Abstract
Systemic sclerosis (SSc) is a life-threatening autoimmune disease characterized by widespread fibrosis in the skin and several internal organs. Nudix Hydrolase 21 (NUDT2 or CFIm25) downregulation in fibroblasts is known to play detrimental roles in both skin and lung fibrosis. This study aims to investigate the upstream mechanisms that lead to NUDT21 repression in skin fibrosis. We identified transforming growth factor β (TGFβ1) as the primary cytokine that downregulated NUDT21 in normal skin fibroblasts. In the bleomycin-induced dermal fibrosis model, consistent with the peak activation of TGFβ1 at the late fibrotic stage, NUDT21 was downregulated at this stage, and delayed NUDT21 knockdown during this fibrotic phase led to enhanced fibrotic response to bleomycin. Further investigation suggested TGFβ downregulated NUDT21 through microRNA (miRNA) 181a and 181b induction. Both miR-181a and miR-181b were elevated in bleomycin-induced skin fibrosis in mice and primary fibroblasts isolated from SSc patients, and they directly targeted NUDT21 and led to its downregulation in skin fibroblasts. Functional studies demonstrated that miR-181a and miR-181b inhibitors attenuated bleomycin-induced skin fibrosis in mice in association with decreased NUDT21 expression, while miR-181a and miR-181b mimics promoted bleomycin-induced fibrosis. Overall, these findings suggest a novel role for miR-181a/b in SSc pathogenesis by repressing NUDT21 expression.
Keywords: alternative polyadenylation, cleavage factor Im 25, dermal fibrosis, fibroblasts, miR-181a, miR-181b, systemic sclerosis, transforming growth factor β
1 |. INTRODUCTION
Systemic Sclerosis (SSc, scleroderma) is a complex and multifaceted autoimmune disease that affects various organs including skin, lungs, gastrointestinal tract, heart, and other internal organs, leading to significant functional impairment and complications. The hallmark of disease is widespread fibrosis, which involves excessive accumulation of collagen and other connective tissue components.
With a standardized mortality ratio of 3.5,1 SSc has the highest mortality among major rheumatic diseases.2,3 This high disease burden is in part driven by only modest responses of its fibrotic features to immunosuppressive agents.4,5 While some treatments have been approved for specific manifestations of SSc, such as nintedanib6 and tocilizumab7 for SSc-related interstitial lung disease (SSc-ILD), there is currently no FDA-approved medication that effectively targets both lung and skin fibrosis in SSc.8 Despite the extensive studies carried out to investigate the biomarkers and pathways leading to the dysregulated fibroblasts and immune cells, the mechanisms involved in uncontrolled fibrosis remain elusive. This fragmented understanding of pathogenesis poses a significant obstacle to developing a disease-modifying therapy for SSc. A deeper understanding of the underlying mechanisms driving fibrosis in SSc is essential for advancing our knowledge to manage and treat this complex disease. Moreover, understanding the fibrotic mechanisms in a prototypic systemic fibrotic disease such as SSc can provide insights into other fibrotic diseases that are a common cause of mortality.
Previous studies from our group suggest that Nudix Hydrolase 21 (NUDT21, also named CFIm25) mediated alternative polyadenylation (APA) plays a role in SSc pathogenesis. APA is a posttranscriptional RNA regulation that is implicated in several cellular and biological processes. APA allows attachment of poly(A) tails at alternative sites of genes containing more than one polyadenylation site (PAS), including many profibrotic mediators, and encodes transcripts with different lengths of 3′UTR or in rare situations, even different transcripts.9,10 NUDT21 is a key component of cleavage factor Im that our group and others identified as a master APA regulator whose depletion induced 3′UTR shortening of target genes. Transcripts with shorter 3′UTR escape miRNA or 3′UTR binding protein-mediated repression and generally have enhanced protein translation. Previous studies from our group show that NUDT21 is downregulated in myofibroblasts of SSc skin and fibrotic lungs, and its fibroblast-specific repression promotes bleomycin-induced skin and lung fibrosis in mice by targeting genes involved in important fibrotic pathways including transforming growth factor receptor (TGFBR1), collagen I alpha 1 (COL1A1) and Secreted Protein Acidic and Cysteine Rich (SPARC).11,12 These findings highlight the potential role of NUDT21-loss in amplifying tissue fibrosis. Therefore, understanding the upstream pathways leading to decreased NUDT21 expression in skin fibrosis is important to target NUDT21 for therapeutic interventions. Previous studies from our group have demonstrated miR-203 as a TGFβ1-inducible miRNA targeting NUDT21 for downregulation in lung fibroblasts.13 However, mechanisms by which NUDT21 is downregulated in skin fibrosis are not reported. The primary goal of this study is to identify the upstream factors involved in NUDT21 downregulation and demonstrate the therapeutic potential of targeting these factors for the treatment of skin fibrosis in an animal model of skin fibrosis through NUDT21 rescue.
2 |. MATERIALS AND METHODS
2.1 |. Materials
Wild-type C57BL/6J and Col1a1Cre (B6.Cg-Tg(Col1a1cre/ERT2)1Crm/J) mice were purchased from Jackson laboratories. Nudt21f/f (also named CFIm25f/f) mouse line that has floxed alleles designed to delete exons 2 and 3 of the Nudt21 gene was a gift from Dr. Michael R. Blackburn and was originally generated by Ozgene. Antibodies against beta-actin (ACTB) and fibronectin (FN1) were purchased from Sigma-Aldrich. Antibodies against NUDT21 and COL1A1 were from Proteintech. Control, miR-181a/-181b mimics, and inhibitors were obtained from Qiagen. RT-qPCR kit and primers for reverse transcription and detection of transcript levels were purchased from Gendepot. RT-qPCR kit and primers for reverse transcription and detection of miRNA levels were purchased from Qiagen. 293T cells were purchased from ATCC. Dulbecco’s Modified Eagle Medium (DMEM) was purchased from Cytiva. Fetal bovine serum and antibiotics for cell culture were from Gendepot. Recombinant TGFβ1, IL4, IL6, IL6, and PDGF were purchased from R&D systems.
2.2 |. Human samples
All human skin biopsy samples and dermal fibroblasts were obtained in the Rheumatology Clinic at UTHealth-Houston (Table 1 and Supplemental Table S1). All patients included in this study fulfilled the 2012 ACR/EULAR Classification Criteria14 and had diffuse cutaneous involvement. The healthy control subjects had no personal or family medical history of autoimmune diseases. Skin biopsies were obtained from the arm of the study participants. Biopsy samples were collected using a 3-mm punch. Written informed consent was obtained from all subjects, and the study protocol was approved by the institutional review boards at UTHealth-Houston.
Table 1.
Demographic and clinical characteristics of participants
| Systemic sclerosis (n=27) | Control (n=20) | |
|---|---|---|
| Age, mean (SD) | 49.63 (13.9) | 49.5 (12.89) |
| Female, n (%) | 13 (48.1%) | 10 (50%) |
| Race/ethnicity | ||
| White | 15 (55.6%) | 14 (70%) |
| African American | 7 (25.9%) | 5 (25%) |
| Hispanic | 5 (18.5%) | 1 (5%) |
| Disease duration in years, mean (SD) | 2.16 (1.07) | NA |
| Diffuse cutaneous involvement, n (%) | 27 (100%) | NA |
| Anti-topoisomerase I antibodies | 6 (24%) | N/A |
| Anti-RNA polymerase III | 10 (37%) | N/A |
| Anticentromere antibodies | 0 (0%) | NA |
| Skin score at the site of biopsy, mean (SD) | 1.51 (0.85) | NA |
2.3 |. Mouse experiments
All mice used in this study are on the C57BL/6J background. Mice were housed in pathogen-free conditions at UTHealth McGovern Medical School, Houston, TX. All experiments performed here were approved by the UTHealth Animal Welfare Committee. Col1a1Cre-Nudt21f/f mice were generated by crossing the Col1a1Cre mice with Nudt21f/f mice that have floxed alleles designed to delete exons 2 and 3 of the Nudt21 gene. For the delayed Cre activation experiment, 6–8-week-old female Col1a1Cre or Col1a1Cre-Nudt21f/f mice were injected with repeated s.c. bleomycin (0.02 U/mice/day, injected five times a week for 4 weeks). One week after the initial bleomycin injection, mice were i.p. injected with 75 mg/kg/day tamoxifen for 5 days to induce Cre recombination and NUDT21 depletion. On day 28 after the first bleomycin injection, mice were euthanized by carbon dioxide asphyxiation followed by cervical dislocation, and skin samples were collected for RNA, protein, and histological analysis.
For the miRNA experiments, 6–8 week-old female WT C57BL/6J were injected with subcutaneous (s.c.) bleomycin (0.02 U/mice/day) 6 days a week for 4 weeks. Ten days after the initial bleomycin injection, mice were s.c. injected with LNA control or miR-181a or miR-181b mimic/inhibitors (0.5 nmole in 100 μL PBS per injection per mouse) at the same bleomycin-injected spots twice a week until day 28 for analysis.
2.4 |. Masson’s trichrome staining
Skin biopsy samples collected from mice (6 mm punch) or humans (3 mm punch) were dehydrated, paraffin-embedded, and sectioned (4 μm). For Masson’s trichrome staining, paraffin-embedded skin sections were rehydrated in a series of alcohol and stained with Masson’s Trichrome Kit (Sigma-Aldrich). Dermal thickness was defined as the distance between the epidermal–dermal junction to the dermal–adipose layer junction and was measured blindly at six randomly selected sites/microscopic fields in each skin sample.15 Data were presented as average skin thickness ± Mean square error.
