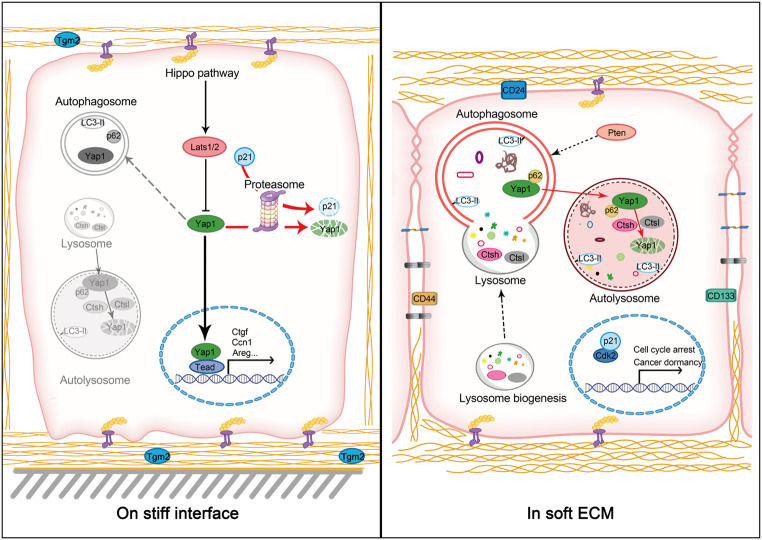
Abstract
The progression and malignancy of many tumors are associated with increased tissue stiffness. Conversely, the oncogenically transformed cells can be confined in soft stroma. Yet, the underlying mechanisms by which soft matrix confines tumorigenesis and metastasis remain elusive. Here, we show that pancreatic cancer cells are suppressed in the soft extracellular matrix, which is associated with YAP1 degradation through autophagic-lysosomal pathway rather than Hippo signal mediated proteasome pathway. In the soft stroma, PTEN is upregulated and activated, which consequently promotes lysosomal biogenesis, leading to the activation of cysteine-cathepsins for YAP1 degradation. In vitro, purified cathepsin L can directly digest YAP1 under acidic conditions. Lysosomal stress, either caused by chloroquine or overexpression of cystatin A/B, results in YAP1 accumulation and malignant transformation. Likewise, liver fibrosis induced stiffness can promote malignant potential in mice. Clinical data show that down-regulation of lysosomal biogenesis is associated with pancreatic fibrosis and stiffness, YAP1 accumulation, and poor prognosis in PDAC patients. Together, our findings suggest that soft stroma triggers lysosomal flux-mediated YAP1 degradation and induces cancer cell dormancy.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-024-05466-y.
Keywords: Soft matrix, Pancreatic ductal adenocarcinoma, Tumor dormancy, Yes-associated protein 1, Autophagy
Introduction
Even tumor cells with multiple oncogenic transformation events can remain dormant for years or decades without metastatic dissemination, indicating a mechanism of niche-dependent tumor suppression [1–3]. Cancer cell dormancy is characterized as cell cycle arrest, cellular size compaction, acquisition of cellular stemness, and chemotherapeutic resistance [4]. Conversely, chronic infection and toxin exposure can drive tissue fibrosis and stiffness which can awaken dormant cancer cells for malignant dissemination. Pancreatic ductal adenocarcinoma (PDAC) often occurs in elderly with a median onset age of 65–70 years old. The age-delayed onset of PDAC is likely associated with the nature of extreme softness of the pancreatic tissue, with an elastic modulus of approximately 1.0-1.5 kPa for the healthy pancreas. On the other hand, the liver is a stiff organ with an elastic modulus of 6 kPa, and liver cancer often occurs at middle age, consistent with cirrhosis and stiffness, which can reach 8–12 kPa [5]. In fact, many risk factors for pancreatic cancer, such as tobacco smoking, alcohol abuse, and chronic pancreatitis, all can cause pancreatic fibrosis and stiffness [6], suggesting that tissue stiffness may awaken cancer dormancy [7]. However, the mechanism for cancer dormancy in soft tissues remains poorly understood. Likewise, it is unknown as to how fibrotic stiffness awakens dormant cells for malignant transformation.
