Abstract
Background
Human papillomavirus (HPV) has emerged as a significant contributor to cancer incidence globally, particularly in the context of oropharyngeal squamous cell carcinoma (OPSCC) and cancer of unknown primary (HNCUP). This study aimed to develop and validate droplet digital PCR (ddPCR) assays for the detection of circulating tumor HPV DNA (ctHPV-DNA) in plasma, focusing on high-risk HPV genotypes associated with these cancers.
Methods
ddPCR assays for HPV16, 18, 33, 35, 56, and 59 were developed and tested using gBlocks, HPV cell-free DNA, fragmented tumor HPV+ DNA, and plasma samples from patients with HPV+ OPSCC (n = 110) and HNCUP (n = 9).
Results
Assays demonstrated robust technical sensitivity across all tested HPV genotypes. Clinical application of the assays on a cohort of patients with HPV+ OPSCC and HNCUP revealed high sensitivity (91.6%) and wide variability in ctHPV-DNA levels. Analyses revealed correlations between ctHPV-DNA levels and TNM stage and tumor viral load. The association between ctHPV-DNA and tumor viral load persisted even after adjusting for TNM stage. At posttreatment, 72.5% of samples had reached undetectable ctHPV-DNA levels. Having detectable ctHPV-DNA posttreatment was associated with a higher ctHPV-DNA level at diagnosis and higher viral load at diagnosis.
Conclusion
The findings underscore the potential of ctHPV-DNA as a biomarker for monitoring HPV+ cancers and offer insights into tumor dynamics. Implementation of these assays in clinical practice could enhance no-invasive treatment monitoring and recurrence detection in HPV-associated cancers.
Clinical Trials
Supplementary Information
The online version contains supplementary material available at 10.1007/s40291-024-00743-9.
Key Points
| Human papillomavirus can be detected with high sensitivity in plasma from patients with oropharyngeal cancer and cancer of unknown primary in the head and neck. |
| Human papillomavirus levels in plasma could be correlated to both TNM stage as well as tumor viral load. |
Introduction
Due to the significant increase in human papilloma virus (HPV)-induced cancers over recent decades, the role of DNA diagnostics has become of great importance. Almost 0.7 million new HPV cancer cases were diagnosed in 2018, equal to 10% of the total cancer incidence worldwide [1]. HPV, a DNA virus that infects squamous cells, is widely recognized for its potential to cause genital cancer through persistent infections. However, HPV+ head neck cancer, such as oropharyngeal squamous cell carcinoma (OPSCC) and cancer of unknown primary (HNCUP), has become one of the most frequent HPV driven cancer types worldwide. The prevalence varies by region, but HPV+ OPSCC has been reported to have a pooled global burden of 33%, with a higher prevalence in the Nordic countries [2]. High-risk HPV genotypes, including 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 59, are considered carcinogenic due to their expression of E6 and E7 oncoproteins, along with their ability to integrate genomic components into the human genome [3]. Consequently, circulating tumor DNA (ctDNA) released from HPV-induced tumors contains fragments of HPV DNA sequences (ctHPV-DNA).
Ultrasensitive detection of ctDNA holds great potential as an important tool in molecular diagnostics for cancer, including molecular profiling, detection of residual disease, treatment monitoring, and early detection of recurrence [4–7]. These applications currently rely on invasive procedures to obtain tumor tissue, whereas ctDNA has been identified in various body fluids, such as plasma, serum, urine, cerebrospinal fluid, and saliva [8]. Consequently, ctDNA would offer a noninvasive alternative for these clinical applications [9].
However, the identification of low levels of ctDNA among circulating cell-free DNA (cfDNA), generated by normal cell turn-around, necessitates ultra-sensitive detection methods. While next generation sequencing (NGS) is commonly used for comprehensive ctDNA characterization, it presents drawbacks such as high cost, complex analysis, and a lengthy turnaround time. Droplet digital PCR (ddPCR), on the other hand, overcomes these disadvantages, exhibiting higher sensitivity for the analysis of known ctDNA biomarkers.
Studies have successfully linked the detection of ctHPV-DNA fragments to treatment response and prognosis in OPSCC [10–13]. Notably, Chera et al. showed a 100% negative predictive value and a 94% positive predictive value for recurrent disease in a cohort of OPSCC using ctHPV-DNA. Additionally, Siravegna et al. investigated the health economic benefits of ctHPV-DNA, demonstrating a higher diagnostic accuracy and a shorter diagnostic interval compared with standard clinical workup [14]. These findings underscore the potential of ctHPV-DNA as a biomarker for OPSCC.
This study’s primary objective was to develop a set of highly sensitive ddPCR assays for use as a surveillance tool in patients with HPV+ tumors after radiotherapy or radiotherapy combined with chemotherapy. The sensitivity and specificity of the assays were validated using fragmented tumor genomes diluted in cfDNA. Subsequently, the assays were applied to a cohort of patients with HPV+ OPSCC and HNCUP, included in the CIRCulating biomarkers in Oropharyngeal CancerS (CIRCOS) study (ClinicalTrials.gov: NCT05904327), for which the relationship between ctHPV-DNA levels and tumor characteristics relevant to the shedding process of cfDNA were explored.
Materials and Methods
Assay Development
ddPCR assays for ctDNA targeting the E7 gene in HPV genotypes 16, 18, 33, 35, 56, and 59 were developed and validated at SAGA Diagnostics (Lund, Sweden). The primer and probe sequences are proprietary to SAGA Diagnostics and assays can be ordered through contacting SAGA Diagnostics. Initially, assays for HPV16, 18, and 33 were developed based on known prevalence in OPSCC. Additional genotypes, HPV35, 56, and 59, were then detected during inclusion in the CIRCOS study; consequently, assays for these genotypes were also developed. The assay chemistry is of dual nature where the first phase of the amplification is linear through inhibition of the primers targeting one of the template strands. This generates only one single stranded copy directly from the original template molecule for each cycle. During the second phase, all primers and probes are active and exponential amplification is performed. Through this, generation of copies-of-copies and propagation of base misincorporation errors are absent during the first phase and can later be distinguishable by a lower fluorescence intensity in the readout as compared to true positive droplets. Assays were tested using gBlocks (Integrated DNA Technologies, Coralville, IA, USA) specific for the different HPV types and a pool of normal cfDNA (n ~ 30 healthy donors). In addition, HPV16, 18, and 33 were tested for linearity by serial 1:2 dilution of gBlocks. Each assay contained a human control sequence located on 12q24.32, which was used as amplification control.
