Abstract
During herpesvirus egress, capsids bud through the inner nuclear membrane. Underlying this membrane is the nuclear lamina, a meshwork of intermediate filaments with which it is tightly associated. Details of alterations to the lamina and the inner nuclear membrane during infection and the mechanisms involved in capsid transport across these structures remain unclear. Here we describe the fate of key protein components of the nuclear envelope and lamina during herpes simplex virus type 1 (HSV-1) infection. We followed the distribution of the inner nuclear membrane protein lamin B receptor (LBR) and lamins A and B2 tagged with green fluorescent protein (GFP) in live infected cells. Together with additional results from indirect immunofluorescence, our studies reveal major morphologic distortion of nuclear-rim LBR and lamins A/C, B1, and B2. By 8 h p.i., we also observed a significant redistribution of LBR-GFP to the endoplasmic reticulum, where it colocalized with a subpopulation of cytoplasmic glycoprotein B by immunofluorescence. In addition, analysis by fluorescence recovery after photobleaching reveals that LBR-GFP exhibited increased diffusional mobility within the nuclear membrane of infected cells. This is consistent with the disruption of interactions between LBR and the underlying lamina. In addition to studying stably expressed GFP-lamins by fluorescence microscopy, we studied endogenous A- and B-type lamins in infected cells by Western blotting. Both approaches reveal a loss of lamins associated with virus infection. These data indicate major disruption of the nuclear envelope and lamina of HSV-1-infected cells and are consistent with a virus-induced dismantling of the nuclear lamina, possibly in order to gain access to the inner nuclear membrane.
Herpesvirus genomes are packaged within preformed capsids inside the nuclei of infected cells. The subsequent processes of assembly and egress involve the acquisition of tegument proteins and an envelope incorporating viral glycoproteins, followed by release of the mature virion by membrane fusion at the cell surface. The precise route of egress and the source of mature envelope remain the subject of much debate (reviewed by Enquist et al. [16]), but a feature common to all proposed models is the exit of nucleocapsids from the nucleus by budding through the inner nuclear membrane (INM). This event is referred to as primary envelopment and has been widely documented in electron microscopy (EM) studies (12, 37).
Several viral proteins involved in the process have been characterized, and in herpes simplex virus type 1 (HSV-1) infection these include the products of the UL11, UL31, UL34, and UL53 genes (3, 4, 9, 19, 28, 42, 46, 57). However, the involvement of cellular proteins in the exit of nucleocapsids from the nucleus has not been well defined. The nuclear envelope consists of two lipid bilayers and a proteinaceous meshwork known as the nuclear lamina. The envelope is studded with nuclear pore complexes, and the inner and outer nuclear membranes meet at these sites. The outer nuclear membrane is a continuation of the endoplasmic reticulum (ER), while the INM has a unique composition and contains specific resident proteins, including lamin B receptor (LBR), lamin-associated polypeptides (LAPs), emerin, and nurim (reviewed by Chu et al. [10] and Dechat et al. [13]). While there are exceptions, many of these proteins are thought to localize to the INM by a diffusion-and-retention mechanism. They are first targeted by their transmembrane domains to the continuous ER-nuclear envelope compartment. Within this compartment, they subsequently diffuse laterally, transiting the nuclear pore within the membrane. They are then retained at the INM by interactions between their exposed nucleoplasmic domains and the underlying lamina and/or chromatin (21, 40, 47, 52).
The main component of the lamina lining the INM are the lamins, members of the intermediate filament family of cytoskeletal proteins (recently reviewed by Gruenbaum et al. [27] and Moir et al. [34]). Like cytoplasmic intermediate filaments, they comprise an α-helical rod domain flanked by less well conserved N- and C-terminal domains. In addition, nuclear lamins contain a nuclear localization signal (31) and a C-terminal target site for posttranslational modification (isoprenylation) which increases hydrophobicity and is required for correct targeting of newly synthesized lamins to the lamina (29). In vitro studies show lamins dimerizing via coiled-coil interactions between rod domains and then forming head-to-tail associations to form polymers which bundle into filaments (reviewed by Nigg [35]). The lamina in situ has been described most clearly by Aebi and coworkers (1), whose EM observations of Xenopus oocytes revealed a regular meshwork with an average crossover spacing of 52 nm. Although some reports have suggested that in higher eukaryotes this fibrillar network may be discontinuous (5, 24, 41), this structure is likely to present significant restrictions to the access of 100-nm-diameter herpesvirus nucleocapsids to the INM (Fig. 1),
FIG. 1.
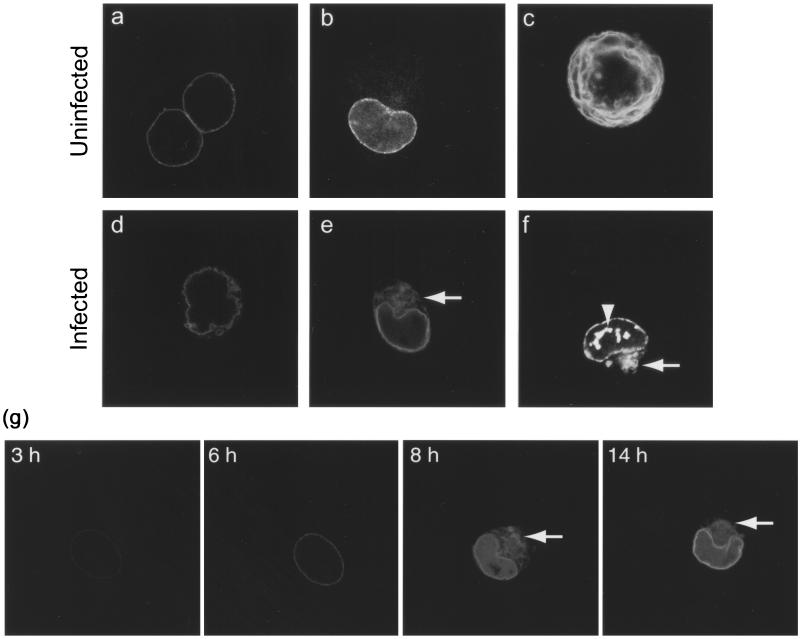
Live COS-1 cells transiently expressing LBR-GFP viewed by confocal microscopy. (a and b) Uninfected interphase cells show nuclear-rim fluorescence and some fluorescence in the ER (b). (c) Uninfected mitotic cell shows fluorescence in the ER. (d to g) Cells infected with HSV-1 at 10 PFU/cell (e, 8 h; d and f, 24 h) show a distorted nuclear-rim pattern (d) in a membranous cytoplasmic compartment (e and f [arrows]) and in intranuclear domains (f [arrowhead]). (g) A single COS-1 cell transiently expressing LBR-GFP and infected with HSV-1 at 10 PFU/cell was tracked throughout the course of infection. Live confocal microscope images are shown, taken between 3 and 14 h p.i. Between 6 and 8 h p.i., the nuclear-rim LBR-GFP partially redistributes to a cytoplasmic compartment (arrows) which is stable over several hours.
Direct interactions of lamins both with chromatin and with INM proteins have been demonstrated, and it is proposed that each component of the INM and lamina interact with multiple partners to form a complex web of links between these structures. In this way, lamins provide structural support to the interphase nuclear envelope and play a role in its disassembly and reassembly at mitosis (18, 22). They also play roles in spatial organization within the nucleus, including positioning of nuclear pore complexes, chromatin, and DNA replication factors (5, 25, 53). Although full-scale dismantling of the nuclear envelope does not occur in HSV-1 infection, it is possible that processes normally involved in INM-lamina dynamics are invoked in order for the exiting nucleocapsid to traverse the lamina and achieve envelopment at the INM. To date, our understanding of these events has come largely from EM studies (reviewed by Nii [36]) where individual protein components of the nuclear envelope were not studied in detail. In this study, we have used live-cell imaging of green fluorescent protein (GFP)-tagged proteins in conjunction with biochemical techniques to monitor an INM protein (LBR) and lamins in the context of HSV-1 infection.
MATERIALS AND METHODS
Cells and viruses.
COS-1, HeLa, and BHK-21 cells were grown in Dulbecco's modified Eagle medium with 10% newborn calf serum. To maintain VLP4/BHK and VLP28/BHK stable lines, the growth medium was supplemented with 250 μg of Geneticin/ml. For virus infections, HSV-1 (strain 17) was applied to cells at 10 PFU/cell in serum-free medium. After 1 h of incubation at 37°C, the inoculum was replaced with complete medium.
