Abstract
Objectives
Given substantial obstacles surrounding health data acquisition, high-quality synthetic health data are needed to meet a growing demand for the application of advanced analytics for clinical discovery, prediction, and operational excellence. We highlight how recent advances in large language models (LLMs) present new opportunities for progress, as well as new risks, in synthetic health data generation (SHDG).
Materials and Methods
We synthesized systematic scoping reviews in the SHDG domain, recent LLM methods for SHDG, and papers investigating the capabilities and limits of LLMs.
Results
We summarize the current landscape of generative machine learning models (eg, Generative Adversarial Networks) for SHDG, describe remaining challenges and limitations, and identify how recent LLM approaches can potentially help mitigate them.
Discussion
Six research directions are outlined for further investigation of LLMs for SHDG: evaluation metrics, LLM adoption, data efficiency, generalization, health equity, and regulatory challenges.
Conclusion
LLMs have already demonstrated both high potential and risks in the health domain, and it is important to study their advantages and disadvantages for SHDG.
Keywords: large language models, generative artificial intelligence, synthetic data, health equity, responsible AI
Introduction
The recent release of generative large language models (LLMs), such as OpenAI’s GPT models1 and Google’s PaLM2 has generated both robust enthusiasm as well as significant concern related to the use of generative artificial intelligence (AI) in healthcare.3–5 Numerous potential applications for healthcare have been documented, including processing of administrative data, such as discharge summary generation, interfacing as a chatbot with doctors for diagnosis or treatment determination, interfacing as a chatbot with patients for mental healthcare delivery, producing clinical trial documentation, intelligent tagging of patient images (eg, radiology or pathology images), and creation of educational health material.6–14 General-purpose LLMs have been found to achieve high performance on clinical licensing exams and comprehensive medical Q&A benchmarks,15–17 and LLMs trained on medical data have successfully augmented clinician diagnostic performance.18
In this perspective, we focus on 1 particularly promising avenue for LLMs: the creation of synthetic health data. There is a significant need for augmented datasets, as health data are often limited in size, may be costly to collect, not representative of diverse populations, and privacy concerns limit its sharing.19,20 Machine learning models for prediction and classification often require large datasets, particularly for deep learning models common in health.21 Yet datasets can be insufficiently large due to the rapid evolution of diseases, such as COVID-19,22 rarity of disease,23 or the myriad obstacles to sharing and acquiring existing health data, including ethical, legal, political, economic, cultural, and technical barriers.24,25 Synthetic data provide a unique opportunity for health dataset expansion or creation, by addressing privacy concerns and other barriers. We build on existing research on the state of the art in synthetic health data generation (SHDG)20,23,26–32 and broad exploration of the potential risks and opportunities for LLMs in healthcare.6 We contribute to the literature with a focused assessment of LLMs for SHDG, including a review of early research in the area and recommendations for future research directions (see Figure 1 for a summary of the paper’s key concepts).
Figure 1.
An overview of the concepts and research directions discussed in the article.
Synthetic health data generation
Synthetic data can be characterized by a combination of their resemblance to and distance from real data—they aim to mimic the statistical distribution and usability of real data while restricting reidentification of original data points (ie, individuals). Common characteristics for assessing synthetic datasets include data realism, the extent to which the synthetic datasets resemble and reflect patterns in real datasets, utility, measured by the performance on predictive tasks, and privacy, evaluated by the risk of identification of patients or attributes in the original data.20
Non-LLM approaches and unresolved challenges
Standard synthetic data generation methods seek to simulate the generating process of the original data through an estimation of the original data distributions. Classical statistical approaches include kernel density estimation33 and Markov Chain Monte Carlo.34 However, these methods often impose limiting assumptions on the data distribution, which precludes estimation of the complex correlation structure typically found in medical data.20 Popular state-of-the-art methods include Generative Adversarial Networks (GANs)35 and variational autoencoders.36 GANs are composed of 2 neural networks, a “generator” and a “discriminator,” trained in tandem. The generator’s objective is to create synthetic data indistinguishable from the real data, and the discriminator’s objective is to differentiate between the generated and real data. Variational autoencoders are also composed of 2 neural network models: the “encoder” aims to compress an original dataset into a latent representation and the “decoder” aims to decompress the latent representation back to the original data. The decoder can then be used to generate synthetic data. These methods have been used to generate a variety of health data types, including both snapshot and longitudinal electronic health record (EHR) data, sociodemographic data, lab/measurement data, and image data.30,37–41 We provide illustrative applications of these methods with recent examples from the literature.