Control Samples
DNA from tumor tissue samples from different HPV+ tumor types used in clinical diagnostics were used as control samples for genotypes 16, 18, 33, 35, 56, and 59. All control samples were anonymized before analysis.
Extracted cfDNA originating from anonymized healthy donors (n ~ 10) was used as technical control material of the method as described in Sect. 2.3. “Assay validation.” All donors gave oral informed consent and cfDNA was pooled prior to analysis. Blood was collected in Cell-Free DNA BCT® tube (Streck, La Vista, NE, USA) and underwent two consecutive centrifugations: 1600g for 10 min followed by 3000g for 10 min. Plasma was transferred to a new tube without disrupting the cell layer. The clean plasma was stored in – 80 °C until DNA extraction. Plasma was brought to room temperature before manual DNA extraction using QIAamp circulating nucleic acid kit (QIAGEN) according to manufacturers’ instruction. Extracted DNA was stored in – 20 °C until further analysis.
Assay Validation
To validate the assays, controls consisting of fragmented tumor DNA diluted in cfDNA were prepared. One tumor DNA sample from each of the six different HPV genotypes (16, 18, 33, 35, 56, and 59) was therefore diluted to 20 ng/µl and cleaved using BamH1 20,000 units/ml (New England Biolabs). The fragmented tumor DNA was then diluted 1:10 before being analyzed with the developed HPV specific ddPCR assays to determine baseline positivity. The prereaction (20 µl), including 1 µl assay, 5 µl 4× ddPCR™ Multiplex Supermix (BIORAD), and 14 µl template, was prepared according to instructions provided by the manufacturer (SAGA diagnostics). Droplets were generated according to instructions, using QX200 droplet generator™ (BIO-RAD) and PCR was performed on a Veriti 96-well thermal cycler (Applied Biosystems) according to instructions provided by the manufacturer (SAGA Diagnostics). QX200 Droplet Reader™ (BIO-RAD) was used to read droplets, and data was analyzed using QuantaSoft version 1.0.596. Each PCR was run in duplicate on both the specific HPV genotypes and the human control sequence located on 12q24.32. Based on the baseline positivity, the fragmented tumor DNA samples from each HPV genotype were serially diluted six times in cfDNA from healthy donors as described in Sect. 2.2. “Control samples.” The first dilution was calculated to generate ten positive droplets and the last dilution to reach undetectable levels.
Based on the results from the serial dilutions, two HPV+ controls were prepared: one with low positivity and one with high positivity. Preparation of the controls was performed as for the sensitivity tests. The controls were used to set thresholds for positivity, and to monitor the assays over time. The assay performance was analyzed over time by observing copies/µl for both HPV and the human control sequence in the positive controls at each analysis point. An assay was considered having high precision if (1) no single measurement fell outside three standard deviations (SD) and (2) measurements did not fall outside two SD in two consecutive runs in accordance with Westgard multi-rule approach [15].
Clinical Application
The assays were applied on clinical plasma samples (n = 119) from the cohort of CIRCOS study (ClinicalTrials.gov: NCT05904327). CIRCOS is a prospective multicenter study aiming to monitor ctHPV-DNA after treatment and during surveillance in HPV+ OPSCC and HNCUP. Inclusion criteria were patients with OPSCC or HNCUP having a HPV+ tumor as defined by DNA genotyping, and a blood sample retrieved prior to start of treatment to be used for ctHPV-DNA analysis. Patients with distant metastases or a previous history of OPSCC or HNCUP were excluded. All participants were given oral and written information about the study and provided written in-formed consent upon inclusion. The study was approved by the ethics review board (2019-0656/2022-02405-02).
The diagnostic tissue sample for each patient was retrieved for determination of HPV genotype using the Anyplex TM II HPV28 detection (Seegene). Detection was performed according to instructions provided by the manufacturer. In brief, 100 ng of DNA underwent melt curve analysis with data collection at 30 (+++), 40 (++), and 50 (+) cycles on a CFX96TM Real-time PCR system (Bio-Rad). A ++/+++ result was considered positive and a + result was reanalyzed to confirm positivity.
All patients were p16+, as determined by immunohistochemistry. Tumors were staged and histologically classified according to the UICC TNM classification 8th edition [16].
Patients underwent radiotherapy with 68 Gy in 35 fractions (2.0 Gy/fraction) for 7 weeks for target lesions and 54 Gy for elective targets. Selected patients with stage III/IV were also given weekly intravenous Cisplatin of 40 mg/m2.
Pretreatment and posttreatment blood samples were drawn in a Cell-Free DNA BCT® tube (Streck, La Vista, NE, USA) and prepared as for the cfDNA used in the controls. cfDNA was extracted from 4 mL of plasma. The cfDNA was analyzed for its specific genotype, predetermined on paired FFPE tissue as described under the preparation of controls section, using ddPCR. Mastermix was made according to instructions provided by the manufacturer (SAGA diagnostics) and ddPCR was performed as described for the controls in the assay verification. Each run included a negative template control and two HPV+ controls: one low and one high, as in the assay validation. A sample with more than one positive droplet was considered positive. A sample with one positive droplet was reanalyzed, if positivity could be repeated the result was interpreted as positive. The number of copies per mL plasma was calculated as:
Viral Load
Additional diagnostic FFPE tumor tissue material was available for 63 patients and used to determine the HPV copy number in the tumor, i.e., tumor viral load. HPV viral load for HPV16, 18, 33, and 35 was measured using ddPCR as previously described [17]. DNA was extracted, cleaved, and genotyped as described for the FFPE control samples.
All HPV assays were run in separate reactions together with probes targeting the HBB gene, which was used as internal control and for normalization of input amount. Tumor viral load was calculated as the ratio between HPV copies and HBB copies and subsequently normalized to tumor cell content according to:
Tumor cell content was assessed on p16 stained slides by AQ and MK. Slides were analyzed in imageJ (version 1.54d) by quantification of the proportion of p16 stained area. Cell density was calculated by marking one area with a perimeter of 500 µm in a representative area of the tumor and one area in the surrounding tissue in CaseViewer (version 2.4). The proportion of cells in the respective areas was then calculated. If the tissue had a high cell density, e.g., lymphocyte-rich, the proportion of p16 stained area was corrected with a factor of two.