Expression vectors and transfection.
phLBR1TMGFP was provided by Jan Ellenberg, National Institutes of Health (15). Transient transfections of this plasmid were performed using the calcium phosphate precipitation method modified by using N,N-bis(2-hydroxethyl)-2-amino-ethanesulfonic acid-buffered saline (pH 7.06) in place of HEPES-buffered saline. The day before transfection, COS-1 cells were seeded into six-well cluster dishes containing 13-mm-diameter coverslips at 2 × 105 cells/well. Transfections were done with 1 μg of phLBR1TMGFP plus 1 μg of pUC19 carrier DNA per well. The cells were superinfected 18 h after transfection.
Plasmids pVLP4 and pVLP28 were provided by Tom Rapoport, Harvard Medical School (47). The stable cell lines VLP4/BHK and VLP28/BHK were generated by transfecting BHK-21 cells with these plasmids. The cells (5 × 105) were transfected with 2.5 μg of pVLP4 or pVLP28 and 2.5 μg of pUC19 using Lipofectamine (Gibco-BRL) according to the manufacturer's instructions. From 24 h posttransfection, the growth medium of these cells was supplemented with 250 μg of Geneticin/ml.
Antibodies, indirect immunofluorescence, and live fluorescence microscopy.
Calreticulin antibody (rabbit polyclonal) was supplied by Calbiochem, and grp94 antibody (rat monoclonal) was supplied by Affinity Bioreagents. Both antibodies were diluted 1:200 for immunofluorescence studies. The mouse monoclonal antibody against HSV-1 and -2 glycoprotein B (gB) (Virusys Corp.) was diluted 1:500. Mouse monoclonal antibodies against lamin A/C and lamin B2 (Novocastra) were diluted 1:100 for immunofluorescence. For Western blotting, lamin A/C antibody was diluted 1:200 and lamin B2 antibody was diluted 1:500. The rabbit polyclonal antibody against lamin B1 was a generous gift from Murray Stewart, University of Cambridge, and was diluted 1:50 for immunofluorescence studies. For Western blotting, mouse monoclonal antibodies against ICP0 (clone 5H7; Virusys Corp.) and VP16 (LP1; a generous gift of Tony Minson, University of Cambridge) were diluted 1:8,000 and 1:4,000, respectively.
For indirect immunofluorescence studies, live cells on coverslips were rinsed with phosphate-buffered saline (PBS) and then fixed in ice-cold methanol for 20 min. The coverslips were blocked with 10% newborn calf serum in PBS for 20 min. They were incubated with 40 μl of primary antibody diluted in the blocking solution for 20 min and then rinsed three times in PBS for 5 min each time. Secondary antibodies were fluorescently labeled with fluorescein isothiocyanate (FITC) (Vector Laboratories) or Alexa 546 (Molecular Probes). They were also diluted in blocking solution, and 40 μl was applied to the coverslips for 20 min. The coverslips were rinsed three more times in PBS. All of these steps were carried out at room temperature. The coverslips were mounted with Vectashield fluorescent mount when FITC secondary antibodies were used or in equal parts PBS and glycerol when Alexa 546 antibodies were used. Dual-channel fluorescence images were acquired on a Zeiss LSM410 laser scanning confocal microscope using the 63× objective lens.
For live fluorescence imaging, cells expressing GFP fusion proteins were seeded on 40-mm-diameter coverslips (BDH) to fit a heated microscope chamber in two-well chamber cover glasses (Nunc) or in 35-mm gridded coverslip chambers (MatTek Corp., Ashland, Mass.) and viewed with the confocal microscope described above.
Fluorescence recovery after photobleaching (FRAP) analysis.
Live COS-1-1 cells transiently expressing LBR-GFP were maintained at 37°C on the heated stage of the microscope described above. A portion of the cell, including a section of the nuclear rim, was defined using the LSM software zoom facility and bleached by a 10-s scan using a 200-mW Ar laser at 100% transmission. Pre- and postbleach images of the whole cell were acquired by averaging two laser scans at 3% transmission for each time point. These low-intensity scans were repeated at 10-s intervals for 400 s following the bleaching. LSM software was used to generate a time series from these images and to plot the pixel intensity in defined regions of interest (ROIs) over the duration of the experiment. At each time point, the intensity of an equivalent background area was subtracted and values were normalized for total loss of fluorescence. The percentage fluorescence recovery for LBR-GFP was calculated as follows: (fluorescence intensity at recovery plateau − intensity immediately postbleaching)/(initial intensity − intensity immediately postbleaching) × 100.
Western blotting and cell fractionation.
For Western blotting, cells were seeded in six-well cluster dishes at 5 × 105/well the day before infection. To collect total cell lysates, the cells were rinsed in cold PBS at an appropriate time postinfection (p.i.) and then harvested in 250 μl of sodium dodecyl sulfate (SDS) loading buffer. Samples were boiled for 10 min and briefly sonicated before 10-μl samples were fractionated by polyacrylamide gel electrophoresis and transferred to Hybond-C nitrocellulose membranes. The membranes were blotted with primary antibodies at the dilutions given above. Secondary antibodies carried a horseradish peroxidase tag and were detected by enhanced chemiluminescence (Pierce).
To prepare soluble and insoluble protein samples, cells in cluster dishes as described above were rinsed in cold PBS and then scraped into cold PBS and pelleted in a microcentrifuge. The cells were resuspended in ice-cold low-salt buffer (0.5% NP-40, 10 mM HEPES–KOH [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride) or high-salt buffer (same as low-salt buffer but with 500 mM NaCl) for 2 min. Insoluble proteins were pelleted at 12,000 × g (in a microcentrifuge) for 15 min and resuspended in SDS loading buffer. An equal volume of 2× SDS loading buffer was added to the supernatant to give the soluble fraction. These fractions were processed for Western blotting as described above.
RESULTS
Nuclear-rim LBR-GFP is distorted in HSV-1-infected cells and partially redistributes to an altered ER.
The lamin B receptor is a well-defined component of the INM. To examine the distribution of an LBR-GFP fusion protein as a marker for the INM, COS-1 cells were transiently transfected with the construct phLBR1TMGFP, which expresses the N-terminal nucleoplasmic tail and first transmembrane domain of LBR fused to a C-terminal GFP tag (LBR-GFP). Subcellular distribution of the fusion protein was then studied in live cells by confocal microscopy. In uninfected interphase cells, LBR-GFP exhibited a predominantly nuclear-rim pattern of fluorescence (Fig. 1a), although minor amounts localized to the ER could be seen in cells expressing high levels (Fig. 1b). In uninfected mitotic cells, LBR-GFP was in a continuous membranous compartment (Fig. 1c) previously characterized as the ER (15). These observations are consistent with previously published data using this construct (15).
In cells infected with HSV-1, the nuclear-rim fluorescence of LBR-GFP became distorted, assuming a thickened and/or crenellated appearance (Fig. 1). Such alterations were observed in virtually all infected cells over the time course of infection. In addition, we observed a population of infected cells (approximately 20%) in which LBR-GFP was partially redistributed to a membranous cytoplasmic compartment (Fig. 1e and f), and occasionally LBR-GFP was also observed in intranuclear domains (Fig. 1f). Fluorescence imaging of live individual LBR-GFP-expressing cells during the course of infection revealed that cytoplasmic redistribution could occur as early as 8 h p.i.. It occurred in cells that previously showed normal nuclear-rim fluorescence, and cytoplasmic LBR-GFP was retained in this compartment as the infection progressed over at least 6 h (Fig. 1).
In an attempt to identify the cytoplasmic compartment containing LBR-GFP in infected cells, we undertook a series of immunofluorescence colocalization studies and used drug treatments to disrupt known cellular compartments. The ER-Golgi intermediate compartment, the Golgi complex, the trans-Golgi network, and the microtubule network were identified by using antibodies against β-COP, the 58K Golgi protein, γ-adaptin, and α-tubulin. Both Golgi and microtubule proteins in these cells were found to have an altered distribution in HSV-1-infected COS-1 cells, as had been previously reported for other cell types (2, 8). Although LBR-GFP was found closely juxtaposed to the altered Golgi compartment, there was no significant colocalization with any of the Golgi markers or with α-tubulin (data not shown). Since the altered Golgi and cytoskeletal compartments in infected cells made these colocalization experiments relatively difficult to interpret, we also used Brefeldin A or Nocodazol treatment, respectively, to disrupt these structures. However neither treatment had any effect on the compartment containing cytoplasmic LBR-GFP (data not shown).