As a cellular degradation and recycling center, the lysosome is recognized as a metabolic signaling hub regulating metabolism, cell growth, and differentiation [8]. The roles of lysosomal dysfunction in tumorigenesis is emerging. One study showed that Drosophila harboring a human H-Ras gene was insufficient to induce tumor growth, but additional disruption of a gene for lysosomal biogenesis, called dor, or feeding the H-Ras transgenic fly with chloroquine to create lysosomal stress resulted in massive tumor growth and metastasis in the fly, suggesting that lysosomal flux may delay the malignant transformation [9]. Likewise, knockout of lysosomal cathepsin L gene (CtsL) exacerbated tumor growth in the epidermis of mice expressing human papillomavirus oncogene K14–HPV16[10]. Moreover, high expression of cystatins, the endogenous inhibitors of lysosomal cathepsins, is associated with a poor cancer prognosis [11, 12]. Lysosome-based degradation relies on autophagic flux. Indeed, impairment of autophagy by genetic disruption can promote tumorigenesis in animal models [13] and knockdown of Atg5 promotes KRas gene mediated PDAC [14]. It is unknown how stromal softness promotes lysosomal biogenesis and autophagic flux to maintain cancer cell dormancy.
YAP, yes-associated protein 1 or YAP1, is a transcription factor which is often amplified in various human cancers for cell proliferation and metastatic dissemination [15]. YAP is critical for progression from pancreatic intraepithelial lesion (PanIN) to the malignant PDAC, either through direct transformation of pancreatic epithelial cells or indirect influence on the pancreatic stellate cells [16]. As a mechano-sensor and transcriptional activator that senses of stromal rigidity, YAP can orchestrate tissue fibrosis and stiffness through various mechanisms [17]. However, it is unknown as to whether and how stromal softness can promote YAP degradation. In this study, we demonstrated the critical roles of stromal softness in maintaining cancer cell dormancy and explored its underlying mechanisms.