Li et al42 developed a GAN-based model to simultaneously generate multiple types of clinical time series data. After training on 141 488 unique patients’ intensive care unit (ICU) data, they were able to synthesize sequences of patients’ health indicators, including oxygen level, blood pressure, and heart rate. They demonstrate that augmenting their real training data with synthetic data to increase training data size improves performance on a downstream task of predicting patients’ need for mechanical ventilation or vasopressors. However, this is achieved through significant data pre-processing, including imputation, smoothing, and truncation of time series. Biswal et al43 proposed a variational autoencoder model to generate patients’ clinical encounters and their corresponding features, including diagnoses, medications, and procedures. The authors were able to generate longitudinal encounter sequences for a chosen disease. The paper highlighted that complex longitudinal clinical interactions can be generated, but with the limitation of not being able to account for comorbidities from more than 1 disease. Collectively, the literature suggests that previous approaches can generate a wide variety of health data types, can focus on generation for specific health conditions, and incorporate privacy into model training.44 A variety of other models have also been developed to handle varying time periods between clinical visits,45 missing or incomplete original data,46 and incorporation of disease-specific domain knowledge.47 However, these models each have their drawbacks, such as their limited focus on 1 data modality or on 1 disease, or their inflexible data pre-processing requirements.
However, many technical challenges remain. Hernandez et al26 and Ghosheh et al31 observe that current GAN-based approaches are tailored to specific data structures and contexts, making generalization and transfer between contexts difficult. Ghosheh et al31 also point out GAN’s lack of ability to generate complex multimodal data, which has been shown to improve predictive performance.48 Augmentation of real data with synthetic data has the potential to improve multimodality models’ performance, yet current methods cannot generate data across data types. Murtaza et al20 similarly point out that while existing methods have shown proficiency at synthesizing disease-specific longitudinal EHR data, “generating comprehensive longitudinal records with co-morbidities remains an open challenge.” Another issue is the difficulty of incorporating expert knowledge into generation methods, whether in the form of disease progression models or constraints on clinical knowledge violation.20 Yan et al49 found in their benchmarking study that all tested models made mistakes in assigning gender-specific disease codes to varying degrees. LLMs have the potential to overcome these current limitations.
Application of LLMs for SHDG
Although several studies have explored the use of LLMs for SHDG, both in the context of text-based tasks, such as generation or augmentation of clinical language,50,51 and tabular EHR data tasks,52–54 developments in this area are still nascent, focusing largely on proof-of-concept rather than field applications. Table 1 summarizes key recent studies investigating SHDG with LLMs. One of these studies, Yuan et al, 50 addressed the issue of matching patients, based on their EHR data, to clinical trials, accounting for the trials’ inclusion and exclusion criteria. Existing models generally have had limited success due to terminology discordance across the datasets, which renders matching more challenging. Thus, Yuan et al50 used ChatGPT to augment inclusion and exclusion criteria descriptions for clinical trials in order to facilitate improved matching. Tang et al51 focused on the challenge of acquiring labeled data for 2 text classification tasks: recognition of biomedical vocabulary, or “entities,” and extraction of relationships between those entities. They used ChatGPT to generate a synthetic dataset for these 2 tasks, incorporating a combination of prompt engineering (prompt engineering is the process of optimizing prompts given to interactive LLMs, such as ChatGPT, to achieve a certain task without having to further train the LLM) and human-labeled seed examples into their workflow. Borisov et al52 investigated the ability of LLMs to generate tabular data. They converted rows of heterogeneous features (both categorical and numerical) into sentence-like textual representation, fine-tuned (fine-tuning refers to a strategy of adapting pre-trained models to specific tasks, by using a dataset of labeled examples to update either the whole model, or by adding and training a relatively small number of additional layers) GPT-2 to generate similar synthetic text data, and then converted the synthesized text back into tabular data. Borisov et al,52 however, considered just 1 tabular health dataset, and did not evaluate privacy preservation characteristics. Seedat et al53 examined the capabilities of GPT4 to generate tabular data out-of-the-box, using a 3-section prompt with data context, data examples, and generation instructions and post-generation filtering for data quality. They found that GPT4 generates high-utility data from few examples, holding promise for health applications with low data availability (eg, rare diseases). Finally, Kim et al54 focus on synthetic data generation with LLMs in settings of outcome class imbalance, common in health settings. Using out-of-the-box generation with GPT4, Llama 2 58 and Mistral,59 they find that prompts specifically identifying and partitioning examples from each class produce synthetic data that boost model performance for a minority class. Collectively, these studies highlight the potential of LLMs for multimodal synthetic data generation and generation from few training examples.
Table 1.
