Abstract
Successful antimicrobial therapy depends on achieving optimal drug concentrations within individual patients. Inter-patient variability in pharmacokinetics (PK) and differences in pathogen susceptibility (reflected in the minimum inhibitory concentration, [MIC]) necessitate personalised approaches. Dose individualisation strategies aim to address this challenge, improving treatment outcomes and minimising the risk of toxicity and antimicrobial resistance. Therapeutic drug monitoring (TDM), with the application of population pharmacokinetic (popPK) models, enables model-informed precision dosing (MIPD). PopPK models mathematically describe drug behaviour across populations and can be combined with patient-specific TDM data to optimise dosing regimens. The integration of machine learning (ML) techniques promises to further enhance dose individualisation by identifying complex patterns within extensive datasets. Implementing these approaches involves challenges, including rigorous model selection and validation to ensure suitability for target populations. Understanding the relationship between drug exposure and clinical outcomes is crucial, as is striking a balance between model complexity and clinical usability. Additionally, regulatory compliance, outcome measurement, and practical considerations for software implementation will be addressed. Emerging technologies, such as real-time biosensors, hold the potential for revolutionising TDM by enabling continuous monitoring, immediate and frequent dose adjustments, and near patient testing. The ongoing integration of TDM, advanced modelling techniques, and ML within the evolving digital health care landscape offers a potential for enhancing antimicrobial therapy. Careful attention to model development, validation, and ethical considerations of the applied techniques is paramount for successfully optimising antimicrobial treatment for the individual patient.
Key Points
| Personalised approaches are necessary for successful antimicrobial therapy due to interpatient variability in the pharmacokinetic parameters and differences in pathogen susceptibility. |
| Therapeutic drug monitoring (TDM), combined with population pharmacokinetic (popPK) models and machine learning (ML) techniques, offers a promising strategy for individualising antimicrobial drug dosing. |
| The implementation of TDM and advanced modelling techniques poses challenges, including rigorous model selection and validation, understanding the relationship between drug exposure and clinical outcomes, and ensuring regulatory compliance. |
Introduction
For the treatment of infectious diseases, the application of antimicrobial drugs is of paramount importance [1]. Their efficacy depends on achieving optimal drug concentrations in the body, or rather, at the site of infection [2]. However, these concentrations can vary significantly among individuals due to differences such as age, body weight, renal function, co-morbidities, having an infection and co-administered medication [3]. These differences may be the result of differences in the individual’s drug exposure [4], which are described by the individual pharmacokinetic (PK) parameters. Beyond patient-specific factors, the pharmacodynamic (PD) parameters of the targeted microbe also play a significant role. A key PD parameter is the minimum inhibitory concentration (MIC), a quantitative measure of the antimicrobial potency. Understanding the MIC is essential for selecting an appropriate dosing regimen—microbes with an MIC above their breakpoint are considered resistant and may necessitate higher doses or an alternative antimicrobial agent [5]. By considering both the individual patient’s PK profile and the PD characteristics of the microbe, clinicians can individualise dosing regimens for improved therapeutic outcomes.
Dose individualisation involves tailoring dosing regimens based on a patient’s unique characteristics. This ensures achieving optimal drug exposure while minimising side effects, the emergence of resistance and toxicity [6]. Traditional dosing approaches often rely on fixed dosing regimens, which may not account for inter-patient variability in drug exposure. This may result in sub-therapeutic or supra-therapeutic drug exposure and, consequently, treatment failure, or side effects and toxicity. To overcome these limitations, doses can be tailored based on the individual patient’s characteristics. In addition, therapeutic drug monitoring (TDM) can be applied by measuring drug concentrations in blood, plasma, or urine, and subsequently guiding dose adjustments to achieve a predetermined target concentration or range [7].
Healthcare’s ongoing digital transformation, characterised by the digitisation of hospitals, the vast availability of patient data, and ever-increasing computing power, opened many new possibilities that were previously unfeasible. This has led to significant advancements in methodologies used to perform dose individualisation, including the application of population PK (popPK) models in model-informed precision dosing and the utilisation of machine learning (ML) techniques. Therefore, this review aims to explore the current state and future potential of dose individualisation for antimicrobial drugs. The focus will be on the integration of TDM, popPK models, and the transformative potential of ML methods in optimising and individualising antibiotic treatment.
Therapeutic Drug Monitoring/Model Informed Precision Dosing
Therapeutic drug monitoring allows for the adjustment of the dosing regimen based on the measured concentration of a drug [7]. However, optimal dose adjustments are difficult to assess. Therefore, TDM can be applied in conjunction with subsequent drug exposure predictions for different dosing regimen provided by a popPK model, referred to as model informed precision dosing (MIPD) [6]. These modalities, applied to individualise antimicrobial drug dosing, are elaborated upon below.
Application of Therapeutic Drug Monitoring
Therapeutic drug monitoring, which measures drug concentrations in biological matrices (e.g., blood, plasma, urine), offers clinical utility when four key conditions are satisfied. First, the drug must exhibit significant interindividual variability (IIV) in its pharmacokinetics. Second, alternative non-invasive methods to assess drug efficacy must be lacking. Third, a validated assay for precise quantification of drug concentrations must be readily available. Finally, a well-defined exposure-response relationship, encompassing both therapeutic and toxic thresholds, must be established. The latter includes knowledge about the MIC of the pathogen and is, therefore, referred to as the PK/PD index.
For antimicrobial drugs, the relationship between drug concentration and its clinical efficacy and bacterial kill characteristics is best described by three distinct PK/PD indices, depending on the drug’s activity pattern [8]. Antimicrobials with concentration-dependent activity are described by either the ratio of the unbound (free) maximal concentration (fCmax) and the MIC (fCmax/MIC) or the ratio between the 24-hour area under the free concentration-time curve and the MIC (fAUC0h-24h/MIC) [9]. For time-dependent antimicrobials, effectiveness is determined by the percentage of time the free drug concentration exceeds the MIC (%fT>MIC) throughout the dosing interval [10]. Finally, antimicrobials exhibiting both concentration-dependent and time-dependent effects are best described using the fAUC0h–24h/MIC ratio. The magnitude of the PK/PD index correlating best with a specific antimicrobial effect is referred to as the PK/PD target. The values used as the PK/PD targets are usually the median values, thereby ignoring the variability in the magnitude values.
To apply TDM effectively, it is important to obtain accurate information regarding the times of dosing prior to and after the concentration measurement. Moreover, the time of sampling is also crucial. In the case of a trough sample, it may be required to sample during steady-state, which may differ widely between antimicrobial drugs and clinical situations such as renal insufficiency. Multiple samples between subsequent doses may be required if an AUC has to be obtained. A peak concentration may also be required, in the case of application of an antimicrobial with a small volume of distribution, such as with aminoglycosides. When the volume of distribution alters, this may result in a large change in the antimicrobial concentration. Therefore, a peak measurement may be necessary in the latter case.