Results
Soft matrix maintains pancreatic cancer cell dormancy in association with autophagic flux, lysosomal biogenesis, and YAP1 degradation
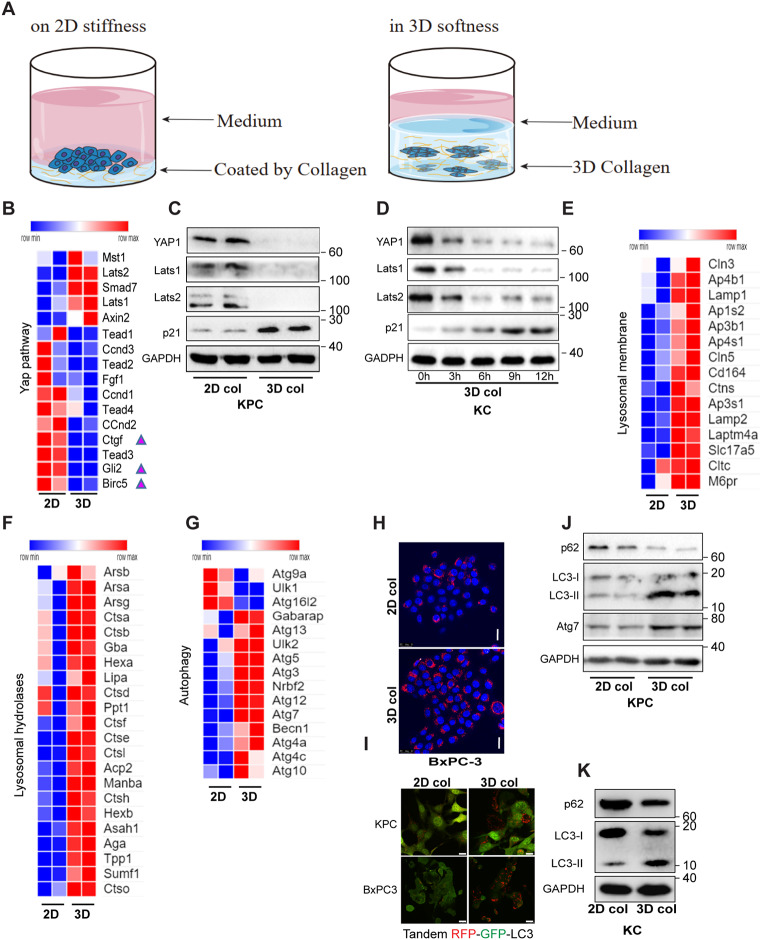
Gain-of-function mutation of KRas and loss-of-function mutation of p53 are critical for the malignancy of PDAC and the characteristics of human PDAC are recapitulated in transgenic KrasG12D/Trp53R172H mice [18]. We reasoned that even such oncogenically transformed cells might remain dormant if a soft niche is imposed, which may explain the delayed onset of PDAC in light of the nature of pancreatic softness. Tumor cellular dormancy is characterized by slow proliferation and G1 cell cycle arrest, reduced cell size, presence of tight junctions, and acquisition of stem cell markers [19, 20]. To simulate the softness of pancreatic stroma, murine KPC (UN-KPC961, Pdx1 driving KRas [12] and Trp53R172H) and Bxpc-3 cells were embedded in 3D collagen lattices (approximately 0.5 kPa) or seeded on collagen-coated 2D surface to mimic the fibrotic stiffness [21] (Fig. 1A). On the 2D stiff interface, the KPC and BxPC-3 cells swiftly went through growth cycle with a larger cell volume. However, in 3D collagen, the growth of cells was substantially slowed down and the cell cycle was arrested at the G0/1 phase, along with smaller cell size (Fig. S1A-F). Similarly, epithelial-to-mesenchymal transition (EMT) associated factors such as β-catenin, Twist1 and Vimentin were down-regulated, whereas E-cadherin was up-regulated in cancer cells embedded in the soft stroma (Fig. S1G-H). We also found that stem cell markers including Sox2, Sox4, Sox9, Notch1 and Jag1, as well as putative pancreatic cancer stem cell markers (CD24, CD44 and CD133) were all up-regulated in KPC cells embedded in soft 3D compared to those cultured on stiff 2D (Fig. S1I).
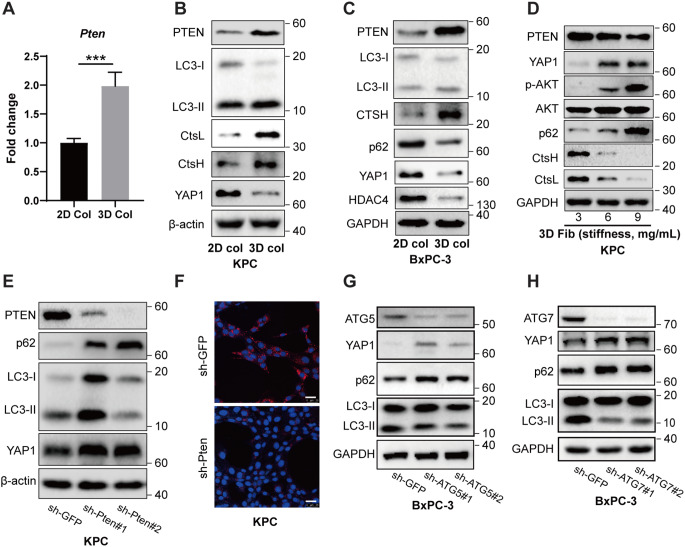
Fig. 1.