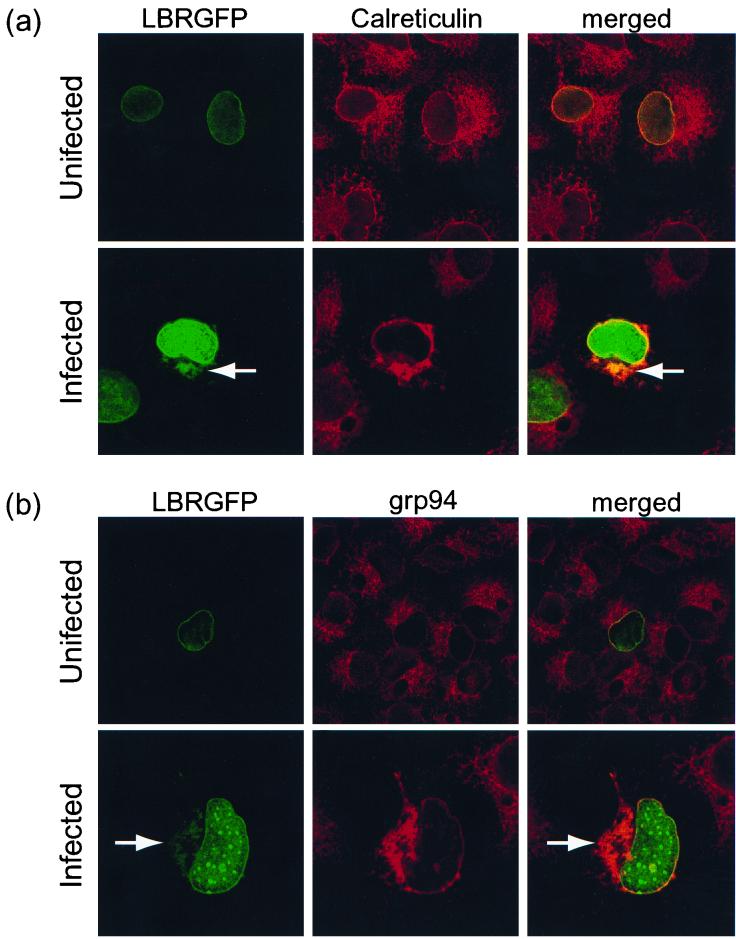
We next compared the localization of LBR-GFP to the position of the ER markers calreticulin and grp94. Antibodies against these proteins labeled a fine network of membranes throughout the cytoplasm of uninfected cells, consistent with ER staining (Fig. 2a and b). In HSV-1-infected cells, the ER appeared collapsed, being composed of broader structures generally limited to a juxtanuclear position (Fig. 2a and b). Cytoplasmic LBR-GFP was found to colocalize with both calreticulin (Fig. 2a) and grp94 (Fig. 2b, high magnification), indicating that it had redistributed to an altered ER in HSV-1-infected cells.
FIG. 2.
COS-1 cells transiently expressing LBR-GFP were either infected with HSV-1 at 10 PFU/cell or left uninfected and then fixed with methanol at 8 h p.i. Fixed cells were treated with antibodies to calreticulin (a) or grp94 (b) followed by secondary antibodies tagged with the fluorochrome Alexa 546 (red). Confocal dual-channel fluorescence images reveal some colocalization (yellow) of cytoplasmic LBR-GFP (arrows) with these ER markers.
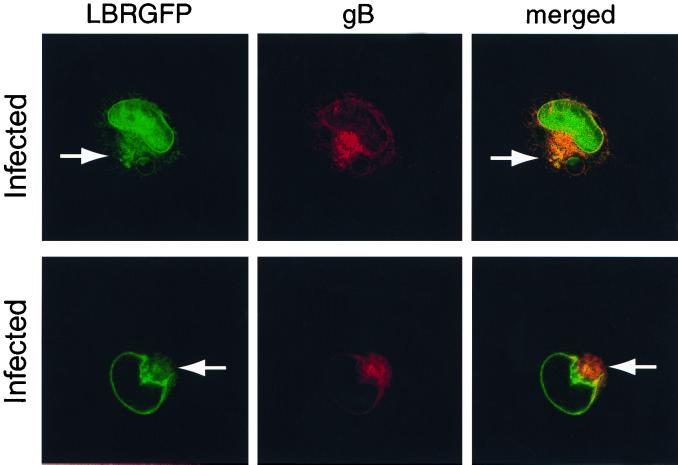
We also compared the localization of LBR-GFP and the viral gB in HSV-1-infected cells at 8 h p.i. gB localized to the nuclear rim as well as to membranous cytoplasmic structures (Fig. 3). This is consistent with previous reports describing gB in the nuclear envelope, ER, Golgi, and plasma membrane in infected cells (23, 30, 55). Although LBR-GFP showed a higher ratio of nuclear-rim to cytoplasmic fluorescence than gB, these proteins did colocalize at the nuclear rim and in a subset of gB-positive cytoplasmic membranes (Fig. 3). Taking this together with the Golgi and ER studies described above, we believe cytoplasmic LBR-GFP and gB are colocalizing within the ER.
FIG. 3.
COS-1 cells transiently expressing LBR-GFP were infected with HSV-1 at 10 PFU/cell and then fixed in methanol at 8 h p.i. Fixed cells were treated with antibody to HSV-1 and -2 gB followed by secondary antibodies tagged with Alexa 546 (red) and were imaged as for Fig. 4. The arrows indicate cytoplasmic LBR-GFP. LBR-GFP colocalized with gB at the nuclear rim and in a subset of the cytoplasmic membranes they occupy (yellow).
LBR-GFP shows increased lateral mobility within the nuclear envelope of HSV-1-infected cells.
FRAP analysis is a microscopy-based technique used to track the movement of fluorescently tagged proteins in live cells (56). A subpopulation of tagged proteins within the cellular structure of interest is irreversibly photobleached, and then the recovery of fluorescence within the bleached region is monitored. Recovery corresponds to the movement of intact fluorescent proteins from outside the bleached area into the region; the speed of recovery relates to the mobility of the protein within the cellular compartment. FRAP analysis has been used to demonstrate that LBR-GFP is immobilized at the inner nuclear membrane in interphase cells but is laterally mobile within the interphase ER and within mitotic membranes (15). We used FRAP analysis to compare the lateral mobilities of LBR-GFP within the inner nuclear membranes of uninfected and HSV-1-infected cells.
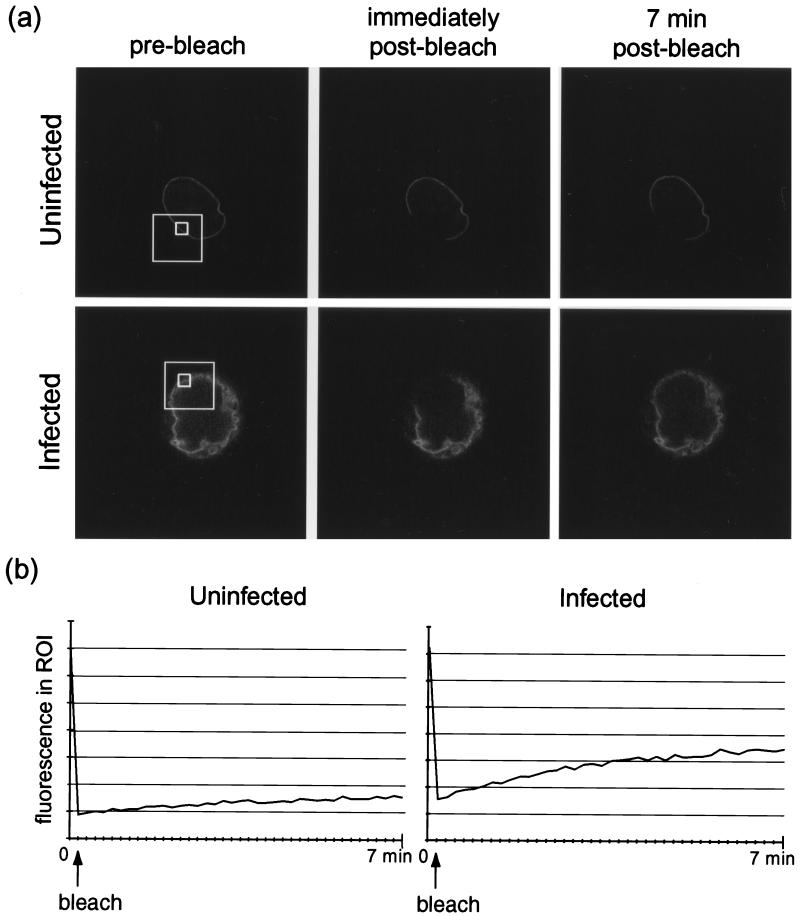
The distribution of LBR-GFP was examined in living cells by confocal microscopy using a low-intensity laser scan, and then a subpopulation of LBR-GFP at the nuclear rim was irreversibly photobleached by a high-intensity laser pulse applied to only a limited region (Fig. 4a, large white frame). Recovery of fluorescence in this region was monitored by a series of whole-cell low-intensity scans over a 7-min period following the bleaching.