Depending on the use of PK models, TDM can be initiated early to simulate steady-state concentrations and adjust the dose accordingly. If models are not utilised, a sample can be taken at steady state, and the dose can be adjusted after analysis. This approach is influenced by the drug’s characteristics, the laboratory’s turnaround time, the use of dosing software, and whether efficacy and/or toxicity are specifically monitored. Ideally, dose optimisation should occur as soon as possible. The timing of TDM is expected to be of importance for clinical cure and it is expected that early TDM might improve clinical outcome. However, in clinical patients many factors influence clinical outcome complicating the analysis. In a retrospective study in critically ill patients, it has been shown that patients who obtained clinical cure and microbial eradication had beta-lactam drug concentrations measured earlier [11]. However, more research is required to assess the clinical outcome of early TDM measurements.
Studies have been performed on the superiority as it comes to clinical outcome of applying TDM. In a systemic review and meta-analysis of randomised controlled trials by Sanz-Codina et al, it was demonstrated that TDM and MIPD do have benefit with regard to reducing nephrotoxicity and treatment failure and improving target attainment [12]. Also, an improvement in mortality, clinical cure or microbiological outcome was described, although not statistically significant. This is probably because there exist so many factors influencing these outcomes. More research should go into defining realistic clinical outcomes and targeting specific target groups.
Application of Pharmacokinetic Models
Pharmacokinetic modelling has a long and established history in drug research. Pharmacokinetic models offer a powerful way to describe the behaviour of a drug within a population, mathematically capturing the relationship between drug concentrations and individual patient characteristics [13, 14]. These models have diverse applications, ranging from early pre-clinical drug development to post-marketing studies. Their insights aid in understanding disease processes, simulating drug dosing scenarios, and ultimately guiding treatment decisions. It is important to notice that each model has its own unique strengths and limitations. Models inherently simplify real-world complexity, relying on assumptions to provide valuable approximations under specific conditions.
Compartmental analysis is mostly used for models applied in TDM, which comprises the construction of both popPK and physiologically-based pharmacokinetic (PBPK) models [15]. PopPK analyses follows a 'top-down' approach, beginning with observed PK data and fitting increasingly complex models; these models don’t always directly represent physiological compartments. Conversely, PBPK utilises a ‘bottom-up’ approach, combining models of physiological and chemical processes until they accurately simulate observed PK data [16]. The key advantage of PBPK models lies in their ability to extrapolate beyond initial populations and conditions and allow for prediction of drug concentrations within specific organs or tissues [17]. However, to our knowledge no PKPB models have yet been applied for TDM purposes in clinical practice [18].
In MIPD, dose tailoring using a popPK model can be applied with and without the drug concentrations obtained from an individual patient. When drug concentrations are unavailable, a priori dosing utilises a popPK model’s median parameter estimates and covariate relationships to personalise dosing [19–21]. If drug concentrations have been obtained for a patient, maximum a posteriori (MAP) Bayesian analysis provides even greater precision by blending prior knowledge (PK parameter distributions) with observed patient data (drug concentrations and individual characteristics) to estimate the patient’s individual PK parameters [22]. These individual PK parameters enable optimised dosing regimens that target a specific concentration range, ultimately enhancing therapeutic outcomes and minimising side effects, toxicity and resistance [3]. However, it is important to remember that the accuracy of MAP Bayesian analysis hinges on the validity of the underlying popPK model.
Application of Machine Learning Techniques
In recent years, ML techniques have gained significant attention in various fields, including health care [23, 24]. Machine learning algorithms can analyse large amounts of data and uncover complex patterns and relationships that might be overlooked by traditional popPK analyses [25]. This opens doors for enhanced precision in dose individualisation. Within the field of pharmacometrics, ML and artificial intelligence (AI) applications span from data handling (e.g., imputing missing values) to model selection. Researchers are exploring hybrid models that merge PK, ML and AI, as well as pure ML/AI-based prediction models for tasks like determining antimicrobial target attainment [26].
Classical popPK analyses and newer ML approaches are both valuable tools in drug development, but their performance and outcomes differ. Classical PK analyses, based on nonlinear mixed-effects modelling, provide a well-established framework for characterising drug behaviour in populations and have been successfully used in dose optimisation studies. Machine learning approaches, on the other hand, offer potential advantages in terms of handling large datasets, identifying complex relationships and automating model building processes. While promising results have been reported with ML in PK modelling, direct comparisons are limited and often context-specific. A study by Destere et al found that an ML model in combination with a population PK model, referred to as hybrid approach, outperformed the application of a popPK model in predicting drug concentrations in a specific patient population, but the generalisability of this finding remains uncertain [27]. Moreover, a study conducted by Li et al showed that this hybrid approach improved individual prediction of vancomycin clearance. However, the latter was obtained in simulated patients and not using real-world patient data [28].
The limitations of ML in popPK analyses include the need for large and high-quality datasets, potential overfitting of models, and the “black box” nature of some algorithms, which can make interpretation and regulatory acceptance challenging. Additionally, while ML can identify complex relationships, it may not always provide the mechanistic insights offered by classical PK models.
Challenges
Applying MIPD involves numerous challenges, particularly when aiming for innovative methods. The challenges currently faced in the dose individualisation of antimicrobials will be discussed in the following sections.
Choosing the Correct Model
The use of a proper model is of utmost importance. Implementing a model into clinical practice that is not suitable for the target population can lead to patient harm due to unpredictable and unwanted concentration-dose relations. Hence, it is important that the model is validated on the target population prior to its use [29]. Models developed using seemingly similar populations can even result in a poor fit [21, 30]. Therefore, careful model selection and external validation of model performance using an independent dataset is key [31].
To ensure the reliability and predictability of models used for TDM and MIPD, pharmacometric model validation is a critical step. It involves assessing the model’s ability to accurately describe observed data and its performance in predicting drug PK in new scenarios. Common validation approaches include internal validation (e.g., cross-validation, bootstrapping) and external validation using independent datasets. Model validation guidelines, such as those published by the European Medicines Agency (EMA), provide a framework for best practices in this area. Moreover, Taylor et al have provided a step-by-step guide for clinical implementation of models into MIPD software [32].
Additionally, choosing multiple models over a single model might be beneficial. Different techniques are available for choosing the most suitable model for clinical use, such as model averaging or model ensembling [33–35]. With these methods, the models are weighted based on their ability to describe the data, demographics of the population used to construct the model, or other features to select the most optimal model or (weighted) combination of models for obtaining individual PK parameters used for dose individualisation. Pharmacometric model averaging and ensembling are techniques used to combine predictions from multiple models, aiming to improve overall accuracy and robustness. Model averaging typically involves taking a weighted average of predictions from different models, with weights determined based on factors such as model performance, complexity or prior knowledge [34]. Model ensembling, on the other hand, is a more sophisticated approach that combines predictions from diverse models to leverage their strengths while mitigating their weaknesses. This can be implemented using various methods, such as stacking, boosting or bagging. The benefits of these techniques in pharmacometrics include improved predictive accuracy, reduced overfitting, and increased robustness. However, challenges such as model selection, weighting schemes and interpretability need to be carefully considered. Recent advancements like Synthetic Model Combination (SMC) have shown promise in incorporating demographic information for improved predictions in PK and PD settings [33].