Summary of current research applying LLMs to synthetic data generation.
| Modality | Downstream application | LLM(s) used | Reference(s) |
|---|---|---|---|
| Text | Clinical trial-patient matching | GPT3 | Yuan et al50 |
| Biomedical term comprehension | GPT3 | Tang et al51 | |
| Radiology report generation | GPT4 | Xie et al55 | |
| Alzheimer’s detection from EHR notes | GPT4 | Li et al56 | |
| Clinical NLP tasks (general purpose) | GPT3.5 | Xu et al57 | |
| Tabular | Binary classification (general purpose) | GPT2 | Borisov et al52 |
| GPT3.5, GPT4 | a Seedat et al53 | ||
| GPT3.5, LLaMa-2-7b, Mistral-7b | a Kim et al54 |
Not currently peer reviewed.
Abbreviation: LLM, large language model.
Open research directions
Given the limited research to date on LLMs for SHDG, many important questions remain around the potential opportunities and risks. Building on and extending systematic reviews of synthetic data generation in healthcare20,23,32–36 and benchmarking studies,49,60–63 we describe useful avenues for further research below.
Evaluation metrics
To fully understand the value potential of LLMs, it is important to establish a portfolio of metrics to evaluate the quality of the generated data and to compare synthetic data generating LLMs to the current state of the art. There is already a lack of standardization of metrics in the broader field of health data generation20,26,28 and metrics for LLM performance on clinical prediction tasks.64 Dimensions that are critical to measure include the aforementioned data realism, utility, and privacy, token usage, computational cost (including power consumption), and diversity. Researchers should aim to present results on benchmark datasets and metrics, as well as assessments specific to a potential deployment context.
Some evaluation criteria have established metrics, such as the predictive performance of a downstream machine learning (ML) model after augmentation with synthetic data, which is both common28 and considers a likely setting for EHR data use. Computational cost metrics have recently become more prevalent26 and are particularly important in the LLM context, given the high costs of training and prompting, whether monetary or environmental.65 Efficiency metrics, such as generation time, algorithmic complexity, and computing power required, can help organizations make economic assessments of synthetic data generation alternatives. When evaluating privacy, metrics should focus on risks specific to healthcare data, such as re-identification, using measurements like the ability to predict whether a real data point belonged to the generator’s training set.28 One must also consider unique vulnerabilities of the LLMs. For example, ChatGPT is vulnerable to prompt injection, where users design prompts to reveal sensitive data that ChatGPT is intended not to expose.66 Thus, a new privacy metric could evaluate whether the generating LLMs reveal private medical information across a range of prompts. Furthermore, given the breadth of LLM training data, new categories of risks must be anticipated, including inadvertent infringement of intellectual property or generation of toxic or harmful language.67
Given existing research questioning the value of general-purpose models over tailored clinical models,68 a comprehensive comparison of LLM-based methods to tailored methods for synthetic data generation across a portfolio of relevant metrics is critical.
LLM adoption
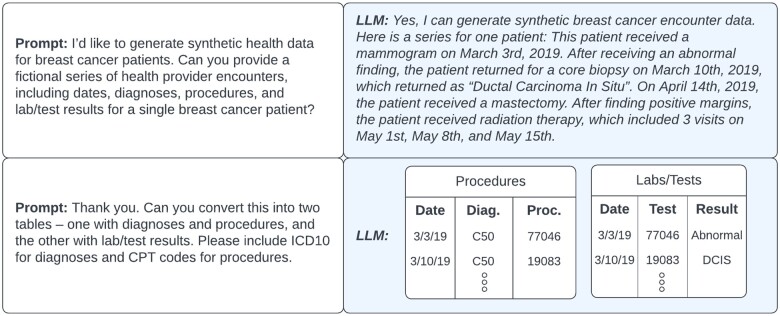
Choices related to the specific LLM model to be deployed as well as the generation approach are critical to understand. In the context of synthetic data, both prompt engineering and fine-tuning have already been applied.51,52 Prompt engineering can involve a variety of prompt templates and include either few or zero examples as part of the prompt69—an example of a zero-example prompt is shown in Figure 2. Similarly, one can explore or develop multiple different fine-tuning approaches.70 Furthermore, there are many existing LLMs, each of which may be more appropriate for certain approaches—fine-tuning may be easier with “smaller” LLMs such as Llama71 whereas prompt engineering is better suited for “larger” LLMs such as GPT-4. These choices can also be framed in the tradeoff between “buy” versus “build”—do the benefits of fine-tuning LLM models over direct application of out-of-the-box models outweigh the fine-tuning development cost? Extensive exploration of such strategic choices in adoption is necessary to make informed decisions for a specific context and task.
Figure 2.
An example of synthetic health data generation with prompts to a large language model (LLM).