FIG. 4.
COS-1 cells transiently expressing LBR-GFP were infected with HSV-1 at 10 PFU/cell or left uninfected and viewed live on a confocal microscope at 24 h p.i. (a) A restricted region (large frames) of the nuclear-rim fluorescence was irreversibly photobleached by a 10-s, 100% intensity laser pulse. Images of the whole cell were recorded before bleaching and at 10-s intervals for 7 min following the bleaching; selected images are shown. (b) LSM software was used to record fluorescence (in arbitrary units) in ROIs within the bleached area (panel a, small frames). These readings from the cells shown in panel a are plotted over time.
Qualitative analysis revealed that within this time there was little recovery of fluorescence in the bleached region of the nuclear rims of uninfected cells (Fig. 4a). Similar observations have previously been reported and indicate that LBR-GFP in INM regions flanking the bleach box is tightly tethered there by interactions with underlying chromatin and lamins. It is therefore not free to diffuse laterally into the bleached region (15, 47).
By way of semiquantitative analysis, we measured the fluorescence in defined ROIs in each time series of whole-cell images, and we present these representative plots in Fig. 4b. Data from at least five such plots from each condition were normalized to allow direct comparison (see Materials and Methods) and were used to calculate the mean recovery of fluorescence as a percentage of initial fluorescence (Table 1). As a positive control, we have included measurements of LBR-GFP fluorescence recovery in mitotic cells, where lateral diffusion of this protein is known to be higher than in interphase cells (15).
TABLE 1.
Mean recovery of fluorescence as a percentage of initial fluorescence
| Experimental condition | Mean recovery of fluorescence (%) | SE |
|---|---|---|
| Interphase uninfected | 13.1 | 1.7 |
| Mitotic uninfected | 26.0a | 3.8 |
| Infected (8 h) | 17.2 | 1.3 |
| Infected (24 h) | 22.4a | 3.9 |
Values significantly different from interphase uninfected cells by two-tailed t test.
In line with our qualitative conclusions, there was little recovery of fluorescence in a bleached ROI at the nuclear rim of an uninfected interphase cell (Fig. 4a, small frame). In contrast, bleached nuclear-rim regions of HSV-1-infected cells at 24 h p.i. showed significant recovery of fluorescence within the same 7-min time span (Fig. 4a). This is highlighted by the traces in Fig. 4b and data in Table 1, where significantly more recovery was observed in the nuclear rim of an infected cell compared to that of an uninfected cell. These data suggest that LBR-GFP becomes increasingly free to diffuse laterally within the INMs of infected cells. One explanation for this could be a virus-induced release of tethering at the INM, which in turn would be consistent with our observations of LBR-GFP in the ER of infected cells, as the untethered protein would be able to diffuse back into this compartment.
Nuclear lamina is also distorted and endogenous lamin levels are reduced in HSV-1-infected cells.
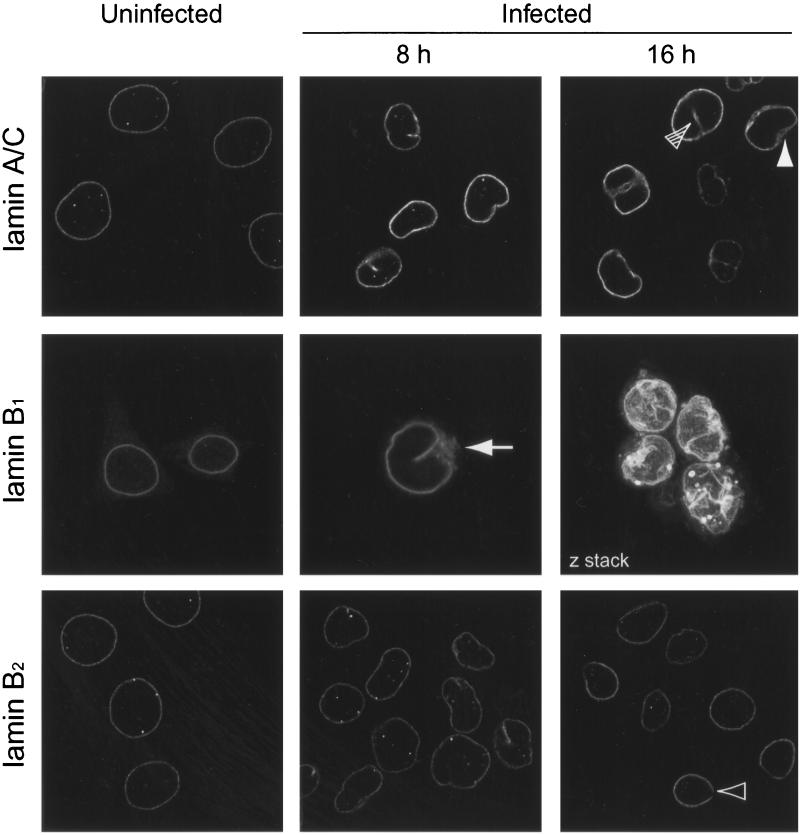
One plausible explanation for the release of LBR tethering could be the loss or modification of lamins within the underlying lamina. We have therefore studied the fate of endogenous lamins during HSV-1 infection both by indirect immunofluorescence and by Western blotting of cell lysates. Uninfected HeLa cells and HSV-1-infected cells fixed at 8 or 16 h p.i. were processed for immunofluorescence using antibodies against lamin A/C, B1, or B2. Lamins are classified as type A or type B, based primarily on sequence homology but also reflecting differences in their expression patterns, biochemical properties, and behavior at mitosis. Lamins A and C (type A) are alternative splice products of the LmnA gene, while lamin B1 and lamin B2 are encoded by separate genes.
Confocal microscope images of uninfected cells showed the expected nuclear-rim pattern of fluorescence for these lamins (Fig. 5). In addition, we observed some spots of lamin staining within the nuclei. These are likely to be intranuclear lamin structures which run perpendicular to the growth substrate and have previously been described (6, 20).
FIG. 5.
HeLa cells infected with HSV-1 at 10 PFU/cell were fixed in methanol at 8 or 16 h p.i. These and control uninfected cells were treated with antibodies against lamins A/C, B1, and B2 and with the appropriate FITC-tagged secondary antibody. Confocal images of staining by all three antibodies show nuclear-rim staining. This nuclear rim becomes increasingly distorted as the infection progresses. The solid arrowhead indicates an example of thickened lamina, the open arrowhead indicates a gap in lamina staining, and the hatched arrowhead indicates intranuclear staining in an infected cell. Lamin B1 antibody reveals partial redistribution of lamin to the cytoplasm (8 h p.i.; arrow). The lamin B1 image taken at 16 h p.i. is compiled from serial z sections through four infected cells.
In contrast to the relatively uniform nuclear-rim staining in uninfected cells, lamin rim staining in infected cells at 8 and 16 h p.i. appeared uneven; in some places it was thickened, and in others it was discontinuous. Representative examples of these features are highlighted in Fig. 5 (thickening, solid arrowhead; discontinuity, empty arrowhead). The path of the lamina followed the same pattern of gross nuclear distortions noted above for LBR-GFP in infected cells (seen best as combined z sections in Fig. 5, lamin B1 at 16 h) which also matched distortions of the nuclear envelope seen by phase-contrast microscopy (data not shown). In addition to the spots noted in uninfected cells, lines of intranuclear lamins were found in infected cells. These may correspond to invaginated furrows of the lamina and seem to be structures distinct from the lamins within uninfected nuclei, which are only seen as spots. It was noteworthy that the lamin B1 antibody also exhibited cytoplasmic staining in some HSV-1-infected cells (Fig. 5, 8 h, arrowhead). This occurred at a time p.i., and at a frequency, similar to the redistribution of LBR-GFP to the ER.