2.5 |. Fibroblast culture and transfection
Primary human dermal fibroblasts were isolated using an outgrowth model from skin punch from healthy donors and SSc patients. Isolated fibroblasts were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Sigma-Aldrich) containing 10% fetal bovine serum (FBS) and 1% antibiotics to avoid contamination, and mycoplasma infection was tested using the MycoAlert™ Mycoplasma Detection Kit (Lonza). Cell culture was maintained at 37°C in a humidified 5% carbon dioxide atmosphere.
For miRNA overexpression or inhibition experiment, fibroblasts were transfected with 50 nM control or miR-181a/181b mimic or inhibitors (Qiagen) using Lipofectamine® RNAiMAX (ThermoFisher Scientific) on day 0 and day 1, and RNA and protein were collected on day 2 for analysis.
2.6 |. MiRNA sequencing (miR-seq) and data analysis
A 3-millimeter punch biopsy was immersed in RNAlater solution (Qiagen). RNA was extracted using miRNeasy Mini kits (Qiagen). RNA integrity was assessed using the Bioanalyzer 2100 (Agilent). Small RNA sequencing was performed by Novogene at a minimum of 10 million reads per sample.
2.7 |. Real-time qRT PCR
For mRNA PCR, total RNA was extracted using RNeasy Mini Kit (Qiagen) and reverse-transcribed using the Gendepot amfiRivert cDNA Synthesis Platinum Master Mix. Real-time PCR was performed using the Gendepot SybrGreen Master Mix, and under the CFX 384 Real-Time PCR system (BioRad), and data were quantified using the relative Ct method and presented as a mean ratio to β-actin or 18s rRNA.
The APA of target genes was determined using a previously described PCR-based method.16 For each candidate gene, two pairs of primers were designed with one targeting the open reading frame to represent the total transcript level and the other targeting sequences just before the dPAS to detect long transcripts that used the dPAS. The percentage of dPAS usage was calculated as . Data were presented as fold changes normalized to control by calculating . A negative value indicates that the mRNA has 3′UTR shortening compared to controls, and this approach has been used to quantify APA.16 The primers used for this study are listed in Table 2.
Table 2:
Primer sequences
| Gene Name | Forward | Reverse |
|---|---|---|
| Human Actin | CATGTACGTTGCTATCCAGGC | CTCCTTAATGTCACGCACGAT |
| Human COL1A1 | GTGCGATGACGTGATCTGTGA | CGGTGGTTTCTTGGTCGGT |
| Human COL1A1 Long | GTGAGGGAGACAGACACCTG | TTGCTAGTTTACCGTTCAGAAGT |
| Human NUDT21 | GGTCACTCAGTTCGGCAACAA | CTCATGCGCTGAAATCTGGC |
| Mouse Actin | GGCTGTATTCCCCTCCATCG | CCAGTTGGTAACAATGCCATGT |
| Mouse Col1a1 | GCTCCTCTTAGGGGCCACT | CCACGTCTCACCATTGGGG |
| Mouse Col1a1 Long | GGCAATGCTGAAATGTCCCA | ACAGTCCAAGAACCCCATGT |
| Mouse Col1a2 | AAGGGTGCTACTGGACTCCC | TTGTTACCGGATTCTCCTTTGG |
| Mouse Fn1 | GCTCAGCAAATCGTGCAGC | CTAGGTAGGTCCGTTCCCACT |
| Mouse Nudt21 | AGGAGGCTGACATGGATTGTT | TCTTCTCCTGGGTTAAGTTCCC |
| Mouse Tgfbr1 | TCTGCATTGCACTTATGCTGA | AAAGGGCGATCTAGTGATGGA |
| Mouse Tgfbr1 Long | AGATAGGTCATGGTTAATGTGGC | ATACTACACTTTGGGGCTGGG |
| Mouse/Human 18S | GTAACCCGTTGAACCCCATT | CCATCCAATCGGTAGTAGCG |
For miRNA PCR, total RNA was reversed-transcribed using the Qiagen miRCURY LNA RT Kit. PCR was carried out using the miRCURY LNA miRNA PCR primers targeting mouse or human miR-203, miR-181a, miR181b, miR-539, and miR-216. MiRCURY LNA SYBR Green PCR kit was used for preparing the reaction mix, and all PCR was performed on a CFX 384 Real-Time PCR system. Data were normalized to the levels of U6 snRNA or RNU1A1.
2.8 |. MiRNAscope
The miRNAscope staining was employed using probes that recognize both miR-181a and miR-181b to visualize miR-181 in the tissue samples. Initially, the tissue sections were deparaffinized by sequential washes in xylene and graded ethanol solutions. Samples are subsequently post-fixed by submerging in 12% formaldehyde for 120 min. Once samples have been fixed, a 10 min H202 incubation at RT is performed. Antigen retrieval was then achieved by incubating the sections in proprietary target retrieval reagents at 98°C for 15 min. The tissue sections were then washed with H20 and ethanol and allowed to dry overnight. The tissue sections were then incubated with a protease solution at 40°C for 30 min in a humidity control tray. Samples were washed with H20 and then hybridized with an RNAscope control probe or probe specific to miR-181a for 120 min at 40°C in a humidity control tray. After hybridization, the sections were washed to remove unbound probes. Signal amplification was achieved by sequential incubation in a humidity control tray with three preamplifiers at 40°C for the following time intervals: 15′, 15′, 30′. This is followed by sequential incubation in a humidity control tray with three AP-labeled probes. The first incubation was at 40°C for 15 min, followed by two incubations at RT for 30′ and 15′. Signal detection was achieved using the chromogenic substrate FAST RED for 10′ at RT in a humidity control tray. Finally, the sections were counterstained with 50% hematoxylin for 120 s, washed, dehydrated, and coverslipped. Images were captured using a Nikon microscope equipped with an appropriate camera. The % of miR-181a positively stained cells was blindly counted.
2.9 |. Plasmids construction and dual-luciferase assay
The psi-CHECK2 plasmids containing the wild-type NUDT21 3′UTR (psi-NUDT21) were gifted from Dr. Eric Wagner’s lab. Briefly, the human NUDT21 3′UTR were PCR amplified from 293 T cDNA and digested with XhoI and NotI. Digested PCR products were gel purified and inserted into an empty psi-CHECK2 plasmid between the Xho1 and Not1 sites. The miR-181a/b binding site mutations were introduced to the NUDT21 3′UTR using the Q5® Site-Directed Mutagenesis Kit (New England Biolabs). The miR-181a/b binding site at 328–334 was mutated from TGAATGT to TCCATGT, and at 1946–1953 was mutated from TGAATGTA to TGAGGGTA. All of the constructed plasmids were sequenced by GENEWIZ to confirm the sequence of WT and 181m NUDT21 3′UTR.
For dual-luciferase analysis, HEK293T cells were cultured in DMEM supplemented with 10% FBS and 1% antibiotics and plated in a 96-well plate. When cells reached ~60%–80% confluence, cells were transfected with 50 ng/well psi-CHECK2, psi-NUDT21 WT or psi-NUDT21 181mut plasmids, together with 50 nM control, miR-181a or miR-181b mimics using Lipofectamine 2000 according to the manufacturer’s instruction. Forty-eight hours after transfection, the Firefly and Renilla luciferase activities were measured using the Promega Dual-Glo® Luciferase Assay System and a Tecan Infinite® 200 PRO plate reader.
2.10 |. Sircol collagen assay
Freshly collected mouse skin was weighed and homogenized in 0.5M acetic acid. For each milligram of mouse skin, 0.1 mg of pepsin (Sigma-Aldrich) were added. Collagen was released by rocking the skin samples in Pepsin overnight at room temperature. Digested skin samples were centrifuged, and the supernatant was collected to determine the collagen concentration using the Sircol Soluble Collagen Assay kit (Biocolor). The final data were normalized to the wet skin weight.
2.11 |. Statistical analysis
The statistical analysis was performed using GraphPad Prism 9. Results are expressed as the mean ± mean standard error (MSE). For comparison between the two groups, data were analyzed using the student’s t-test. For comparisons among more than two groups, one-way or two-way ANOVA followed by Dunnett’s multiple comparisons test was used to determine whether there was any statistical difference among groups. The number of asterisks represents the degree of significance for the p-value. p-values less than .05 were considered significant
3 |. RESULTS
3.1 |. TGFβ1 downregulates NUDT21 in dermal fibroblasts
To identify the upstream factors leading to NDUT21 downregulation, we first screened several cytokines and growth factors that are known to induce fibrotic activity in fibroblasts including TGFβ1, interleukin 4, −6, and −13 (IL4, IL6, and IL13), and platelet-derived growth factor (PDGF) (Figure 1A). TGFβ1 most strongly downregulated NUDT21 expression in normal dermal fibroblasts. Consistent with previously reported decreased NUDT21 in skin fibroblasts from SSc patients compared to normal donors,11 TGFβ1 cytokine levels were significantly elevated in the supernatant of primary dermal fibroblasts from SSc patients (n = 9) than those from healthy controls (Figure 1B and Supplemental Table S1). Therefore, the upregulation of TGFβ may account for the aforementioned NUDT21 downregulation in SSc fibroblasts.13 For these reasons, we focused on the TGFβ1 pathway for further investigation. To study the dynamics of TGFβ1-mediated NUDT21 repression, we exposed normal human dermal fibroblasts to TGFβ1 for various lengths of time. Specifically, TGFβ1 downregulated NUDT21 protein (Figure 1C) and mRNA (Figure 1D) expression after 48-h exposure; and the levels of NUDT21 remained low through 96-h exposure. We have previously identified COL1A1 as a NUDT21 target and shown its 3′UTR shortening in fibroblasts with NUDT21 repression.11 Interestingly, 3′UTR shortening of COL1A1 was also observed in a time-dependent manner in response to TGFβ1 treatment (Figure 1E). This result is consistent with decreased NUDT21 expression upon TGFβ treatment. Taken together, these findings suggest that TGFβ1 upregulation in SSc fibroblasts suppresses NUDT21 expression.