Data efficiency
While the promise of synthetic data is clear, the practical feasibility of creating quality data must address the question of how much real data are necessary to generate high-fidelity synthetic observations. Given the large number of rare diseases and conditions (McDuff et al., 2023),23 well-documented difficulties of acquiring health data, the privacy risks involved with sharing increasing patient data with models, and the potential costs of fine-tuning LLMs with increasing data,72 there is great interest in maximizing the data efficiency of synthetic data generating LLMs. However, providing fewer examples can lead to decreased generating ability,42 particularly of rare real cases that are all the more important to have represented in synthetic data. Yet, LLMs have a crucial advantage in this environment. As shown by Seedat et al,53 LLMs can leverage prior knowledge from training to generate synthetic tabular data from very few examples. Application of their and related methods to health data holds great promise for the many low data contexts in healthcare, that should be explored.
Generalization
As previously noted, non-LLM-based models for synthetic data generation struggle to generalize, whether in handling multiple data modalities, transferring between health contexts, or incorporating domain knowledge.26,31 Because LLMs are general purpose models trained on diverse knowledge bases, they are well-equipped to handle each of these challenges. Borisov et al52 showed that LLMs can simultaneously generate different data modalities with little tailoring, whether discrete, continuous, or categorical text data. Gruver et al73 demonstrated that LLMs can generate time series forecasts, while also handling missing data and creating textual explanations of predictions. Additionally, there are many existing LLMs trained specifically on medical text corpuses such as ClinicalBERT, Med-PaLM, and GatorTron which have the potential to automatically incorporate their domain knowledge into data generation.74–76 Thus, an important question for future work is a deeper understanding of if and how LLMs can convert knowledge from their training data into generalization across data modalities and contexts.
Health equity
Another critical area that warrants further research is the risks and opportunities of synthetic data generation with LLMs in the domain of health equity. One clear risk is the perpetuation of existing biases in the data used to train LLMs and biases in health data77,78—Bhanot et al79 have documented violations of fairness metrics in existing synthetic data generation methods. For instance, they found that the realism of synthetic sleep data was not equal across age groups.
However, as noted by McDuff et al,23 synthetic data generation also provides opportunities to correct existing health equity issues. For instance, machine learning models often underperform for minority groups due to a lack of training data.80 Racial minorities are underrepresented in clinical trials and in health data81—synthetic data generation can help increase representation of smaller groups in augmented datasets and boost performance of these models for underrepresented groups. Thus, 1 valuable research direction is to compare the boost in performance for minority groups that LLM-based synthetic data provide over other common ML fairness methods aimed at improving performance for small groups.
Regulatory challenges
Moving forward, the regulatory environment for generative AI will likely evolve rapidly. Regulation of LLMs in general, and synthetic health data in particular, is not clearly delineated in current data protection guidelines (eg, The General Data Protection Regulation [GDPR]).82 However, privacy preservation is a key policy requirement in emerging legislation on AI such as the EU AI Act83 and the Executive Order issued in the United States.84 Giuffrè et al85 discuss the difficulty of proving whether synthetic data are fully anonymized, as required by GDPR, as recent methods claiming to achieve de-identification were shown to retain vulnerability to re-identification attacks. One must also consider the privacy policies of LLM providers. For example, OpenAI’s policies around storing user data, training future models on user data, and sharing data with third parties may preclude researchers from providing sensitive patient data in input prompts.86 There are also considerations for the data used to validate the performance and safety of medical devices, which regulators must consider; relatedly, the Food and Drug Administration (FDA) and the Advanced Research Projects Agency for Health (ARPA-H) recently launched a program to help facilitate access to broader diversity in training and test data to better empower FDA pre-market submissions.87 The intersection between the regulatory landscape and the use of LLMs for SHDG represents a final opportunity for further work.
Conclusion
LLMs have already shown great promise in a variety of healthcare applications, and SHDG is a logical next high-impact application area of LLMs. These methods can provide opportunities to address persistent challenges such as fairness in health modeling. However, we must ensure that potential drawbacks to LLM-based models are carefully examined as well. Each of these concerns—privacy, data efficiency, or bias perpetuation—is an important area of research as we develop new LLM-based synthetic health generation approaches.
Contributor Information
Daniel Smolyak, Department of Computer Science, University of Maryland, College Park, College Park, MD 20742, United States.
Margrét V Bjarnadóttir, Robert H. Smith School of Business, University of Maryland, College Park, College Park, MD 20740, United States.
Kenyon Crowley, Accenture Federal Services, Arlington, VA 22203, United States.
Ritu Agarwal, Center for Digital Health and Artificial Intelligence, Carey Business School, Johns Hopkins University, Baltimore, MD 21202, United States.
Author contributions
Concept and design (Ritu Agarwal, Margrét V. Bjarnadóttir, and Kenyon Crowley), drafting of manuscript (Daniel Smolyak, Ritu Agarwal, and Margrét V. Bjarnadóttir), critical revision of manuscript for important intellectual content (Daniel Smolyak, Ritu Agarwal, Margrét V. Bjarnadóttir, and Kenyon Crowley), statistical analysis (N/A), and obtained funding (N/A).