PK/PD Targets
The PK/PD target is, classically, studied using neutropenic murine thigh and lung infection models [36]. Three different values for the antibacterial effect are usually described: stasis, 1-log10 kill and 2-log10 kill. In general, the 1-log10 kill and 2-log10 kill values are preferred over a static effect. Especially in infections where a high bacterial load is presumed to be present at the site of the infection, such as with pneumonia, the 1-log10 kill target is preferred [37]. However, the 1-log10 kill or 2-log10 kill cannot be achieved for all antimicrobials. For the beta-lactam antimicrobials, the correlation between animal-derived PK/PD targets and the outcome of treatment in human infections has been reviewed [38]. It was concluded that most clinical studies in human suggests a PK/PD target that is congruent with the target from pre-clinical murine studies. Importantly, infection-site specific PK/PD targets might differ from the classical PK/PD targets often based on blood taken from systemic circulation [39]. Therefore, concentrations taken at the site of infection cannot be automatically interpreted using classical PK/PD targets.
In clinical studies, other PK/PD targets such as the 100%fT>MIC or fT>4xMIC are also used for guiding antimicrobial dosing [40]. The 100%fT>MIC is commonly used in critically ill patients because of the severity of their condition [41]. These patients often have altered PK parameters, which poses a risk for target non-attainment [42]. For many antimicrobials, the bacterial killing does not increase above a threshold value. Consequently, a PK/PD target of 80%fT>MIC might be equally effective to 100%fT>MIC. In pre-clinical studies a near-maximal killing can be reached at lower values than 100%fT>MIC, indicating that the additional effect of increasing the %fT>MIC above a certain value to 100% might be limited. As can be seen for example for Klebsiella pneumoniae and cefotaxime, in which at 65%fT>MIC near maximal killing is achieved [43]. In addition, a systematic literature review demonstrated that there is no compelling clinical evidence showing superior efficiency of bactericidal as compared with bacteriostatic agents in treatment of severe infections [44]. Therefore, it is questionable whether high PK/PD targets are required in cases where alterations in the PK parameters are accounted for by TDM-guided dosing. The latter may also result in supra-therapeutic or toxic doses. However, it is unclear what concentration is needed at the target site. It can be argued that for difficult-to-reach infections, a higher systemic concentration may be necessary to achieve the correct concentration at the target site. Dependent on the characteristics of the drug (chance of toxicity), a dosing advise should be given balancing between sufficient efficacy and risk of toxicity.
Although MIC values used to determine PK/PD targets are obtained by repeated measurement using a gold standard method, measuring MICs multiple times in clinical practice is not feasible. Due to the inherent variability of the MIC measurement, the use of the epidemiological cut-off (ECOFF) value is recommended for TDM instead [45]. However, the %fT>ECOFF value might be close to the %fT>4xMIC value depending on the wild-type distribution for several antimicrobial-pathogen combinations.
Another challenge for determining an accurate PK/PD index is the target that needs to be adhered to when combination therapy (multiple antimicrobials) is applied [46]. Future research should focus on integrating the population PK and PD of multiple drugs, mechanisms of synergy and resistance and computational models to optimise combination therapies.
Antimicrobial Resistance
With the increasing resistance to antimicrobials, assessment of adequate dosing regimen is required. For different pathogens and antimicrobials, there is considerable variation between the antimicrobial exposure needed to suppress the emergence of resistance. Notwithstanding, the antimicrobial exposure required to suppress the emergence of resistance generally exceeds that of clinical efficacy [47]. This highlights a complex and evolving challenge for clinicians and researchers. Furthermore, the optimal exposure to prevent resistance can vary significantly across different pathogens and antimicrobial agents. This is further complicated by the imprecise measurement of bacterial susceptibility, showing high variability [48]. Understanding the relationship between antimicrobial concentration and the development of resistance is crucial. Currently, these relationships only consider the emergence of resistance in the targeted pathogen and not in commensal microorganisms. The latter poses a major limitation.
If a more accurate method for MIC testing is developed, all PD analyses have to be reassessed. This also goes for current MIC testing, included in the determination of PK/PD targets. When treatment with a specific antibiotic is needed for isolates with MIC values above the clinical breakpoint, it has been suggested to perform TDM based on the MIC measured with additional 2-fold dilutions added to the measurement. For isolates with a wide wild-type distribution it might be necessary to add more than a 2-fold dilution to the measured MIC [45].
Practical Implementation
Regulatory Guidelines
To apply models clinically, compliance with medical device regulations (MDR) is essential, as models and the software where they are implemented in are considered to be medical devices. Validation of this software is required, and certification confirming data storage outside the user’s jurisdiction is important. Additionally, data transfer must adhere to the General Data Protection Regulation (GDPR) [49]. In addition, as proposed doses by the MIPD software may exceed the approved dose ranges for a specific antimicrobial, an approval by a Medical Research Ethics Committee (MREC) may also be required.
Model Complexity and User Experience
To facilitate seamless adoption and application of PK/PD models in real-world clinical settings, user-friendly interfaces and training programmes for health care professionals are essential [50]. Increasing model complexity may hamper its application in clinical practice as it is less clear how the model results have been obtained. As for ML models, one is generally unaware of the underlying processes driving the model results. Therefore, clinicians may be hesitant to adopt the use of these models in clinical practice. Unlike many AI/ML techniques, popPK and PBPK models offer transparency in their underlying calculations. Additionally, high model complexity may lead to other issues such as unavailability of required data to apply the model (discussed below). Moreover, as the calculations performed by such models are complex and computationally demanding, specialised computer software and hardware and IT support are also required, especially for integration and implementation in the current hospital infrastructure.
Outcome Measures
Although clinical trials evaluating the impact of MIPD on antimicrobial outcomes are increasing, the existing evidence base remains limited. A meta-analysis of randomised controlled trials (RCTs) noted that MIPD potentially reduces mortality. Although statistically significant reductions in treatment failure and nephrotoxicity were obtained, the reductions obtained in mortality were not statistically significant [12]. Direct comparison of dose optimisation strategies across clinical trials is challenging due to complexities in non-inferiority trial designs and large sample size requirements [51]. The Desirability of Outcome Ranking (DOOR) and Response Adjusted for Duration of Antibiotic Risk (RADAR) methodologies are transforming clinical trial practices, particularly in antimicrobial optimisation. These approaches establish a two-tiered evaluation system. The DOOR innovates by ranking trial participants based on their overall outcomes, considering benefits, harms, and potentially quality of life. This patient-centric approach contrasts with traditional methods that focus on analysing outcomes rather than patients. The RADAR, a specialised DOOR application for antimicrobial trials, integrates clinical outcomes with drug use, prioritising reduced antimicrobial exposure without compromising patient results.