FIGURE 1.
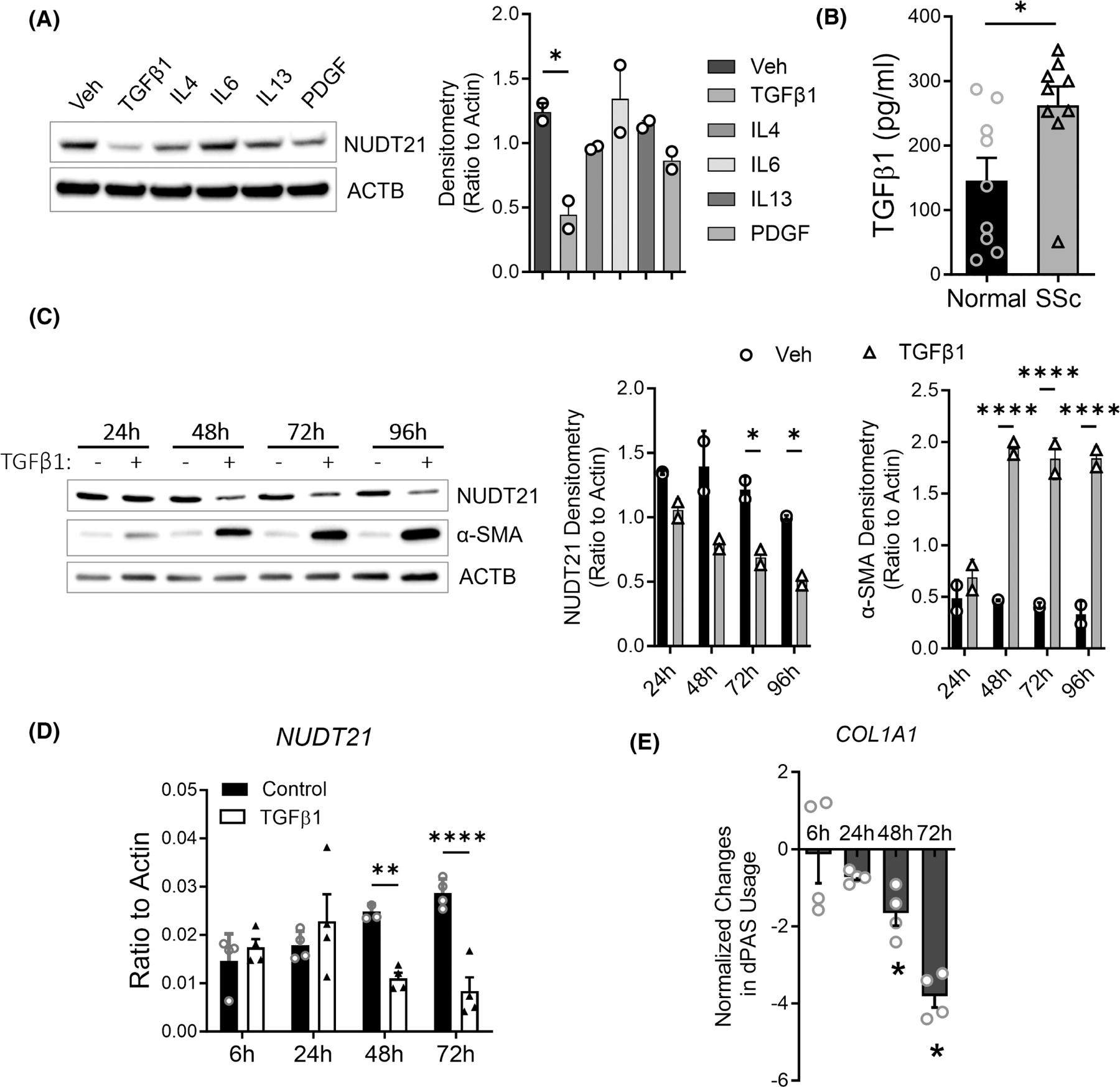
Transforming growth factor beta 1 (TGFβ1) downregulates NUDT21 in dermal fibroblasts. (A) Left panel: Western blot of NUDT21 and β-Actin (ACTB) was performed using the protein lysate of normal human dermal fibroblasts treated with PBS, 5 ng/mL TGFβ1, 10 ng/mL interleukin 4 (IL4), 60 ng/mL IL6 + interleukin 6 receptor (IL6R), 10 ng/mL IL13, or 30 μL/mL platelet-derived growth factor (PDGF) for 2 days. Right panel: Densitometry analysis of the Western blot image. (B) The levels of TGFβ1 in the supernatant of cultured normal or SSc dermal fibroblasts were determined using ELISA. (C) Left panel: The protein levels of NUDT21 were determined in normal human dermal fibroblasts treated with 5 ng/mL TGFβ1 for different hours. Right panels: Densitometry analysis of the Western blot image. (D) The transcript levels of NUDT21 in normal human dermal fibroblasts treated with 5 ng/mL TGFβ1 for different hours. (E) The normalized percentage of distal polyadenylation site (dPAS) usage for COL1A1 was determined using PCR methods, and data were presented as Log (% long transcript in TGFβ1 cells/% long transcript in control) ± mean square error (MSE). p-values were calculated using a two-tailed student t-test for panel B, ANOVA followed by Dunnett’s multiple comparisons test for panels A, C, and D, and one sample t-test versus 0 for panel C. *p < .05; **p < .01; ****p < .0001.
3.2 |. Identification of microRNAs targeting NUDT21
Next, we investigated the signaling cascade by which TGFβ1 induces NUDT21 downregulation. For direct targets, TGFβ1 can normally induce transcript changes as fast as 3–6 h (h).17 However, the transcript levels of NUDT21 were not decreased until 2 days after TGFβ1 treatment (Figure 1D), suggesting that TGFβ1 suppresses NUDT21 expression by an indirect mechanism other than directly regulating its promoter activity. NUDT21 has a very long 3′UTR (~3.5 KD), and thus it is likely to be targeted by miRNAs. TargetScan identified several miRNAs (e.g., miR-181a, miR-181b, miR-23a, miR-203, miR-27b, miR-539) having at least two binding sites within the NUDT21 3′UTR. Previous studies from our group have demonstrated miR-203 as a TGFβ1-inducible miRNA targeting NUDT21 for downregulation in lung fibroblasts.13 To identify the miRNAs that are involved in TGFβ1-mediated NDUT21 repression in skin fibroblasts, we first performed miRNA sequencing of dermal fibroblasts obtained from 27 patients with early diffuse SSc and 20 age-, race-, and gender-matched controls. The demographic and clinical characteristics of the study participants are shown in Table 1. MiR-181b-5p was identified as the only significantly upregulated NUDT21-binding miRNA in SSc fibroblasts. Of note, miR-181a-5p was also numerically elevated but the difference did not reach statistical significance (p-value = .11) (Figure 2A), while the other NUD21 binding miRNAs based TargetScan search including miR-203a-3p, miR-539-5p, and miR-216a-5p were not differentially expressed (Supplemental Figure S1). Next, we performed RNAscope using a probe that recognizes both miR-181a and miR-181b. Consistent with SSc fibroblast results, RNAscope also demonstrated a significant increase in the percentage of miR-181-positive fibroblasts in SSc skin (Figure 2B). To confirm whether similar miRNA upregulation could be observed in a murine model of skin fibrosis, we measured the levels of these Nudt21 targeting miRNAs in a repetitive subcutaneous (s.c.) bleomycin-induced skin fibrosis model. In this model, wild-type C57/BL6 mice were s.c. injected at the back of the skin with 0.02 U/day bleomycin 6 days a week for 4 weeks. And the skin was analyzed on day 28. MiR-181a-5p, miR-181b-5p (Figure 2C), and miR-539-5p (Supplemental Figure S1) were significantly increased in day-21 bleomycin-treated murine skin. However, only miR-181a and miR-181b (Figure 2D) but not the other NUDT21 binding miRNAs (Supplemental Figure S1) were also increased in normal skin fibroblasts (n = 5) treated with TGFβ1 for 6 hours. Because miR-181a and miR-181b are consistently upregulated in both human and mouse fibrotic skin and by TGFβ1, we focused on these two miRNAs for the follow-up studies.
FIGURE 2.
