kanehisa et al. 2017, 2021).
Results
Determination of the PAO1 acetylome reveals 1076 unique acetylation sites
We analysed the acetylome of P. aeruginosa PAO1 using stable isotope labeling by amino acids in cell culture (SILAC) in combination with LC-MS/MS. Next, we processed the raw MS files with MaxQuant (1% FDR threshold) (Cox and Mann 2008, Cox et al. 2011), and sites with a site localization probability above 0.75 and a posterior error probability (PEP) smaller than 0.05 were selected (Birhanu et al. 2017). These filtering steps resulted in a total of 1076 unique identified sites in 508 proteins (Supplementary Table S1).
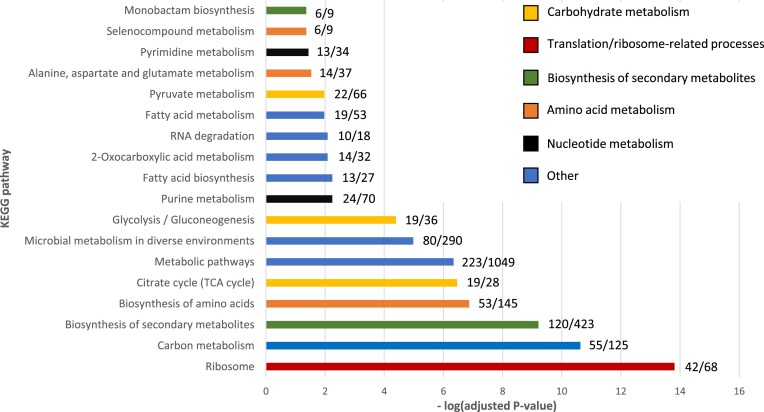
Functional enrichment analysis of the acetylated proteins provides a perspective on their biological relevance (Ashburner et al. 2000, Huang et al. 2009, The Gene Ontology Consortium et al. 2021, Sherman et al. 2022) (Fig. 1, Supplementary Figure S1, Supplementary Table S2). Here, the enrichment analysis results in several GO terms related to translation—such as tRNA binding, ribosomal subunit, and ribosome binding—being overrepresented in the acetylome (Supplementary Figure S1). In addition, we find proteins involved in two metabolic pathways, the citrate cycle (TCA) and glycolysis, in our list of acetylated proteins (Supplementary Figure S1). Analysis of enrichment of KEGG metabolic pathways confirms that the TCA cycle and glycolysis are enriched (Fig. 1). Additional metabolic pathways include pathways related to fatty acid metabolism, amino acid metabolism, and the biosynthesis of secondary metabolites (e.g. monobactam). Overall, these enrichment analyses show that acetylated proteins are spread across different pathways and cellular processes, but are predominantly found in proteins involved in translation and carbon metabolism.
Figure 1.
KEGG enrichment analysis of the identified acetylated proteins. The numbers next to each pathway represent the number of acetylated proteins and total number of proteins in a pathway, respectively. To highlight similarities between the different enriched terms and the GO enrichment analyses in supplementary Figure S1, terms related to the same process are displayed in the same colour. Overall, terms associated with translation, the carbohydrate and amino acid metabolism are enriched.
To assess whether the acetylation sites play a regulatory role in the aforementioned processes, we performed a two-step analysis. First, we compiled a list of experimentally confirmed acetylation sites with a functional effect in other bacterial species (Ramakrishnan et al. 1998, Crosby et al. 2010, Thao et al. 2010, Wang et al. 2010, Li et al. 2011, 2017, 2020a, b, 2021, Liang et al. 2011, Lima et al. 2011, 2012, 2016, Liang and Deutscher 2012, Hu et al. 2013, Tucker and Escalante-Semerena 2013, Vergnolle et al. 2013, Zhang et al. 2013b, 2016, 2020, 2022, Xu et al. 2014, Fraiberg et al. 2015, Tu et al. 2015, Xie et al. 2015, Carabetta et al. 2016, 2019, Qin et al. 2016, Ren et al. 2016, 2019, Song et al. 2016, Sun et al. 2016, 2019, You et al. 2016, Baron and Eisenbach 2017, Chen et al. 2017, Ishigaki et al. 2017, Liao et al. 2017, Nakayasu et al. 2017, Sang et al. 2017, Wang et al. 2017, Wei et al. 2017, Ye et al. 2017, Venkat et al. 2017a, b, 2018, 2019, Bi et al. 2018, Davis et al. 2018, Hockenberry et al. 2018, Reverdy et al. 2018, Sakatos et al. 2018, Umehara et al. 2018, VanDrisse and Escalante-Semerena 2018, Yang et al. 2018, Gao et al. 2019, Kim et al. 2020, Komine-Abe et al. 2021, Singh et al. 2021, Barlow and Tsai 2022, Cai et al. 2022, Fang et al. 2022, Luu et al. 2022, Di et al. 2023) (Supplementary Table S3). Second, we mapped these sites to PAO1 homologs using pairwise sequence alignments. The mapping results in 12 sites for which acetylation was shown to have an impact (Table 1). Two functional sites are conserved at the residue level in proteins involved in translation; for the leucine–tRNA ligase LeuS residue K632 (EC 6.1.1.4) and arginine–tRNA ligase ArgS residue K131 (EC 6.1.1.19) (Ye et al. 2017), three for a protein involved in S-adenosylmethonionine biosynthesis; S-adenosylmethionine synthase MetK (K267, K285, and K386) (EC 2.5.1.6) (Sun et al. 2016), and one for a protein from the TCA cycle; isocitrate dehydrogenase Icd K57 (EC 1.1.1.42) (Venkat et al. 2018). Four additional sites—one in the RNA polymerase protein RpoA K291 (EC 2.7.7.6) (Lima et al. 2016) and the other in the DNA-binding protein HU-beta HupB (K18, K86, and K86) (Barlow and Tsai 2022), were found to influence transcription and are present in the corresponding PAO1 proteins as well. Overall, mutational experiments on these acetylation sites in the original bacterial species showed mostly negative effects—such as decreased enzymatic activity and decreased DNA compaction (Supplementary Table S3). Based on the conservation it is likely that the acetylation sites have the same effect on the P. aeruginosa proteins. Moreover, the conservation of both the acylation of the protein and the acetylated residue suggests an important function. Indeed, Lima et al. (2016) showed that transcription in E. coli depends on RpoA acetylation, the conservation of this acetylation among distant species suggests that this is universal among bacteria.
Table 1.
Acetylation sites with a functional effect also present in the P. aeruginosa PAO1 acetylome.
| Publication | Organism | Protein (Kac site) | P. aeruginosa homolog (matched Kac site) | P. aeruginosa protein description | Gene name |
|---|---|---|---|---|---|
| Lima et al. (2016) | E. coli | P0A7Z4 (K291) | O52760 (K290) | DNA-directed RNA polymerase subunit alpha | rpoA |
| Singh et al. (2021) | M. tuberculosis | P9WGM7 (K207) | Q9HW21 (K203) | Probable two-component response regulator | PA4381 |
| Ye et al. (2017) | E. coli | P07813 (K619) | Q9HX33 (K632) | Leucine–tRNA ligase | leuS |
| Ye et al. (2017) | E. coli | P11875 (K126) | Q9HUC8 (K131) | Arginine–tRNA ligase | argS |
| Barlow and Tsai (2022) | E. coli | P0ACF4 (K86) | P05384 (K86) | DNA-binding protein HU-beta | hupB |
| Luu et al. (2022) | B. subtilis | P08821 (K18, K80, and K86) | P05384 (K18, K80, and K86) | DNA-binding protein HU-beta | hupB |
| Venkat et al. (2018) | E. coli | P08200 (K55) | Q9I0L5 (K57) | Isocitrate dehydrogenase (NADP) | icd |
| Sun et al. (2016) | E. coli | A0A140N627 (K266, K284, and K373) | Q9I5Z0 (K267, K285, and K386) | S-adenosylmethionine synthase | metK |
| Carabetta et al. (2019) | B. subtilis | P08821 (K18, K80, and K86) | P05384 (K18, K80, and K86) | DNA-binding protein HU-beta | hupB |
Note: A list of sites from 67 publications was compared to our PAO1 acetylome. Further details regarding the biological effect of the sites in this list, how they are acetylated and more can be found in supplementary Table S3 (Ramakrishnan, Schuster and Bourret 1998; Crosby et al. 2010; Thao et al. 2010; Wang et al. 2010; Liang, Malhotra and Deutscher 2011; Li et al. 2011, 2017, 2020a, 2020b, 2021; Lima et al. 2011, 2012, 2016; Liang and Deutscher 2012; Hu et al. 2013; Tucker and Escalante-Semerena 2013; Vergnolle, Xu and Blanchard 2013; Zhang et al. 2013b, 2016, 2020, 2022; Xu et al. 2014; Fraiberg et al. 2015; Tu et al. 2015; Xie et al. 2015; Carabetta et al. 2016, 2019; Qin et al. 2016; Ren et al. 2016, 2019; Song et al. 2016; Sun et al. 2016, 2019; You et al. 2016; Baron and Eisenbach 2017; Chen et al. 2017; Ishigaki et al. 2017; Liao et al. 2017; Nakayasu et al. 2017; Sang et al. 2017; Venkat et al. 2017b, 2017a, 2018, 2019; You et al 2016; Wei et al. 2017; Ye et al. 2017; Bi et al. 2018; Davis et al. 2018; Hockenberry et al. 2018; Reverdy et al. 2018; Sakatos et al. 2018; Umehara et al. 2018; VanDrisse and Escalante-Semerena 2018; Yang et al. 2018; Gao et al. 2019; Kim et al. 2020; Komine-Abe et al. 2021; Singh et al. 2021; Barlow and Tsai 2022; Cai et al. 2022; Fang et al. 2022; Luu et al. 2022; Di et al. 2023).