Multiple studies demonstrate the value of DOOR and RADAR across diverse medical settings. Examples include the CASA RELAX study comparing appendectomy antibiotic strategies [52], procalcitonin-guided algorithms in burn patients [53], urinary tract infections [54], and intra-abdominal infections [55]. Studies in paediatric pneumonia and the DigiSep-Trial for sepsis patients, further illustrate the potential of DOOR and RADAR to improve clinical responses and optimise cost-effective diagnostic strategies [56, 57]. These studies highlight the significant utility of DOOR and RADAR for enhancing antimicrobial use and evaluating comprehensive clinical outcomes.
Model-Informed Precision Dosing Software
While MIPD software offers the potential to streamline antimicrobial dose optimisation, their widespread clinical adoption faces several challenges [49]. Factors hindering implementation include limited robust evidence demonstrating the utility, impact, and cost effectiveness of MIPD, as well as concerns around user-friendliness, high costs, privacy regulations, and the technical expertise required [49, 58]. Additionally, ensuring the availability of validated popPK models within the software is crucial for accurate dosing recommendations. To assist clinicians, a comparative evaluation of ten available MIPD software tools has been conducted, analysing features like user-interface design, popPK models, user-support, and costs [49]. Careful consideration of factors such as model validation, covariate availability, appropriate patient selection and clinician training is essential for successful MIPD software implementation in clinical practice [59]. Ideally, seamless integration with electronic patient record (EPR) systems would increase transferability of patient data and, thus, optimise workflow.
To accurately measure drug concentrations for MIPD, the choice of analytical method is crucial. For certain antimicrobials (like aminoglycosides and vancomycin), immunoassays offer a convenient option. However, many other antimicrobials require more sensitive techniques like liquid chromatography–mass spectrometry/MS or high-performance liquid chromatography. When interpreting MIPD results, it is essential to consider both the analytical method and the biological matrix where the sample was taken, as these factors can significantly influence measured drug concentrations [60]. Additionally, precise documentation of dose administration and sample collection times is important for obtaining accurate MIPD results. Uncertainties in timing can result in biased and imprecise prediction when using popPK models, potentially leading to suboptimal supra-therapeutic dose recommendations [61].
Target-Site Concentrations Versus Plasma Concentrations
Therapeutic drug monitoring is standardly performed in blood and the reference values for TDM in most cases are based on plasma concentrations. This is in line with the preclinical studies on the PD-target in which the PK exposure is determined in murine blood while the effect is measured in murine tissues. However, the latter is often not performed at the site of infection. It would be of added value to also perform TDM directly in tissues. However, in order to interpretate the results obtained from TDM we also need the PD-target based on concentrations in the corresponding tissues. In our research group, we have performed TDM for antibiotics in cerebrospinal fluid (CSF) in clinical practice. Nevertheless, TDM performed at other target sites is still only performed in research [62].
In bone and joint infections, for example, the literature is sparse for most antibiotics. In addition, many variable and total antibiotic concentrations are published. More extensive research on the PK/PD of antibiotics in bone and joint infections is required [63].
Cost Effectiveness and Logistics
Cost-effectiveness evaluations of individualisation strategies in pharmacometrics are crucial to assess their economic viability and sustainability within health care systems. These evaluations should consider both the direct costs associated with model development, validation, and implementation, as well as the indirect costs and benefits related to improved patient outcomes and resource utilisation. Additionally, the logistic requirements for successful individualisation, such as data collection, infrastructure, and personnel training, need to be thoroughly examined. For instance, the implementation of MIPD may require investment in software and hardware infrastructure, as well as training healthcare professionals on model interpretation and application [64].
Future
Data Required for Machine Learning Techniques
Providing data for ML in TDM involves several challenges, which are outlined in the following sections.
Data Quality and Completeness
TDM data can be collected using various approaches, including sparse, opportunistic, or scavenged sampling. However, limited data availability poses a significant challenge for training and deploying effective ML models, as these techniques thrive on vast datasets. When essential data are scarce, it becomes difficult to build reliable models, and missing data points can further compromise accuracy. Imputing missing values requires careful strategies to minimise bias [65–67]. Fortunately, the landscape is changing with the rise of automated PK data recording modalities, such as infusion systems connected to EPRs and the increasing use of methods like dried blood spots (DBS) for sample collection. These advancements have the potential to offer new ways of collecting data needed to power dose optimisation.
Heterogeneity of Data
Patient populations vary in demographics, genetics, and comorbidities. Incorporating this heterogeneity into ML models is challenging but essential for generalisability across different patient groups. Integrating heterogeneous data from multiple sources requires clear guidelines that adhere to high ethical and legal standards [68].
Temporal Dynamics
Drug concentrations in the body change over time, influenced by factors such as metabolism, renal function, and adherence to medication. Rapid fluctuations in physiological functions, such as augmented renal clearance (ARC), impaired kidney or liver function, and altered drug distribution volume are experienced especially by critically ill patients [10]. These dynamic changes can significantly impact antimicrobial exposure. Although adding complexity to the modelling process, accurately capturing these temporal dynamics is crucial for applying TDM effectively. Moreover, an ample number of data points is required to predict such dynamic processes [69]. In clinical practice, this may be hampered by the turn-around time of the applied analytical assay. Accounting for rapidly changing temporal dynamics is important for optimising dosing regimens in critically ill patients.
Temporal dynamics can also occur in the inoculum. Currently, in all in vitro methods (such as MIC testing) and the in vivo preclinical studies, the used inocula are standardised. This makes it easier to compare results. On the other hand, it has been shown that in a mouse model higher drug exposures are required to achieve stasis or 1-log10 killing against a higher (107 CFU/mL) than a lower (105 CFU/mL) inoculum of multiple Staphylococcus aureus strains for four classes of antibiotics [70, 71]. Although the clinical relevance of experimental inoculum effects is not fully understood, it is noted that in infections in which a high inoculum can be expected, such as in ventilator-associated pneumonia, it may be more desirable to achieve the more stringent PD-targets [70].
Validation and Generalisation
Validating models, especially ML models, for TDM is challenging due to the limited availability of independent datasets. Rigorous validation is crucial to ensure the reliable model performance in different real-world clinical scenarios [29, 72]. Furthermore, models trained on specific patient populations or data sources may not adequately predict other populations. Generalising model results across diverse clinical contexts is a common challenge, and careful extrapolation of parameter estimates is warranted, if at all possible.
Regulatory Compliance
Population PK models, obtained using non-linear mixed effect modelling or ML, may be subject to regulatory scrutiny. Ensuring that data and models comply with regulatory standards is essential for their acceptance in clinical practice [73, 74].
