Abstract
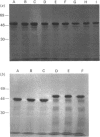
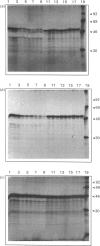
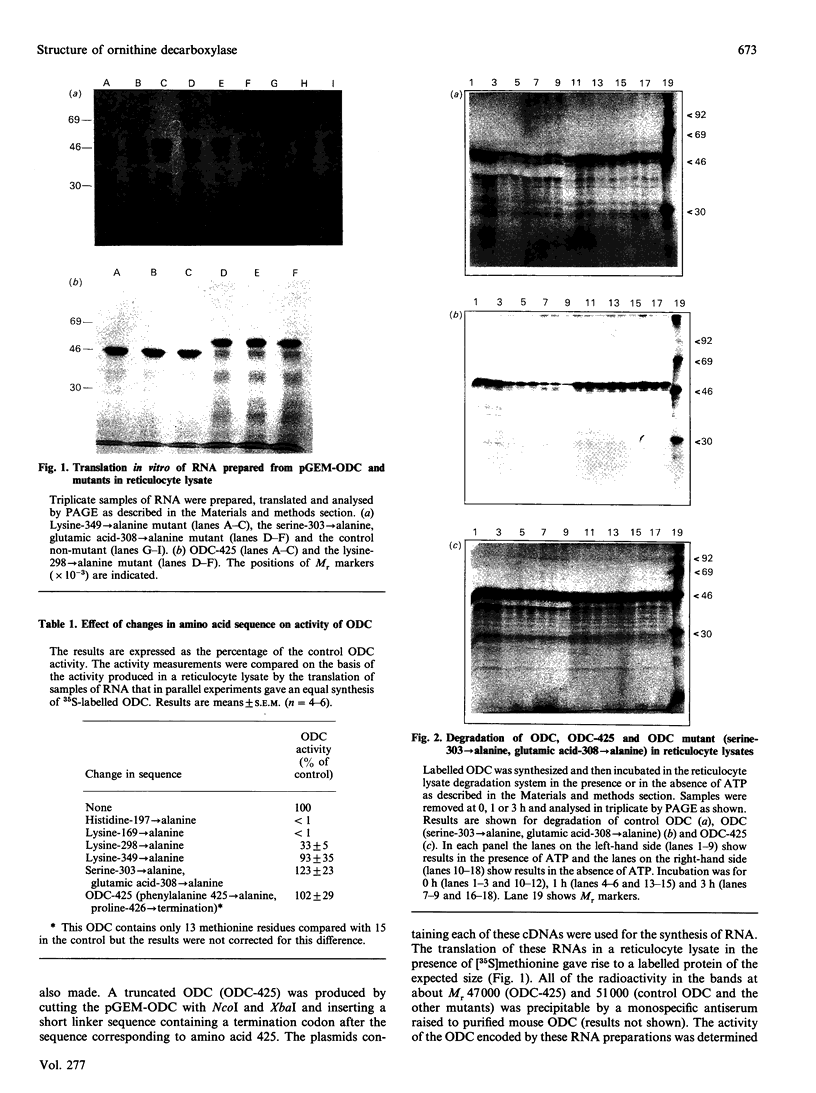
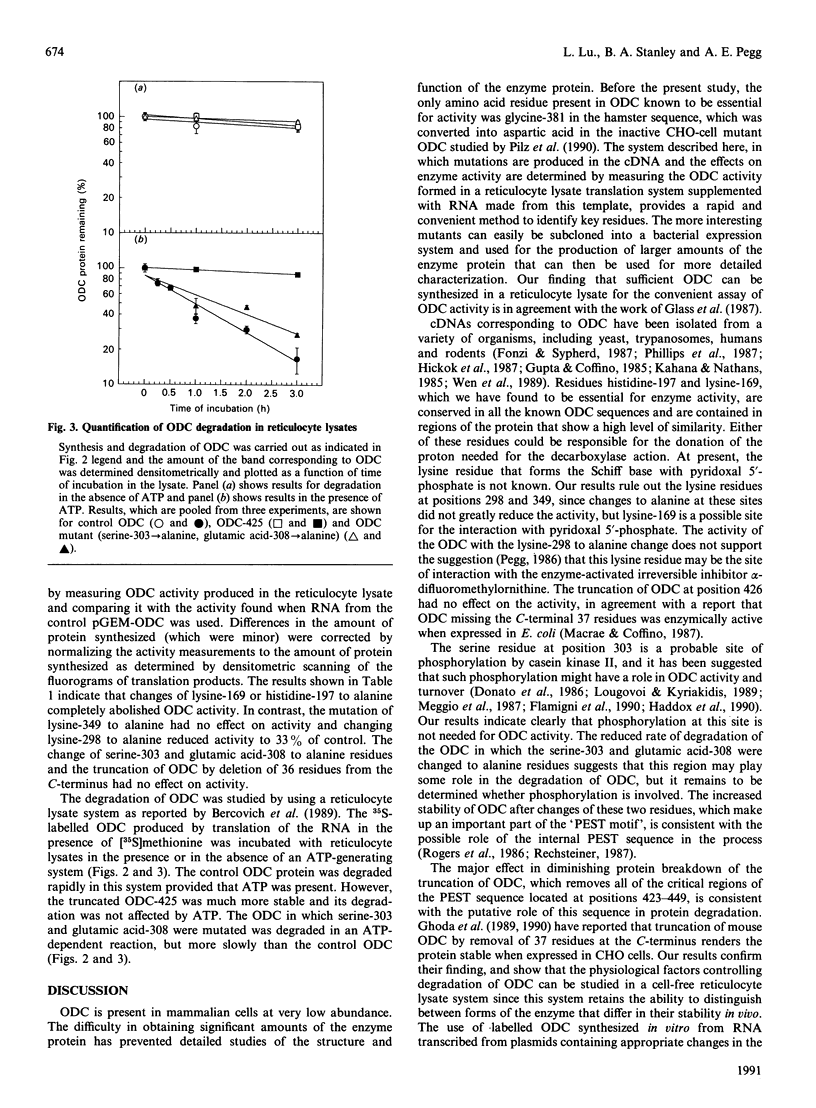
The importance of certain amino acid residues in mammalian ornithine decarboxylase activity and degradation was studied by site-specific mutagenesis. Changes were made to the mouse ornithine decarboxylase cDNA in a plasmid containing a T7 RNA polymerase promoter. The plasmid was then used for the synthesis of RNA, which was translated in a reticulocyte lysate system. The activity of the ornithine decarboxylase formed and the stability of the protein to degradation in a reticulocyte lysate system were determined. Changes of lysine-169 or of histidine-197 to alanine completely abolished enzyme activity, indicating that these residues are essential for enzyme activity. The removal of the C-terminal 36 residues, the mutation of lysine-349 to alanine, of lysine-298 to alanine or the double change of serine-303 and glutamic acid-308 to alanine residues still resulted in an active enzyme. The last-mentioned finding indicates that the phosphorylation of serine-303 does not play an essential role in the catalytic activity of ornithine decarboxylase. The control ornithine decarboxylase protein was degraded rapidly in a reticulocyte lysate provided that ATP was added. The truncated protein missing the 36 residues from the C-terminus was much more stable in this system, and the protein containing the double change of serine-303 and glutamic acid-308 to alanine residues was slightly more stable than control ornithine decarboxylase protein. These results indicate that the altered residues may play a role in interaction with factors responsible for the rapid turnover of ornithine decarboxylase.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bercovich Z., Rosenberg-Hasson Y., Ciechanover A., Kahana C. Degradation of ornithine decarboxylase in reticulocyte lysate is ATP-dependent but ubiquitin-independent. J Biol Chem. 1989 Sep 25;264(27):15949–15952. [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffino P. Probable cloning artefacts previously interpreted as unusual leader sequences of rodent ornithine decarboxylase mRNAs--a cautionary tale. Gene. 1988 Sep 30;69(2):365–368. doi: 10.1016/0378-1119(88)90448-9. [DOI] [PubMed] [Google Scholar]
- Donato N. J., Ware C. F., Byus C. V. A rat monoclonal antibody which interacts with mammalian ornithine decarboxylase at an epitope involved in phosphorylation. Biochim Biophys Acta. 1986 Nov 19;884(2):370–382. doi: 10.1016/0304-4165(86)90186-8. [DOI] [PubMed] [Google Scholar]