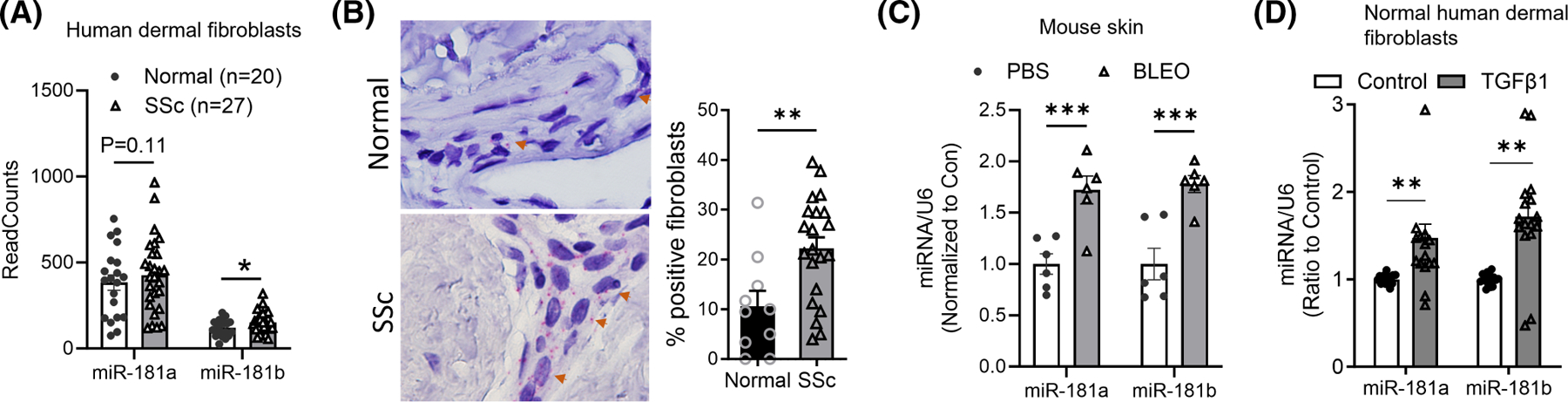
Identification of microRNAs targeting NUDT21 in skin fibrosis. (A) MicroRNA sequencing was performed using 20 normal and 27 SSc primary human dermal fibroblasts. The sequencing results of predicted miR-181a and miR-181b were presented. (B) RNAscope was performed to determine the levels of miR-181a and miR-181b in the skin of 10 normal and 20 SSc donors (left panel). The percentage of miR-181-positive fibroblasts was blindly counted (right panel). (C) Six–eight-week-old female WT C57/BL6 mice were s.c. injected with PBS or 0.02 U/day bleomycin five times a week for 3 weeks, skin samples were collected on day 21 for analysis. The levels of miRNAs were determined using the miRCURY LNA miRNA PCR system and data were normalized to the levels of U6 snRNA. N = 6. (D) MiRNA levels were determined in primary normal human dermal fibroblasts treated with vehicle or 5 ng/mL TGFβ1 for 6 h (n = 5). The graph represents means ± MSE. p-value was calculated using two-tailed student t-tests and adjusted with the Bonferroni method. *p < .05; **p < .01; ***p < .001.
3.3 |. MiR-181a and miR-181b target NUDT21 for downregulation
To determine whether miR-181a and miR-181b directly repress NUDT21 expression in dermal fibroblasts, we first overexpressed miR-181a and miR-181b by transfecting the normal human dermal fibroblasts with miR-181a and miR-181b mimics. Previously, miR-203 was demonstrated to repress NUDT21 expression in lung fibroblasts,13 and thus we included miR-203 as a positive control. NUDT21 protein levels were repressed by all three miRNAs but to the highest extent by miR-181b (Figure 3A). Next, to understand whether miR-181a and miR-181b suppress NUDT21 by directly binding to NUDT21 3′UTR, the NUDT21 WT 3′UTR or NUDT21 3′UTR with miR-181a and miR-181b binding site mutation (181mut) were cloned into the dual-luciferase plasmid psi-check-2 (Promega) that contains both Firefly luciferase and Renilla luciferase cDNAs in the same reporter system (Figure 3B). This design allowed intra-plasmid normalization of transfection and eliminated problems observed with co-transfection of two different plasmids. Then, miRNAs were overexpressed in 293 T cells transfected with psi-check2 or psi-check2-NUDT21 plasmids. Significant repression of NUDT21 wild-type 3′UTR luciferase was detected in miR-181a overexpressed fibroblasts, and to a greater extent, in miR-181b overexpressed fibroblasts (Figure 3C). However, miR-181a and miR-181b mimics failed to repress the luciferase activity in plasmids cloned with mutated NUDT21 3′UTR, suggesting a direct binding of miR-181a/-181b at NUDT21 3′UTR. In addition, miR-181a and miR-181b inhibitors were able to partially attenuate TGFβ1 induced NUDT21 downregulation (Figure 3D), suggesting that their induction was implicated in TGFβ1-mediated NUDT21 repression. Overall, these findings indicate that both miR-181a and miR-181b suppress NUDT21 expression through 3′UTR binding.
FIGURE 3.
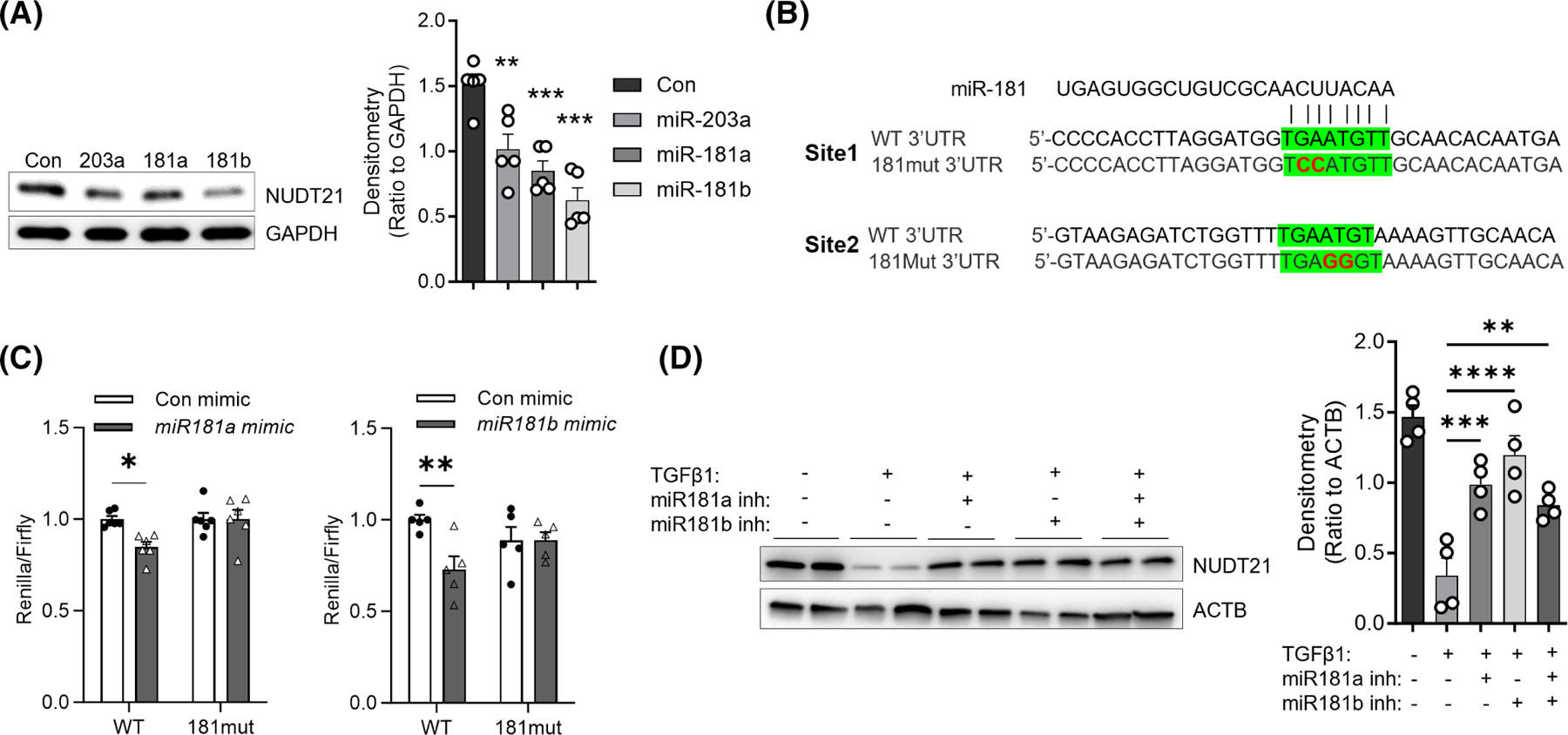
MiR-181a and miR-181b target NUDT21 for downregulation. (A) Left panel: The protein levels of NUDT21 and ACTB were determined using Western blot in normal skin fibroblasts transfected with different miRNA mimics (50 nM in total) for 48 h. Right panel: Densitometry analysis of the Western blot image. n = 5 fibroblast lines from different donors. (B) Sequence of wild-type NUDT21 3′UTR, and mutated NUDT21 3′UTR (181mut 3′UTR) that has both miR-181a/b binding sites mutated (red fonts). Green highlight: MiR-181a/b binding sites. (C) Dual-luciferase assay of WT or mutant NUDT21 3′UTR activities in 293T cells. N ≥ 5. The graph represents means ± MSE. The p-value was determined using a two-tailed student t-test. *p < .05. (D) Left panel: The protein levels of NUDT21 and ACTB in primary human normal dermal fibroblasts transfected with 50 nM control, miR-181a, miR-181b, or both inhibitors on day 0 and day 2 and treated with vehicle or 5 ng/mL TGFβ1 for 4 days. Right panel: Densitometry analysis of the Western blot image. n = 4 fibroblast lines from different donors. p-values were calculated by one-way ANOVA for A and D and two-way ANOVA for C followed by Dunnett’s comparisons test. *p < .05; **p < .01; ***p < .001; ****p < .0001.