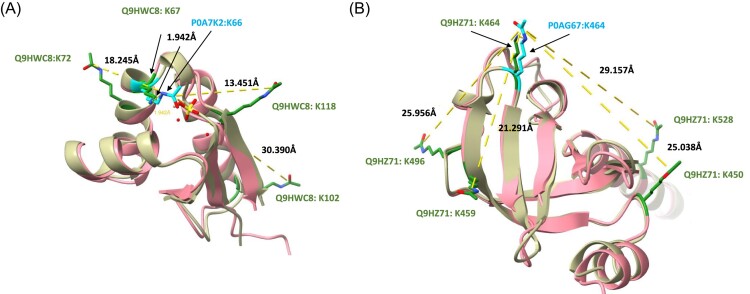
Despite the substantial number of detected acetylation sites in ribosomal proteins, the potentially functional acetylation sites discovered by Zhang et al. (2020)—which influence the chaperone activity of S1 (protein P0AG67, K411, and K464), and potentially EF-TU recruitment and GTP hydrolysis (protein P0A7K2 and K66) in E. coli—were not present in our acetylome (Supplementary Table S3). In total, the PAO1 acetylome contains 91 acetylation sites in 42 out of 56 structural ribosomal proteins (Supplementary Table S4). Seven of these sites are located relatively close to these previously described sites (Fig. 2) (Leijonmarck et al. 1987, Goddard et al. 2018, Berman et al. 2000, Jumper et al. 2021, Pettersen et al. 2021, Varadi et al. 2022, Meng et al. 2023). However, upon closer inspection, we can observe that the acetylated sidechains are oriented away from the sidechains of the sites described by Zhang et al. (2020). Nevertheless, due to their proximity, the detected acetylation sites could still play a regulatory role.
Figure 2.
Lysine acetylation of the ribosomal proteins, Q9HWC8 and Q9HZ71, in P. aeruginosa surrounding two sites that were shown to have potential regulatory role - via mutational studies — in E. coli by Zhang et al. (2020). In this figure, acetylation sites that were measured in this study are highlighted in green on the Pseudomonas AlphaFold structures (Jumper et al. 2021, Varadi et al. 2022) (beige) using ChimeraX (Goddard et al. 2018, Pettersen et al. 2021, Meng et al. 2023). Two sites described by Zhang et al. (2020), K66 in protein P0A7K2 and K464 in protein P0AG67, are highlighted in cyan on the E. coli structures (pink). The P0A7K2 structure originates from the PDB database (1CTF) (Leijonmarck and Liljas 1987, Berman et al. 2000) and P0AG67 from the AlphaFold database (Varadi et al. 2022). Seven of the measured acetylation sites are located relatively closely to these previously characterised sites, thereby suggesting a possible regulatory role of these sites as well.
Comparison of the PAO1 and PA14 acetylomes indicates protein- and site- but not pathway-level differences
We studied the interstrain acetylome variation in P. aeruginosa PAO1 and PA14, by comparing overlapping acetylated proteins and lysine residues. The number of acetylated proteins and sites between the PAO1 acetylome studied here and Gaviard et al.’s (2018) PA14 acetylome are generally consistent. We identified 508 acetylated proteins and 1076 sites, while Gaviard et al. (2018) found 522 proteins and 1102 unique sites. Taking into account the smaller PAO1 proteome (5564 versus 5886 proteins; The UniProt Consortium 2023), the acetylomes are even more similar. Indeed, the PAO1 acetylome covers a slightly larger part of the strain's proteome (9.1% of the proteins), compared to the PA14 acetylome (8.7%). Compared to Gaviard et al.’s (2018) acetylome, larger differences could be observed between our and Ouidir et al.’s (2015) acetylome. We identified 188 additional proteins and 646 additional acetylation sites in our study and consequently obtain a higher fraction of acetylated proteins (9.1%) versus Ouidir et al.’s study (2015) (5.4%).
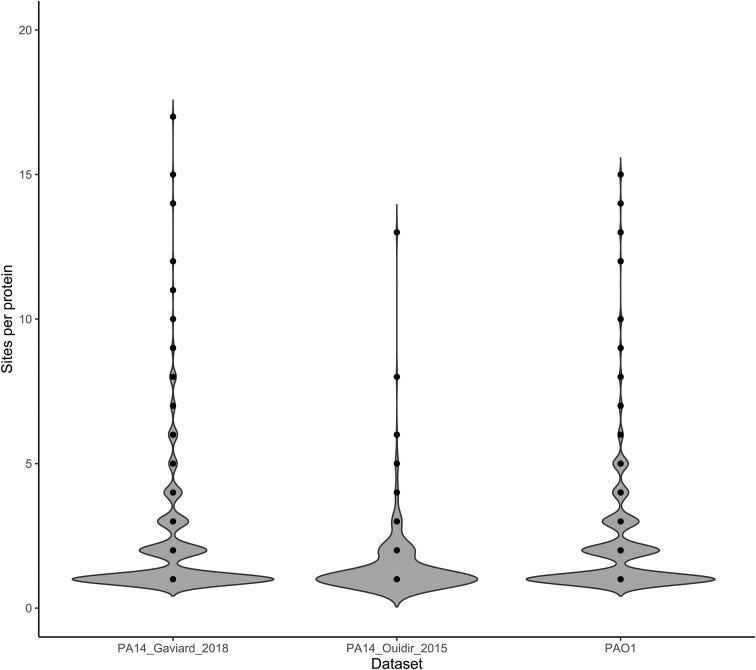
In addition to the acetylome size, the distribution of the number of acetylated sites per protein are similar between the PAO1 acetylome and Gaviard et al.’s (2018) PA14 acetylome (Fig. 3). Most proteins contain one acetylation site. However, some possess many more. For example, Chaperone protein DnaK (Q02FR1) displayed 17 acetylation sites in the study by Gaviard et al. (2018) (Supplementary Table S5). In addition, it is of note that several of these proteins that possess multiple acetylation sites are homologs. For example, Chaperonin GroEL (P30718–EC 5.6.1.7) contains 14 acetylation sites in our acetylome and 11 and 13 sites in its PA14 homolog (Q02H55) in Gaviard et al.’s (2018) and Ouidir et al.’s (2015) acetylome, respectively (Supplementary Table S5). This trend of high numbers of acetylation sites in both strains—and even between the two PA14 datasets—does not hold for the majority of proteins. To illustrate, Cysteine desulfurase IscS (Q02RW8–EC 2.8.1.7) contains seven acetylation sites in Gaviard et al.’s (2018) acetylome, none in Ouidir et al.’s (2015) dataset and only one in its PAO1 homolog in our acetylome (Supplementary Table S5). Within the context of these acetylome comparisons, it is important to keep in mind that the acetylome of Ouidir et al. (2015) is markedly smaller. As such, these similarities with Gaviard’s acetylome and differences with Ouidir’s might be attributable by differences in the experimental setup of the studies in question.
Figure 3.
Site per protein distribution of the two available PA14 acetylomes by Ouidir et al. (2015) and Gaviard et al. (2018) and our PAO1 acetylome. A similar distribution can be observed between our and the Gaviard et al. dataset. Most acetylated proteins contain one acetylation site, however, a few proteins possess many more.
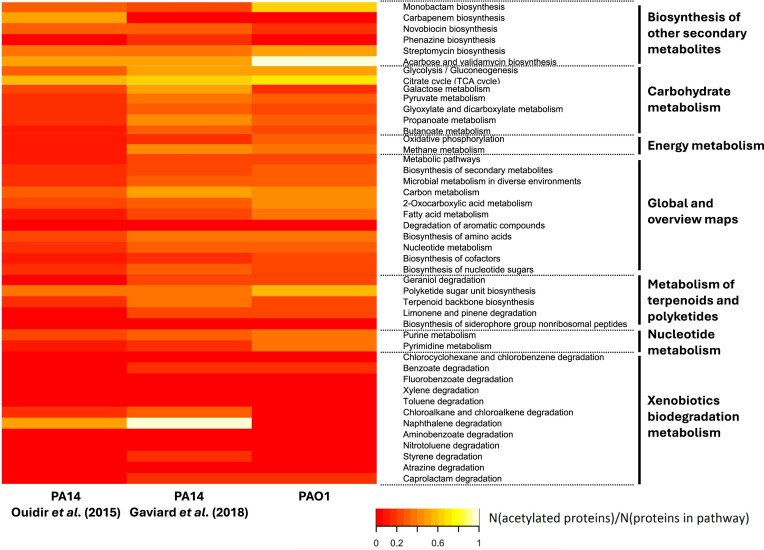
Due to the smaller PA14 acetylome measured by Ouidir et al. (2015), most of the metabolic pathways showed reduced acetylation levels—defined as the fraction of genes involved in a KEGG pathway for which their corresponding proteins were found to be acetylated—compared to those for the two other P. aeruginosa acetylomes (Fig. 4). Despite the similarities regarding the number of acetylated proteins and sites between the PAO1 and PA14 acetylome from Gaviard et al. (2018), the overall acetylation patterns at the pathway level also display several differences. In general, acetylation events are spread throughout the metabolic network. Only 15 of the 102 metabolic pathways lacked any acetylated proteins for all three acetylomes, six of which belong to the ‘pathway group’, which is involved in xenobiotics biodegradation and metabolism. The remaining unacetylated pathways are distributed across the other pathway groups, excluding the amino acid, energy, nucleotide metabolism, and the global and overview maps pathway groups.
Figure 4.