- Dower W. J., Miller J. F., Ragsdale C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988 Jul 11;16(13):6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamigni F., Marmiroli S., Meggio F., Guarnieri C., Pinna L. A. Phosphorylation of ornithine decarboxylase in intact erythroleukemia cells. Biochim Biophys Acta. 1990 May 2;1052(2):345–347. doi: 10.1016/0167-4889(90)90232-3. [DOI] [PubMed] [Google Scholar]
- Fonzi W. A., Sypherd P. S. The gene and the primary structure of ornithine decarboxylase from Saccharomyces cerevisiae. J Biol Chem. 1987 Jul 25;262(21):10127–10133. [PubMed] [Google Scholar]
- Ghoda L., Phillips M. A., Bass K. E., Wang C. C., Coffino P. Trypanosome ornithine decarboxylase is stable because it lacks sequences found in the carboxyl terminus of the mouse enzyme which target the latter for intracellular degradation. J Biol Chem. 1990 Jul 15;265(20):11823–11826. [PubMed] [Google Scholar]
- Ghoda L., van Daalen Wetters T., Macrae M., Ascherman D., Coffino P. Prevention of rapid intracellular degradation of ODC by a carboxyl-terminal truncation. Science. 1989 Mar 17;243(4897):1493–1495. doi: 10.1126/science.2928784. [DOI] [PubMed] [Google Scholar]
- Glass J. R., Gerner E. W. Spermidine mediates degradation of ornithine decarboxylase by a non-lysosomal, ubiquitin-independent mechanism. J Cell Physiol. 1987 Jan;130(1):133–141. doi: 10.1002/jcp.1041300119. [DOI] [PubMed] [Google Scholar]
- Glass J. R., MacKrell M., Duffy J. J., Gerner E. W. Ornithine decarboxylase production in vitro by using mouse cDNA. Biochem J. 1987 Jul 1;245(1):127–132. doi: 10.1042/bj2450127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda D. K., Bachmair A., Wünning I., Tobias J. W., Lane W. S., Varshavsky A. Universality and structure of the N-end rule. J Biol Chem. 1989 Oct 5;264(28):16700–16712. [PubMed] [Google Scholar]
- Gupta M., Coffino P. Mouse ornithine decarboxylase. Complete amino acid sequence deduced from cDNA. J Biol Chem. 1985 Mar 10;260(5):2941–2944. [PubMed] [Google Scholar]
- Heby O. Role of polyamines in the control of cell proliferation and differentiation. Differentiation. 1981;19(1):1–20. doi: 10.1111/j.1432-0436.1981.tb01123.x. [DOI] [PubMed] [Google Scholar]
- Hickok N. J., Seppänen P. J., Gunsalus G. L., Jänne O. A. Complete amino acid sequence of human ornithine decarboxylase deduced from complementary DNA. DNA. 1987 Jun;6(3):179–187. doi: 10.1089/dna.1987.6.179. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Ito K., Kashiwagi K., Watanabe S., Kameji T., Hayashi S., Igarashi K. Influence of the 5'-untranslated region of ornithine decarboxylase mRNA and spermidine on ornithine decarboxylase synthesis. J Biol Chem. 1990 Aug 5;265(22):13036–13041. [PubMed] [Google Scholar]
- Jänne J., Pösö H., Raina A. Polyamines in rapid growth and cancer. Biochim Biophys Acta. 1978 Apr 6;473(3-4):241–293. doi: 10.1016/0304-419x(78)90015-x. [DOI] [PubMed] [Google Scholar]