Immunohistochemistry
Conventional hematoxylin and eosin (H&E) stain and immunohistochemical detection of P16INK4a protein (p16) were used on sections of clinical FFPE tumor samples from the included patients to identify tumor characteristics. H&E was stained automatically using Tissue Tek Prisma (Sakura) as follows: two baths of Tissue Clear (Histolab) for 2 min, 2 min ethanol 99.5%, 2 min ethanol 95%, 1 min running tap water, 5 min Mayers Htx (Histolab), 4 min running tap water, 1 min eosin Y 0.2% (Histolab), 1 min running tap water, 1 min 95% ethanol, two baths of 99.5% ethanol for 1 min, and two baths of xylene for 1.5 min. Stained slides were mounted automatically by Tissue Tek Prisma (Sakura).
Immunohistochemistry for p16 was performed using BenchMark ULTRA IHC/ISH System (Roche Diangostics) with the mouse monoclonal antibody E6H4 (Roche Diagnostics). Samples were deparaffinized and rehydrated manually as follows: two baths of xylene for 5 min, two baths of 99.5% ethanol for 1 min, 95% ethanol for 1 min, 70% ethanol for 1 min, and distilled water before transfer to automatic staining in BenchMark ULTRA IHC/ISH. Automatic procedure included cell conditioning using CC1 ULTRA for 48 min in 100 °C, preprimary peroxidase inhibitor, primary antibody for 12 min at 36 °C, hematoxylin II for 8 min, and bluing reagent for 4 min. Slides was manually dehydrated with 70% ethanol for 1 min, 95% ethanol for 1 min, three baths of 99.5% ethanol for 1 min, and xylene for 1 min before automated mounting in Tissue Tek Prisma (Sakura). Representative stainings can be viewed in Online resource 1.
Statistics
Continuous variables were assessed by Mann–Whitney U or Kruskal Wallis tests. The Spearman correlation coefficient was used for analyzing correlations. Multivariate analysis on viral load and TNM stage for ctHPV-DNA levels was performed using linear regression. For correlation and linear regression analysis, ctHPV-DNA levels and viral load were logarithmically transformed and all assumptions for linear regression were met. Statistical analyses were performed using IBM SPSS Statistics Viewer (version 29.0). Figures were done in R version 4.3.2 (R Project for Statistical Computing; http://www.r-project.org) with the ggpubr package.
Results
Assay Development
ddPCR assays targeting HPV16, 18, 33, 35, 56, and 59 were developed. All assays were tested against extracted cfDNA from plasma from healthy donors (n ~ 30). No false positive droplets were observed for any of the assays. All assays were also tested against gBlocks corresponding to the specific HPV genotypes (Online resource 2). All assays showed high positive amplitude and good separation between positive and negative droplets, showing no pronounced raindrop pattern. The threshold amplitude for control channel was set to 4000. Thresholds for HPV16, 18, and 56 was set to 6000, for HPV33 it was set to 5000, for HPV35 it was set to 4000, and for HPV59, it was set to 8500. The cluster of positive droplets was centered around 11,600 in amplitude for HPV16 and 18, 9000 for HPV33, 8300 for HPV35, 13,600 for HPV56, and 15,200 for HPV59.
In addition, HPV16, 18, and 35 were tested for linearity and sensitivity using extracted DNA from FFPE tumor tissue mixed with cfDNA from healthy volunteers (Fig. 1). High linearity (r2 = 1.00) was observed for all assays and the assay sensitivity was 0.08, 0.07, and 0.07 copies/µl for HPV16, 18, and 35, respectively.