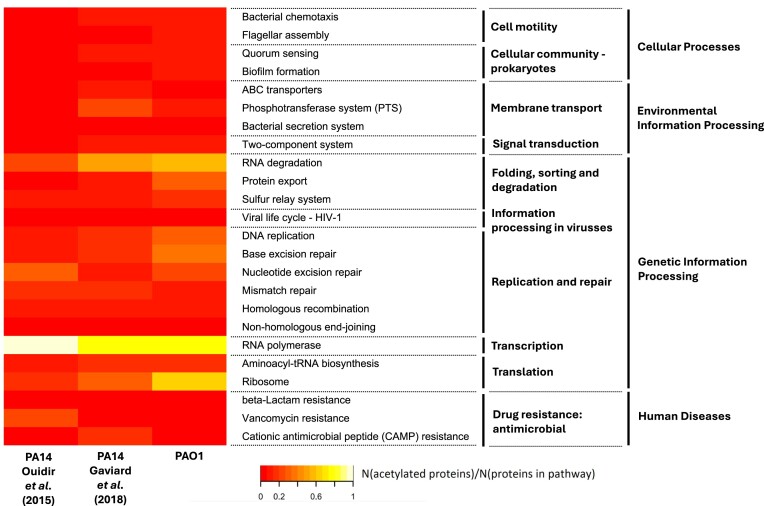
Overview of acetylation levels throughout a subsection of metabolic KEGG pathways in P. aeruginosa strain PAO1 and PA14. The colour of each cell in the heatmap corresponds to the fraction of genes involved in a KEGG pathway for which their corresponding proteins were found to be acetylated. This fraction is also referred to as the acetylation level of a pathway in this paper. Specifically, two acetylomes from P. aeruginosa strain PA14 were compared with our PAO1 acetylome (Ouidir et al. 2015, Gaviard et al. 2018). Several pathways were highly acetylated in all three datasets (e.g. TCA cycle). On the other hand, for some pathways large differences could be observed, e.g. the naphthalene degradation and monobactam biosynthesis pathway. However, some of these differences may be caused by pathways containing a limited number of proteins.
Interestingly, several pathways with relatively high acetylation levels are shared among the three acetylomes (Fig. 4). The two prime examples here are the glycolysis and TCA cycle pathways of the carbohydrate metabolism. They display acetylation levels of between 30.1% (11/36) in Ouidir et al.’s (2015) acetylome and 52.8% (19/36) in PAO1, and between 53.6% (15/28) in Ouidir et al.’s (2015) acetylome in PAO1 and 67.9% (19/28) in PAO1, respectively (Supplementary Table S6). As mentioned previously, these pathways are often found to be acetylated in acetylome studies (Ouidir et al. 2015, Nakayasu et al. 2017, Gaviard et al. 2018). Notably, the acarbose and validamycin biosynthesis pathway show high acetylation levels for all P. aeruginosa acetylomes (50%–100%). However, this pathway only contains two protein coding genes. As such, high acetylation levels can easily be reached.
By contrast, not every pathway, which shows extensive acetylation in one dataset is highly acetylated in another. For example, the monobactam biosynthesis pathway only possesses a high acetylation level in PAO1, whereas the naphthalene degradation pathway has extensive acetylation levels in the PA14 acetylomes. Nevertheless, similar to the acarbose and validamycin pathway, we must keep in mind that the monobactam and naphthalene pathways are small. Missing a single acetylated protein can drastically affect the acetylation level. However, there are some pathways, which consist of more proteins that show differences in acetylation levels. For example, in PAO1 13 out of 34 proteins involved in the pyrimidine metabolism are acetylated, whereas only three and six out of the 36 proteins in the same pathway in the Gaviard et al. (2018) and Ouidir et al. (2015) acetylomes are acetylated, respectively.
Aside from the metabolic pathways, the acetylation levels of the pathways involved in cellular processes, environmental information processing, genetic information processing, and human diseases (that includes antimicrobial resistance) were investigated as well. Here, the common highly acetylated processes are associated with the functioning of RNA polymerase and RNA degradation (Fig. 5). In the former process, homologs of all the proteins that are acetylated in PAO1 (3) are acetylated in PA14, albeit not as many sites per protein. The fourth protein involved in this process according to KEGG, RpoZ (Q9HTM1–EC 2.7.7.6), is only acetylated in the Ouidir et al. (2015) acetylome. In the RNA degradation process, most proteins acetylated in PAO1 are also acetylated in one of the PA14 acetylomes. Two proteins in particular—chaperonin GroEL (P30718) and chaperon protein DnaK (Q9HV43)—are highly acetylated across all three acetylomes (11–14 sites per protein) (Supplementary Table S7). The only protein that is uniquely acetylated in the PAO1 acetylome is Q9×4P2, an RNA pyrophosphohydrolase. Interestingly, the ribosome pathway is more heavily acetylated in PAO1 than PA14. Here, 17 proteins are uniquely acetylated in PAO1. However, here too the most acetylated proteins are acetylated in at least one of the PA14 acetylomes (Supplementary Table S7).
Figure 5.
Overview of acetylation levels across the KEGG pathways — involved in cellular processes, environmental information processing, genetic information processing and human diseases - in P. aeruginosa. The colour of each cell in the heatmap corresponds to fraction of genes involved in a KEGG pathway for which their corresponding proteins were found to be acetylated. This fraction is also referred to as the acetylation level of a pathway in this paper. Specifically, two acetylomes from P. aeruginosa strain PA14 were compared with our PAO1 acetylome (Ouidir et al. 2015, Gaviard et al. 2018). Transcription-related genes were highly acetylated in all three datasets (e.g. TCA cycle). For some pathways differences could be seen (e.g. ribosome). However, no clear separation of the acetylomes based on the strain to which they belong, could be seen.
The distribution of acetylation events in virulence-related processes based on the VFDB database (Liu et al. 2022) revealed that 19 and 22 virulence factors were acetylated in PAO1 and PA14, respectively (Supplementary Table S8). Notably, there is little overlap between the specific proteins that were acetylated in these two strains. Nevertheless, the acetylated proteins are involved in mostly the same processes and subprocesses (e.g. biofilm formation, adherence, and so on), with exception of the virulence factors related to pyocyanin. The proteins in the latter were only acetylated in PA14.
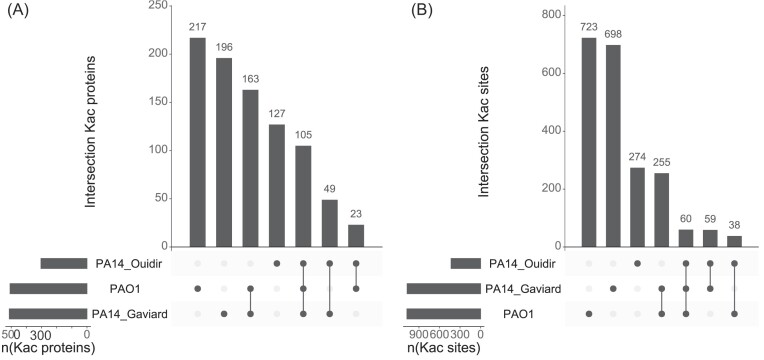
Given the lack of large differences at the pathway level, the natural follow-up question was whether this trend continued at the protein and acetylation site level. To analyse this, we mapped the PA14 acetylation sites to their corresponding sites for their PAO1 orthologs. In this manner, orthologs could be found for respectively 96.5% and 97.7% of the acetylated proteins extracted from the publications of Ouidir et al. (2015) and Gaviard et al. (2018). This corresponds to respectively 7 and 12 proteins for which no orthologs could be found (Ouidir et al. 2015, Gaviard et al. 2018). Notably, only one of these, the CRISPR-associated protein Csy2 (Q02MM0), plays a role in antiviral defense. The sites themselves were also mapped with high efficiency. A matching lysine in a PAO1 ortholog was found for 96.5% of the extracted sites from the Ouidir et al. (2015) acetylome and 97.7% of the extracted sites from Gaviard research team acetylome. In the end, the mapping process revealed a total of 880 proteins and 2107 sites across the three datasets (Fig. 6). In total, our PAO1 acetylome shared 52.8% of the proteins and 29.3% of the sites with the PA14 acetylome of Gaviard et al. (2018), and 25.2% of the proteins and 9.1% of the sites with the PA14 acetylome of Ouidir et al. (2015) Consequently, most of the acetylated proteins and sites are unique to one of the datasets. To be more precise, 42.7% proteins and 67.2% sites in the POA1 acetylome, 38.2% proteins and 65.1% sites in the PA14 acetylome of Gaviard et al. (2018), and 41.8% proteins and 63.6% sites in the PA14 acetylome of Ouidir et al. (2015) were unique in their respective acetylome. Based on the absolute number of overlapping acetylated proteins and sites, our PAO1 acetylome appears to be more similar to the PA14 acetylome of Gaviard et al. (2018) than that of Ouidir et al. (2015), similar to the previous comparisons (number of proteins, sites, sites per protein distribution). However, if we inspect the fractions of shared proteins and sites, this view changes. For example, the Ouidir acetylome shares 50.6% of all its proteins and 27.6% of all its sites with the Gaviard acetylome. This is quite similar compared to our acetylome, which shares 52.8% and 29.2% of its proteins and sites with Gaviard et al.’s (2018) acetylome, respectively. Nevertheless, we can conclude that clear differences exist between the Pseudomonas acetylomes (Ouidir et al. 2015, Gaviard et al. 2018).
Figure 6.