3.4 |. Delayed NUDT21 depletion promotes skin fibrosis in mice
TGFβ1 was previously demonstrated to have peak levels during the late stage of bleomycin-induced skin or lung fibrosis.18,19 Consistently, NUDT21 levels were not significantly decreased in mouse skin until 21 days after bleomycin treatment (Figure 4A), suggesting that it mainly plays a role during the late fibrotic stage. We have previously shown that fibroblast-specific knockdown of NUDT21 in the Col1a1-cre/ERT2-Nudt21f/f strain before bleomycin treatment promotes skin thickening and collagen deposition in the bleomycin-induced dermal fibrosis model.11 To confirm whether NUDT21 was successfully knocked down in Col1a1-cre/ERT2-Nudt21f/f mice, we treated Col1a1-cre/ERT2-Nudt21f/f and control Col1a1-cre mice with 75 mg/kg/day tamoxifen for 5 days, and isolated primary skin fibroblasts 1 week after the last tamoxifen injection (Figure 4B). NUDT21 protein level was significantly downregulated in the fibroblasts of Col1a1-cre/ERT2-Nudt21f/f skin in association with elevated Collagen I levels (Figure 4C). Here, to bypass the NUDT21 depletion during the early inflammatory phase of the bleomycin dermal fibrosis model, we delayed the Cre activation by initiating tamoxifen injection 7 days after the first bleomycin injection in Col1a1-cre/ERT2-Nudt21f/f and control Col1a1-cre/ERT2 mice (Figure 4D). This treatment allows NUDT21 protein knockdown starting 14 days after the initial bleomycin treatment, a time point when fibrosis starts developing. At the skin tissue level, RT-qPCR analysis suggested that Nudt21 transcript levels were not significantly decreased (Figure 4E), but Western blot revealed that its protein levels were significantly decreased (Figure 4H). The transcript levels of Col1a2 and Fn1 were highly elevated in NUDT21 knockdown mice (Figure 4E). Although the transcript level of Col1a1 was not changed (Figure 4E), it had significant 3′UTR shortening in NUDT21 knockout mice compared to control mice (Figure 4F). Consistent with these findings, the protein levels of Collagen I and fibronectin were highly elevated in the skin of Col1a1cre/ERT2-NUDT21f/f mice treated with s.c. bleomycin (Figure 4H), suggesting that COL1A1 was regulated post-transcriptionally by NUDT21-mediated APA. In parallel with these findings, the dermal pepsin-soluble collagen levels (Figure 4G) and skin thickness (Figure 4I,J) were increased in the delayed activation Col1a1-cre/ERT2-NUDT21f/f mice. Taken together, these data suggest that delayed NUDT21 depletion at the late fibrotic stage promotes bleomycin-induced skin fibrosis in mice.
FIGURE 4.
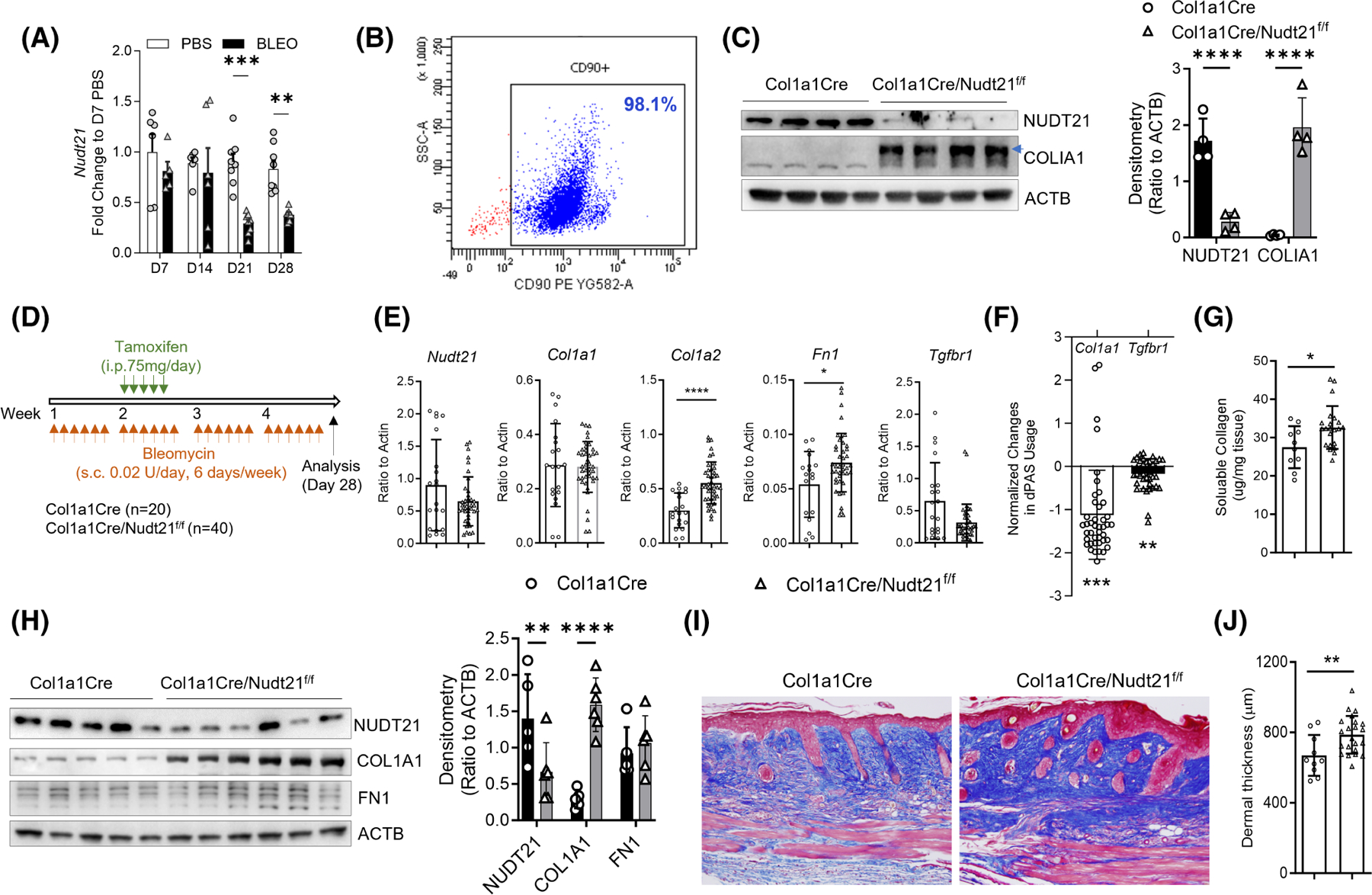
Delayed Nudt21 depletion exaggerates bleomycin-induced skin fibrosis. (A) The transcript levels of Nudt21 at different time points were determined in skin of female WT mice s.c. injected with PBS or 0.02 U/day bleomycin six times a week for 4 weeks. (B) Six–eight-week-old Col1a1Cre or Col1a1Cre/Nudt21f/f female mice were i.p. injected with 75 mg/day tamoxifen daily for 5 days. One week after the last tamoxifen injection, primary mouse fibroblasts were isolated for analysis. Flow cytometry of primary isolated mouse skin fibroblasts showing more than 95% of cells are CD90 positive. (C) Left panel: Western blot of primary isolated skin fibroblasts showing NUDT21 and COLIA1 levels. Right panel: Densitometry analysis of the Western blot image. n = 4 fibroblast lines from different mice. (D–J) Female Col1a1Cre or Col1a1Cre/Nudt21f/f mice were injected with repeated s.c. bleomycin six times a week for 4 weeks. One week after the first bleomycin injection, mice were i.p. injected with 75 mg/day tamoxifen daily for 5 days to induce cre recombination. Skin was analyzed on day 28. (D) Scheme of experimental design. (E) Real-time qRT PCR was performed to determine the transcript levels of Nudt21, Col1a1 Col1a2, Fn1, and Tgfbr1 in the skin. (F) Real-time qRT PCR was performed to determine the APA of Col1a1 in the skin. Data were presented as . (G) Sircol assay was performed to determine the pepsin-soluble collage levels in the skin. (H) Left panel: Western blot was performed to determine the protein levels of NUDT21, COL1A1, FN1, and ACTB. Right panel: Densitometry analysis of the Western blot image. n ≥ 5 skin from different mice. (I) Representative Masson’s Trichrome staining images. Scale bar =50 μm. (J) Histological dermal thickness. The graph represents means ± MSE. The p-value was determined using a two-tailed student t-test, and the p-values for panels A, C, and G were Bonferroni corrected. *p < .05; **p < .01; ***p < .001; ****p < .0001.
3.5 |. MiR-181a and miR-181b inhibitors attenuate bleomycin-induced skin fibrosis in association with NUDT21 rescue
Our in vitro studies have demonstrated that miR-181a and miR-181b inhibitors can rescue NDUT21 levels in TGFβ1-treated dermal fibroblasts, suggesting that miR-181a and miR-181b inhibitors may attenuate bleomycin-induced dermal fibrosis in vivo by preventing NUDT21 downregulation. To test this hypothesis, 6–8-week-old female wild-type mice were treated with s.c. bleomycin five times a week for 4 weeks. Starting on day 10 after the first bleomycin injection, when dermal fibrosis starts to develop, LNA control, miR-181a, or miR-181b inhibitors (10 mg/kg body weight in 50 μL PBS) were administered s.c. at the bleomycin injection site twice weekly for 2 weeks (Figure 5A). The skin was collected on day 28 for analysis. Although the transcript levels of Nudt21 are not affected (Figure 5B), both miR-181a and miR-181b effectively increased NUDT21 protein levels (Figure 5C) and reduced the expression of collagen I (COL1) and fibronectin (FN1) at both the protein and transcript levels (Figure 5D). Notably, the miR-181b inhibitor exhibited stronger effects. Consistently, Masson’s Trichrome staining indicated decreased skin collagen accumulation (Figure 5E) and dermal thickness (Figure 5F) in miR-181a or miR-181b inhibitors treated mice. Overall, these data indicate that miR-181a and miR-181b inhibitors can attenuate skin fibrosis by restoring NDUT21 expression.
FIGURE 5.