Fig. 1.
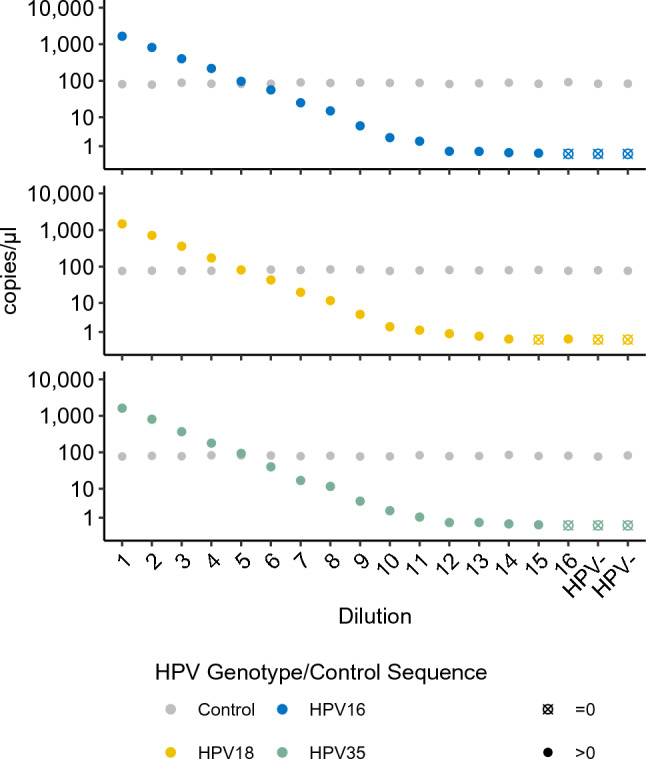
Serial dilution of ctHPV-DNA assays on tumor tissue. Tumor DNA positive for HPV16 (A), 18 (B) and 35 (C) was fragmented and underwent twofold serial dilutions in cfDNA extracted from healthy donors. The concentration of the undiluted control sequence on 12q24.32 is shown in gray. All measurements were performed in singleton
Assay Validation
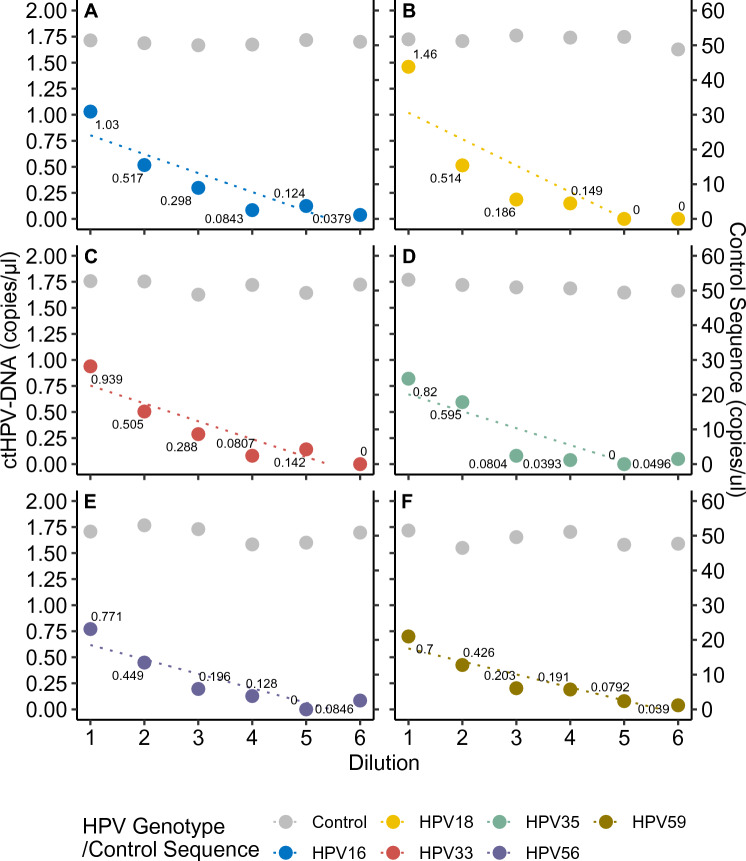
To further test the sensitivity of the assays, controls consisting of fragmented HPV+ tumor DNA diluted in HPV-cfDNA were analyzed for HPV genotypes 16, 18, 33, 35, 56, and 59. Controls underwent twofold serial dilutions around the previously validated sensitivity threshold. Total input, as measured by copy number of control sequence, was kept consistent and showed a stable result for all genotypes, with a variation from 46.5 copies/µl to 53.1 copies/µl. As for HPV, a linear relationship was found in the four first dilutions, while the targets became undetectable during the two final dilutions. The lowest detected number of copies for the different assays were 0.038 copies/µl for HPV16 (r2 = 0.81), 0.149 for HPV18 (r2 = 0.81), 0.081 for HPV33 (r2 = 0.84), 0.040 for HPV35 (r2 = 0.92), 0.085 for HPV56 (r2 = 0.93), and 0.039 for HPV59 (r2 = 0.88; fig. 2). The collective limit of detection of ctHPV-DNA was 0.082 copies/µl (95% CI: 0.029–0.135 copies/µl).
Fig. 2.
Serial dilution of ctHPV-DNA assays in plasma cfDNA. Concentration of ctHPV-DNA in progressive twofold serial dilutions for six different HPV genotypes: A HPV16, B HPV18, C HPV33, D HPV35, E HPV56, and F HPV59. The concentration of the undiluted control sequence on 12q24.32 is shown in gray. All measurements were performed in duplicate
The performance of each assay was also tested over time during a period of 7–23 months at 3–21 different timepoints. This was performed using two different dilutions of HPV+ tumor DNA, named HPV low and HPV high, fragmented and diluted in HPV plasma cfDNA for each assay. The lower control contained between 0.151 and 0.922 copies/µl and the higher control contained between 0.654 and 2.26 copies/µl across all six assays (Online resource 3). Each assay was readily quantifiable and met the criteria for precision in both controls, except for two consecutive observations in the HPV33 low control that exceeded two standard deviations (SDs). The limit of quantification is potentially lower than the results obtained for the low HPV control, but this was not further investigated. No deterioration in detectable ctHPV-DNA and amplitude levels was observed (data not shown).
Clinical Application
To test the assay in a clinical setting, pretreatment plasma samples from patients with OPSCC (n = 110) and HNCUP (n = 9) were analyzed. The patients had a mean age of 63.1 years (SD 9.0) and a male predominance (76.6 %, Table 1). A clear majority of the diagnostic tissue samples were positive for HPV16 (n = 98) followed by HPV33 (n = 11), HPV18 (n = 3), HPV35 (n = 3), HPV59 (n = 3), and HPV56 (n = 1). The majority of patients had a stage I disease (58.8%). Further characteristics are presented in Online resource 4.
Table 1.