Clinical Integration
Implementation of models into health information systems (HIS) comprising EPRs has multiple advantages. It reduces the human factor of manually typing information from one system into the other. Moreover, the models may be able to autonomically tailor the dose of an antimicrobial to the needs of the individual patient. Furthermore, models integrated in a HIS may continuously learn from new incoming real-world data (RWD) as well as on feedback of their own performance. Therefore, autonomic updating of the model may continuously improve the model performance and provide more accurate adjustments of a dosing regimen.
Real-Time Monitoring
When performing TDM, biological samples (typically plasma) are transported to an analysis laboratory, with a turn-around time ranging from approximately 30 minutes to 48 hours. This timeframe varies depending on the specific drug and the availability of the assay. Recognising the importance of promptly initiating effective antimicrobial therapy, reducing the turnaround time becomes crucial for timely adjustments of antimicrobial dosing regimen [75].
Another limitation of current TDM practices revolves around the minimal number of available antimicrobial assays in most laboratories [76]. Considering these challenges, there is a necessity to create innovative technologies that address the limitations of traditional TDM processes. In this context, the exploration of real-time TDM sensor monitoring emerges as a promising solution [77, 78]. Prerequisites for this technology encompass: transduction, involving the identification of targets in human matrices [79]; ongoing sample collection via, for instance, interstitial fluid sampling using microneedle biosensors [80]; and instantaneous signal processing of assay outcomes [81]. Therefore, biosensors could facilitate target site concentration measurement.
Numerous biosensor techniques are currently undergoing testing, including aptamer-based electrochemical and electronic sensors [82]. Among these technologies is one that originates from particle mobility, utilising a 'competition assay' [83]. The assay identifies a binding event through a sudden reduction in particle mobility, attributed to interactions between a particle and a sensor surface. As these interactions are facilitated by weak biological forces, they are reversible, allowing continuous monitoring. Bright field optical microscopy records the transitions between unbound and bound states over time for numerous particles concurrently, facilitating precise concentration determination. Critical factors for enhancing the robustness of biosensors encompass selectivity, sensitivity, reproducibility, reusability, and long-term stability. Additionally, there is a necessity to validate the new technology against the gold standard of mass-spectrometry (MS). Given the extensive data captured, processed, and managed during clinical use, it is imperative to prioritise data confidentiality.
Alongside novel techniques, biosensors could also be measuring novel blood or target site markers. For example, Gastine et al investigated the use of the galactomannan index (GMI) for determining the dynamic endpoint of antifungal response of invasive pulmonary aspergillosis (IPA) in neutropenic patients [84]. It was demonstrated that a PK/PD index could be adequately established using the GMI (30 mg*h/L). The latter shows that other PK/PD parameters such as the MIC can also be applied for providing optimised antibiotic treatment.
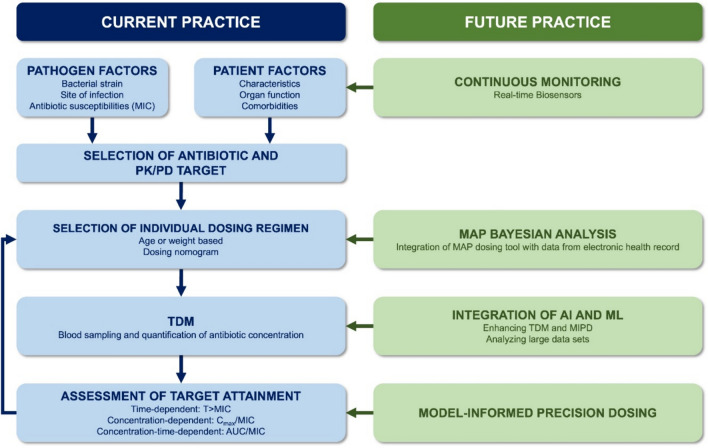
Looking ahead, the integration of AI may prove instrumental in facilitating data analysis and exposure prediction, potentially playing a substantial role in the future implementation of real-time biosensors, ultimately leading to continuous dose optimisation [85]. Applying real-time measurements may only be beneficial for a particular group of patients, such as critically ill patients. It should be further investigated what type of patients could benefit from this modality. Moreover, clinical trials need to be performed to obtain evidence for cost effectiveness as well as improved antimicrobial efficiency (Fig. 1).
Fig. 1.
Differences between current practice of MIPD and future practice. The left panel delineates the conventional methodology for optimising antibiotic dosing in current clinical practice, whereas the right panel outlines the proposed advancements for future practice. AI artificial intelligence, AUC/MIC the ratio of the area under the concentration-time curve to MIC, Cmax/MIC the ratio of maximum drug concentration to MIC, MAP maximum a posteriori, MIC minimum inhibitory concentration, MIPD model-informed precision dosing, ML machine learning, PK/PD pharmacokinetic/pharmacodynamic, TDM therapeutic drug monitoring, T>MIC the duration of time that the drug concentration remains above the MIC during a dosing interval
Conclusion
Dose individualisation of antimicrobial drugs is crucial to optimise therapeutic outcomes and minimise the risk of adverse effects. Population PK models, combined with MAP Bayesian analysis, have been valuable tools in this context, allowing for the estimation of individual PK parameters and the prediction of drug concentrations. Machine learning and AI techniques show promise in enhancing the accuracy of dose individualisation by capturing complex relationships that may not be captured by traditional popPK models. However, further research is needed to fully explore the potential of ML and AI in this field. Careful model selection, validation, and consideration of ethical and regulatory aspects are essential to ensure the reliability and applicability of the results obtained using these methodologies.
Data availability
Not applicable.
Declarations
Funding
For this review, no funding was obtained.
Conflict of interest
TP, AM, AA, BdW, SS, and BK have no conflict of interest to report.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
Not applicable.
Code availability
Not applicable.
Author contributions
All authors contributed equally to the conception, writing, and revision of the review.
Footnotes
Tim Preijers, Anouk E. Muller, Alan Abdulla, Brenda C. M. de Winter, Birgit C. P. Koch, and Sebastiaan D. T. Sassen contributed equally.