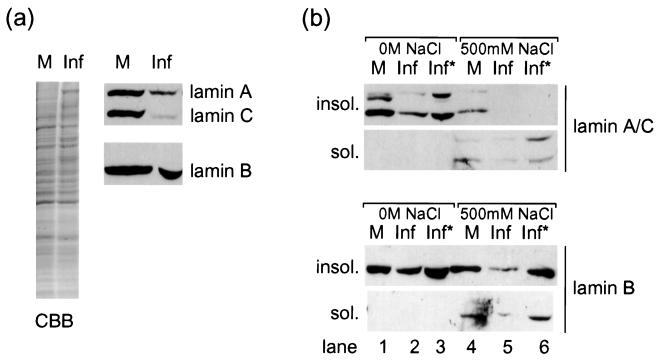
We also examined the fate of the total lamin population in infected cells by Western blotting. HSV-1-infected and mock-infected COS-1 cells were harvested at 24 h p.i., and total cell lysates were separated on an 8% polyacrylamide gel. The gel was then stained with Coomassie brilliant blue to confirm equal loading or transferred to nitrocellulose filters and blotted with antibodies against lamin A/C or lamin B2 (Fig. 6a). The results show that there was a significant decrease in lamin levels in infected compared to uninfected cell lysates. This was more pronounced for lamins A/C than for lamin B2. Although formally the decrease could be due to modification of the epitopes recognized by these antibodies, we believe this reflects loss of lamins in the infected cells (see below).
FIG. 6.
(a) Total cell lysates of COS-1 cells infected with HSV-1 at 10 PFU/cell or of control uninfected cells, stained with Coomassie brilliant blue (CBB) or blotted for lamins A/C or B2. Levels of lamin B2 and, more strikingly, lamins A/C are reduced in infected-cell lysates (Inf) compared to uninfected-cell lysates (M). (b) Soluble and insoluble fractions of COS-1 cells infected or uninfected as for panel a blotted for lamins A/C or B2. Lamins are insoluble (insol.) in a low-salt buffer (lanes 1 to 3). As in panel a, there is a loss of lamins in infected COS-1 cells which is most pronounced for lamins A/C. In high-salt buffers, lamins are partially soluble (sol.) (lanes 4 to 6). Loading double quantities (Inf*) reveals that the only detectable lamin A/C in infected cells is in the soluble fraction, while lamin B2 retains the predominantly insoluble profile seen in uninfected cells.
Endogenous A-type lamins from HSV-1-infected cell extracts show increased solubility compared to those from uninfected cells.
We also examined any change in the solubility of A- and B-type lamins during HSV-1 infection. Uninfected and infected cells were extracted in a low- or high-salt buffer, and each sample was separated into soluble and insoluble fractions for analysis by Western blotting as described above. As expected, lamins were largely insoluble in the low-salt buffer, and the blots of the insoluble fractions show essentially the same result as that described above for the total cell lysates. Thus, the insoluble fraction from infected cells contained lower levels of lamins A/C and a less pronounced but reproducible decrease in the level of lamin B2 than an uninfected control fraction (Fig. 6b, lanes 1 and 2). By loading double quantities of the infected cell fractions, it can be seen that this constitutes no more than a twofold fall in lamin A/C levels (compare lanes 1 and 3). It is also noteworthy that by this method of sample preparation the lamin A band (upper) was resolved into a doublet, and that in uninfected cells the majority of lamin A existed in the higher-mobility form (Fig. 6b, lane 1). In lane 3, both forms were still detectable, but most of the remaining lamin A was in the lower-mobility form, i.e., the faster form had been preferentially lost.
Lamins were more soluble in the high-salt buffer containing 500 mM NaCl (Fig. 6b, lane 4), and in uninfected cells, lamins A/C were distributed approximately evenly between soluble and insoluble fractions. In contrast, in fractions from infected cells, lamins A/C were predominantly lost from the insoluble fractions, where they were undetectable even when double quantities were loaded (Fig. 6b, lane 6). Under these conditions, lamin B2 levels were again lower in infected cell fractions (compare lanes 4 and 5), but compared to lamins A/C, lamin B2 retained the predominantly insoluble profile seen in uninfected cells (compare lanes 4 and 6).
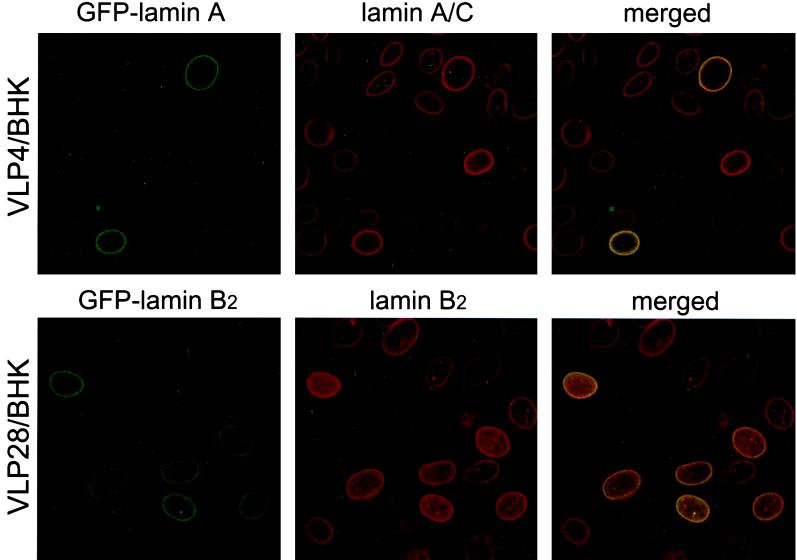
Live-cell analysis of HSV-1 infection in GFP-lamin stable cell lines reveals nuclear lamina distortion and lamin loss.
We wished to visualize the fate of lamins at the level of individual infected cells and to establish whether the fall in lamin levels described above represents reduced epitope accessibility by the antibodies used or true loss of lamin protein. To this end, we have generated and characterized cell lines stably expressing GFP-tagged lamins. BHK-21 cells were transfected with plasmids encoding GFP-lamin A (pVLP4) or GFP-lamin B2 (pVLP28) and grown in culture medium containing 250 μg of Geneticin/ml to select for expression from these vectors. These cell lines, referred to as VLP4/BHK and VLP28/BHK, both exhibited nuclear-rim fluorescence which colocalized with endogenous lamin staining by indirect immunofluorescence (Fig. 7). GFP-lamin levels were heterogeneous within the population and were only detected in a subpopulation of the cells. Before investigating the effect of HSV-1 infection on GFP-lamin fate, we compared the infectivity of the virus in these cells and in the parental BHK-21 cell line. The virus showed comparable titer on VLP4/BHK, VLP28/BHK, and BHK-21 cells by plaque assay (data not shown). Due to the heterogeneity of the GFP-lamin expression, we cannot unequivocally conclude that the presence of the GFP-lamin has no effect on virus replication. However, attempts to obtain more-homogeneous lines were unsuccessful, and our present lines offer the best opportunity to study the fate of lamins after infection in living cells.
FIG. 7.
BHK-21 cells stably expressing GFP-lamin A (VLP4/BHK) or GFP-lamin B2 (VLP28/BHK) were fixed in methanol and treated with antibodies for lamin A/C and B2, respectively, followed by Alexa 546-tagged secondary antibodies. The locations of the GFP-lamin or the appropriate native lamin were then recorded separately by confocal microscopy. Dual-channel fluorescence images reveal colocalization (yellow) between GFP-lamins and endogenous lamins at the nuclear rim.