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflicts of interest
The authors have no competing interests to declare.
Data availability
There are no new data associated with this article.
References
- 1. GPT-4. Accessed October 9, 2023. https://openai.com/gpt-4
- 2. Google AI PaLM 2. Google AI. Accessed October 9, 2023. https://ai.google/discover/palm2/
- 3. The Lancet Digital Health. ChatGPT: friend or foe? Lancet Digit Health. 2023;5:e102. 10.1016/S2589-7500(23)00023-7 [DOI] [PubMed] [Google Scholar]
- 4. Asch DA. An interview with ChatGPT about health care. Catalyst. 2023;4. [Google Scholar]
- 5. Lee P, Bubeck S, Petro J.. Benefits, limits, and risks of GPT-4 as an AI chatbot for medicine. N Engl J Med. 2023;388:1233-1239. 10.1056/NEJMsr2214184 [DOI] [PubMed] [Google Scholar]
- 6. Thirunavukarasu AJ, Ting DSJ, Elangovan K, et al. Large language models in medicine. Nat Med. 2023;29:1930-1940. 10.1038/s41591-023-02448-8 [DOI] [PubMed] [Google Scholar]
- 7. Cascella M, Montomoli J, Bellini V, et al. Evaluating the feasibility of ChatGPT in healthcare: an analysis of multiple clinical and research scenarios. J Med Syst. 2023;47:33. 10.1007/s10916-023-01925-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lai T, Shi Y, Du Z, et al. Psy-LLM: scaling up global mental health psychological services with AI-based large language models. 2023. Accessed October 6, 2023. http://arxiv.org/abs/2307.11991
- 9. Lim S, Schmälzle R.. Artificial intelligence for health message generation: an empirical study using a large language model (LLM) and prompt engineering. Front Commun. 2023;8:1129082. [Google Scholar]
- 10. Sallam M. ChatGPT utility in healthcare education, research, and practice: systematic review on the promising perspectives and valid concerns. Healthcare (Basel). 2023;11:887. 10.3390/healthcare11060887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu J, Wang C, Liu S.. Utility of ChatGPT in clinical practice. J Med Internet Res. 2023;25:e48568. 10.2196/48568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Javaid M, Haleem A, Singh RP.. ChatGPT for healthcare services: an emerging stage for an innovative perspective. TBench. 2023;3:100105. 10.1016/j.tbench.2023.100105 [DOI] [Google Scholar]
- 13. Morley J, DeVito NJ, Zhang J.. Generative AI for medical research. BMJ. 2023;382:1551. 10.1136/bmj.p1551 [DOI] [PubMed] [Google Scholar]
- 14. Liu S, Wright AP, Patterson BL, et al. Using AI-generated suggestions from ChatGPT to optimize clinical decision support. J Am Med Inform Assoc. 2023;30:1237-1245. 10.1093/jamia/ocad072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Strong E, DiGiammarino A, Weng Y, et al. Chatbot vs medical student performance on free-response clinical reasoning examinations. JAMA Intern Med. 2023;183:1028-1030. 10.1001/jamainternmed.2023.2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Singhal K, Azizi S, Tu T, et al. Large language models encode clinical knowledge. Nature. 2023;620:172-180. 10.1038/s41586-023-06291-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liévin V, Hother CE, Winther O. Can large language models reason about medical questions? 2023. Accessed October 6, 2023. http://arxiv.org/abs/2207.08143 [DOI] [PMC free article] [PubMed]
- 18. McDuff D, Schaekermann M, Tu T, et al. Towards accurate differential diagnosis with large language models. 2023. Accessed January 13, 2024. http://arxiv.org/abs/2312.00164
- 19. Chen RJ, Lu MY, Chen TY, et al. Synthetic data in machine learning for medicine and healthcare. Nat Biomed Eng. 2021;5:493-497. 10.1038/s41551-021-00751-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Murtaza H, Ahmed M, Khan NF, et al. Synthetic data generation: state of the art in health care domain. Comput Sci Rev. 2023;48:100546. 10.1016/j.cosrev.2023.100546 [DOI] [Google Scholar]
- 21. Abdel-Jaber H, Devassy D, Al Salam A, et al. A review of deep learning algorithms and their applications in healthcare. Algorithms. 2022;15:71. 10.3390/a15020071 [DOI] [Google Scholar]
- 22. Waheed A, Goyal M, Gupta D, et al. CovidGAN: data augmentation using auxiliary classifier GAN for improved COVID-19 detection. IEEE Access. 2020;8:91916-91923. 10.1109/ACCESS.2020.2994762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McDuff D, Curran T, Kadambi A. Synthetic data in healthcare. 2023. Accessed September 26, 2023. http://arxiv.org/abs/2304.03243