References
- 1.Gould K. Antibiotics: from prehistory to the present day. J Antimicrob Chemother. 2016;71(3):572–5. [DOI] [PubMed] [Google Scholar]
- 2.Radlinski L, Conlon BP. Antibiotic efficacy in the complex infection environment. Curr Opin Microbiol. 2018;42:19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wicha SG, Martson AG, Nielsen EI, Koch BCP, Friberg LE, Alffenaar JW, et al. From therapeutic drug monitoring to model-informed precision dosing for antibiotics. Clin Pharmacol Ther. 2021;109(4):928–41. [DOI] [PubMed] [Google Scholar]
- 4.Adembri C, Novelli A. Pharmacokinetic and pharmacodynamic parameters of antimicrobials: potential for providing dosing regimens that are less vulnerable to resistance. Clin Pharmacokinet. 2009;48(8):517–28. [DOI] [PubMed] [Google Scholar]
- 5.Zhang L, Xie H, Wang Y, Wang H, Hu J, Zhang G. Pharmacodynamic parameters of pharmacokinetic/pharmacodynamic (PK/PD) integration models. Front Vet Sci. 2022;9: 860472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darwich AS, Polasek TM, Aronson JK, Ogungbenro K, Wright DFB, Achour B, et al. Model-informed precision dosing: background, requirements, validation, implementation, and forward trajectory of individualizing drug therapy. Annu Rev Pharmacol Toxicol. 2021;61:225–45. [DOI] [PubMed] [Google Scholar]
- 7.Kang JS, Lee MH. Overview of therapeutic drug monitoring. Korean J Intern Med. 2009;24(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez-Gascon A, Solinis MA, Isla A. The role of PK/PD analysis in the development and evaluation of antimicrobials. Pharmaceutics. 2021;13(6):833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen EI, Cars O, Friberg LE. Pharmacokinetic/pharmacodynamic (PK/PD) indices of antibiotics predicted by a semimechanistic PKPD model: a step toward model-based dose optimization. Antimicrob Agents Chemother. 2011;55(10):4619–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koch BCP, Muller AE, Hunfeld NGM, de Winter BCM, Ewoldt TMJ, Abdulla A, Endeman H. Therapeutic drug monitoring of antibiotics in critically ill patients: current practice and future perspectives with a focus on clinical outcome. Ther Drug Monit. 2022;44(1):11–8. [DOI] [PubMed] [Google Scholar]
- 11.Al-Shaer MH, Rubido E, Cherabuddi K, Venugopalan V, Klinker K, Peloquin C. Early therapeutic monitoring of beta-lactams and associated therapy outcomes in critically ill patients. J Antimicrob Chemother. 2020;75(12):3644–51. [DOI] [PubMed] [Google Scholar]
- 12.Sanz-Codina M, Bozkir H, Jorda A, Zeitlinger M. Individualized antimicrobial dose optimization: a systematic review and meta-analysis of randomized controlled trials. Clin Microbiol Infect. 2023;29(7):845–57. [DOI] [PubMed] [Google Scholar]
- 13.Mould DR, Upton RN. Basic concepts in population modeling, simulation, and model-based drug development-part 2: introduction to pharmacokinetic modeling methods. CPT Pharmacometrics Syst Pharmacol. 2013;2(4): e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duffull SB, Wright DF, Winter HR. Interpreting population pharmacokinetic-pharmacodynamic analyses—a clinical viewpoint. Br J Clin Pharmacol. 2011;71(6):807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rescigno A. The rise and fall of compartmental analysis. Pharmacol Res. 2001;44(4):337–42. [DOI] [PubMed] [Google Scholar]
- 16.Suri A, Chapel S, Lu C, Venkatakrishnan K. Physiologically based and population PK modeling in optimizing drug development: a predict-learn-confirm analysis. Clin Pharmacol Ther. 2015;98(3):336–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsamandouras N, Rostami-Hodjegan A, Aarons L. Combining the “bottom up” and “top down” approaches in pharmacokinetic modelling: fitting PBPK models to observed clinical data. Br J Clin Pharmacol. 2015;79(1):48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira A, Lapa R, Vale N. PBPK modeling and simulation and therapeutic drug monitoring: possible ways for antibiotic dose adjustment. Processes. 2021;9(11):2087. [Google Scholar]
- 19.Heus A, Uster DW, Grootaert V, Vermeulen N, Somers A, In’t Veld DH, et al. Model-informed precision dosing of vancomycin via continuous infusion: a clinical fit-for-purpose evaluation of published PK models. Int J Antimicrob Agents. 2022;59(5): 106579. [DOI] [PubMed] [Google Scholar]
- 20.Duong A, Simard C, Williamson D, Marsot A. Tobramycin a priori dosing regimens based on PopPK model simulations in critically ill patients: are they transferable? Ther Drug Monit. 2023;45(5):616–22. [DOI] [PubMed] [Google Scholar]
- 21.Yang N, Wang J, Xie Y, Ding J, Wu C, Liu J, Pei Q. External evaluation of population pharmacokinetic models to inform precision dosing of meropenem in critically ill patients. Front Pharmacol. 2022;13: 838205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo T, van Hest RM, Zwep LB, Roggeveen LF, Fleuren LM, Bosman RJ, et al. Optimizing predictive performance of bayesian forecasting for vancomycin concentration in intensive care patients. Pharm Res. 2020;37(9):171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Javaid M, Haleem A, Pratap Singh R, Suman R, Rab S. Significance of machine learning in healthcare: features, pillars and applications. Int J Intell Netw. 2022;3:58–73. [Google Scholar]
- 24.Stankevičiūtė K, Woillard J-B, Peck RW, Marquet P, van der Schaar M. Bridging the worlds of pharmacometrics and machine learning. Clin Pharmacokinet. 2023;62(11):1551–65. [DOI] [PubMed] [Google Scholar]
- 25.Huang Z, Denti P, Mistry H, Kloprogge F. Machine learning and artificial intelligence in PK-PD modeling: fad, friend, or foe? Clin Pharmacol Ther. 2024;115(4):652–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang BH, Zhang JY, Allegaert K, Hao GX, Yao BF, Leroux S, et al. Use of machine learning for dosage individualization of vancomycin in neonates. Clin Pharmacokinet. 2023;62(8):1105–16. [DOI] [PubMed] [Google Scholar]
- 27.Destere A, Marquet P, Labriffe M, Drici MD, Woillard JB. A hybrid algorithm combining population pharmacokinetic and machine learning for isavuconazole exposure prediction. Pharm Res. 2023;40(4):951–9. [DOI] [PubMed] [Google Scholar]
- 28.Li G, Sun Y, Zhu L. Application of machine learning combined with population pharmacokinetics to improve individual prediction of vancomycin clearance in simulated adult patients. Front Pharmacol. 2024;15:1352113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherwin C, Kiang T, Spigarelli M, Ensom M. Fundamentals of population pharmacokinetic modelling: validation methods. Clin Pharmacokinet. 2012;51:573–90. [DOI] [PubMed] [Google Scholar]
- 30.Guo T, van Hest RM, Roggeveen LF, Fleuren LM, Thoral PJ, Bosman RJ, et al. External evaluation of population pharmacokinetic models of vancomycin in large cohorts of intensive care unit patients. Antimicrob Agents Chemother. 2019;63(5):e02543-e2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baklouti S, Marolleau S, Chavanet P, Bonnet E, Concordet D, Gandia P. Why is it desirable to do the external evaluation of a population pharmacokinetic model? Antimicrob Agents Chemother. 2022;66(1): e0149321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor ZL, Poweleit EA, Paice K, Somers KM, Pavia K, Vinks AA, et al. Tutorial on model selection and validation of model input into precision dosing software for model-informed precision dosing. CPT Pharmacometrics Syst Pharmacol. 2023;12(12):1827–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan A, Peck R, Gibbs M, van der Schaar M. Synthetic model combination: a new machine-learning method for pharmacometric model ensembling. CPT Pharmacometrics Syst Pharmacol. 2023;12(7):953–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uster DW, Stocker SL, Carland JE, Brett J, Marriott DJE, Day RO, Wicha SG. A model averaging/selection approach improves the predictive performance of model-informed precision dosing: vancomycin as a case study. Clin Pharmacol Ther. 2021;109(1):175–83. [DOI] [PubMed] [Google Scholar]
- 35.Sibieude E, Khandelwal A, Girard P, Hesthaven JS, Terranova N. Population pharmacokinetic model selection assisted by machine learning. J Pharmacokinet Pharmacodyn. 2022;49(2):257–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adembri C, Novelli A, Nobili S. Some suggestions from PK/PD principles to contain resistance in the clinical setting-focus on ICU patients and gram-negative strains. Antibiotics (Basel). 2020;9(10):676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guideline on the use of pharmacokinetics and pharmacodynamics in the development of antimicrobial medicinal products. EMA/CHMP/594085/2015. 2017.