Soft matrix confines pancreatic cancer cells in cellular dormancy in association with autophagic flux, lysosomal biogenesis, and YAP1 degradation. (A) Schematic illustrating cell culture on 2D stifness or in 3D softness. (B) RNA-seq analysis of genes involved in YAP pathway in KPC cells cultured on 2D monolayers or embedded in 3D collagen (N = 2). (C) Western blot analysis of KPC cells cultured on 2D monolayers or embedded in 3D collagen. (D) Western blot analysis of KPC cells embedded in 3D collagen at the indicated time points. (E-G) RNA-seq analysis of genes involved in lysosome biogenesis and autophagy pathways in KPC cells cultured on 2D monolayers or embedded in 3D collagen (N = 2). (H) Lysosome biogenesis was visualized through confocal microscopy analysis of Lysotracker/Hoechst staining in BxPC3 cells cultured on 2D monolayers or embedded in 3D collagen. (I) Confocal microscopy analysis of autophagic flux of murine or human pancreatic cancer cells expressing LC3-tandemRFP-GFP in cells cultured on 2D monolayers or embedded in 3D collagen. The experiments were repeated 2–3 times, and typical results were presented. (J-K) Western blot analysis of KPC and KC cells cultured on 2D monolayers or embedded in 3D collagen. Scale bar = 25 μm
Interestingly, the transcriptome analysis revealed that several target genes of the mechano-sensor YAP1, such as Ctgf, Gli2 and Birc5, were markedly down-regulated in 3D soft stroma (Fig. 1B). Moreover, both the total and phosphorylated levels of YAP1 were found to be markedly down-regulated in both KC and KPC cells embedded in soft 3D collagen (Fig. 1C-D, Fig. S1J), whereas the mRNA levels of Yap1 remained unchanged (Fig. S1K), indicating that YAP1 is regulated at the post-transcriptional level by the cells in 3D soft stroma. The Hippo kinase pathway is well known for its negative regulation of YAP1 though phosphorylation and subsequent ubiquitination and proteasome-mediated degradation [22]. Here, the mammalian counterparts of the Hippo kinases (Lats1 and Lats2), and the proteasome substrate p21 were markedly down-regulated in cells cultured in soft substrates, suggesting that a Hippo-independent mechanism is involved in stromal softness-mediated YAP1 degradation (Fig. 1C-D).
There are two main mechanisms of protein degradation in eukaryotic cells, including ubiquitin-proteasome system (UPS) and autophagy-lysosome pathway (ALP) [23]. Transcriptome expression profiling showed that mRNA levels of many lysosomal hydrolases (such as Ctsh, Ctsl, Ctsb, Ctsf, and Lipa), lysosomal membrane proteins (such as Lamp1, Lamp2, and M6pr) and autophagic genes were up-regulated in cells embedded in the soft 3D collagen lattices compared to those in cells cultured on 2D stiffed interface (Fig. 1E-G). Lysosomal biogenesis was further evaluated by Lysotracker-Red staining in human pancreatic cancer cells (BxPC-3), where increased number and size of lysosomal puncta were observed in cells embedded in the soft 3D collagen lattices compared to those in cells cultured on 2D interface (Fig. 1H). To monitor the impact of stromal rigidity on the autophagic flux of cancer cells, we transfected KPC and BxPC-3 cells with the tandem-fluorescence-tagged LC3 (tfLC3) constructs, followed by fluorescence imaging of GFP and RFP. As shown in Fig. 1I, the formation of acidic autophagic-lysosomal puncta network, indicated by red fluorescence and quenching of green fluorescence, was significantly increased in cells embedded in the soft 3D collagen lattices compared to that in cells cultured on 2D interface. Autophagic flux levels in BxPC-3 cells were further quantified by FACS, where a significant higher red/green fluorescence ratio was observed in cells cultured on soft substrates compared to that in cells cultured on stiff substrates (Fig. S1L). We further performed Western blot analysis to examine the autophagic flux in KC and KPC cells embedded in soft 3D type-I collagen lattices or seeded on plastic coated with collagen. In soft 3D matrix, p62/Sqstm1, a cargo protein that mediates autophagic-lysosomal degradation, underwent prompt turnover (Fig. 1J-K). Phospholipid conjugated LC3 (LC3-II) and Atg7, the markers of phagophore formation and autophagic flux, were increased in cells cultured in soft stroma (Fig. 1J-K). These data suggested that lysosomal biogenesis and autophagic flux levels were up-regulated by the cancer cells in reponse to stromal softness. Taken together, these lines of evidence indicate that stromal softness can promote autophagic-lysosomal flux, attenuate the YAP pathway, and confine tumor cell growth.