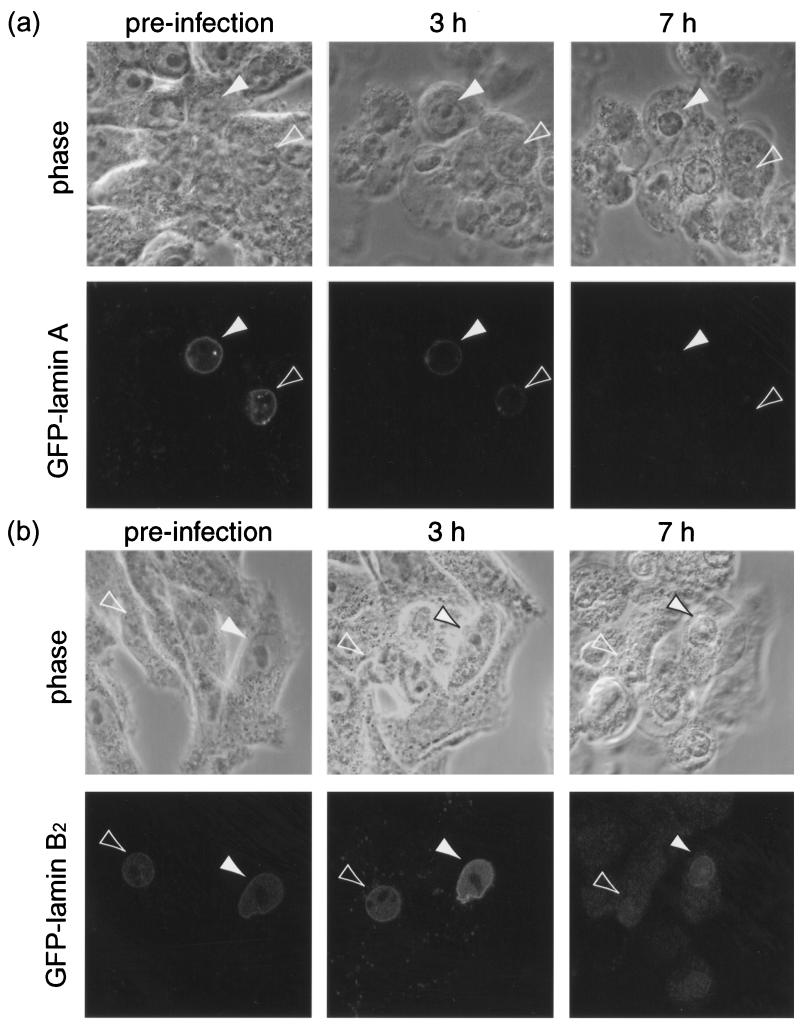
We next studied the fate of GFP-lamins during the course of infection. VLP4/BHK and VLP28/BHK cells were seeded on gridded coverslips, and live confocal images (both fluorescent and phase) of cells expressing GFP-lamin A or GFP-lamin B2 were obtained. The cells were then infected, and further live images of the same fields of view were captured during the course of infection. We used identical imaging settings at each time point to monitor changes in both the intensity and the distribution of GFP-lamin fluorescence during the study.
In all cases, whether tracking GFP-tagged viral components using recombinant virus or following unmarked virus in cells expressing GFP-tagged cellular components, some heterogeneity in the progression of infection is observed. For each cell type, numerous individual cells were therefore tracked throughout the course of infection. Of the individual infected VLP4/BHK cells expressing GFP-lamin A which were tracked (n = 14), more than half showed cytopathic effects (CPE) by 3 h p.i. which was accompanied in all cases by a significant drop in GFP-lamin A fluorescence, in some cases to virtually undetectable levels (Fig. 8a). Nuclear-rim fluorescence also became distorted in several infected cells, although in many the GFP signal had faded by the first time point p.i., meaning any such distortion was not recorded. By contrast, in the control study of uninfected VLP4/BHK cells, only 2 of 18 cells showed significant fading or distortion of GFP-lamin A fluorescence over a 24-h period.
FIG. 8.
Examples of individual BHK-21 cells stably expressing GFP-lamin A (a) or GFP-lamin B2 (b) and infected with HSV-1 at 10 PFU/cell were followed by phase and fluorescence confocal microscopy during the course of infection. The arrowheads indicate cells expressing GFP-lamin which exhibit CPE and reduced nuclear-rim fluorescence as the infection progresses.
In a parallel study tracking individual infected VLP28/BHK cells (n = 48), the majority of GFP-lamin B2-positive cells that showed relatively early CPE (noted at 3- or 8-h p.i. time points) subsequently showed reduced nuclear-rim fluorescence (15 of 22 cells) (Fig. 8b). A smaller proportion of cells that were slower to show CPE also exhibited reduced nuclear-rim GFP-lamin B2 (7 of 26 cells). Over half of these infected cells had distorted nuclear-rim fluorescence as we have noted above for endogenous lamins and for GFP-LBR. Again, this contrasted with the uninfected VLP28/BHK cells, which showed little variation in GFP-lamin B2 intensity or distribution over a 24-h period.
Thus, the reduction in lamin levels (approximately twofold) in HSV-1-infected cells, observed by Western blotting, was confirmed by live-cell analysis of GFP-lamins. The overall reduction in the population seen by blotting probably reflects different levels of reduction in individual cells, and together the data indicate that the observed reduction in lamin levels constitutes true loss of these proteins.
DISCUSSION
The interactions of herpesviruses with the nuclear lamina and envelope at the beginning of viral egress are poorly understood. Published ultrastructural studies to date have provided a detailed but static picture of the process. We have attempted to complement these findings with observations of HSV-1 infection in live cells carrying GFP-tagged lamina and envelope components. We have expanded on these using immunofluorescence microscopy in fixed cells and biochemical analysis. Several lines of evidence from our work taken together provide a more detailed picture of an HSV-1-induced dismantling of the nuclear lamina.
Alterations in the distribution of the INM protein LBR in HSV-1-infected cells was our initial indicator that envelope and possibly lamina components were perturbed. LBR underwent a partial redistribution from a predominantly INM localization to a significant proportion accumulating in the ER. The timing of LBR redistribution, seen from 8 h p.i., is consistent with a reorganization of nuclear envelope structure as egress of nucleocapsids begins. This is accompanied by an increase in the lateral mobility of LBR at the nuclear rims of infected cells as determined by FRAP analysis. LBR has been shown to interact with several partners (32, 43, 51, 58, 59), and of these, the lamins and chromatin have been proposed as fixed, structural nucleoplasmic ligands capable of immobilizing LBR at the INM (15). Disruption of these interactions accounts for its release at the onset of mitosis, and we propose that a similar disruption could occur prior to egress in HSV-1 infection. In mitosis, the severance of INM proteins from lamina and chromatin is comprehensive, resulting in the total dismantling of the nuclear envelope. In infection, the process appears to be more limited, so that the envelope remains intact but distorted and at least some of its components are released to the ER.
Immunofluorescence studies of endogenous lamins provided direct evidence of lamina disruption during HSV-1 infection. In contrast to the uniform nuclear-rim distribution seen in uninfected cells, lamin staining at the nuclear peripheries of infected cells was of irregular thickness and was even discontinuous in some cells. In addition, the gross structure of the lamina itself appeared distorted, in a manner similar to that of the INM distortions seen with LBR-GFP. While lamins showed clear qualitative differences in their distribution by immunofluorescence, heterogeneity of lamin levels across the population and experimental variation in staining levels made it impossible to confirm quantitative changes by this method. Analysis of total lamin levels in the whole cell population by Western blotting showed a decrease in infected cells which was more pronounced for lamin A/C but also present for lamin B2. Furthermore, the generation of cell lines stably expressing GFP-tagged lamins allowed us to follow the fates of these proteins in individual cells during the course of infection. These observations were consistent with the Western blotting data in that a significant proportion of infected cells showed a decrease in nuclear-rim GFP-lamin fluorescence, often to undetectable levels.
While LBR tethering is altered in virus infection, the question still remains as to which binding partner LBR is dissociating from and what is the precise mechanism involved. As its name suggests, LBR's first binding partner to be characterized was lamin B (50). However, this interaction is maintained following dismantling of the nuclear envelope during mitosis (33). Our observation that both LBR and lamin B1 are present in a cytoplasmic compartment at 8 h p.i. suggests that these two components may also remain associated during infection. In this case, the release of LBR tethering could be an indirect consequence of a disruption between lamin B and lamin A/C components of the lamina, such that the LBR-lamin B would be released as a complex. This scenario, in which lamina disassembly and envelope disruption are driven primarily by lamin A/C loss, would be consistent with our data showing that levels of A-type more than B-type lamins are reduced in infected cells and that A-type lamins become more soluble. It is noteworthy that A-type lamins are also more soluble than the B type during mitosis (22). The latter remain membrane associated by virtue of the hydrophobic C terminus which A-type lamins have lost following a posttranslational cleavage.
A high-resolution EM study of nuclear lamina structure, albeit in Xenopus oocytes, describes a regular meshwork with an average crossover spacing of 52 nm, which could be expected to restrict nucleocapsid access to the INM (1). Even a small-scale thinning of this meshwork, following depolymerization and loss of some lamin components, would be predicted to dramatically increase this access. The effects of lamin loss in mammalian cells have been described: a knock-out mouse lacking the lamin A gene displays muscular dystrophy (54). At the cellular level, nuclear envelope structure is perturbed and several envelope components are mislocalized, including the lamin A/C INM receptor emerin. In cells lacking lamin A/C, emerin shows partial cytoplasmic accumulation similar to that of LBR-GFP in HSV-1 infection.