The overlapping acetylated proteins (A) and sites (B) between our PAO1 acetylome and the publicly available PA14 acetylomes. To start, the gene IDs provided by the two PA14 publications were converted to UniProt IDs in order to know which proteins to align in a later phase. Unfortunately, not every gene ID could be converted. As such, a few acetylated proteins were not included in this analysis. In total, 94.4% of Ouidir et al’s acetylated proteins and 94.9% of the sites were successfully mapped to PAO1. Moreover, 97.9% of Gaviard et al.’s acetylated proteins and 98.2% of their sites were successfully mapped to PAO1. Next, the overlap was determined after mapping the PA14 sites to their corresponding positions in PAO1 orthologs. Overall, we can see that many of the acetylated proteins and sites are unique to each dataset.
Some conservation can also be observed; 105 proteins and 60 sites are shared between the three acetylomes. Several of these shared proteins are part of earlier discussed processes, such as the TCA cycle, RNA polymerase related processes and RNA degradation. The latter possesses the protein, which shares the largest number of sites (six) with the PA14 acetylomes, called chaperonin GroEL (P30718). Other processes that are featured in this overlapping region are: ribosomal processes (e.g. small ribosomal subunit protein uS2 K128–O82850), glycolysis (e.g phosphoglycerate kinase K120 –Q9I5Y4–EC 2.7.2.3), and reaction to oxidative stresses (e.g. superoxide dismutase K128–P53641–EC 1.15.1.1). Notably, some of the potentially functional sites in our PAO1 acetylome (Table 1) were also found in the PA14 acetylomes (e.g. HupB K67, S-adenosylmethionine synthase K35 and K285). This conservation further suggests that these acetylation events play a functional role in P. aeruginosa. Finally, one site in a Cysteine–tRNA ligase (Q9I2U7 K73–EC 6.1.1.16) was present in the three acetylomes. As acetylation events were shown to have an effect in other tRNA ligases, this could be an interesting site to study (Venkat et al. 2017a).
Interspecies comparison reveals high acetylation levels in proteins involved in transcription
Since the publication of the first bacterial acetylome in 2008, several bacterial acetylomes have been mapped and published (Yu et al. 2008). The publications that passed our criteria (acetylation site information and an available UniProt (or other) proteome) were selected for comparison with our PAO1 acetylome (see methods) (28/50 publications). In total, 30 acetylomes were found, covering 23 bacterial species (Kim et al. 2013, Lee et al. 2013, Okanishi et al. 2013, Castaño-Cerezo et al. 2014, Pan et al. 2014, Kosono et al. 2015, Ouidir et al. 2015, Xie et al. 2015, Carabetta et al. 2016, Chen et al. 2016, 2017, Kentache et al. 2016, Birhanu et al. 2017, Weinert et al. 2017, Gaviard et al. 2018, Jers et al. 2018, Liu et al. 2018, Ravikumar et al. 2018, Türkowsky et al. 2018, Xu et al. 2018, 2020, Yang et al. 2018, 2021, Sun et al. 2019, Wang et al. 2019, Marakasova et al. 2020, Novak et al. 2020, Lei et al. 2021). After mapping all protein identifiers to their respective UniProt Accession numbers—these acetylomes covered between 0.8% and 80.5% of the proteomes of these bacterial species. However, the extensive coverages (80.5% and 58.0%) were only measured for two genome-reduced bacteria, Mycoplasma pneumoniae and M. genitalium, in the same study (Chen et al. 2016). The remaining acetylomes, including the P. aeruginosa PAO1 acetylome (9.1%), covered less than 31% of the bacterial proteomes (Supplementary Table S9).
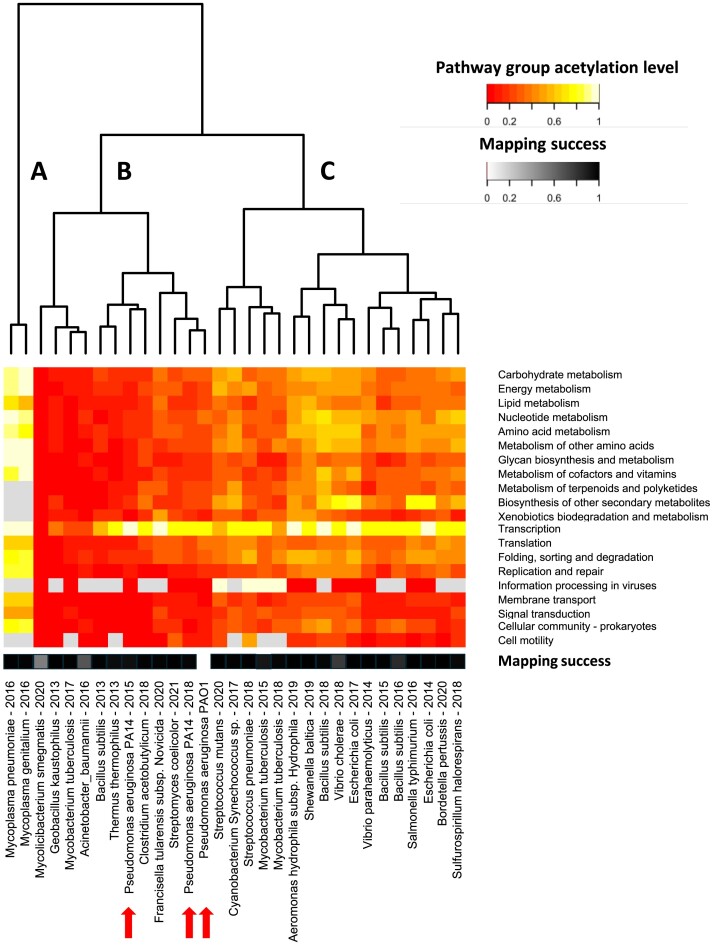
Similar to the PAO1–PA14 comparison, we calculated the acetylation levels in biological processes for the selected datasets. However, instead of focusing on all the individual KEGG pathways, we opted to rely on the ‘pathway groups’ defined by KEGG (Supplementary Table S10). Based on the hierarchical clustering of the acetylation patterns, the datasets were grouped in three main clusters (Fig. 7), with cluster B and C having more similar acetylation levels. Cluster A is comprised of two Mycoplasma species—M. pneumoniae and M. genitalium—which display high acetylation levels for all pathway groups (Fig. 7, cluster A). The remaining two clusters (clusters B and C) have lower overall acetylation levels for the different pathway groups, with cluster B possessing on average lower acetylation levels than group C. One interesting exception is the ‘transcription’ pathway group. Here, extensive acetylation levels can be seen for the majority of datasets contained in clusters B and C.
Figure 7.
Distribution of acetylation events throughout the KEGG ‘pathways groups’ across 31 bacterial acetylomes. The colour of each cell in the heatmap corresponds to the fraction of genes involved in a KEGG pathway group for which their corresponding proteins were found to be acetylated. This fraction is also referred to as the acetylation level of the pathway group in this paper. Cells were coloured grey if no annotations were found in a particular species. If almost no annotations were available for any of the bacterial species in this comparison, the pathway group was left out of this figure. At the bottom, one row highlights the mapping success. This grayscale represents the fraction of acetylated proteins that were successfully mapped to UniProt identifiers from the original datasets. The datasets are clustered in three main clusters (A, B, C). Cluster A contains both Mycoplasma species and is characterized by high acetylation levels across all pathway groups. The remaining acetylomes are clustered in B and C. Notably, high acetylation levels for the transcription pathway group can be observed.
Notably, the Pseudomonas datasets are all part of cluster B (red arrows Fig. 7). However, datasets pertaining to the same species were not always grouped together. For example, two M. tuberculosis acetylomes were grouped in cluster C, while another belongs to cluster B. This could be due to a myriad of factors, e.g. growth phase, carbon source (Christensen et al. 2017, 2019, Greiner-Haas et al. 2021), mass spectrometry search algorithms, or technological improvements. The explanatory power of the parameters regarding the clustering pattern was investigated (Supplementary Table S11). Notably, the majority of acetylomes measured during the stationary phase are grouped in cluster B, whereas most acetylomes measured in either the exponential growth phase or a mix of growth phases are grouped in cluster C. Nevertheless, a few datasets measured during the exponential phase are also present in group B and vice versa. As such, none of the factors were able to fully explain the grouping. Hence, other or a combination of factors must be at play.
Discussion
Over the past two decades, it has become more and more apparent that PTMs in bacteria, including Nε-lysine acetylation, play a vital role in responding to changes in the environment. Here, we present a snapshot of one potential regulatory layer in P. aeruginosa strain PAO1, the acetylome. We identified 1076 unique acetylated lysine positions in 508 proteins. A further enrichment analysis of the acetylated proteins revealed that many acetylated proteins are involved in translation, and more specifically the ribosomes. In addition, GO and KEGG terms associated with the central carbon metabolism (e.g. TCA cycle) were also identified. This corresponds to findings in other acetylome studies (Ashburner et al. 2000, Kanehisa and Goto 2000, Kanehisa et al. 2017, 2021, The Gene Ontology Consortium et al. 2021). Several acetylation sites in these pathways were shown to be widely conserved (Ouidir et al. 2015, Nakayasu et al. 2017, Gaviard et al. 2018). Furthermore, the functioning of multiple proteins involved in these pathways [TCA (Venkat et al. 2017b, 2018, 2019, Komine-Abe et al. 2021) and glycolysis (Xu et al. 2014, Fang et al. 2022)] has been shown to be affected by acetylation, for example by modulation of enzymatic activity. Similarly, potentially functional sites have been found in aminoacyl-tRNA synthetases (Ye et al. 2017, Venkat et al. 2017a, Umehara et al. 2018), an elongation factor (Zhang et al. 2020) and ribosomal proteins (e.g. ribosomal protein S1 in E. coli) (Zhang et al. 2020, Feid et al. 2022). In case of the latter, one study reported that acetylation slowed down translation (elongation) (Zhang et al. 2020), while another reported acetylation to influence subunit association (Feid et al. 2022). Notably, several of these sites were found to be acetylated in PAO1 homologs in our acetylome. Altogether, this suggests that acetylation could play a functional and even a regulatory role in P. aeruginosa PAO1 in the aforementioned processes.