Clinical cohort characteristics. HPV type according to diagnostic tumor tissue samples
| Characteristic | All genotypes n = 119 |
HPV16 n = 98 |
HPV18 n = 3 |
HPV33 n = 11 |
HPV35 n = 3 |
HPV56 n = 1 |
HPV59 n = 3 |
|---|---|---|---|---|---|---|---|
| Age | |||||||
| Mean (SD) | 63.1 (9.0) | 62.7 (8.7) | 64.7 (12.5) | 65.0 (8.6) | 56.0 (12.1) | 71.0 (0.0) | 73.3 (4.5) |
| Sex | |||||||
| Female, n (%) | 29 (24.4) | 21 (21.4) | 0 (0.0) | 6 (54.5) | 2 (66.7) | 0 (0.0) | 0 (0.0) |
| Male, n (%) | 90 (76.6) | 77 (78.6) | 3 (100.0) | 5 (45.5) | 1 (33.3) | 1 (100.0) | 3 (100.0) |
| Diagnosis | |||||||
| OPSCC, n (%) | 110 (92.4) | 90 (91.8) | 2 (66.7) | 11 (100.0) | 3 (100.0) | 1 (100.0) | 3 (100.0) |
| HNCUP, n (%) | 9 (7.6) | 8 (8.2) | 1 (33.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Stage | |||||||
| I, n (%) | 70 (58.8) | 57 (58.2) | 0 (0.0) | 8 (72.7) | 1 (33.3) | 1 (100.0) | 3 (100.0) |
| II, n (%) | 34 (28.6) | 29 (29.6) | 2 (66.7) | 2 (18.2) | 1 (33.3) | 0 (0.0) | 0 (0.0) |
| III, n (%) | 15 (12.6) | 12 (12.2) | 1 (33.3) | 1 (9.1) | 1 (33.3) | 0 (0.0) | 0 (0.0) |
SD standard deviation, OPSCC oropharyngeal squamous cell carcinoma, HNCUP cancer of unknown primary in the head and neck
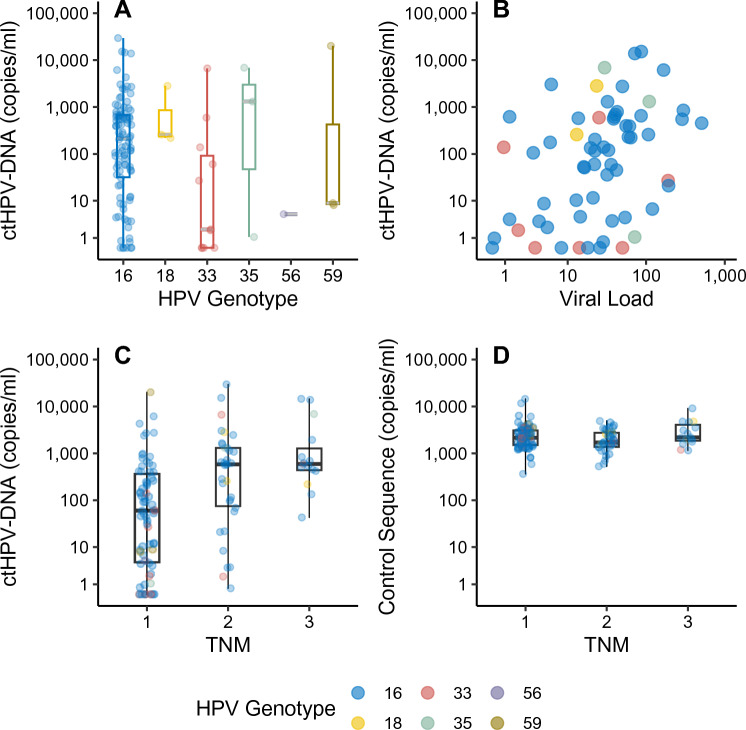
For the entire cohort, tissue HPV status was detected and confirmed in plasma in 91.6% of the cases (109/119). The ten samples with undetected ctHPV-DNA belonged to HPV16+ (n = 6) and HPV33+ (n = 4) tumors, corresponding to nine cases of OPSCC and one case of HNCUP. The median level of ctHPV-DNA across all samples was 146.0 copies/ml plasma (range 0.0–29600.0). Due to the small number of HPV+ cases of genotypes other than HPV16, they were considered as one group for statistical analyses. There were no significant differences in ctHPV-DNA levels between HPV16+ tumors and tumors positive for other genotypes (190.0 copies/ml versus 27.0 copies/ml, p = 0.307, fig. 3A).
Fig. 3.
ctHPV-DNA levels and clinical variables. A ctHPV-DNA across different HPV types. B Normalized tumor viral load and ctHPV-DNA concentration. C TNM-stage and ctHPV-DNA concentration. D TNM stage and circulating control DNA concentrations
The levels of ctHPV-DNA copies were weakly correlated with the total amount of cfDNA in the plasma (r = 0.229 and p = 0.012, Online resource 5). There was no association between ctHPV-DNA negativity and total amount of cfDNA in plasma (2157 copies/ml, range 365–11,700 copies/ml, versus 1712.5 copies/ml, range 810–14,464.3 copies/ml, p = 0.206).
Association of ctHPV-DNA with Clinical Variables
To further study the large variation of ctHPV-DNA levels in the cohort, we investigated the association between the levels of ctHPV-DNA and clinical variables. Interestingly, ctHPV-DNA levels correlated to the TNM stage (r = 0.414, p < 0.001; fig. 3c), while the total cfDNA amount did not (r = −0.012, p = 0.896; fig. 3d). In 52.9% (n = 63) of cases, including 7 ctHPV-DNA-negative cases, tumor tissue was available for further analysis of tumor viral load. Tumor viral load had a median of 15,740 copies/reaction (range 70.4–153,580.0). After normalization for input amount and tumor cell content, there were no significant differences in tumor viral load between TNM stages (p = 0.512) or between HPV16 and other genotypes (p = 0.674). The number of ctHPV-DNA copies was significantly correlated to normalized tumor viral load (r = 0.434, p < 0.001; fig. 3B). In a multivariate analysis with TNM stage, this association persisted [β = 0.362, p = 0.001, 95% CI: 0.268–1.070].
Tumor viral load was available for seven of the ten ctHPV-DNA negative patients. An association between low tumor viral load and ctHPV-DNA negativity was observed, but the association was not statistically significant (median 14.2 versus 32.2, p = 0.06, Online resource 6).
For 113 of the cases, posttreatment measurements of ctHPV-DNA were available. The clear majority of samples (72.5%, n = 82) presented with undetectable levels of ctHPV-DNA at posttreatment. The detectable samples ranged from 0.7 to 341.1 copies/ml plasma (median 3.0 copies/ml plasma, Online resource 7). Two samples had higher ctHPV-DNA levels at posttreatment. Samples with detectable ctHPV-DNA at posttreatment also had significantly higher ctHPV-DNA levels at diagnosis (99.5 versus 587.5 copies/ml plasma, p = 0.011). A higher normalized viral load was significantly associated with having detectable ctHPV-DNA at posttreatment (median 22.3 versus 41.3 copies/ml plasma, p = 0.031) while a higher stage was not (p = 0.158). Patients with undetectable ctHPV-DNA at diagnosis remained undetectable posttreatment.
Discussion
In this study, we developed and validated a series of ultrasensitive ddPCR assays tailored for the detection of ctHPV-DNA in plasma. All assays demonstrated robust sensitivity and specificity when tested, both on tumor tissue and plasma samples. Subsequently, the assays were evaluated on a clinical cohort of patients diagnosed with HPV+ OPSCC and HNCUP as part of the CIRCOS study, consistently exhibiting high sensitivity. Importantly, our investigation uncovered a wide variability in ctHPV-DNA levels among patients, prompting a thorough exploration of correlations with various tumor characteristics. Our findings unveiled significant associations with TNM stage and correlations with tumor viral load, providing valuable insights into the complex dynamics underlying HPV-related cancers.