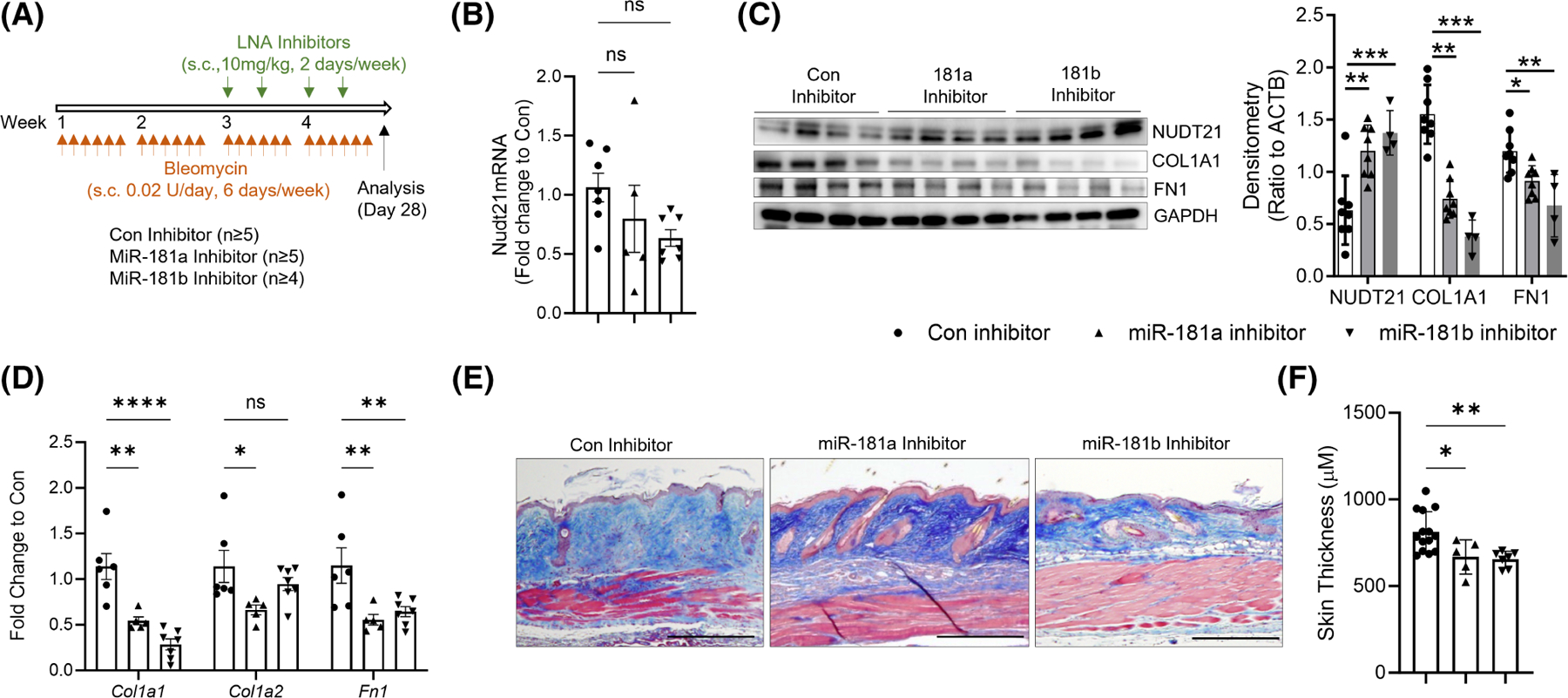
Protective roles of miR-181a and miR-181b inhibitors in skin fibrosis. (A) Scheme of experimental design. Eight-week-old female wild-type C57/BL6 mice were injected with repeated s.c. bleomycin six times a week for 4 weeks. Two weeks after the first bleomycin injection, mice were s.c. injected with 10 mg/kg/day LNA control, miR-181a, or miR-181b inhibitors twice a week at the bleomycin-injected spot. The skin was collected 28 days after the first bleomycin injection for analysis. (B, D) Real-time qRT PCR was performed to determine the transcript levels of (B) Nudt21, and (D) Col1a1, Col1a2, and Fn1. (C) Left panel: Western blot was used to determine the protein levels of NUDT21, COLIA1, FN1, and ACTB. Right panel: Densitometry analysis of the Western blot image. n ≥ 4 fibroblast lines from different mice. (E) Masson’s Trichrome staining was performed to show dermal fibrosis in different treatment groups. Scale bar = 50 μm. (F) Quantification of dermal thickness. N ≥ 5. The graphs represent means ± MSE. p-value was determined using one-way ANOVA (C, F) or two-way ANOVA (D) followed by Dunnett’s multiple comparisons tests. *p < .05; **p < .01; ***p < .001; and ****p < .0001.
3.6 |. MiR-181a and miR-181b mimics exaggerated bleomycin-induced skin fibrosis
In addition, we have also investigated whether the fibrotic phenotype can be exaggerated by miR-181a and miR-181b mimics. MiR-181a and miR-181b mimics were s.c. injected starting on day 10 after the first bleomycin injection, and the skin was analyzed on day 28. Opposite to what we have observed in the miR-181a and miR-181b inhibitors treated skin, miR-181a and miR-181b mimics, particularly miR-181b mimics suppressed NUDT21 and promoted COL1 and FN1 protein expression in bleomycin-treated skin (Figure 6A). Additionally, both miR-181a and miR-181b mimics were able to promote collagen deposition at the dermal layer (Figure 6B) and increased dermal thickness (Figure 6C). Taken together, these findings indicate that miR-181a and miR-181b mimics may exaggerate skin fibrosis through NUDT21 repression in vivo.
FIGURE 6.
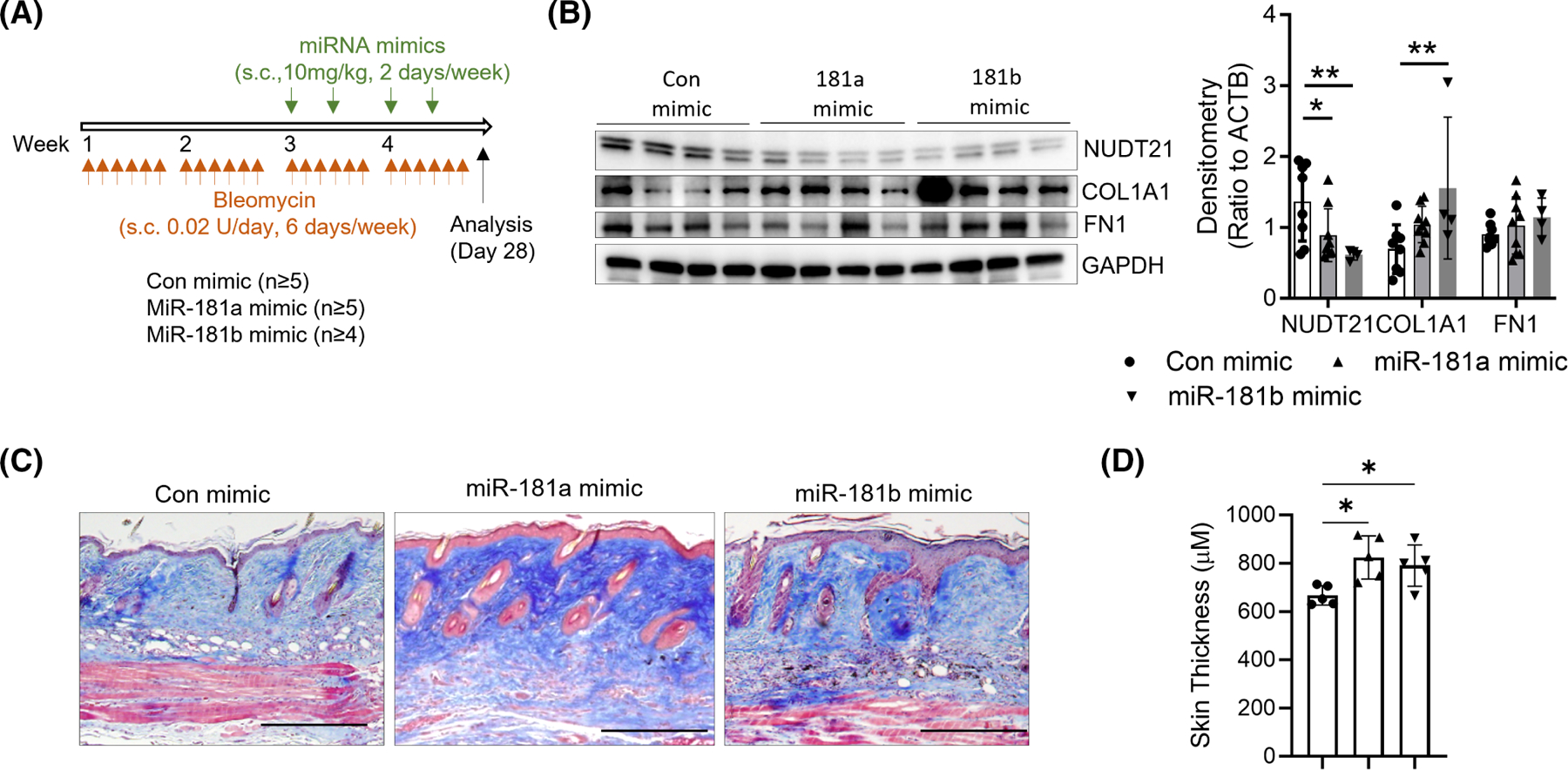
MiR-181a/b mimics worsened bleomycin-induced skin fibrosis. (A) Scheme of mouse treatment. Six–eight-week-old female WT C57/BL6 mice were injected with repeated s.c. bleomycin five times a week. Two weeks after the first bleomycin injection, mice were s.c. injected with 10 mg/kg/day LNA control, miR-181a, or miR-181b mimics twice a week at the same injection spot. The skin was collected 28 days after the first bleomycin injection for analysis. (B) Left panel: The protein levels of NDUT21, COL1A1, FN1, and ACTB in the skin were determined using Western blot. Right panel: Densitometry analysis of the Western blot image. n ≥ 4 fibroblast lines from different donors. N ≥ 4. (C) Collagen deposition in skin samples from different treatment groups was determined using Masson’s Trichrome. Scale bar=50 μm. (D) Dermal thickness was blindly examined and quantitated. N = 5. The graphs represent means ± MSE. p-value was calculated using one-way ANOVA followed by Dunnett’s multiple comparison tests. *p < .05; **p < .01.