Nevertheless, the functionality of most of these identified sites in PAO1 remains unknown. Interestingly, the monobactam biosynthesis pathway was also identified using the enrichment analysis. Six out of nine proteins that have been assigned to this pathway in P. aeruginosa PAO1 in the KEGG database were acetylated (Kanehisa and Goto 2000, Kanehisa et al. 2017, 2021). Two of these genes, 4-hydroxy-tetrahydrodipicolinate synthase (Q9I4W3–EC 4.3.3.7) and Aspartate-semialdehyde dehydrogenase (Q51344–EC 1.2.1.11), are essential for P. aeruginosa growth and have been put forward as potential drug targets (Moore et al. 2002, Kaur et al. 2011)
In the second part of this study, we compared our PAO1 acetylome to the available PA14 acetylomes (Ouidir et al. 2015, Gaviard et al. 2018). The initial comparisons at the level of acetylome size and site per protein distribution showed extensive similarity between our PAO1 acetylome and the PA14 acetylome by Gaviard et al. (2018). However, a more detailed analysis at the pathway and especially specific protein and acetylation site levels, revealed large differences. Indeed, the majority of the sites identified in each P. aeruginosa acetylome (>63%) was unique to that acetylome. As mentioned in the introduction, this is consistent with the study in A. baumannii, where only limited acetylome overlap was found between an antibiotic resistant and sensitive strain (Kentache et al. 2016). These interstrain differences likely contribute to strain-specific attributes—similar to those found in E. coli, between an antibiotic sensitive and several resistant strains (Fang et al. 2022). Nonetheless, despite their differences, our PAO1 acetylome resembled the Gaviard et al. (2018) PA14 acetylome as much or even much more closely than the other PA14 acetylome (also at the pathway level), depending on the perspective (e.g. fraction of shared sites vs absolute number of overlapping sites) (Ouidir et al. 2015, Gaviard et al. 2018). However, one should be cautious with the interpretation of these results, as the effect of strain-specific factors could be masked by technical parameters that differ between the P. aeruginosa acetylome studies (e.g. type of antibodies used for peptide enrichment, growth phase, growth medium, and search engine) (Ouidir et al. 2015, Christensen et al. 2017, 2019, Gaviard et al. 2018, Greiner-Haas et al. 2021). Moreover, this study determined the acetylome during exponential growth in MMP medium, whereas the Gaviard acetylome measured acetylation sites after growth on four different media (citrate, glutamate, succinate, and glucose). This wider range of growth conditions may have increased the overlap between the two acetylomes (Christensen et al. 2017, 2018, Gaviard et al. 2018, Greiner-Haas et al. 2021). Notwithstanding these limitations, our results suggest that the overall acetylome patterns in P. aeruginosa are not primarily driven by strain-specific factors.
Finally, we placed our P. aeruginosa PAO1 acetylome in a broader context by comparing it to 30 other acetylomes from 22 different bacterial species, extracted from literature. As such, comparison between the different acetylomes proved more difficult, because they were determined using different MS techniques for different conditions. Further analysis was also confounded by the fact that the versions of proteomes used during the peptide identifying step in these publications were often not clearly specified (led to the loss of some sites after extraction). Aware of these restrictions, we critically compared the bacterial acetylomes. At the pathway group level, the bacterial acetylomes split in three clusters based on their acetylation patterns. Cluster A possessed high acetylation levels for all pathways, in contrast to clusters B and C. These two clusters displayed slightly lower acetylation levels with the differences being the relatively higher acetylation levels for cluster C in pathways related to carbohydrate metabolism, energy metabolism, and the biosynthesis of secondary metabolites. We attempted to explain this clustering taking into account several parameters (mass spectrometry search algorithms, carbon source, growth phase, and technological improvements), but none could fully explain it. Similar to the PAO1-PA14 comparison, other factors—such as the type of antibody used—could have an impact. One potential limitation of this analysis is the heavy reliance on the pathway definitions provided by KEGG. These are quite large compared to some other databases (e.g. EcoCyc) (Karp et al. 2021). As a result, finer nuances in acetylation patterns could be missed. Notably, high acetylation levels for transcription-related proteins were shared between most acetylomes. The enrichment in this pathway could be due to the high level of protein abundance of proteins belonging to transcription and translation. Several acetylation sites that can impact transcription—for example via influencing transcription factors (Thao et al. 2010, Hu et al. 2013, Qin et al. 2016, Ren et al. 2016, Bi et al. 2018, Koo et al. 2020, Li et al. 2021, Singh et al. 2021), a sigma factor (Kim et al. 2020), the RNA polymerase (Lima et al. 2011, 2012, 2016), or histone-like and nucleoid-associated proteins (Ghosh et al. 2016, Barlow and Tsai 2022, Luu et al. 2022)—have been identified in bacterial species outside P. aeruginosa. However, most acetylation sites remain understudied. The conservation of acetylated pathways, proteins and residues between strains and bacterial species, can help prioritize the functional characterization of the tens of thousands of acetylation sites.
Supplementary Material
Acknowledgements
The authors thank Professor Jorge C. Escalante-Semerena and Professor Ute Römling for their constructive comments during the peer review process.
Notes
Reviewer: Ute Römling
Contributor Information
Nand Broeckaert, Computational Systems Biology, Department of Microbial and Molecular Systems, KU Leuven, Kasteelpark Arenberg 20 box 2460, 3001 Heverlee, Belgium; Laboratory of Gene Technology, Department of Biosystems, KU Leuven, Kasteelpark Arenberg 21 box 2462, 3001 Heverlee, Belgium.
Hannelore Longin, Computational Systems Biology, Department of Microbial and Molecular Systems, KU Leuven, Kasteelpark Arenberg 20 box 2460, 3001 Heverlee, Belgium; Laboratory of Gene Technology, Department of Biosystems, KU Leuven, Kasteelpark Arenberg 21 box 2462, 3001 Heverlee, Belgium.
Hanne Hendrix, Laboratory of Gene Technology, Department of Biosystems, KU Leuven, Kasteelpark Arenberg 21 box 2462, 3001 Heverlee, Belgium.
Jeroen De Smet, Research Group for Insect Production and Processing, Department of Microbial and Molecular Systems (M²S), KU Leuven, Kleinhoefstraat 4, 2440 Geel, Belgium.
Mirita Franz-Wachtel, Proteome Center Tuebingen, Institute of Cell Biology, University of Tübingen, Auf d. Morgenstelle 15, D-72076 Tübingen, Germany.
Boris Maček, Proteome Center Tuebingen, Institute of Cell Biology, University of Tübingen, Auf d. Morgenstelle 15, D-72076 Tübingen, Germany.
Vera van Noort, Computational Systems Biology, Department of Microbial and Molecular Systems, KU Leuven, Kasteelpark Arenberg 20 box 2460, 3001 Heverlee, Belgium; Institute of Biology, Leiden University, Sylviusweg 72, 2333 Leiden, the Netherlands.
Rob Lavigne, Laboratory of Gene Technology, Department of Biosystems, KU Leuven, Kasteelpark Arenberg 21 box 2462, 3001 Heverlee, Belgium.
Conflict of interest
None declared.
Funding
This research was funded through the KU Leuven project C1 ‘ACES’ (C16/20/001) (R.L. and V.N.). Jeroen De Smet holds a postdoctoral fellowship (12V5222N) of the Flanders Research Foundation.
Data Availability
The PAO1 acetylomics dataset discussed in this paper has been submitted to the ProteomeXchange Consortium via the PRIDE database. You can find the data using the identifier: PXD051454.
The list of experimentally assessed acetylation sites was compiled from sources in the public domain. You can find the data in the following references: Ramakrishnan et al. (1998), Crosby et al. (2010), Thao et al. (2010), Wang et al. (2010), Li et al. (2011, 2017, 2020a, b, 2021), Liang et al. (2011), Lima et al. (2011, 2012, 2016), Liang and Deutscher (2012), Hu et al. (2013), Tucker and Escalante-Semerena (2013), Vergnolle et al. (2013), Zhang et al. 2013b, 2016, 2020, 2022), Xu et al. (2014), Fraiberg et al. (2015), Tu et al. (2015), Xie et al. (2015), Carabetta et al. (2016, 2019), Qin et al. (2016), Ren et al. (2016, 2019), Song et al. (2016), Sun et al. (2016, 2019), You et al. (2016), Baron and Eisenbach (2017), Chen et al. (2017), Ishigaki et al. (2017), Liao et al. (2017), Nakayasu et al. (2017), Sang et al. (2017), Wang, Ye et al. (2017), You et al. (2016) , Wei et al. (2017), Venkat et al. 2017a, b, 2018, 2019), Bi et al. (2018), Davis et al. (2018), Hockenberry et al. (2018), Reverdy et al. (2018), Sakatos et al. (2018), Umehara et al. (2018), VanDrisse and Escalante-Semerena (2018), Yang et al. (2018), Gao et al. (2019), Ren et al. (2019), Kim et al. (2020), Komine-Abe et al. (2021), Singh et al. (2021), Barlow and Tsai (2022), Cai et al. (2022), Fang et al. (2022), Luu et al. (2022), Di et al. (2023).