Several studies have investigated the use of ctHPV-DNA as a clinical biomarker for OPSCC. However, the focus has largely been on the most prevalent types, HPV16 and 18. Only a limited number of studies have tested assays for several HPV types in oropharyngeal cancer [14, 18, 19], and only one included assays for six HPV genotypes as in this study [20]. Given the diverse range of high-risk HPV types associated with oropharyngeal cancer, it is essential to analyze multiple HPV types for future implementation in the clinic [21]. To our knowledge, our study is the first to have successfully developed and tested assays for HPV56 and HPV59 for use in this clinical context.
The technical validation tests, conducted using serially diluted ctHPV-DNA from patient plasma, consistently demonstrated excellent sensitivities across all six HPV assays, ranging from 0.038 to 0.149 copies/µl. Technical sensitivity, as explored in this study, has not previously been thoroughly investigated and focus has instead been directly on clinical sensitivity. Such studies have reported variable clinical sensitivity ranging from 56 to 98.4% detection rate of ctHPV-DNA [10, 14, 18, 22, 23]. Following our rigorous technical validation tests, we further investigated the performance in a clinical setting. Our developed assays showed high clinical sensitivity when employed on the CIRCOS cohort, detecting 91.6% of the cases.
We observed ten HPV+ tumors with undetectable ctHPV-DNA belonging to HPV16 and HPV33 genotypes. The HPV33 E6 and E7 genes have been shown to have single nucleotide polymorphisms (SNPs) resulting in three lineages, including two sublineages, in cervical cancer [24]. In addition, multiple different splice variants in E6/E7 have been observed in tonsillar carcinoma, indicating a biological variation in HPV33. Leung et al. compared ctHPV-DNA detection success rate depending on the fraction of HPV genome analyzed. They achieved 100% accuracy when analyzing whole-genome HPV compared with 94–99% accuracy when analyzing only the E6/E7 genes for HPV16 and HPV18 [25]. This difference could be attributed to the aforementioned factors or to an uneven secretion of ctHPV-DNA across the HPV genome. An uneven secretion of cfDNA across the human genome has previously been observed [26]. The fragmentation pattern of cfDNA has been linked to in vivo gene regulation through chromatin protection; an open chromatin sequence yields more fragmented DNA during apoptosis compared with tightly histone-wrapped DNA. This restricts access of nucleases during apoptosis, affecting the fragmentation pattern of cfDNA [26]. Consequently, the active E6/E7 genes of integrated HPV may undergo similar nuclease exposure as expressed human genes, while episomal HPV may exhibit a different fragmentation pattern, influencing detection ability.
For tumor tissue testing in this study, a HPV genotype specific assay was used in combination with p16 immunohistochemistry, decreasing the risk of an unreliable result. Through this approach, the correct genotype for ctHPV-DNA analysis was directly obtained, which can decrease turnaround time and save the limited amount of cfDNA. Although discrepancies between HPV+ and p16 staining have been reported previously [27, 28], in our cohort, there was a complete overlap between p16+ and HPV+ in tissue.
We found a wide range of levels of ctHPV-DNA in clinical plasma samples, aligning with findings from multiple prior studies [14, 18, 19]. This variability has been linked to tumor burden, a correlation our study confirmed by identifying an association between ctHPV-DNA levels and TNM stage [12, 14, 19, 29]. While several clinical tumor characteristics have been investigated for an association with ctHPV-DNA levels, less is known about the molecular characteristics of the tumor. When investigating the relationship between tumor viral load and ctHPV-DNA, we demonstrated a significant correlation between the two. Despite the association of ctHPV-DNA with tumor stage, both in this study and in others, the association of ctHPV-DNA and viral load persisted even after adjusting for TNM stage. Indeed, Chera et al. also observed that the copy number of HPV in tumor tissue, as measured by next generation sequencing (NGS), was significantly higher in patients with more than 200 copies/ml plasma of ctHPV-DNA compared with patients with below 200 copies/ml plasma [12].
Chera et al. found evidence of HPV integration in 40% of patients with OPSCC [12]. Furthermore, tumors with an integrated HPV had a lower copy number of HPV in the tumor and a trend of having lower ctHPV-DNA. Anayannis et al. detected 32% integration in their cohort as well as a significantly lower HPV viral load in tumors with integrated HPV [30]. In our study, tumor viral load was measured by targeting the E6 gene, which is often intact upon HPV integration into the human genome. Therefore, our assay would detect all copies of HPV, irrespective of integration status. However, our assay would not distinguish between episomal and integrated HPV, raising interest in investigating if the correlation between integration and viral load could be replicated in our cohort.
After treatment, 72.5% of the cases in this study had reached undetectable levels of ctHPV-DNA. This is in accordance with Chera et al., who observed an 80% clearance of ctHPV-DNA posttreatment in patients treated with chemotherapy as in this study [12]. A clearance of 74% was observed by Rothman et al. after surgery and a high clearance of 94% was observed by Ferrier et al. in a treatment-mixed group of cases [18, 20]. We further found that having detectable levels of ctHPV-DNA posttreatment was associated with higher ctHPV-DNA levels at diagnosis as well as with higher tumor viral load at diagnosis but not with TNM stage. The association between tumor viral load at diagnosis and slower clearance of ctHPV-DNA has not been previously reported to our knowledge. This could be explained by a simple kinetic effect, where a higher viral load in the tissue requires a longer time to be eliminated after treatment. Viral load could therefor add a dimension to the favorable clearance profile proposed by Chera et al. to distinguish patients that are highly responsive to definitive chemoradiotherapy [12].
A limitation in this study was that we did not screen for ongoing HPV infections at other anatomical sites that might lead to false positive results. The highest risk group for this being women with HPV infections in the cervix, including cases where the infection may have resulted in an undiagnosed intraepithelial lesion. However, multiple studies have demonstrated ctHPV-DNA negativity in plasma among patients with premalignant cervical lesions [31–33]. Nonetheless, one study reported low positivity among a few younger females in the control group and they were unable to investigate if those females had any malignancy [12]. Thus, low abundance positivity should be interpreted with caution due to the high sensitivity of the ddPCR method. To mitigate the risk of a false positive result, we propose a tumor HPV type informed analysis, i.e., performing a targeted ctHPV-DNA analysis for the genotype found in the primary tumor of interest, as utilized in this study. The assay can then by extension be used to complement the clinical picture and help guide treatment interventions. To validate these assays for a nontumor, viral informed application, such as early detection or standalone diagnostic testing, further specificity testing would be required. For such applications, it would be highly beneficial to multiplex the used assays. Since all assays showed great separation between positive and negative droplets and PCR settings are identical, multiplexing has a high probability of success. As a tissue sample is currently necessary to diagnose OPSCC, HPV genotyping of the obtained tissue could easily be performed during the diagnostic workup, enabling subsequent analysis of ctHPV-DNA to be initiated directly. Hence, ctHPV-DNA can be clinically applied as a potential prognostic marker at diagnosis; furthermore, as a treatment evaluation tool and during surveillance.