We envisage that during virus infection a reduction in lamin levels results in a dismantling of the lamina, either as a uniform thinning or in discrete foci. This is particularly pronounced for A-type lamins, whose depolymerization from lamina fibers is inferred from their increased solubility in infected cells. The consequences of this dismantling would include the release of lamin B and its partner, LBR, allowing them to diffuse into the ER, and the distortion of both the lamina and nuclear membrane. Crucially, nucleocapsid access to the INM for primary envelopment would be increased.
Our data match these predictions of lamin and LBR release and of nuclear envelope distortion. They are also reminiscent of the fates of lamina and INM proteins in the processes of mitosis and apoptosis. These programs both involve lamina disassembly but by different mechanisms. In mitosis, lamin depolymerization is triggered by phosphorylation at sites flanking the conserved lamin rod domain by kinases, including p34 (also called cdc2) (17). In apoptosis, lamins, as well as a subset of INM and nuclear matrix proteins, are cleaved by the cysteine proteases caspase-3 and -6 (7, 14, 26, 39). More recently, lamin hyperphosphorylation has also been observed prior to lamin cleavage in apoptosis. Candidate kinases include p34, PKC-α, and PKC-δ (11, 48, 49), although their involvement has been disputed (38).
Our data are more consistent with the mitosis model, as we have not detected significant levels of lamin cleavage products during HSV-1 infection. Also, the selective loss of the higher-mobility form of lamin A could be the result of lamin hyperphosphorylation, as seen in mitosis. Interestingly, lamin dephosphorylation has been observed in human cytomegalovirus infection (44), although this was reported not to occur in HSV-1 infection (45). If lamina disassembly in HSV-1 infection does share early steps with mitotic nuclear envelope breakdown, then the question might also arise as to how the virus restricts the process to a partial dismantling. The mechanism behind HSV-1-induced lamina disassembly is the subject of ongoing investigation.
ACKNOWLEDGMENTS
This work was funded by the Marie Curie Cancer Care.
We are very grateful to Jan Ellenberg and Tom Rapoport for provision of plasmids, Murray Stewart for the lamin B1 antibody, and Tony Minson for VP16 antibody.
REFERENCES
- 1.Aebi U, Cohn J, Buhle L, Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 1986;323:560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- 2.Avitabile E, Gaeta S D, Torrisi M R, Ward P, Roizman B, Campadelli-Fiume G. Redistribution of microtubules and golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J Virol. 1995;69:7472–7482. doi: 10.1128/jvi.69.12.7472-7482.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines J D, Jacob R J, Simmerman L, Roizman B. The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J Virol. 1995;69:825–833. doi: 10.1128/jvi.69.2.825-833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baines J D, Roizman B. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J Virol. 1992;66:5168–5174. doi: 10.1128/jvi.66.8.5168-5174.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belmont A S, Zhai Y, Thilenius A. Lamin B distribution and association with peripheral chromatin revealed by optical sectioning and electron microscopy tomography. J Cell Biol. 1993;123:1671–1685. doi: 10.1083/jcb.123.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broers J, Nachiels B, van Eys G, Kuijpers H, Manders E, van Driel R, Ramaekers F. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J Cell Sci. 1999;112:3463–3475. doi: 10.1242/jcs.112.20.3463. [DOI] [PubMed] [Google Scholar]
- 7.Buendia B, Santa-Maria A, Courvalin J C. Caspase-dependent proteolysis of integral and peripheral proteins of nuclear membranes and nuclear pore complex proteins during apoptosis. J Cell Sci. 1999;112:1743–1753. doi: 10.1242/jcs.112.11.1743. [DOI] [PubMed] [Google Scholar]
- 8.Campadelli G, Brandimarti R, Di Lazzaro C, Ward P L, Roizman B, Torrisi M R. Fragmentation and dispersal of Golgi proteins and redistribution of glycoproteins and glycolipids processed through the Golgi apparatus after infection with herpes simplex virus 1. Proc Natl Acad Sci USA. 1993;90:2798–2802. doi: 10.1073/pnas.90.7.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang Y E, Roizman B. The product of the UL31 gene of herpes simplex virus 1 is a nuclear phosphoprotein which partitions with the nuclear matrix. J Virol. 1993;67:6348–6356. doi: 10.1128/jvi.67.11.6348-6356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu A, Rassadi R, Stochaj U. Velcro in the nuclear envelope: LBR and LAPs. FEBS Lett. 1998;441:165–169. doi: 10.1016/s0014-5793(98)01534-8. [DOI] [PubMed] [Google Scholar]
- 11.Cross T, Griffiths G, Deacon E, Sallis R, Gough M, Watters D, Lord J M. PKC-delta is an apoptotic lamin kinase. Oncogene. 2000;19:2331–2337. doi: 10.1038/sj.onc.1203555. [DOI] [PubMed] [Google Scholar]
- 12.Darlington R W, Moss L H d. Herpesvirus envelopment. J Virol. 1968;2:48–55. doi: 10.1128/jvi.2.1.48-55.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dechat T, Vlcek S, Foisner R. Review: lamina-associated polypeptide 2 isoforms and related proteins in cell cycle-dependent nuclear structure dynamics. J Struct Biol. 2000;129:335–345. doi: 10.1006/jsbi.2000.4212. [DOI] [PubMed] [Google Scholar]
- 14.Duband-Goulet I, Courvalin J C, Buendia B. LBR, a chromatin and lamin binding protein from the inner nuclear membrane, is proteolyzed at late stages of apoptosis. J Cell Sci. 1998;111:1441–1451. doi: 10.1242/jcs.111.10.1441. [DOI] [PubMed] [Google Scholar]
- 15.Ellenberg J, Siggia E, Moreira J, Smith C, Presley J, Worman H, Lippincott-Schwartz J. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enquist L W, Husak P J, Banfield B W, Smith G A. Infection and spread of alphaherpesviruses in the nervous system. Adv Virus Res. 1998;51:237–347. doi: 10.1016/s0065-3527(08)60787-3. [DOI] [PubMed] [Google Scholar]
- 17.Fields A P, Thompson L J. The regulation of mitotic nuclear envelope breakdown: a role for multiple lamin kinases. Prog Cell Cycle Res. 1995;1:271–286. doi: 10.1007/978-1-4615-1809-9_22. [DOI] [PubMed] [Google Scholar]
- 18.Foisner R. Dynamic organisation of intermediate filaments and associated proteins during the cell cycle. Bioessays. 1997;19:297–305. doi: 10.1002/bies.950190407. [DOI] [PubMed] [Google Scholar]
- 19.Foster T P, Kousoulas K G. Genetic analysis of the role of herpes simplex virus type 1 glycoprotein K in infectious virus production and egress. J Virol. 1999;73:8457–8468. doi: 10.1128/jvi.73.10.8457-8468.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fricker M, Hollinshead M, White N, Vaux D. Interphase nuclei of many mammalian cell types contain deep, dynamic, tubular membrane-bound invaginations of the nuclear envelope. J Cell Biol. 1997;136:531–544. doi: 10.1083/jcb.136.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furukawa K, Fritze C E, Gerace L. The major nuclear envelope targeting domain of LAP2 coincides with its lamin binding region but is distinct from its chromatin interaction domain. J Biol Chem. 1998;273:4213–4219. doi: 10.1074/jbc.273.7.4213. [DOI] [PubMed] [Google Scholar]
- 22.Gerace L, Blobel G. The nuclear envelope lamina is reversibly depolymerized during mitosis. Cell. 1980;19:277–287. doi: 10.1016/0092-8674(80)90409-2. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert R, Ghosh H P. Immunoelectron microscopic localization of herpes simplex virus glycoprotein gB in the nuclear envelope of infected cells. Virus Res. 1993;28:217–231. doi: 10.1016/0168-1702(93)90023-g. [DOI] [PubMed] [Google Scholar]
- 24.Goldberg M W, Allen T D. High resolution scanning electron microscopy of the nuclear envelope: demonstration of a new, regular, fibrous lattice attached to the baskets of the nucleoplasmic face of the nuclear pores. J Cell Biol. 1992;119:1429–1440. doi: 10.1083/jcb.119.6.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldberg M W, Allen T D. The nuclear pore complex and lamina: three-dimensional structures and interactions determined by field emission in-lens scanning electron microscopy. J Mol Biol. 1996;257:848–865. doi: 10.1006/jmbi.1996.0206. [DOI] [PubMed] [Google Scholar]