Similarly, the acetylomics datasets, excluding the PAO1 dataset, were derived from sources in the public domain. You can find the data in the following references: Kim et al. (2013), Lee et al. (2013), Okanishi et al. (2013), Castaño-Cerezo et al. (2014), Pan et al. (2014), Kosono et al. 2015), Ouidir et al. (2015), Xie et al. (2015), Carabetta et al. (2016), Chen et al. (2016, 2017), Kentache et al. (2016), Birhanu et al. (2017), Weinert et al. (2017), Gaviard et al. (2018), Jers et al. (2018), Liu et al. (2018), Ravikumar et al. (2018), Türkowsky et al. (2018), Xu et al. (2018, 2020), Yang et al. (2018, 2021), Sun et al. (2019), Wang et al. (2019), Marakasova et al. (2020), Novak et al. (2020), Lei et al. (2021).
The remaining data underlying this article are available in the article and in its online supplementary material.
References
- Ashburner M, Ball CA, Blake JA et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak R, Welch M, Yanovsky A et al. Acetyladenylate or its derivative acetylates the chemotaxis protein CheY in vitro and increases its activity at the flagellar switch. Biochemistry. 1992;34:10099–107. [DOI] [PubMed] [Google Scholar]
- Barlow VL, Tsai Y-H. Acetylation at lysine 86 of Escherichia coli huβ modulates the DNA-binding capability of the protein. Front Microbiol. 2022;12:809030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron S, Eisenbach M. CheY acetylation is required for ordinary adaptation time in Escherichia coli chemotaxis. FEBS Lett. 2017;591:1958–65. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J, Gou Z, Zhou F et al. Acetylation of lysine 182 inhibits the ability of Mycobacterium tuberculosis DosR to bind DNA and regulate gene expression during hypoxia. Emerg Microbes Infect. 2018;7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birhanu AG, Yimer SA, Holm-Hansen C et al. Nε- and O-acetylation in Mycobacterium tuberculosis lineage 7 and lineage 4 strains: proteins involved in bioenergetics, virulence, and antimicrobial resistance are acetylated. J Proteome Res. 2017;16:4045–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontemps-Gallo S, Gaviard C, Richards CL et al. Global profiling of lysine acetylation in Borrelia burgdorferi B31 reveals its role in central metabolism. Front Microbiol. 2018;9:2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert N, Dieterich C, Krug K et al. Proteogenomics of Pristionchus pacificus reveals distinct proteome structure of nematode models. Genome Res. 2010;20:837–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S-S, Zhang L-Q, Zhang Q et al. Acetylation of NarL K188 and K192 is involved in regulating Escherichia coli anaerobic nitrate respiration. Appl Microbiol Biotechnol. 2022;106:7209–21. [DOI] [PubMed] [Google Scholar]
- Carabetta VJ, Greco TM, Cristea IM et al. YfmK is an Nε-lysine acetyltransferase that directly acetylates the histone-like protein HBsu in Bacillus subtilis. Proc Natl Acad Sci. 2019;116:3752–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabetta VJ, Greco TM, Tanner AW et al. Temporal regulation of the Bacillus subtilis acetylome and evidence for a role of MreB acetylation in cell wall growth. mSystems. 2016;1:e00005–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño-Cerezo S, Bernal V, Post H et al. Protein acetylation affects acetate metabolism, motility and acid stress response in Escherichia coli. Mol Syst Biol. 2014;10:762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R, Sadrieh A, Hoffman NJ et al. PhosphOrtholog: a web-based tool for cross-species mapping of orthologous protein post-translational modifications. BMC Genomics. 2015;16:617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W-H, van Noort V, Lluch-Senar M et al. Integration of multi-omics data of a genome-reduced bacterium: prevalence of post-transcriptional regulation and its correlation with protein abundances. Nucleic Acids Res. 2016;44:1192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhang G, Yang M et al. Lysine acetylome analysis reveals photosystem II manganese-stabilizing protein acetylation is involved in negative regulation of oxygen evolution in model Cyanobacterium synechococcus sp. PCC 7002. Mol Cell Proteomics. 2017;16:1297–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary S, Barth S, Verma RS. SNAP-tag technology: a promising tool for ex vivo immunophenotyping. Mol Diagn Ther. 2017;21:315–26. [DOI] [PubMed] [Google Scholar]
- Christensen DG, Meyer JG, Baumgartner JT et al. Identification of novel protein lysine acetyltransferases in Escherichia coli. mBio. 2018;9:e01905–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen DG, Orr JS, Rao CV et al. Increasing growth yield and decreasing acetylation in Escherichia coli by optimizing the carbon-to-magnesium ratio in peptide-based media. Appl Environ Microbiol. 2017;83:e03034–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen DG, Xie X, Basisty N et al. Post-translational protein acetylation: an elegant mechanism for bacteria to dynamically regulate metabolic functions. Front Microbiol. 2019;10:1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock PJA, Antao T, Chang JT et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–805. [DOI] [PubMed] [Google Scholar]
- Crosby HA, Heiniger EK, Harwood CS et al. Reversible N epsilon-lysine acetylation regulates the activity of acyl-CoA synthetases involved in anaerobic benzoate catabolism in Rhodopseudomonas palustris. Mol Microbiol. 2010;76:874–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R, Écija-Conesa A, Gallego-Jara J et al. An acetylatable lysine controls CRP function in E. coli. Mol Microbiol. 2018;107:116–31. [DOI] [PubMed] [Google Scholar]
- Di Y, Xu S, Chi M et al. Acetylation of cyclic AMP receptor protein by acetyl phosphate modulates mycobacterial virulence. Microbiol Spectr. 2023;11:e0400222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–14. [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z, Lai F, Cao K et al. Potential role of lysine acetylation in antibiotic resistance of Escherichia coli. mSystems. 2022;7:e00649–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feid SC, Walukiewicz HE, Wang X et al. Regulation of translation by lysine acetylation in Escherichia coli. mBio. 2022;13:e01224–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraiberg M, Afanzar O, Cassidy CK et al. CheY's acetylation sites responsible for generating clockwise flagellar rotation in Escherichia coli: acetylation sites involved in clockwise rotation. Mol Microbiol. 2015;95:231–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz-Wachtel M, Eisler SA, Krug K et al. Global detection of protein kinase D-dependent phosphorylation events in nocodazole-treated human cells. Mol Cell Proteomics. 2012;11:160–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freschi L, Vincent AT, Jeukens J et al. The Pseudomonas aeruginosa pan-genome provides new insights on its population structure, horizontal gene transfer, and pathogenicity. Genome Biol Evol. 2019;11:109–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Wei W, Hassan BH et al. A single regulator NrtR controls bacterial NAD+ homeostasis via its acetylation. eLife. 2019;8:e51603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaviard C, Broutin I, Cosette P et al. Lysine succinylation and acetylation in Pseudomonas aeruginosa. J Proteome Res. 2018;17:2449–59. [DOI] [PubMed] [Google Scholar]
- Gaviard C, Cosette P, Jouenne T et al. LasB and CbpD virulence factors of Pseudomonas aeruginosa carry multiple post-translational modifications on their lysine residues. J Proteome Res. 2019;18:923–33. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Padmanabhan B, Anand C et al. Lysine acetylation of the Mycobacterium tuberculosis HU protein modulates its DNA binding and genome organization: acetylation of MtHU by Eis. Mol Microbiol. 2016;100:577–88. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace A, Sahu R, Owen DR et al. Pseudomonas aeruginosa reference strains PAO1 and PA14: a genomic, phenotypic, and therapeutic review. Front Microbiol. 2022;13:1023523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner-Haas F, von Bergen M, Sawers G et al. Changes of the proteome and acetylome during transition into the stationary phase in the organohalide-respiring Dehalococcoides mccartyi strain CBDB1. Microorganisms. 2021;9:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentchel KL, Escalante-Semerena JC. Acylation of biomolecules in prokaryotes: a widespread strategy for the control of biological function and metabolic stress. Microbiol Mol Biol Rev. 2015;79:321–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockenberry AM, Post DMB, Rhodes KA et al. Perturbing the acetylation status of the type IV pilus retraction motor, PilT, reduces Neisseria gonorrhoeae viability. Mol Microbiol. 2018;110:677–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu LI, Chi BK, Kuhn ML et al. Acetylation of the response regulator RcsB controls transcription from a small RNA promoter. J Bacteriol. 2013;195:4174–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- Ishigaki Y, Akanuma G, Yoshida M et al. Protein acetylation involved in streptomycin biosynthesis in Streptomyces griseus. J Proteomics. 2017;155:63–72. [DOI] [PubMed] [Google Scholar]
- Jers C, Ravikumar V, Lezyk M et al. The global acetylome of the human pathogen Vibrio cholerae V52 reveals lysine acetylation of major transcriptional regulators. Front Cell Infect Microbiol. 2018;7:537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Sato Y et al. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Midford PE, Caspi R et al. Pathway size matters: the influence of pathway granularity on over-representation (enrichment analysis) statistics. BMC Genomics. 2021;22:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N, Gautam A, Kumar S et al. Biochemical studies and crystal structure determination of dihydrodipicolinate synthase from Pseudomonas aeruginosa. Int J Biol Macromol. 2011;48:779–87. [DOI] [PubMed] [Google Scholar]