Stromal softness induces YAP1 degradation in cancer cells in a lysosomal flux-dependent manner
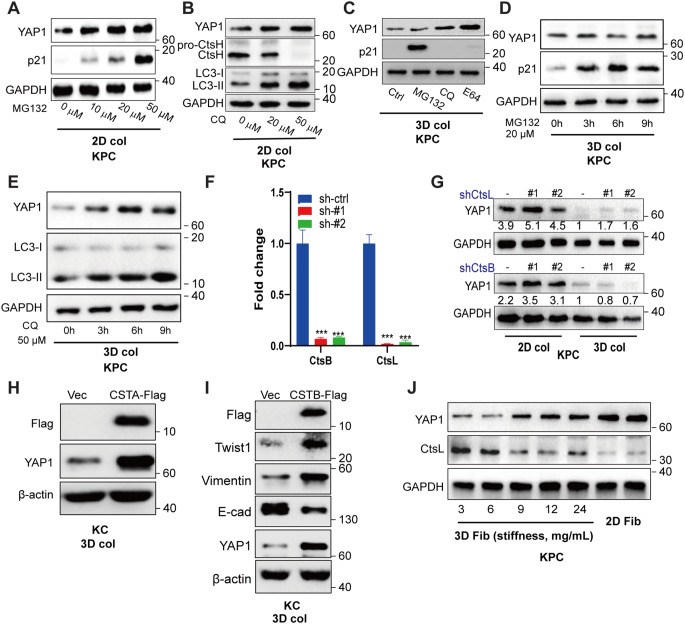
YAP turnover is attributed to Hippo kinase through YAP phosphorylation, followed by ubiquitination and proteasome digestion, while impediment of YAP degradation is related to tumor growth [22]. Here, we confirmed this notion, showing that both YAP1 and p21 proteins were restored by treatment with MG132, a specific proteasome inhibitor, when KPC cells were cultured on stiff interface (Fig. 2A). Chloroquine, a lipophilic amine that impairs lysosomal acidification, also rescued cellular YAP1 in KPC cells grown on stiff interface, which was in line with the absence of activated CtsH and accumulation of its substrate LC3-II (Fig. 2B). However, when the cancer cells were cultured in soft 3D collagen, YAP1 was degraded predominantly through the lysosomal machinery rather than the proteasome, since MG132 failed to rescue YAP1, but could restore p21, showing efficacy of the inhibitor and activation of proteasome pathway by the cells in soft stroma, whereas chloroquine and E64 were capable to rescue YAP1 (Fig. 2C), which was in agreement with the increased expression and activation of lysosomal cathepsins (Fig. 1E). Time course experiments showed that CQ treatment, but not MG132 treatment, was capable to rescue YAP1 in association with autophagy-lysosomal stress and inhibition, as indicated by LC3 accumulation (Fig. 2D-E), which further verified that stromal softness-mediated YAP1 degradation in cancer cells occurred mostly through autophagy-lysosome pathway rather than proteasome-mediated degradation pathway. To define the lysosomal-cysteine proteinases involved in YAP1 degradation, we knocked down the major lysosomal cysteine-type cathepsins, CtsB and CtsL, which were abundantly expressed by the cells in soft stroma. As shown in Fig. 2F and G, knockdown of CtsL resulted in the restoration of YAP1. In contrast, knockdown of CtsB had no effect on YAP1 levels, indicating substrate preference among the lysosomal cathepsins for YAP1 degradation by the cells in soft stroma. Cystatins, the endogenous inhibitors of cysteine cathepsins, are known to be associated with poor prognosis in PDAC patients [24]. Consistently, we found that overexpression of cystatin A (CSTA), known for its inhibition of cathepsin H/L, in KC cells restored YAP1 expression (Fig. 2H), and a similar result was confirmed by overexpression of cystatin B (Fig. 2I). Importantly, the cystatin B promoted lysosomal stress and the expression of EMT markers, as indicated by the increased expression of Twist1 and Vimentin and down-regulation of E-cadherin (Fig. 2I).