4 |. DISCUSSION
The major goal of this study is to identify the upstream pathways leading to NUDT21 downregulation in skin fibrosis and demonstrate whether targeting these pathways could attenuate skin fibrosis by rescuing NUDT21 expression. Here, we identified TGFβ1 as the top cytokine that suppresses NUDT21 expression in human dermal fibroblasts. Moreover, TGFβ1 represses NUDT21 protein expression by inducing miR-181a and miR-181b that directly target NUDT21 through 3′UTR binding. In line with the exaggerated skin fibrosis in NUDT21 fibroblast knockdown mice, miR-181a or miR-181b inhibitors attenuate bleomycin-induced skin fibrosis in mice in association with NUDT21 rescue, while miR-181a and miR-181b mimics suppress NUDT21 expression and promote skin fibrosis. Additionally, genetic deletion of NUDT21 in fibroblasts at a late fibrotic stage leads to increased ECM deposition in bleomycin-induced dermal fibrosis. Taken together, this study revealed a novel role of TGFβ1-mediated miR-181a and miR-181b induction and NUDT21 repression in fibroblasts during SSc pathogenesis.
Several studies have demonstrated the potential role of APA in tissue fibrosis. Studies from our groups have revealed that NUDT21 loss was associated with 3′UTR shortening, increased expression of genes involved in fibrosis (e.g., TGFBR1, COL1A1, and SPARC), and exaggerated bleomycin-induced skin and lung fibrosis in murine fibrosis models.11 Cleavage stimulating factor 64 (CstF64) is a protein involved in polyadenylation machinery whose depletion normally causes 3′UTR lengthening. Consistent with our findings, CstF64 was upregulated in heart failure left ventricular tissues and cardiac fibroblasts, and its knockdown was associated with 3′UTR lengthening and repressed expression of profibrotic genes in cardiac fibroblasts.20 Similarly, cleavage stimulation factor subunit 2 (CSTF2), a gene involved in 3′UTR cleavage, was identified as a TGFβ inducible protein that promoted epithelial–mesenchymal transition (EMT) and extracellular matrix (ECM) overproduction in renal fibrosis by promoting the APA and enhancing the protein expression of fibroblast growth factor 2 (FGF2).21 Moreover, 3′UTR shortening of profibrotic genes (e.g., COL1A, FN1, TGFβ1, TGFβR1, and NFκB) is observed in patients with end-stage right ventricular failure and could be implicated in fibroblast activation and heart fibrosis.22 Together, these findings suggest that fibrosis can be attenuated by preventing 3′UTR shortening of key fibrotic genes, and future studies aiming to investigate the role of other APA factors in dermal fibrosis may provide important molecular mechanisms.
TGFβ1 is a master regulator of fibrotic diseases that drives fibroblast proliferation, myofibroblast differentiation, and ECM synthesis.23 Our in vitro experiments identified TGFβ1 as the top upstream regulator that downregulated NUDT21 expression in dermal fibroblasts. This finding is consistent with our previous unbiased global Ingenuity pathway analysis that predicted TGFβ1 as the upstream regulator of the 971 genes that showed 3′UTR shortening upon NUDT21 depletion.11 Notably, among these 971 genes, approximately 10% (~97 genes) were found to be regulated by TGFβ1, including collagens, Integrins,24 TGFBR1,25 and SPARC.23,26,27 This observation also aligns with our previous gene expression studies, where TGFβ1 emerged as the top predicted regulator in the signature associated with SSc skin.28 Furthermore, our investigations revealed elevated levels of TGFβ1 protein in the supernatant of cultured SSc fibroblasts, suggesting the presence of an autocrine mechanism that contributes to the repression of NUDT21 levels in SSc dermal fibroblasts. As the key fibrotic factor, TGFβ1 was previously demonstrated to have peak levels during the late stage of bleomycin-induced skin or lung fibrosis.18,19 Consistently, NUDT21 was also downregulated at the late fibrotic stage (after 21 days) in the murine skin fibrosis model. Notably, delayed NUDT21 fibroblast depletion at the fibrotic stage still exaggerated bleomycin-induced skin fibrosis. These findings suggest that fibroblast-specific NUDT21 expression played a major role during the fibrotic stage of this murine model. Taken together, these findings highlight the intricate interplay between TGFβ1 and NUDT21 that could contribute to the pathogenesis of skin fibrosis.
While TGFβ1 can directly bind promoters to repress target gene expression, a more common explanation involves its role in inducing miRNA expression. Moreover, TGFβ1 normally induces transcript changes within a relatively short timeframe of 3–6 h.17 Interestingly, the transcript levels of NUDT21 did not show a decrease until 2 days after TGFβ1 treatment, suggesting that TGFβ1 suppresses NUDT21 expression through an indirect mechanism other than directly regulating its promoter activity. To further investigate this, we first treated the TGFβ1-treated fibroblasts with MG132 to suppress NUDT21 transcript synthesis. However, the protein levels of NUDT21 were not affected, suggesting that TGFβ1 did not accelerate the protein degradation of NUDT21 (data not shown). We next screened the miRNAs that have binding sites at both human and mouse NUTDT21 3′UTR and identified miR-181a and miR-181b as potential miRNAs targeting NUDT21 for repression. Consistently, miR-181a and miR-181b were found to be upregulated upon TGFβ1 treatment, in mouse skin fibrosis models, and primary skin fibroblasts from SSc patients. Mechanistically, both miR-181a and miR-181b mimics were able to decrease NUDT21 protein levels in cultured cells and repress luciferase activities in plasmids containing the wild-type NUDT21 3′UTR but not in those with mutated miR-181 binding sites, suggesting a direct binding of miR-181a and miR-181b at NUDT21 3′UTR. Furthermore, inhibition of miR-181a and miR-181b partially rescued the TGFβ1-induced downregulation of NUDT21. Overall, these findings highlight the involvement of miR-181a and miR-181b in mediating the repression of NUDT21 in response to TGFβ1 and linked their function to NUDT21-mediated APA.
In this study, we also investigated the function of miR-181a and miR-181a in bleomycin-induced skin fibrosis and their link with NUDT21. Our study demonstrated that miR-181a and miR-181b inhibitors attenuated bleomycin-induced skin fibrosis in association with elevated NUDT21 protein levels compared to control inhibitor-treated skin, while miR-181a and miR-181b mimics promoted bleomycin-induced skin fibrosis. These findings are consistent with our previous studies showing mice with NUDT21 depletion in fibroblasts had further enhanced ECM expression and fibrosis in bleomycin-induced skin or lung fibrosis models.11,12 These results are also in alignment with the findings from other tissues.29,30 For example, miR-181b was upregulated in hepatic tissues of schistosomiasis-infected individuals and experimental animal models. It exerted its fibrotic effects by directly targeting Smad7, a critical intracellular inhibitor of TGF-β signaling.29 MiR-181b has also been reported as a profibrotic miRNA in liver fibrosis by targeting phosphatase and tensin homolog (PTEN),31 and PTEN’s loss was associated with the progressive development of steatohepatitis and fibrosis.32 Additionally, miR-181b has been induced in TGFβ-treated atrial endocardial endothelial cells and has been shown to decrease atrial fibrosis in TGF-β-transgenic mice by targeting semaphorin 3A.33 Consistently, miR-181a was significantly upregulated in mesangial cells from diabetic nephropathy mice, and miR-181a overexpression promoted extracellular matrix deposition, while miR-181a inhibition alleviated high glucose-induced renal fibrosis.30 In our studies, we identified NUDT21 as a novel target of miR-181a and miR-181b, establishing a new link between these miRNAs and alternative polyadenylation (APA). However, our in vivo studies in mice reveal that while the protein levels of NUDT21 are elevated in the skin of mice injected with miR-181a or miR-181b inhibitors, there is no corresponding change in transcript levels. This discrepancy suggests that miR-181a or miR-181b may bind to NUDT21 mRNA, leading to translational suppression without causing mRNA degradation.34 Additionally, miRNAs can bind to target mRNAs and sequester them into P-bodies or other RNA granules, effectively reducing their availability for translation.35,36 This sequestration does not result in mRNA degradation but prevents their translation into proteins. Cells may also employ compensatory mechanisms to stabilize mRNA levels or enhance transcription rates, thereby maintaining consistent mRNA levels despite translational repression by these miRNAs.34,37 These factors likely contribute to the observed discordance between mRNA and protein levels in our experimental model. Our results also provide an additional explanation of the findings related to the roles of miR-181a and miR-181b in fibrosis. While we focused on fibroblasts in the present study, the miR-181a/b mimics and inhibitors used herein can also target other cell types present in the skin tissue such as epithelial and inflammatory cells. Further studies are needed to elucidate the cell-specific roles of miR-181a and miR-181b and their interactions with NUDT21 in these cell types.