- 24. van Panhuis WG, Paul P, Emerson C, et al. A systematic review of barriers to data sharing in public health. BMC Public Health. 2014;14:1144. 10.1186/1471-2458-14-1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rhodes KL, Echo-Hawk A, Lewis JP, et al. Centering data sovereignty, tribal values, and practices for equity in American Indian and Alaska native public health systems. Public Health Rep. 2024;139:10S-15S. 10.1177/00333549231199477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hernandez M, Epelde G, Alberdi A, et al. Synthetic data generation for tabular health records: a systematic review. Neurocomputing. 2022;493:28-45. 10.1016/j.neucom.2022.04.053 [DOI] [Google Scholar]
- 27. Gonzales A, Guruswamy G, Smith SR.. Synthetic data in health care: a narrative review. PLOS Digit Health. 2023;2:e0000082. 10.1371/journal.pdig.0000082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaabachi B, Despraz J, Meurers T, et al. Can we trust synthetic data in medicine? A scoping review of privacy and utility metrics. 2023. Accessed January 8, 2024. https://www.medrxiv.org/content/10.1101/2023.11.28.23299124v1
- 29. Tsao S-F, Sharma K, Noor H, et al. Health synthetic data to enable health learning system and innovation: a scoping review. Stud Health Technol Inform. 2023;302:53-57. 10.3233/SHTI230063 [DOI] [PubMed] [Google Scholar]
- 30. Georges-Filteau J, Cirillo E. Synthetic observational health data with GANs: from slow adoption to a boom in medical research and ultimately digital twins? 2020. Accessed October 6, 2023. http://arxiv.org/abs/2005.13510
- 31. Ghosheh GO, Li J, Zhu T.. A survey of generative adversarial networks for synthesizing structured electronic health records. ACM Comput Surv. 2024;56:1-34. 10.1145/3636424 [DOI] [Google Scholar]
- 32. Perkonoja K, Auranen K, Virta J. Methods for generating and evaluating synthetic longitudinal patient data: a systematic review. 2023. Accessed January 13, 2024. http://arxiv.org/abs/2309.12380
- 33. Foraker RE, Yu SC, Gupta A, et al. Spot the difference: comparing results of analyses from real patient data and synthetic derivatives. JAMIA Open. 2020;3:557-566. 10.1093/jamiaopen/ooaa060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Park Y, Ghosh J, Shankar M. Perturbed Gibbs Samplers for generating large-scale privacy-safe synthetic health data. In: 2013 IEEE International Conference on Healthcare Informatics, Philadelphia, PA, USA. 2013:493-498.
- 35. Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative adversarial nets. Adv Neural Inf Process Syst. 2014;27. [Google Scholar]
- 36. Kingma DP, Welling M. Auto-encoding variational Bayes. 2022. Accessed November 2, 2023. http://arxiv.org/abs/1312.6114
- 37. Choi E, Biswal S, Malin B, et al. Generating multi-label discrete patient records using generative adversarial networks. In: Proceedings of the 2nd Machine Learning for Healthcare Conference, Boston, MA, USA. PMLR; 2017:286-305.
- 38. Bilici Ozyigit E, Arvanitis TN, Despotou G.. Generation of realistic synthetic validation healthcare datasets using generative adversarial networks. Stud Health Technol Inform. 2020;272:322-325. 10.3233/SHTI200560 [DOI] [PubMed] [Google Scholar]
- 39. Kaur D, Sobiesk M, Patil S, et al. Application of Bayesian networks to generate synthetic health data. J Am Med Inform Assoc. 2021;28:801-811. 10.1093/jamia/ocaa303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koivu A, Sairanen M, Airola A, et al. Synthetic minority oversampling of vital statistics data with generative adversarial networks. J Am Med Inform Assoc. 2020;27:1667-1674. 10.1093/jamia/ocaa127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang Z, Yan C, Malin BA.. Keeping synthetic patients on track: feedback mechanisms to mitigate performance drift in longitudinal health data simulation. J Am Med Inform Assoc. 2022;29:1890-1898. 10.1093/jamia/ocac131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li J, Cairns BJ, Li J, et al. Generating synthetic mixed-type longitudinal electronic health records for artificial intelligent applications. NPJ Digit Med. 2023;6:98. 10.1038/s41746-023-00834-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Biswal S, Ghosh S, Duke J, et al. EVA: generating longitudinal electronic health records using conditional variational autoencoders. In: Proceedings of the 6th Machine Learning for Healthcare Conference. PMLR; 2021:260-282.