Fig. 2.
Stromal softness induces YAP1 degradation in cancer cells through lysosomal flux (A) Western blot analysis of KPC cells, seeded on 2D monolayers and treated with the proteasome inhibitor MG132 at 0, 10, 20 and 50 µM for 6 h. (B) Western blot analysis of KPC cells cultured on 2D monolayers and treated with the lysosome inhibitor chloroquine at 0, 20 and 50 µM for 6 h. (C) Western blot analysis of cells embedded in 3D collagen, treated with DMSO, MG132, E64 and chloroquine for 12 h. (D-E) KPC cells were treated with MG132 (20 µM) or CQ (50 µM) for different periods of time (0, 3, 6–9 h), followed by Western blot analysis. (F) The knockdown efficiency of lenti-shRNA on CtsB and CtsL in KPC cells was determined by RT-qPCR analysis. (G) Western blot analysis of the cells transfected with the indicated shRNA and cultured on 2D monolayers or embedded in 3D collagen for 24 h. (H-I) Western blot analysis of KC cells over-expressing Flag-Cystatin A and Flag-Cystatin B in 3D collagen. (J) Western blot analysis of KPC cells embedded in 3D fibrin gel at 3 (0.6 kPa), 6, 9, 12, and 24 (2.6 kPa) mg/mL or seeded on the fibrin coated plastic for 24 h. The experiments were repeated more than twice, and typical results are presented. The values are presented as mean ± SD; ***p < 0.001 vs. sh-ctrl group; ###p < 0.001 vs. sh-ctrl group
Cancer progression is intimately related to wound healing, by which fibrin clot often occurs as a key component of the tumor micro-environment. Thus, we examined the effects of stromal softness on YAP1 degradation in KPC cells embedded in 3D fibrin lattices, through which the gel stiffness can be adjusted over a wider range. As shown in Fig. 2J, with increasing stiffness from 0.6 kPa (3 mg/ml fibrin) to 2.6 kPa (24 mg/ml), the expression levels of YAP1 were progressively increased to that of the fibrin-coated 2D surface. Strikingly, in response to the increased stiffness of 3D fibrin, expression of matured form of CtsL gradually decreased accompanying with YAP1 accumulation. These results indicated that the protein turnover of YAP1 by the cells in soft stroma is mostly mediated through lysosomal-cathepsins machinery, rather than proteosome machinery.