Taken together, our study highlights a novel role of miR-181a and miR-181b in skin fibrosis. The impact of miR-181a and miR-181b is achieved by mediating TGFβ1-induced NUDT21 downregulation in skin fibroblasts. Together with previous findings showing NUDT21-loss amplifying skin and lung fibrosis, our studies indicate a novel mechanism for NUDT21 repression in skin fibrosis, which contributes to the complexity of APA biology for the field. Furthermore, given the detrimental role of NUDT21-downregulation in skin fibrosis, exploring novel compounds that can inhibit NUDT21 downregulation or prevent 3′UTR shortening holds promise for the treatment of SSc. Moreover, TGFβ1 has been implicated in many other fibrotic diseases, including heart, liver, and kidney fibrosis. Thus, investigating how the TGFβ1/miR-181/NUDT21 pathway may contribute to the pathophysiology of these conditions could be of great interest to the field.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Dr. Michael R Blackburn for providing the Nudt21f/f mice, Ning-Yuan Chen for helping with the mouse maintenance, GENEWIZ for plasmid sequencing, and Novogene for miRNA sequencing.
FUNDING INFORMATION
This work was supported by National Institutes of Health (NIH) National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) grants to T.W. and S.A. (R01AR073284), S.A (R61AR078078), and S.A. and M.W. (R56AR078211), NIH National Institute on Aging (NIA) grant to T.W (1R56AG076144-01A1), and NIH National Heart, Lung, and Blood Institute grant to T.M. (R01HL168128).
Abbreviations:
- 3′UTR
3′ untranslated region
- APA
alternative polyadenylation
- COL1A1, COL1A2
Collagen Type I Alpha 1, 2
- CSTF2
cleavage stimulation factor subunit 2
- DMEM
Dulbecco’s Modified Eagle Medium
- ECM
extracellular matrix
- EMT
epithelial-mesenchymal transition
- FBS
fetal bovine serum
- FGF2
fibroblast growth factor 2
- FN1
fibronectin
- IL4, IL6, IL13
interleukin 4, 6, 13
- LNA
locked nucleic acid
- miRNA
microRNA
- NUDT21/CFIm25
Nudix Hydrolase 21/Cleavage Factor Im 25
- PAS
polyadenylation site
- PDGF
platelet-derived growth factor
- RT-qPCR
real-time quantitative polymerase chain reaction
- SPARC
secreted protein acidic and cysteine rich
- SSc
systemic sclerosis
- TGFBR1
transforming growth factor beta receptor 1
- TGFβ1
transforming growth factor beta 1
- WT
wild type
Footnotes
DISCLOSURES
This study was only funded by federal funds. SA declares Grant support from Janssen, aTyr, and Boehringer Ingelheim. Personal Fees from aTyr, Boehringer Ingelheim, AstraZeneca, TeneoFour, CSL Behring, Merck, Abbvie, and Mitsubishi Tanabe.
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available in the Materials and Methods, Results, and/or Supplemental Material of this article.
REFERENCES
- 1.Elhai M, Meune C, Avouac J, Kahan A, Allanore Y. Trends in mortality in patients with systemic sclerosis over 40 years: a systematic review and meta-analysis of cohort studies. Rheumatology. 2012;51:1017–1026. [DOI] [PubMed] [Google Scholar]
- 2.Thomas E, Symmons DP, Brewster DH, Black RJ, Macfarlane GJ. National study of cause-specific mortality in rheumatoid arthritis, juvenile chronic arthritis, and other rheumatic conditions: a 20 year followup study. J Rheumatol 2003;30:958–965. [PubMed] [Google Scholar]
- 3.Elfving P, Puolakka K, Kautiainen H, Virta LJ, Pohjolainen T, Kaipiainen-Seppanen O. Mortality and causes of death among incident cases of systemic lupus erythematosus in Finland 2000–2008. Lupus. 2014;23:1430–1434. [DOI] [PubMed] [Google Scholar]
- 4.Tashkin DP, Elashoff R, Clements PJ, et al. Cyclophosphamide versus placebo in scleroderma lung disease. N Engl J Med 2006;354:2655–2666. [DOI] [PubMed] [Google Scholar]
- 5.Khanna D, Denton CP, Jahreis A, et al. Safety and efficacy of subcutaneous tocilizumab in adults with systemic sclerosis (faSScinate): a phase 2, randomised, controlled trial. Lancet. 2016;387:2630–2640. [DOI] [PubMed] [Google Scholar]
- 6.Distler O, Highland KB, Gahlemann M, et al. Nintedanib for systemic sclerosis-associated interstitial lung disease. N Engl J Med 2019;380:2518–2528. [DOI] [PubMed] [Google Scholar]
- 7.Khanna D, Lin CJF, Furst DE, et al. Tocilizumab in systemic sclerosis: a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Respir Med 2020;8:963–974. [DOI] [PubMed] [Google Scholar]
- 8.Skaug B, Assassi S. Biomarkers in systemic sclerosis. Curr Opin Rheumatol 2019;31:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 2013;14:496–506. [DOI] [PubMed] [Google Scholar]
- 10.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weng T, Huang J, Wagner EJ, et al. Downregulation of CFIm25 amplifies dermal fibrosis through alternative polyadenylation. J Exp Med 2020;217:e20181384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weng T, Ko J, Masamha CP, et al. Cleavage factor 25 deregulation contributes to pulmonary fibrosis through alternative polyadenylation. J Clin Invest 2019;129:1984–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ko J, Mills T, Huang J, et al. Transforming growth factor beta1 alters the 3′-UTR of mRNA to promote lung fibrosis. J Biol Chem 2019;294:15781–15794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van den Hoogen F, Khanna D, Fransen J, et al. 2013 classification criteria for systemic sclerosis: an American College of Rheumatology/European League against Rheumatism collaborative initiative. Arthritis Rheum 2013;65:2737–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu M, Schneider DJ, Mayes MD, et al. Osteopontin in systemic sclerosis and its role in dermal fibrosis. J Invest Dermatol 2012;132:1605–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masamha CP, Xia Z, Yang J, et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510:412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang WL, Lee MT, Wu LS, et al. CREB coactivator CRTC2/TORC2 and its regulator calcineurin crucially mediate follicle-stimulating hormone and transforming growth factor beta1 upregulation of steroidogenesis. J Cell Physiol 2012;227:2430–2440. [DOI] [PubMed] [Google Scholar]
- 18.Strobel B, Klein H, Leparc G, Stierstorfer BE, Gantner F, Kreuz S. Time and phenotype-dependent transcriptome analysis in AAV-TGFbeta1 and Bleomycin-induced lung fibrosis models. Sci Rep 2022;12:12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto T, Takagawa S, Katayama I, et al. Animal model of sclerotic skin. I: local injections of bleomycin induce sclerotic skin mimicking scleroderma. J Invest Dermatol 1999;112:456–462. [DOI] [PubMed] [Google Scholar]
- 20.Neupane R, Youker K, Yalamanchili HK, et al. Cleavage stimulating factor 64 depletion mitigates cardiac fibrosis through alternative polyadenylation. Biochem Biophys Res Commun 2022;597:109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan Y, Zheng T, Zhang R, et al. Alternative polyadenylation writer CSTF2 forms a positive loop with FGF2 to promote tubular epithelial-mesenchymal transition and renal fibrosis. Biochim Biophys Acta Mol basis Dis 2022;1868:166541. [DOI] [PubMed] [Google Scholar]
- 22.Neupane R, Cieslik KA, Youker K, Palaniyandi SS, Guha A, Thandavarayan RA. 3′UTR shortening of profibrotic genes and reversibility of fibrosis in patients with end-stage right ventricular failure. Clin Transl Med 2022;12:e1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lafyatis R Transforming growth factor beta—at the centre of systemic sclerosis. Nat Rev Rheumatol. 2014;10:706–719. [DOI] [PubMed] [Google Scholar]
- 24.Ray K Connective tissue diseases: Integrins crucial for the onset of fibrosis in systemic sclerosis—a new therapeutic target? Nat Rev Rheumatol 2013;9:637. [DOI] [PubMed] [Google Scholar]
- 25.Pannu J, Nakerakanti S, Smith E, ten Dijke P, Trojanowska M. Transforming growth factor-beta receptor type I-dependent fibrogenic gene program is mediated via activation of Smad1 and ERK1/2 pathways. J Biol Chem 2007;282:10405–10413. [DOI] [PubMed] [Google Scholar]
- 26.Bhattacharyya S, Wei J, Varga J. Understanding fibrosis in systemic sclerosis: shifting paradigms, emerging opportunities. Nat Rev Rheumatol 2011;8:42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou X, Tan FK, Guo X, Arnett FC. Attenuation of collagen production with small interfering RNA of SPARC in cultured fibroblasts from the skin of patients with scleroderma. Arthritis Rheum 2006;54:2626–2631. [DOI] [PubMed] [Google Scholar]
- 28.Assassi S, Swindell WR, Wu M, et al. Dissecting the heterogeneity of skin gene expression patterns in systemic sclerosis. Arthritis Rheum 2015;67:3016–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S, Zhang J, Chen H, et al. MicroRNA-181b promotes schistosomiasis-induced hepatic fibrosis by targeting Smad7. Mol Biochem Parasitol 2022;252:111523. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Wu C, Dong J, Liu J, Wei X. Downregulation of miR-181a alleviates renal fibrosis in diabetic nephropathy mice. Int J Clin Exp Pathol 2018;11:4004–4011. [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng J, Wu C, Xu Z, et al. Hepatic stellate cell is activated by microRNA-181b via PTEN/Akt pathway. Mol Cell Biochem 2015;398:1–9. [DOI] [PubMed] [Google Scholar]
- 32.Horie Y, Suzuki A, Kataoka E, et al. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J Clin Invest 2004;113:1774–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai YJ, Tsai FC, Chang GJ, et al. miR-181b targets semaphorin 3A to mediate TGF-beta-induced endothelial-mesenchymal transition related to atrial fibrillation. J Clin Invest 2022;132:e142548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 2015;16:421–433. [DOI] [PubMed] [Google Scholar]
- 35.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol 2007;27:3970–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol 2005;7:719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available in the Materials and Methods, Results, and/or Supplemental Material of this article.