- Kahana C., Nathans D. Nucleotide sequence of murine ornithine decarboxylase mRNA. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1673–1677. doi: 10.1073/pnas.82.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameji T., Pegg A. E. Inhibition of translation of mRNAs for ornithine decarboxylase and S-adenosylmethionine decarboxylase by polyamines. J Biol Chem. 1987 Feb 25;262(6):2427–2430. [PubMed] [Google Scholar]
- Kopitz J., Rist B., Bohley P. Post-translational arginylation of ornithine decarboxylase from rat hepatocytes. Biochem J. 1990 Apr 15;267(2):343–348. doi: 10.1042/bj2670343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984 Sep 25;12(18):7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lougovoi C. P., Kyriakidis D. A. Interconversion of Tetrahymena pyriformis ornithine decarboxylase from inactive to active form by phosphorylation. Biochim Biophys Acta. 1989 Jun 13;996(1-2):70–75. doi: 10.1016/0167-4838(89)90096-4. [DOI] [PubMed] [Google Scholar]
- Macrae M., Coffino P. Complementation of a polyamine-deficient Escherichia coli mutant by expression of mouse ornithine decarboxylase. Mol Cell Biol. 1987 Jan;7(1):564–567. doi: 10.1128/mcb.7.1.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzella J. M., Blackshear P. J. Regulation of rat ornithine decarboxylase mRNA translation by its 5'-untranslated region. J Biol Chem. 1990 Jul 15;265(20):11817–11822. [PubMed] [Google Scholar]
- Meggio F., Flamigni F., Guarnieri C., Pinna L. A. Location of the phosphorylation site for casein kinase-2 within the amino acid sequence of ornithine decarboxylase. Biochim Biophys Acta. 1987 Jun 15;929(1):114–116. doi: 10.1016/0167-4889(87)90246-1. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Hayashi S. Role of antizyme in degradation of ornithine decarboxylase in HTC cells. Biochem J. 1985 Mar 15;226(3):893–896. doi: 10.1042/bj2260893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y., Nishiyama M., Hayashi S. Involvement of antizyme in stabilization of ornithine decarboxylase caused by inhibitors of polyamine synthesis. Eur J Biochem. 1989 Mar 1;180(1):181–184. doi: 10.1111/j.1432-1033.1989.tb14630.x. [DOI] [PubMed] [Google Scholar]
- Pegg A. E., Madhubala R., Kameji T., Bergeron R. J. Control of ornithine decarboxylase activity in alpha-difluoromethylornithine-resistant L1210 cells by polyamines and synthetic analogues. J Biol Chem. 1988 Aug 5;263(22):11008–11014. [PubMed] [Google Scholar]
- Pegg A. E., McCann P. P. Polyamine metabolism and function. Am J Physiol. 1982 Nov;243(5):C212–C221. doi: 10.1152/ajpcell.1982.243.5.C212. [DOI] [PubMed] [Google Scholar]
- Pegg A. E. Polyamine metabolism and its importance in neoplastic growth and a target for chemotherapy. Cancer Res. 1988 Feb 15;48(4):759–774. [PubMed] [Google Scholar]