- 44. Torfi A, Fox EA, Reddy CK.. Differentially private synthetic medical data generation using convolutional GANs. Inf Sci. 2022;586:485-500. 10.1016/j.ins.2021.12.018 [DOI] [Google Scholar]
- 45. Wang X, Lin Y, Xiong Y, et al. Using an optimized generative model to infer the progression of complications in type 2 diabetes patients. BMC Med Inform Decis Mak. 2022;22:174. 10.1186/s12911-022-01915-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu M, He Y, Raghunathan TE.. A semiparametric multiple imputation approach to fully synthetic data for complex surveys. J Surv Stat Methodol. 2022;10:618-641. 10.1093/jssam/smac016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sood M, Sahay A, Karki R, et al. Realistic simulation of virtual multi-scale, multi-modal patient trajectories using Bayesian networks and sparse auto-encoders. Sci Rep. 2020;10:10971. 10.1038/s41598-020-67398-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Soenksen LR, Ma Y, Zeng C, et al. Integrated multimodal artificial intelligence framework for healthcare applications. NPJ Digit Med. 2022;5:149. 10.1038/s41746-022-00689-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yan C, Yan Y, Wan Z, et al. A multifaceted benchmarking of synthetic electronic health record generation models. Nat Commun. 2022;13:7609. 10.1038/s41467-022-35295-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yuan J, Tang R, Jiang X, et al. Large language models for healthcare data augmentation: an example on patient-trial matching. 2023. Accessed September 26, 2023. http://arxiv.org/abs/2303.16756 [PMC free article] [PubMed]
- 51. Tang R, Han X, Jiang X, et al. Does synthetic data generation of LLMs help clinical text mining? 2023. Accessed September 26, 2023. http://arxiv.org/abs/2303.04360
- 52. Borisov V, Sessler K, Leemann T, et al. Language models are realistic tabular data generators. 2022. Accessed September 26, 2023. https://arxiv.org/abs/2210.06280
- 53. Seedat N, Huynh N, van Breugel B, et al. Curated LLM: synergy of LLMs and data curation for tabular augmentation in ultra low-data regimes. 2023. Accessed January 13, 2024. http://arxiv.org/abs/2312.12112
- 54. Kim J, Kim T, Choo J. Exploring prompting methods for mitigating class imbalance through synthetic data generation with large language models. 2024. Accessed June 25, 2024. http://arxiv.org/abs/2404.12404
- 55. Xie Y, Chen Q, Wang S, et al. PairAug: what can augmented image-text pairs do for radiology? Accessed June 24, 2024. https://arxiv.org/abs/2404.04960v1
- 56. Li R, Wang X, Yu H.. Two directions for clinical data generation with large language models: data-to-label and label-to-data. Proc Conf Empir Methods Nat Lang Process. 2023;2023:7129-7143. 10.18653/v1/2023.findings-emnlp.474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xu R, Cui H, Yu Y, et al. Knowledge-infused prompting improves clinical text generation with large language models. In: NeurIPS 2023 Workshop on Synthetic Data Generation with Generative AI, New Orleans, LA, USA. 2023.
- 58. Touvron H, Martin L, Stone K, et al. Llama 2: open foundation and fine-tuned chat models. 2023. Accessed June 17, 2024. http://arxiv.org/abs/2307.09288
- 59. Jiang AQ, Sablayrolles A, Mensch A, et al. Mistral 7B. 2023. Accessed June 17, 2024. http://arxiv.org/abs/2310.06825
- 60. El Emam K, Mosquera L, Fang X, et al. Utility metrics for evaluating synthetic health data generation methods: validation study. JMIR Med Inform. 2022;10:e35734. 10.2196/35734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. El Kababji S, Mitsakakis N, Fang X, et al. Evaluating the utility and privacy of synthetic breast cancer clinical trial data sets. JCO Clin Cancer Inform. 2023;7:e2300116. 10.1200/CCI.23.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Benaim AR, Almog R, Gorelik Y, et al. Analyzing medical research results based on synthetic data and their relation to real data results: systematic comparison from five observational studies. JMIR Med Inform. 2020;8:e16492. 10.2196/16492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hernadez M, Epelde G, Alberdi A, et al. Synthetic tabular data evaluation in the health domain covering resemblance, utility, and privacy dimensions. Methods Inf Med. 2023;62:e19-e38. 10.1055/s-0042-1760247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wornow M, Xu Y, Thapa R, et al. The shaky foundations of large language models and foundation models for electronic health records. NPJ Digit Med. 2023;6:135. 10.1038/s41746-023-00879-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Strubell E, Ganesh A, McCallum A. Energy and policy considerations for deep learning in NLP. In: Korhonen A, Traum D, Màrquez L, eds. Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, Florence, Italy. Association for Computational Linguistics; 2019:3645-3650.