- Kentache T, Jouenne T, Dé E et al. Proteomic characterization of Nα- and Nε-acetylation in Acinetobacter baumannii. J Proteomics. 2016;144:148–58. [DOI] [PubMed] [Google Scholar]
- Kim D, Yu BJ, Kim JA et al. The acetylproteome of Gram-positive model bacterium Bacillus subtilis. Proteomics. 2013;13:1726–36. [DOI] [PubMed] [Google Scholar]
- Kim J-E, Choi J-S, Kim J-S et al. Lysine acetylation of the housekeeping sigma factor enhances the activity of the RNA polymerase holoenzyme. Nucleic Acids Res. 2020;48:2401–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine-Abe A, Kondo N, Kubo S et al. Characterization of lysine acetylation in the peripheral subunit-binding domain of the E2 subunit of the pyruvate dehydrogenase-2-oxoglutarate dehydrogenase hybrid complex from Corynebacterium glutamicum. Biosci Biotechnol Biochem. 2021;85:874–81. [DOI] [PubMed] [Google Scholar]
- Koo H, Park S, Kwak M-K et al. Regulation of gene expression by protein lysine acetylation in Salmonella. J Microbiol. 2020;58:979–87. [DOI] [PubMed] [Google Scholar]
- Kosono S, Tamura M, Suzuki S et al. Changes in the acetylome and succinylome of Bacillus subtilis in response to carbon source. PLoS One. 2015;10:e0131169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers M. Post-translational lysine Ac(et)ylation in bacteria: a biochemical, structural, and synthetic biological perspective. Front Microbiol. 2021;12:757179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D-W, Kim D, Lee Y-J et al. Proteomic analysis of acetylation in thermophilic Geobacillus kaustophilus. Proteomics. 2013;13:2278–82. [DOI] [PubMed] [Google Scholar]
- Lei L, Zeng J, Wang L et al. Quantitative acetylome analysis reveals involvement of glucosyltransferase acetylation in Streptococcus mutans biofilm formation. Environ Microbiol Rep. 2021;13:86–97. [DOI] [PubMed] [Google Scholar]
- Leijonmarck M, Liljas A. Structure of the C-terminal domain of the ribosomal protein L7/L12 from Escherichia coli at 1.7 A. J Mol Biol. 1987;195:555–79. [DOI] [PubMed] [Google Scholar]
- Li D, Ramanathan S, Wang G et al. Acetylation of lysine 7 of AhyI affects the biological function in Aeromonas hydrophila. Microb Pathog. 2020a;140:103952. [DOI] [PubMed] [Google Scholar]
- Li P, Zhang H, Zhao G-P et al. Deacetylation enhances ParB-DNA interactions affecting chromosome segregation in Streptomyces coelicolor. Nucleic Acids Res. 2020b;48:4902–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu S, Su Y et al. Acetylation of PhoP K88 is involved in regulating Salmonella virulence. Infect Immun. 2021;89:e00588–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Gu J, Chen P et al. Purification and characterization of the acetyl-CoA synthetase from Mycobacterium tuberculosis. Acta Biochim Biophys Sin. 2011;43:891–9. [DOI] [PubMed] [Google Scholar]
- Li S, Zhang Q, Xu Z et al. Acetylation of lysine 243 inhibits the oriC binding ability of DnaA in Escherichia coli. Front Microbiol. 2017;8:699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W, Deutscher MP. Post-translational modification of RNase R is regulated by stress-dependent reduction in the acetylating enzyme pka (YfiQ). RNA. 2012;18:37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W, Malhotra A, Deutscher MP. Acetylation regulates the stability of a bacterial protein: growth stage-dependent modification of RNase R. Mol Cell. 2011;44:160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J-H, Tsai C-H, Patel SG et al. Acetylome of Acinetobacter baumannii SK17 reveals a highly-conserved modification of histone-like protein HU. Front Mol Biosci. 2017;4:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima BP, Antelmann H, Gronau K et al. Involvement of protein acetylation in glucose-induced transcription of a stress-responsive promoter: acetylation of RNA polymerase. Mol Microbiol. 2011;81:1190–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima BP, Lennon CW, Ross W et al. In vitro evidence that RNA polymerase acetylation and acetyl phosphate-dependent CpxR phosphorylation affect cpxP transcription regulation. FEMS Microbiol Lett. 2016;363:fnw011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima BP, Thanh Huyen TT, Bäsell K et al. Inhibition of acetyl phosphate-dependent transcription by an acetylatable lysine on RNA polymerase. J Biol Chem. 2012;287:32147–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zheng D, Zhou S et al. VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022;50:D912–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Yang M, Wang X et al. Acetylome analysis reveals diverse functions of lysine acetylation in Mycobacterium tuberculosis. Mol Cell Proteomics. 2014;13:3352–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-T, Pan Y, Lai F et al. Comprehensive analysis of the lysine acetylome and its potential regulatory roles in the virulence of Streptococcus pneumoniae. J Proteomics. 2018;176:46–55. [DOI] [PubMed] [Google Scholar]
- Luu J, Carabetta VJ. Contribution of Nε-lysine acetylation towards regulation of bacterial pathogenesis. mSystems. 2021;6:e00422–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu J, Mott CM, Schreiber OR et al. Nε-lysine acetylation of the histone-like protein HBsu regulates the process of sporulation and affects the resistance properties of Bacillus subtilis spores. Front Microbiol. 2022;12:782815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macek B, Forchhammer K, Hardouin J et al. Protein post-translational modifications in bacteria. Nat Rev Microbiol. 2019;17:651–64. [DOI] [PubMed] [Google Scholar]
- Marakasova E, Ii A, Nelson KT et al. Proteome wide profiling of N-ε-lysine acetylation reveals a novel mechanism of regulation of the chitinase activity in Francisella novicida. J Proteome Res. 2020;19:1409–22. [DOI] [PubMed] [Google Scholar]
- Meng EC, Goddard TD, Pettersen EF et al. UCSF ChimeraX: tools for structure building and analysis. Protein Sci. 2023;32:e4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, Liu P, Wang J et al. Systematic analysis of the lysine acetylome of the pathogenic bacterium Spiroplasma eriocheiris reveals acetylated proteins related to metabolism and helical structure. J Proteomics. 2016;148:159–69. [DOI] [PubMed] [Google Scholar]
- Meyer JG, D'Souza AK, Sorensen DJ et al. Quantification of lysine acetylation and succinylation stoichiometry in proteins using mass spectrometric data-independent acquisitions (SWATH). J Am Soc Mass Spectrom. 2016;27:1758–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen H, McMullan R, Filloux A. The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS. PLoS One. 2011;6:e29113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RA, Bocik WE, Viola RE. Expression and purification of aspartate β-semialdehyde dehydrogenase from infectious microorganisms. Protein Expr Purif. 2002;25:189–94. [DOI] [PubMed] [Google Scholar]
- Nakayasu ES, Burnet MC, Walukiewicz HE et al. Ancient regulatory role of lysine acetylation in central metabolism. mBio. 2017;8:e01894–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak J, Fabrik I, Jurnecka D et al. Bordetella pertussis acetylome is shaped by lysine deacetylase Bkd1. J Proteome Res. 2020;19:3680–96. [DOI] [PubMed] [Google Scholar]
- Okanishi H, Kim K, Masui R et al. Acetylome with structural mapping reveals the significance of lysine acetylation in Thermus thermophilus. J Proteome Res. 2013;12:3952–68. [DOI] [PubMed] [Google Scholar]
- Ouidir T, Cosette P, Jouenne T et al. Proteomic profiling of lysine acetylation in Pseudomonas aeruginosa reveals the diversity of acetylated proteins. Proteomics. 2015;15:2152–7. [DOI] [PubMed] [Google Scholar]
- Pan J, Ye Z, Cheng Z et al. Systematic analysis of the lysine acetylome in Vibrio parahemolyticus. J Proteome Res. 2014;13:3294–302. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin R, Sang Y, Ren J et al. The bacterial two-hybrid system uncovers the involvement of acetylation in regulating of Lrp activity in Salmonella typhimurium. Front Microbiol. 2016;7:1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Xiao W, Zhou C et al. Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Sig Transduct Target Ther. 2022;7:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan R, Schuster M, Bourret RB. Acetylation at Lys-92 enhances signaling by the chemotaxis response regulator protein CheY. Proc Natl Acad Sci USA. 1998;95:4918–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–906. [DOI] [PubMed] [Google Scholar]
- Ravikumar V, Nalpas NC, Anselm V et al. In-depth analysis of Bacillus subtilis proteome identifies new ORFs and traces the evolutionary history of modified proteins. Sci Rep. 2018;8:17246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Sang Y, Qin R et al. Metabolic intermediate acetyl phosphate modulates bacterial virulence via acetylation. Emerg Microbes Infect. 2019;8:55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Sang Y, Tan Y et al. Acetylation of lysine 201 inhibits the DNA-binding ability of PhoP to regulate Salmonella virulence. PLoS Pathog. 2016;12:e1005458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverdy A, Chen Y, Hunter E et al. Protein lysine acetylation plays a regulatory role in Bacillus subtilis multicellularity. PLoS One. 2018;13:e0204687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakatos A, Babunovic GH, Chase MR et al. Posttranslational modification of a histone-like protein regulates phenotypic resistance to isoniazid in mycobacteria. Sci Adv. 2018;4:eaao1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y, Ren J, Qin R et al. Acetylation regulating protein stability and DNA-binding ability of HilD, thus modulating Salmonella typhimurium virulence. J Infect Dis. 2017;216:1018–26. [DOI] [PubMed] [Google Scholar]