Lysosome destinating CTSL, rather than nuclear cathepsins, digests YAP1 from the N-terminus
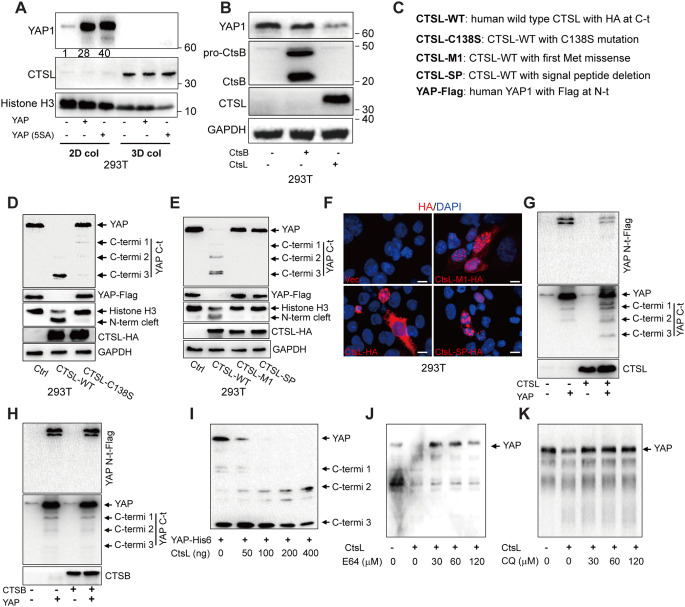
Hippo-kinase-mediated YAP inactivation depends on substrate phosphorylation at multiple serine residues, which collectively serves as signals for the ubiquitination and proteasome-mediated degradation [22]. However, we noticed that a five-serine-to-alanine YAP1 null variant (5SA-YAP: S61A, S109A, S127A, S164A, and S381A) was equally degraded as the wild-type YAP1 in 293T cells embedded in 3D collagen, which again implies a Hippo-independent mechanism for YAP1 degradation by the cells in soft stroma (Fig. 3A). To validate the notion based on loss-of-function analysis as shown in Fig. 2, we also performed gain-of-function analysis. Indeed, forced expression of CTSL but not CTSB, promoted YAP1 degradation in cells (Fig. 3B). Next, 293T cells were co-transfected to express a wild type CTSL or a catalytically null variant (C138S mutant) together with human YAP1 tagged with HA (Human influenza hemagglutinin) at C-terminus (Fig. 3C). As shown, forced expression of the wild-type CTSL, but not its catalytically inactive variant (C138S), led to YAP1 degradation (Fig. 3D). A previous report showed that CTSL was able to cleave the N-terminus of histone H3, which consequently impacted on embryonic stem cell differentiation [25]. Here, using antibodies against the C-terminus of histone H3, we confirmed that CTSL, but not the catalytically null variant, could cleave the N-terminus of histone H3 (Fig. 3D). In addition to lysosome destination, cysteine cathepsins can also be found in the nucleus or extracellular secretion [26]. We then investigated whether the lysosomal-sorted cathepsin L is necessary for the YAP1 degradation. As shown in Fig. 3E, CTSL variants, including M1-CTSL and SP-CTSL, both lacking an N-terminal signal peptide for ER-Golgi-lysosomal sorting, were unable to digest YAP1 and histone H3. This notion was further confirmed by immunofluorescence staining analysis of the transfected cells, revealing that most M1-CTSL and SP-CTSL variants, as well as a portion of wild type CTSL, were found in the nuclear compartment (Fig. 3F). We further captured the intermediate fragments during the course of cathepsin-mediated YAP1 degradation by adding tags at the N- or C-terminus to the substrate YAP1. We noticed that CTSL, but not CTSB, initiated YAP1 degradation starting from its N-terminus (Fig. 3G and H). The notion that the three intermediate fragments of YAP1 being recognized by antibodies against the C-terminus, but not the ones against the N-terminus, indicates that the cathepsin-mediated cleavage of YAP1 begins at the N-terminal region. The specificity of cathepsins for YAP1 degradation according to the gain-of-function analysis agreed well with the results of loss-of-function analysis (Fig. 2G).
Fig. 3.