- 26.Gotzmann J, Vlcek S, Foisner R. Caspase-mediated cleavage of the chromosome-binding domain of lamina-associated polypeptide 2 alpha. J Cell Sci. 2000;113:3769–3780. doi: 10.1242/jcs.113.21.3769. [DOI] [PubMed] [Google Scholar]
- 27.Gruenbaum Y, Wilson K L, Harel A, Goldberg M, Cohen M. Review: nuclear lamins—structural proteins with fundamental functions. J Struct Biol. 2000;129:313–323. doi: 10.1006/jsbi.2000.4216. [DOI] [PubMed] [Google Scholar]
- 28.Hutchinson L, Roop-Beauchamp C, Johnson D C. Herpes simplex virus glycoprotein K is known to influence fusion of infected cells, yet is not on the cell surface. J Virol. 1995;69:4556–4563. doi: 10.1128/jvi.69.7.4556-4563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitten G T, Nigg E A. The CaaX motif is required for isoprenylation, carboxyl methylation, and nuclear membrane association of lamin B2. J Cell Biol. 1991;113:13–23. doi: 10.1083/jcb.113.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koga J, Chatterjee S, Whitley R J. Studies on herpes simplex virus type 1 glycoproteins using monoclonal antibodies. Virology. 1986;151:385–389. doi: 10.1016/0042-6822(86)90059-0. [DOI] [PubMed] [Google Scholar]
- 31.Loewinger L, McKeon F. Mutations in the nuclear lamin proteins resulting in their aberrant assembly in the cytoplasm. EMBO J. 1988;7:2301–2309. doi: 10.1002/j.1460-2075.1988.tb03073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martins S B, Eide T, Steen R L, Jahnsen T, Skalhegg B S, Collas P. HA95 is a protein of the chromatin and nuclear matrix regulating nuclear envelope dynamics. J Cell Sci. 2000;113:3703–3713. doi: 10.1242/jcs.113.21.3703. [DOI] [PubMed] [Google Scholar]
- 33.Meier J, Georgatos S D. Type B lamins remain associated with the integral nuclear envelope protein p58 during mitosis: implications for nuclear reassembly. EMBO J. 1994;13:1888–1898. doi: 10.1002/j.1460-2075.1994.tb06458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moir R D, Spann T P, Lopez-Soler R I, Yoon M, Goldman A E, Khuon S, Goldman R D. Review: the dynamics of the nuclear lamins during the cell cycle—relationship between structure and function. J Struct Biol. 2000;129:324–334. doi: 10.1006/jsbi.2000.4251. [DOI] [PubMed] [Google Scholar]
- 35.Nigg E A. Assembly-disassembly of the nuclear lamina. Curr Opin Cell Biol. 1992;4:105–109. doi: 10.1016/0955-0674(92)90066-l. [DOI] [PubMed] [Google Scholar]
- 36.Nii S. Electron microscopic study on the development of herpesviruses. J Electron Microsc. 1992;41:414–423. [PubMed] [Google Scholar]
- 37.Nii S, Morgan C, Rose H M. Electron microscopy of herpes simplex virus. II. Sequence of development. J Virol. 1968;2:517–536. doi: 10.1128/jvi.2.5.517-536.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oberhammer F A, Hochegger K, Froschl G, Tiefenbacher R, Pavelka M. Chromatin condensation during apoptosis is accompanied by degradation of lamin A+B, without enhanced activation of cdc2 kinase. J Cell Biol. 1994;126:827–837. doi: 10.1083/jcb.126.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orth K, Chinnaiyan A M, Garg M, Froelich C J, Dixit V M. The CED-3/ICE-like protease Mch2 is activated during apoptosis and cleaves the death substrate lamin A. J Biol Chem. 1996;271:16443–16446. [PubMed] [Google Scholar]
- 40.Ostlund C, Ellenberg J, Hallberg E, Lippincott-Schwartz J, Worman H J. Intracellular trafficking of emerin, the Emery-Dreifuss muscular dystrophy protein. J Cell Sci. 1999;112:1709–1719. doi: 10.1242/jcs.112.11.1709. [DOI] [PubMed] [Google Scholar]
- 41.Paddy M, Belmont A, Saumweer H, Aard D, Sedat J. Interphase nuclear envelope lamins form a discontinuous network that interacts with only a fraction of the chromatin in the nuclear periphery. Cell. 1990;62:89–106. doi: 10.1016/0092-8674(90)90243-8. [DOI] [PubMed] [Google Scholar]
- 42.Purves F C, Spector D, Roizman B. UL34, the target of the herpes simplex virus U(S)3 protein kinase, is a membrane protein which in its unphosphorylated state associates with novel phosphoproteins. J Virol. 1992;66:4295–4303. doi: 10.1128/jvi.66.7.4295-4303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pyrpasopoulou A, Meier J, Maison C, Simos G, Georgatos S D. The lamin B receptor (LBR) provides essential chromatin docking sites at the nuclear envelope. EMBO J. 1996;15:7108–7119. [PMC free article] [PubMed] [Google Scholar]
- 44.Radsak K, Brucher K, Georgatos S. Focal nuclear envelope lesions and specific nuclear lamin A/C dephosphorylation during infection with human cytomegalovirus. Eur J Cell Biol. 1991;54:299–304. [PubMed] [Google Scholar]
- 45.Radsak K, Schneider D, Jost E, Brucher K H. Alteration of nuclear lamina protein in human fibroblasts infected with cytomegalovirus (HCMV) Arch Virol. 1989;105:103–112. doi: 10.1007/BF01311120. [DOI] [PubMed] [Google Scholar]
- 46.Roller R J, Zhou Y, Schnetzer R, Ferguson J, DeSalvo D. Herpes simplex virus type 1 U(L)34 gene product is required for viral envelopment. J Virol. 2000;74:117–129. doi: 10.1128/jvi.74.1.117-129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rolls M, Stein P, Taylor S, Ha E, McKeon F, Rapoport T. A visual screen of a GFP-fusion library identifies a new type of nuclear envelope membrane protein. J Cell Biol. 1999;146:29–43. doi: 10.1083/jcb.146.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi L, Nishioka W K, Th'ng J, Bradbury E M, Litchfield D W, Greenberg A H. Premature p34cdc2 activation required for apoptosis. Science. 1994;263:1143–1145. doi: 10.1126/science.8108732. [DOI] [PubMed] [Google Scholar]
- 49.Shimizu T, Cao C X, Shao R G, Pommier Y. Lamin B phosphorylation by protein kinase calpha and proteolysis during apoptosis in human leukemia HL60 cells. J Biol Chem. 1998;273:8669–8674. doi: 10.1074/jbc.273.15.8669. [DOI] [PubMed] [Google Scholar]
- 50.Simos G, Georgatos S D. The inner nuclear membrane protein p58 associates in vivo with a p58 kinase and the nuclear lamins. EMBO J. 1992;11:4027–4036. doi: 10.1002/j.1460-2075.1992.tb05496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simos G, Maison C, Georgatos S D. Characterization of p18, a component of the lamin B receptor complex and a new integral membrane protein of the avian erythrocyte nuclear envelope. J Biol Chem. 1996;271:12617–12625. doi: 10.1074/jbc.271.21.12617. [DOI] [PubMed] [Google Scholar]
- 52.Soullam B, Worman H J. Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J Cell Biol. 1995;130:15–27. doi: 10.1083/jcb.130.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spann T, Moir R, Goldman A, Stick R, Goldman R. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J Cell Biol. 1997;136:1201–1212. doi: 10.1083/jcb.136.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart C, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–919. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Torrisi M R, Di Lazzaro C, Pavan A, Pereira L, Campadelli-Fiume G. Herpes simplex virus envelopment and maturation studied by fracture label. J Virol. 1992;66:554–561. doi: 10.1128/jvi.66.1.554-561.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White J, Stelzer E. Photobleaching GFP reveals protein dynamics inside living cells. Trends Cell Biol. 1999;9:61–65. doi: 10.1016/s0962-8924(98)01433-0. [DOI] [PubMed] [Google Scholar]
- 57.Ye G J, Roizman B. The essential protein encoded by the UL31 gene of herpes simplex virus 1 depends for its stability on the presence of UL34 protein. Proc Natl Acad Sci USA. 2000;97:11002–11007. doi: 10.1073/pnas.97.20.11002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye Q, Worman H. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J Biol Chem. 1996;271:14653–14656. doi: 10.1074/jbc.271.25.14653. [DOI] [PubMed] [Google Scholar]
- 59.Ye Q, Worman H. Primary structure analysis and lamin B and DNA binding of human LBR, an integral protein of the nuclear envelope inner membrane. J Biol Chem. 1994;269:11306–11311. [PubMed] [Google Scholar]