- 38.Berry AV, Kuti JL. Pharmacodynamic thresholds for beta-lactam antibiotics: a story of mouse versus man. Front Pharmacol. 2022;13: 833189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farrington N, McEntee L, Johnson A, Unsworth J, Darlow C, Jimenez-Valverde A, et al. Pharmacodynamics of meropenem and tobramycin for neonatal meningoencephalitis: novel approaches to facilitate the development of new agents to address the challenge of antimicrobial resistance. Antimicrob Agents Chemother. 2022;66(4): e0218121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts JA, Paul SK, Akova M, Bassetti M, De Waele JJ, Dimopoulos G, et al. DALI: defining antibiotic levels in intensive care unit patients: are current beta-lactam antibiotic doses sufficient for critically ill patients? Clin Infect Dis. 2014;58(8):1072–83. [DOI] [PubMed] [Google Scholar]
- 41.Abdul-Aziz MH, Alffenaar JC, Bassetti M, Bracht H, Dimopoulos G, Marriott D, et al. Antimicrobial therapeutic drug monitoring in critically ill adult patients: a position paper. Intensive Care Med. 2020;46(6):1127–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wieringa A, Ewoldt TMJ, Gangapersad RN, Gijsen M, Parolya N, Kats C, et al. Predicting beta-lactam target non-attainment in ICU patients at treatment initiation: development and external validation of three novel (machine learning) models. Antibiotics (Basel). 2023;12(12):1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Craig WA. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing of mice and men. Clin Infect Dis. 1998;26(1):1–10 (quiz 1-2). [DOI] [PubMed] [Google Scholar]
- 44.Wald-Dickler N, Holtom P, Spellberg B. Busting the myth of “static vs cidal”: a systemic literature review. Clin Infect Dis. 2018;66(9):1470–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mouton JW, Muller AE, Canton R, Giske CG, Kahlmeter G, Turnidge J. MIC-based dose adjustment: facts and fables. J Antimicrob Chemother. 2018;73(3):564–8. [DOI] [PubMed] [Google Scholar]
- 46.Bulman ZP, Wicha SG, Nielsen EI, Lenhard JR, Nation RL, Theuretzbacher U, et al. Research priorities towards precision antibiotic therapy to improve patient care. Lancet Microbe. 2022;3(10):e795–802. [DOI] [PubMed] [Google Scholar]
- 47.Sumi CD, Heffernan AJ, Lipman J, Roberts JA, Sime FB. What antibiotic exposures are required to suppress the emergence of resistance for gram-negative bacteria? A Systematic Review Clin Pharmacokinet. 2019;58(11):1407–43. [DOI] [PubMed] [Google Scholar]
- 48.Malmberg C, Torpner J, Fernberg J, Öhrn H, Johansson C, Tängdén T, Kreuger J. Evaluation of the speed, accuracy and precision of the QuickMIC rapid antibiotic susceptibility testing assay in a clinical setting. bioRxiv. 2021:2021.08.11.455925. [DOI] [PMC free article] [PubMed]
- 49.Kantasiripitak W, Van Daele R, Gijsen M, Ferrante M, Spriet I, Dreesen E. Software tools for model-informed precision dosing: how well do they satisfy the needs? Front Pharmacol. 2020;11:620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fuchs A, Csajka C, Thoma Y, Buclin T, Widmer N. Benchmarking therapeutic drug monitoring software: a review of available computer tools. Clin Pharmacokinet. 2013;52(1):9–22. [DOI] [PubMed] [Google Scholar]
- 51.Evans SR, Rubin D, Follmann D, Pennello G, Huskins WC, Powers JH, et al. Desirability of outcome ranking (DOOR) and response adjusted for duration of antibiotic risk (RADAR). Clin Infect Dis. 2015;61(5):800–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeh DD, Hatton GE, Pedroza C, Pust G, Mantero A, Namias N, Kao LS. Complex and simple appendicitis: REstrictive or Liberal postoperative Antibiotic eXposure (CASA RELAX) using desirability of outcome ranking (DOOR) and response adjusted for duration of antibiotic risk (RADAR): study protocol for a randomized controlled trial. Trauma Surg Acute Care Open. 2022;7(1): e000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zbyrak V, Reveron SL, Smoke S, Mehta A, Marano MA, Lee R. Antibiotic usage after procalcitonin-guided therapy algorithm implementation in a burn intensive care unit. Ann Burns Fire Disasters. 2020;33(4):317–21. [PMC free article] [PubMed] [Google Scholar]
- 54.Sherwood K, Ikuta K. 2241. Methenamine for the prevention of recurrent UTIs: a desirability of outcome ranking (DOOR) analysis for the alternative to prophylactic antibiotics for the treatment of recurrent urinary tract infections in women (ALTAR) Trial. Open Forum Infect Dis. 2022;9:Supplement_2. [Google Scholar]
- 55.Celestin AR, Odom SR, Angelidou K, Evans SR, Coimbra R, Guidry CA, et al. Novel method suggests global superiority of short-duration antibiotics for intra-abdominal infections. Clin Infect Dis. 2017;65(9):1577–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams DJ, Creech CB, Walter EB, Martin JM, Gerber JS, Newland JG, et al. Short- vs standard-course outpatient antibiotic therapy for community-acquired pneumonia in children: the SCOUT-CAP randomized clinical trial. JAMA Pediatr. 2022;176(3):253–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brenner T, Skarabis A, Stevens P, Axnick J, Haug P, Grumaz S, et al. Optimization of sepsis therapy based on patient-specific digital precision diagnostics using next generation sequencing (DigiSep-Trial)-study protocol for a randomized, controlled, interventional, open-label, multicenter trial. Trials. 2021;22(1):714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Darwich AS, Ogungbenro K, Vinks AA, Powell JR, Reny JL, Marsousi N, et al. Why has model-informed precision dosing not yet become common clinical reality? Lessons from the past and a roadmap for the future. Clin Pharmacol Ther. 2017;101(5):646–56. [DOI] [PubMed] [Google Scholar]
- 59.Oliver MB, Boeser KD, Carlson MK, Hansen LA. Considerations for implementation of vancomycin Bayesian software monitoring in a level IV NICU population within a multisite health system. Am J Health Syst Pharm. 2023;80(11):670–7. [DOI] [PubMed] [Google Scholar]
- 60.Oyaert M, Peersman N, Kieffer D, Deiteren K, Smits A, Allegaert K, et al. Novel LC-MS/MS method for plasma vancomycin: comparison with immunoassays and clinical impact. Clin Chim Acta. 2015;441:63–70. [DOI] [PubMed] [Google Scholar]