- Pegg A. E. Recent advances in the biochemistry of polyamines in eukaryotes. Biochem J. 1986 Mar 1;234(2):249–262. doi: 10.1042/bj2340249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R., Jackson R. J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem. 1976 Aug 1;67(1):247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- Phillips M. A., Coffino P., Wang C. C. Cloning and sequencing of the ornithine decarboxylase gene from Trypanosoma brucei. Implications for enzyme turnover and selective difluoromethylornithine inhibition. J Biol Chem. 1987 Jun 25;262(18):8721–8727. [PubMed] [Google Scholar]
- Pilz R. B., Steglich C., Scheffler I. E. Molecular and genetic characterization of an ornithine decarboxylase-deficient Chinese hamster cell line. J Biol Chem. 1990 May 25;265(15):8880–8886. [PubMed] [Google Scholar]
- Rechsteiner M. Regulation of enzyme levels by proteolysis: the role of pest regions. Adv Enzyme Regul. 1988;27:135–151. doi: 10.1016/0065-2571(88)90014-3. [DOI] [PubMed] [Google Scholar]
- Rogers S., Wells R., Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986 Oct 17;234(4774):364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Rosenberg-Hasson Y., Bercovich Z., Ciechanover A., Kahana C. Degradation of ornithine decarboxylase in mammalian cells is ATP dependent but ubiquitin independent. Eur J Biochem. 1989 Nov 6;185(2):469–474. doi: 10.1111/j.1432-1033.1989.tb15138.x. [DOI] [PubMed] [Google Scholar]
- Russell D. H. Ornithine decarboxylase as a biological and pharmacological tool. Pharmacology. 1980;20(3):117–129. doi: 10.1159/000137355. [DOI] [PubMed] [Google Scholar]
- Seely J. E., Pegg A. E. Effect of 1,3-diaminopropane on ornithine decarboxylase enzyme protein in thioacetamide-treated rat liver. Biochem J. 1983 Dec 15;216(3):701–707. doi: 10.1042/bj2160701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seely J. E., Pösö H., Pegg A. E. Purification of ornithine decarboxylase from kidneys of androgen-treated mice. Biochemistry. 1982 Jul 6;21(14):3394–3399. doi: 10.1021/bi00257a023. [DOI] [PubMed] [Google Scholar]
- Stanley B. A., Pegg A. E., Holm I. Site of pyruvate formation and processing of mammalian S-adenosylmethionine decarboxylase proenzyme. J Biol Chem. 1989 Dec 15;264(35):21073–21079. [PubMed] [Google Scholar]
- Tabor C. W., Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- Toneguzzo F., Glynn S., Levi E., Mjolsness S., Hayday A. Use of a chemically modified T7 DNA polymerase for manual and automated sequencing of supercoiled DNA. Biotechniques. 1988 May;6(5):460–469. [PubMed] [Google Scholar]
- Wen L., Huang J. K., Blackshear P. J. Rat ornithine decarboxylase gene. Nucleotide sequence, potential regulatory elements, and comparison to the mouse gene. J Biol Chem. 1989 May 25;264(15):9016–9021. [PubMed] [Google Scholar]