- Sherman BT, Hao M, Qiu J et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50:W216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, Athira PJ, Bhardwaj N et al. Acetylation of response regulator protein MtrA in M. tuberculosis regulates its repressor activity. Front Microbiol. 2021;11:516315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Wang G, Malhotra A et al. Reversible acetylation on Lys501 regulates the activity of RNase II. Nucleic Acids Res. 2016;44:1979–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi B, Macek B. Stable isotope labeling by amino acids applied to bacterial cell culture. Methods Mol Biol. 2014;1188:9–22. [DOI] [PubMed] [Google Scholar]
- Sun L, Yao Z, Guo Z et al. Comprehensive analysis of the lysine acetylome in Aeromonas hydrophila reveals cross-talk between lysine acetylation and succinylation in LuxS. Emerg Microbes Infect. 2019;8:1229–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Guo H, Lu G et al. Lysine acetylation regulates the activity of Escherichia coli S-adenosylmethionine synthase. Acta Biochim Biophy Sin. 2016;48:723–31. [DOI] [PubMed] [Google Scholar]
- Thao S, Chen C-S, Zhu H et al. Nε−lysine acetylation of a bacterial transcription factor inhibits its DNA-binding activity. PLoS One. 2010;5:e15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium, Carbon S, Douglass E et al. The Gene ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49:D325–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium . UniProt: the Universal Protein knowledgebase in 2023. Nucleic Acids Res. 2023;51:D523–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S, Guo S-J, Chen C-S et al. YcgC represents a new protein deacetylase family in prokaryotes. eLife. 2015;4:e05322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker AC, Escalante-Semerena JC. Acetoacetyl-CoA synthetase activity is controlled by a protein acetyltransferase with unique domain organization in Streptomyces lividans. Mol Microbiol. 2013;87:152–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Türkowsky D, Esken J, Goris T et al. A retentive memory of tetrachloroethene respiration in Sulfurospirillum halorespirans—involved proteins and a possible link to acetylation of a two-component regulatory system. J Proteomics. 2018;181:36–46. [DOI] [PubMed] [Google Scholar]
- Umehara T, Kosono S, Söll D et al. Lysine acetylation regulates alanyl-tRNA synthetase activity in Escherichia coli. Genes. 2018;9:473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDrisse CM, Escalante-Semerena JC. In Streptomyces lividans, acetyl-CoA synthetase activity is controlled by O-serine and Nɛ -lysine acetylation. Mol Microbiol. 2018;107:577–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi M, Anyango S, Deshpande M et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkat S, Chen H, McGuire P et al. Characterizing lysine acetylation of Escherichia coli type ii citrate synthase. FEBS J. 2019;286:2799–808. [DOI] [PubMed] [Google Scholar]
- Venkat S, Chen H, Stahman A et al. Characterizing lysine acetylation of isocitrate dehydrogenase in Escherichia coli. J Mol Biol. 2018;430:1901–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkat S, Gregory C, Gan Q et al. Biochemical characterization of the lysine acetylation of tyrosyl-tRNA synthetase in Escherichia coli. ChemBioChem. 2017a;18:1928–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkat S, Gregory C, Sturges J et al. Studying the lysine acetylation of malate dehydrogenase. J Mol Biol. 2017b;429:1396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnolle O, Xu H, Blanchard JS. Mechanism and regulation of mycobactin fatty acyl-AMP ligase FadD33. J Biol Chem. 2013;288:28116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virág D, Dalmadi-Kiss B, Vékey K et al. Current trends in the analysis of post-translational modifications. Chromatographia. 2020;83:1–10. [Google Scholar]
- Wang M-M, You D, Ye B-C. Site-specific and kinetic characterization of enzymatic and nonenzymatic protein acetylation in bacteria. Sci Rep. 2017;7:14790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang F, Bao X et al. Systematic analysis of lysine acetylome reveals potential functions of lysine acetylation in Shewanella baltica, the specific spoilage organism of aquatic products. J Proteomics. 2019;205:103419. [DOI] [PubMed] [Google Scholar]
- Wei W, Liu T, Li X et al. Lysine acetylation regulates the function of the global anaerobic transcription factor FnrL in Rhodobacter sphaeroides. Mol Microbiol. 2017;104:278–93. [DOI] [PubMed] [Google Scholar]
- Weinert BT, Satpathy S, Hansen BK et al. Accurate quantification of site-specific acetylation stoichiometry reveals the impact of sirtuin deacetylase CobB on the E. coli acetylome. Mol Cell Proteomics. 2017;16:759–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Wang X, Zeng J et al. Proteome-wide lysine acetylation profiling of the human pathogen Mycobacterium tuberculosis. Int J Biochem Cell Biol. 2015;59:193–202. [DOI] [PubMed] [Google Scholar]
- Xu J-Y, Xu Z, Liu X et al. Protein acetylation and butyrylation regulate the phenotype and metabolic shifts of the endospore-forming Clostridium acetobutylicum. Mol Cell Proteomics. 2018;17:1156–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J-Y, Zhao L, Xu Y et al. Dynamic characterization of protein and posttranslational modification levels in mycobacterial cholesterol catabolism. mSystems. 2020;5:e00424–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Li F, Lv L et al. Oxidative stress activates SIRT2 to deacetylate and stimulate phosphoglycerate mutase. Cancer Res. 2014;74:3630–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Sha W, Liu Z et al. Lysine acetylation of DosR regulates the hypoxia response of Mycobacterium tuberculosis. Emerg Microbes Infect. 2018;7:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhang H, Guo Z et al. Global insights into lysine acylomes reveal crosstalk between lysine acetylation and succinylation in Streptomyces coelicolor metabolic pathways. Mol Cell Proteomics. 2021;20:100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Ji Q-Q, Yan W et al. Acetylation of lysine ϵ-amino groups regulates aminoacyl-tRNA synthetase activity in Escherichia coli. J Biol Chem. 2017;292:10709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You D, Yin B-C, Li Z-H et al. Sirtuin-dependent reversible lysine acetylation of glutamine synthetases reveals an autofeedback loop in nitrogen metabolism. Proc Natl Acad Sci USA. 2016;113:6653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B-J, Kim J-A, Moon J-H et al. The diversity of lysine-acetylated proteins in Escherichia coli. J Microbiol Biotechnol. 2008;18:1529–36. [PubMed] [Google Scholar]
- Zhang B-Q, Bu H-L, You D et al. Acetylation of translation machinery affected protein translation in E. coli. Appl Microbiol Biotechnol. 2020;104:10697–709. [DOI] [PubMed] [Google Scholar]
- Zhang J, Sprung R, Pei J et al. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Proteomics. 2009;8:215–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Zheng S, Yang JS et al. Comprehensive profiling of protein lysine acetylation in Escherichia coli. J Proteome Res. 2013a;12:844–51. [DOI] [PubMed] [Google Scholar]
- Zhang Q-F, Gu J, Gong P et al. Reversibly acetylated lysine residues play important roles in the enzymatic activity of Escherichia coli N-hydroxyarylamine O-acetyltransferase. FEBS J. 2013b;280:1966–79. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yao Z, Tang H et al. The lysine acetylation modification in the porin Aha1 of Aeromonas hydrophila regulates the uptake of multidrug antibiotics. Mol Cell Proteomics. 2022;21:100248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhou A, Li S et al. Reversible lysine acetylation is involved in DNA replication initiation by regulating activities of initiator DnaA in Escherichia coli. Sci Rep. 2016;6:30837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PAO1 acetylomics dataset discussed in this paper has been submitted to the ProteomeXchange Consortium via the PRIDE database. You can find the data using the identifier: PXD051454.
The list of experimentally assessed acetylation sites was compiled from sources in the public domain. You can find the data in the following references: Ramakrishnan et al. (1998), Crosby et al. (2010), Thao et al. (2010), Wang et al. (2010), Li et al. (2011, 2017, 2020a, b, 2021), Liang et al. (2011), Lima et al. (2011, 2012, 2016), Liang and Deutscher (2012), Hu et al. (2013), Tucker and Escalante-Semerena (2013), Vergnolle et al. (2013), Zhang et al. 2013b, 2016, 2020, 2022), Xu et al. (2014), Fraiberg et al. (2015), Tu et al. (2015), Xie et al. (2015), Carabetta et al. (2016, 2019), Qin et al. (2016), Ren et al. (2016, 2019), Song et al. (2016), Sun et al. (2016, 2019), You et al. (2016), Baron and Eisenbach (2017), Chen et al. (2017), Ishigaki et al. (2017), Liao et al. (2017), Nakayasu et al. (2017), Sang et al. (2017), Wang, Ye et al. (2017), You et al. (2016) , Wei et al. (2017), Venkat et al. 2017a, b, 2018, 2019), Bi et al. (2018), Davis et al. (2018), Hockenberry et al. (2018), Reverdy et al. (2018), Sakatos et al. (2018), Umehara et al. (2018), VanDrisse and Escalante-Semerena (2018), Yang et al. (2018), Gao et al. (2019), Ren et al. (2019), Kim et al. (2020), Komine-Abe et al. (2021), Singh et al. (2021), Barlow and Tsai (2022), Cai et al. (2022), Fang et al. (2022), Luu et al. (2022), Di et al. (2023).
Similarly, the acetylomics datasets, excluding the PAO1 dataset, were derived from sources in the public domain. You can find the data in the following references: Kim et al. (2013), Lee et al. (2013), Okanishi et al. (2013), Castaño-Cerezo et al. (2014), Pan et al. (2014), Kosono et al. 2015), Ouidir et al. (2015), Xie et al. (2015), Carabetta et al. (2016), Chen et al. (2016, 2017), Kentache et al. (2016), Birhanu et al. (2017), Weinert et al. (2017), Gaviard et al. (2018), Jers et al. (2018), Liu et al. (2018), Ravikumar et al. (2018), Türkowsky et al. (2018), Xu et al. (2018, 2020), Yang et al. (2018, 2021), Sun et al. (2019), Wang et al. (2019), Marakasova et al. (2020), Novak et al. (2020), Lei et al. (2021).
The remaining data underlying this article are available in the article and in its online supplementary material.