- 61.Alihodzic D, Broeker A, Baehr M, Kluge S, Langebrake C, Wicha SG. Impact of inaccurate documentation of sampling and infusion time in model-informed precision dosing. Front Pharmacol. 2020;11:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bilal H, Bergen PJ, Tait JR, Wallis SC, Peleg AY, Roberts JA, et al. Clinically relevant epithelial lining fluid concentrations of meropenem with ciprofloxacin provide synergistic killing and resistance suppression of hypermutable pseudomonas aeruginosa in a dynamic biofilm model. Antimicrob Agents Chemother. 2020;64(7):e00469-e520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koch BCP, Zhao Q, Oosterhoff M, van Oldenrijk J, Abdulla A, de Winter BCM, et al. The mysteries of target site concentrations of antibiotics in bone and joint infections: what is known? A narrative review. Expert Opin Drug Metab Toxicol. 2022;18(9):587–600. [DOI] [PubMed] [Google Scholar]
- 64.Kluwe F, Michelet R, Mueller-Schoell A, Maier C, Klopp-Schulze L, van Dyk M, et al. Perspectives on model-informed precision dosing in the digital health era: challenges, opportunities, and recommendations. Clin Pharmacol Ther. 2021;109(1):29–36. [DOI] [PubMed] [Google Scholar]
- 65.Emmanuel T, Maupong T, Mpoeleng D, Semong T, Mphago B, Tabona O. A survey on missing data in machine learning. Journal of Big Data. 2021;8(1):140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang H, Tang J, Wu M, Wang X, Zhang T. Application of machine learning missing data imputation techniques in clinical decision making: taking the discharge assessment of patients with spontaneous supratentorial intracerebral hemorrhage as an example. BMC Med Inform Decis Mak. 2022;22(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Irby DJ, Ibrahim ME, Dauki AM, Badawi MA, Illamola SM, Chen M, et al. Approaches to handling missing or “problematic” pharmacology data: pharmacokinetics. CPT Pharmacometr Syst Pharmacol. 2021;10(4):291–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Collin CB, Gebhardt T, Golebiewski M, Karaderi T, Hillemanns M, Khan FM, et al. Computational models for clinical applications in personalized medicine-guidelines and recommendations for data integration and model validation. J Pers Med. 2022;12(2):166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kang D, Schwartz JB, Verotta D. Sample size computations for PK/PD population models. J Pharmacokinet Pharmacodyn. 2005;32(5):685–701. [DOI] [PubMed] [Google Scholar]
- 70.Bulitta JB, Hope WW, Eakin AE, Guina T, Tam VH, Louie A, et al. Generating robust and informative nonclinical in vitro and in vivo bacterial infection model efficacy data to support translation to humans. Antimicrob Agents Chemother. 2019;63(5):e02307-e2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee DG, Murakami Y, Andes DR, Craig WA. Inoculum effects of ceftobiprole, daptomycin, linezolid, and vancomycin with Staphylococcus aureus and Streptococcus pneumoniae at inocula of 10(5) and 10(7) CFU injected into opposite thighs of neutropenic mice. Antimicrob Agents Chemother. 2013;57(3):1434–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaminwar SR, Goschenhofer J, Thomas J, Thon I, Bischl B. Structured verification of machine learning models in industrial settings. Big Data. 2023;11(3):181–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dykstra K, Mehrotra N, Tornøe CW, Kastrissios H, Patel B, Al-Huniti N, et al. Reporting guidelines for population pharmacokinetic analyses. J Pharmacokinet Pharmacodyn. 2015;42(3):301–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun H, Fadiran EO, Jones CD, Lesko L, Huang SM, Higgins K, et al. Population pharmacokinetics. A regulatory perspective. Clin Pharmacokinet. 1999;37(1):41–58. [DOI] [PubMed] [Google Scholar]
- 75.Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, Ferrer R, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Crit Care Med. 2017;45(3):486–552. [DOI] [PubMed] [Google Scholar]
- 76.Ewoldt TMJ, Abdulla A, van den Broek P, Hunfeld N, Bahmany S, Muller AE, et al. Barriers and facilitators for therapeutic drug monitoring of beta-lactams and ciprofloxacin in the ICU: a nationwide cross-sectional study. BMC Infect Dis. 2022;22(1):611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mishi RD, Stokes MA, Campbell CA, Plaxco KW, Stocker SL. Real-time monitoring of antibiotics in the critically ill using biosensors. Antibiotics (Basel). 2023;12(10):1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ates HC, Roberts JA, Lipman J, Cass AEG, Urban GA, Dincer C. On-site therapeutic drug monitoring. Trends Biotechnol. 2020;38(11):1262–77. [DOI] [PubMed] [Google Scholar]
- 79.Barrett JF, Hoch JA. Two-component signal transduction as a target for microbial anti-infective therapy. Antimicrob Agents Chemother. 1998;42(7):1529–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kiang TKL, Ranamukhaarachchi SA, Ensom MHH. Revolutionizing therapeutic drug monitoring with the use of interstitial fluid and microneedles technology. Pharmaceutics. 2017;9(4):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rawson TM, O’Hare D, Herrero P, Sharma S, Moore LSP, de Barra E, et al. Delivering precision antimicrobial therapy through closed-loop control systems. J Antimicrob Chemother. 2018;73(4):835–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zambry NS, Ahmad Najib M, Awang MS, Selvam K, Khalid MF, Bustami Y, et al. Aptamer-based electrochemical biosensors for the detection of salmonella: a scoping review. Diagnostics (Basel). 2022;12(12):3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Visser EWA, Yan J, van Ijzendoorn LJ, Prins MWJ. Continuous biomarker monitoring by particle mobility sensing with single molecule resolution. Nat Commun. 2018;9(1):2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gastine S, Hope W, Hempel G, Petraitiene R, Petraitis V, Mickiene D, et al. Pharmacodynamics of posaconazole in experimental invasive pulmonary aspergillosis: utility of serum galactomannan as a dynamic endpoint of antifungal efficacy. Antimicrob Agents Chemother. 2021;65(2):e01574-e1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Heil EL, Nicolau DP, Farkas A, Roberts JA, Thom KA. Pharmacodynamic target attainment for cefepime, meropenem, and piperacillin-tazobactam using a pharmacokinetic/pharmacodynamic-based dosing calculator in critically ill patients. Antimicrob Agents Chemother. 2018;62(9):e01008-e1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.