- 66. Gupta M, Akiri C, Aryal K, et al. From ChatGPT to ThreatGPT: impact of generative AI in cybersecurity and privacy. IEEE Access. 2023;11:80218-80245. 10.1109/ACCESS.2023.3300381 [DOI] [Google Scholar]
- 67. Solaiman I, Talat Z, Agnew W, et al. Evaluating the social impact of generative AI systems in systems and society. 2023. Accessed October 6, 2023. http://arxiv.org/abs/2306.05949
- 68. Lehman E, Hernandez E, Mahajan D, et al. Do we still need clinical language models? 2023. Accessed October 8, 2023. http://arxiv.org/abs/2302.08091
- 69. Wang J, Shi E, Yu S, et al. Prompt engineering for healthcare: methodologies and applications. 2023. Accessed October 12, 2023. http://arxiv.org/abs/2304.14670
- 70. Sun C, Qiu X, Xu Y, et al. How to fine-tune BERT for text classification? In: Sun M, Huang X, Ji H, et al. , eds. Chinese Computational Linguistics. Springer International Publishing; 2019:194-206. [Google Scholar]
- 71. Gema AP, Daines L, Minervini P, et al. Parameter-efficient fine-tuning of LLaMA for the clinical domain. 2023. Accessed October 17, 2023. http://arxiv.org/abs/2307.03042
- 72. Zhao WX, Zhou K, Li J, et al. A survey of large language models. 2023. Accessed October 9, 2023. http://arxiv.org/abs/2303.18223
- 73. Gruver N, Finzi M, Qiu S, et al. Large language models are zero-shot time series forecasters. 2023. Accessed January 14, 2024. http://arxiv.org/abs/2310.07820
- 74. Huang K, Altosaar J, Ranganath R. ClinicalBERT: modeling clinical notes and predicting hospital readmission. 2020. Accessed January 14, 2024. http://arxiv.org/abs/1904.05342
- 75. Singhal K, Tu T, Gottweis J, et al. Towards expert-level medical question answering with large language models. 2023. Accessed January 14, 2024. http://arxiv.org/abs/2305.09617
- 76. Yang X, Chen A, PourNejatian N, et al. GatorTron: a large clinical language model to unlock patient information from unstructured electronic health records. 2022. Accessed January 14, 2024. http://arxiv.org/abs/2203.03540
- 77. Bender EM, Gebru T, McMillan-Major A, et al. On the dangers of stochastic parrots: can language models be too big? In: Proceedings of the 2021 ACM Conference on Fairness, Accountability, and Transparency. ACM; 2021:610-623.
- 78. Chen IY, Pierson E, Rose S, et al. Ethical machine learning in healthcare. Annu Rev Biomed Data Sci. 2021;4:123-144. 10.1146/annurev-biodatasci-092820-114757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bhanot K, Qi M, Erickson JS, et al. The problem of fairness in synthetic healthcare data. Entropy (Basel). 2021;23:1165. 10.3390/e23091165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Buolamwini J, Gebru T. Gender shades: intersectional accuracy disparities in commercial gender classification. In: Proceedings of the 1st Conference on Fairness, Accountability and Transparency, New York City, NY, USA. PMLR; 2018:77-91.
- 81. Nazha B, Mishra M, Pentz R, et al. Enrollment of racial minorities in clinical trials: old problem assumes new urgency in the age of immunotherapy. Am Soc Clin Oncol Educ Book. 2019;39:3-10. 10.1200/EDBK_100021 [DOI] [PubMed] [Google Scholar]
- 82. Meskó B, Topol EJ.. The imperative for regulatory oversight of large language models (or generative AI) in healthcare. NPJ Digit Med. 2023;6:120-126. 10.1038/s41746-023-00873-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. The Act Texts | EU Artificial Intelligence Act. Accessed June 18, 2024. https://artificialintelligenceact.eu/the-act/
- 84. The White House. Executive order on the safe, secure, and trustworthy development and use of artificial intelligence. The White House; 2023. Accessed June 18, 2024. https://www.whitehouse.gov/briefing-room/presidential-actions/2023/10/30/executive-order-on-the-safe-secure-and-trustworthy-development-and-use-of-artificial-intelligence/
- 85. Giuffrè M, Shung DL.. Harnessing the power of synthetic data in healthcare: innovation, application, and privacy. NPJ Digit Med. 2023;6:186-188. 10.1038/s41746-023-00927-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wu X, Duan R, Ni J.. Unveiling security, privacy, and ethical concerns of ChatGPT. J Inf Intell. 2024;2:102-115. 10.1016/j.jiixd.2023.10.007 [DOI] [Google Scholar]
- 87.ARPA-H Announces Medical Imaging Data Partnership with FDA | ARPA-H. 2024. Accessed June 24, 2024. https://arpa-h.gov/news-and-events/arpa-h-announces-medical-imaging-data-partnership-fda
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are no new data associated with this article.