Conclusions
The results of this study show that a tumor-specific assay could easily be implemented in clinical routine for non-invasive detection ctHPV-DNA at diagnosis and by extension for follow up during treatment. We could also show associations between tumor viral load and ctHPV-DNA, both at diagnosis and posttreatment, opening up for a new dimension of HPV clearance profiles.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We express our gratitude to all the patients whose participation in this study has been instrumental in furthering our understanding of ctHPV-DNA and its potential for improving cancer care.
Declarations
Author contributions
Conceptualization: G.H., A.O.A., and A.Q.; methodology: A.Q., E.A., and G.H..; validation: A.Q., E.A., and G.H.; formal analysis: A.Q., E.A., A.O.A., M.W., and G.H; investigation: A.Q., E.A., A.O.A., and M.K.; resources: G.H.; data curation: E.A., A.Q., A.O.A., M.W., and M.K.; writing—original draft preparation: A.Q.; writing—review and editing: A.Q., E.A., A.O.A., M.W., B.S., C.K., and G.H.; visualization: M.W. and A.Q.; supervision: G.H.; project administration: A.Q. and G.H.; funding acquisition: G.H. and A.O.A. All authors have read and agreed to the published version of the manuscript.
Funding
Open access funding provided by Örebro University. This work was funded by the Örebro County Council Research committee, Nyckelfonden-Örebro University Hospital Research Foundation, Lions fund for cancer research Uppsala-Örebro, and Uppsala-Örebro Regional research council.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Review Board (2019-0656/2022-02405-02).
Informed Consent Statement
Written informed consent was obtained from all research subjects involved in the study. For cfDNA samples used as technical controls, oral informed consent was obtained from all donors and samples were then anonymized and pooled before analysis. For FFPE samples used as technical controls, residual extracted DNA from clinical routine was anonymized before analysis. .
Data Availability Statement
The majority of data used in this study is presented in Online resource 4. Raw data is available upon reasonable request.
Code availability
Not applicable.
Conflicts of Interest
The raw data used for the data analysis under Sect. 2.1 Assay development was generated at SAGA Diagnostics. SAGA Diagnostics had no role in design of the study, interpretation of data, in the writing of the manuscript, or in the decision to publish the results. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results. The authors declare no conflict of interest.
References
- 1.de Martel C, Georges D, Bray F, Ferlay J, Clifford GM. Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Glob Health. 2020;8(2):e180–90. 10.1016/S2214-109X(19)30488-7. [DOI] [PubMed] [Google Scholar]
- 2.Carlander AF, Jakobsen KK, Bendtsen SK, Garset-Zamani M, Lynggaard CD, Jensen JS, et al. A contemporary systematic review on repartition of HPV-positivity in oropharyngeal cancer worldwide. Viruses. 2021. 10.3390/v13071326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghittoni R, Accardi R, Chiocca S, Tommasino M. Role of human papillomaviruses in carcinogenesis. Ecancermedicalscience. 2015;9:526. 10.3332/ecancer.2015.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359(6378):926–30. 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vu P, Khagi Y, Riviere P, Goodman A, Kurzrock R. Total Number of alterations in liquid biopsies is an independent predictor of survival in patients with advanced cancers. JCO Precis Oncol. 2020. 10.1200/PO.19.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavallone L, Aguilar-Mahecha A, Lafleur J, Brousse S, Aldamry M, Roseshter T, et al. Prognostic and predictive value of circulating tumor DNA during neoadjuvant chemotherapy for triple negative breast cancer. Sci Rep. 2020;10(1):14704. 10.1038/s41598-020-71236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lipson EJ, Velculescu VE, Pritchard TS, Sausen M, Pardoll DM, Topalian SL, et al. Circulating tumor DNA analysis as a real-time method for monitoring tumor burden in melanoma patients undergoing treatment with immune checkpoint blockade. J Immunother Cancer. 2014;2(1):42. 10.1186/s40425-014-0042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ofiara LM, Navasakulpong A, Beaudoin S, Gonzalez AV. Optimizing tissue sampling for the diagnosis, subtyping, and molecular analysis of lung cancer. Front Oncol. 2014;4:253. 10.3389/fonc.2014.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tivey A, Church M, Rothwell D, Dive C, Cook N. Circulating tumour DNA - looking beyond the blood. Nat Rev Clin Oncol. 2022;19(9):600–12. 10.1038/s41571-022-00660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka H, Suzuki M, Takemoto N, Fukusumi T, Eguchi H, Takai E, et al. Performance of oral HPV DNA, oral HPV mRNA and circulating tumor HPV DNA in the detection of HPV-related oropharyngeal cancer and cancer of unknown primary. Int J Cancer. 2022;150(1):174–86. 10.1002/ijc.33798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka H, Uno A, Takenaka Y, Suzuki M, Seo Y, Takemoto N, et al. Clearance profile of circulating tumor human papillomavirus DNA during radiotherapy predicts clinical outcomes in human papillomavirus-related oropharyngeal cancer. JCO Precis Oncol. 2023;7: e2200494. 10.1200/PO.22.00494. [DOI] [PubMed] [Google Scholar]
- 12.Chera BS, Kumar S, Beaty BT, Marron D, Jefferys S, Green R, et al. Rapid clearance profile of plasma circulating tumor HPV type 16 DNA during chemoradiotherapy correlates with disease control in HPV-associated oropharyngeal cancer. Clin Cancer Res. 2019;25(15):4682–90. 10.1158/1078-0432.CCR-19-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chera BS, Kumar S, Shen C, Amdur R, Dagan R, Green R, et al. Plasma circulating tumor HPV DNA for the surveillance of cancer recurrence in HPV-associated oropharyngeal cancer. J Clin Oncol. 2020;38(10):1050–8. 10.1200/JCO.19.02444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siravegna G, O’Boyle CJ, Varmeh S, Queenan N, Michel A, Stein J, et al. Cell-free HPV DNA provides an accurate and rapid diagnosis of HPV-associated head and neck cancer. Clin Cancer Res. 2022;28(4):719–27. 10.1158/1078-0432.CCR-21-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westgard JO, Barry PL, Hunt MR, Groth T. A multi-rule Shewhart chart for quality control in clinical chemistry. Clin Chem. 1981;27(3):493–501. [PubMed] [Google Scholar]
- 16.Brierley JD, Gospodarowicz MK, Wittekind C. TNM classification of malignant tumours. 8th ed. Oxford: Wiley-Blackwell; 2016. [Google Scholar]
- 17.Malin K, Louise BM, Gisela H, Mats KG, Gabriella LL. Optimization of droplet digital PCR assays for the type-specific detection and quantification of five HPV genotypes, including additional data on viral loads of nine different HPV genotypes in cervical carcinomas. J Virol Methods. 2021;294: 114193. 10.1016/j.jviromet.2021.114193. [DOI] [PubMed] [Google Scholar]
- 18.Routman DM, Kumar S, Chera BS, Jethwa KR, Van Abel KM, Frechette K, et al. Detectable postoperative circulating tumor human papillomavirus DNA and association with recurrence in patients with HPV-associated oropharyngeal squamous cell carcinoma. Int J Radiat Oncol Biol Phys. 2022;113(3):530–8. 10.1016/j.ijrobp.2022.02.012. [DOI] [PubMed] [Google Scholar]
- 19.O’Boyle CJ, Siravegna G, Varmeh S, Queenan N, Michel A, Pang KCS, et al. Cell-free human papillomavirus DNA kinetics after surgery for human papillomavirus-associated oropharyngeal cancer. Cancer. 2022;128(11):2193–204. 10.1002/cncr.34109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrier ST, Tsering T, Sadeghi N, Zeitouni A, Burnier JV. Blood and saliva-derived ctDNA is a marker of residual disease after treatment and correlates with recurrence in human papillomavirus-associated head and neck cancer. Cancer Med. 2023;12(15):15777–87. 10.1002/cam4.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garset-Zamani M, Carlander AF, Jakobsen KK, Friborg J, Kiss K, Marvig RL, et al. Impact of specific high-risk human papillomavirus genotypes on survival in oropharyngeal cancer. Int J Cancer. 2022;150(7):1174–83. 10.1002/ijc.33893. [DOI] [PubMed] [Google Scholar]
- 22.Akashi K, Sakai T, Fukuoka O, Saito Y, Yoshida M, Ando M, et al. Usefulness of circulating tumor DNA by targeting human papilloma virus-derived sequences as a biomarker in p16-positive oropharyngeal cancer. Sci Rep. 2022;12(1):572. 10.1038/s41598-021-04307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rutkowski TW, Mazurek AM, Snietura M, Hejduk B, Jedrzejewska M, Bobek-Billewicz B, et al. Circulating HPV16 DNA may complement imaging assessment of early treatment efficacy in patients with HPV-positive oropharyngeal cancer. J Transl Med. 2020;18(1):167. 10.1186/s12967-020-02330-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen AA, Heideman DA, Boon D, Chen Z, Burk RD, De Vuyst H, et al. Human papillomavirus 33 worldwide genetic variation and associated risk of cervical cancer. Virology. 2014;448:356–62. 10.1016/j.virol.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leung E, Han K, Zou J, Zhao Z, Zheng Y, Wang TT, et al. HPV sequencing facilitates ultrasensitive detection of HPV circulating tumor DNA. Clin Cancer Res. 2021;27(21):5857–68. 10.1158/1078-0432.CCR-19-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164(1–2):57–68. 10.1016/j.cell.2015.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nauta IH, Rietbergen MM, van Bokhoven A, Bloemena E, Lissenberg-Witte BI, Heideman DAM, et al. Evaluation of the eighth TNM classification on p16-positive oropharyngeal squamous cell carcinomas in the Netherlands and the importance of additional HPV DNA testing. Ann Oncol. 2018;29(5):1273–9. 10.1093/annonc/mdy060. [DOI] [PubMed] [Google Scholar]
- 28.Mehanna H, Taberna M, von Buchwald C, Tous S, Brooks J, Mena M, et al. Prognostic implications of p16 and HPV discordance in oropharyngeal cancer (HNCIG-EPIC-OPC): a multicentre, multinational, individual patient data analysis. Lancet Oncol. 2023;24(3):239–51. 10.1016/S1470-2045(23)00013-X. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Springer S, Mulvey CL, Silliman N, Schaefer J, Sausen M, et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med. 2015;7(293):293ra104. 10.1126/scitranslmed.aaa8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anayannis NV, Schlecht NF, Ben-Dayan M, Smith RV, Belbin TJ, Ow TJ, et al. Association of an intact E2 gene with higher HPV viral load, higher viral oncogene expression, and improved clinical outcome in HPV16 positive head and neck squamous cell carcinoma. PLoS ONE. 2018;13(2): e0191581. 10.1371/journal.pone.0191581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bryan SJ, Lee J, Gunu R, Jones A, Olaitan A, Rosenthal AN, et al. Circulating HPV DNA as a biomarker for pre-invasive and early invasive cervical cancer: a feasibility study. Cancers (Basel). 2023. 10.3390/cancers15092590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonlokke S, Stougaard M, Sorensen BS, Booth BB, Hogdall E, Nyvang GB, et al. The diagnostic value of circulating cell-free HPV DNA in plasma from cervical cancer patients. Cells. 2022. 10.3390/cells11142170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galati L, Combes JD, Le Calvez-Kelm F, McKay-Chopin S, Forey N, Ratel M, et al. Detection of circulating HPV16 DNA as a biomarker for cervical cancer by a bead-based HPV genotyping assay. Microbiol Spectr. 2022;10(2): e0148021. 10.1128/spectrum.01480-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The majority of data used in this study is presented in Online resource 4. Raw data is available upon reasonable request.