Lysosome destinating CTSL digests YAP1 from the N-terminus (A) Western blot analysis of 293T cells embedded in 3D collagen or seeded on plastic for 24 h for expression of wild-type human YAP1 or the Hippo phosphorylation null variant (5SA) of YAP1. (B) Western blot analysis of expression of CTSB, CTSL and YAP1 in 293T cells embedded in 3D collagen. (C) Schematic representation of YAP1 and CTSL variant constructs. (D) CTSL-mediated degradation of YAP1 was analyzed by Western blot analysis of 293T cells expressing wild type CTSL or catalytic inactive CTSL (C138S) together with FLAG-YAP1 (tagged at the N-terminus). The C-terminal fragments of YAP1 and N-terminus cleaved histone H3 are indicated by arrows. (E) Western blot analysis of 293T cells expressing wild-type or the signal-peptide-deletion variants of CTSL together with Flag-YAP1. (F) Confocal immunofluorescence microscopy of HA tag in 293T cells expressing HA-CTSL, HA-M1-CTSL and HA-ΔSP-CTSL. Nuclei are marked by DAPI. (G and H) Western blot analysis of 293T cells expressing CTSL or CTSB together with YAP1-FLAG. C-terminal fragments of YAP1 are indicated by arrows. (I) In vitro degradation assay of the CTSL-mediated digestion of YAP1 was carried out by incubating purified His-tagged-YAP1 with increasing doses of purified human CTSL at pH 4.0 and 37 ℃ for 30 min. The levels of C-terminal fragments of YAP1 were determined by Western blot analysis. (J and K) In vitro degradation of YAP1 by CTSL was inhibited by E64 and chloroquine. Scale bar = 10 μm. Representative images of experiments repeated 2–3 times are shown
We then conducted an in vitro degradation assay by incubating of purified His-tagged YAP1 as a substrate with CTSL as a proteinase. At acidic condition of pH4.0 that is closed to lysosomal pH, YAP1 was completely digested by CTSL in a dose-dependent manner (Fig. 3I). Interestingly, three C-terminus fragments of YAP1 appeared as intermediate products, which are closely match to the YAP1 fragments observed in the cells (Fig. 3D, E). We further tested the effect of the lipophilic amine inhibitors, which can cause lysosomal stress, on CTSL-mediated YAP1 degradation in vitro. Indeed, the CTSL-mediated degradation of YAP1 in vitro could be inhibited by E64 or chloroquine (Fig. 3J and K) at the similar concentrations as those observed in cells (Fig. 2). Thus, the results from in vitro degradation and in vivo transfection experiments explicitly demonstrated that YAP1 is a bona fide substrate for the lysosomal cysteine-cathepsins. These results also demonstrate the roles of lysosomal-cathepsin and autophagic flux in regulating YAP1 degradation.
Stromal softness up-regulates PTEN and promotes autophagic flux for YAP degradation
Phosphatase and tensin homologue (PTEN), a phospholipid and phosphor-protein phosphatase, is known for its role in maintaining somatic stem cells and cancer stem cells [27]. Loss of PTEN is often associated with the dissemination of PDAC [28]. PTEN is also known to promote autophagic flux via inhibiting AKT/mTORC pathway [29]. In an animal model, disruption of PTEN accelerates the KRas-induced pancreatic cancer development [30]. Our RNA expression profiling revealed that PTEN was up-regulated in KPC cells cultivated in 3D collagen lattices, which was confirmed by RT-qPCR and Western blot analysis (Fig. 4A-B). In agreement with the elevated expression of PTEN, autophagic-lysosomal flux was consequently accelerated, as indicated by increased formation of LC3-II and expression of CtsH and CtsL, which are further related to YAP1 degradation (Fig. 4B). Likewise, up-regulation of PTEN was also observed in human BxPC-3 cells cultured in 3D collagen, in agreement with the acceleration of autophagic flux, as indicated by increased turnover of p62/Sqstm1 and expression of CTSH for the degradation of YAP1, and HDAC4 (Fig. 4C), which is in line with our previous report showing CTSH mediated degradation of HDAC4 in the context of stromal softness [31]. Along with the increase in stiffness caused by embedding of KPC cells in 3D fibrin lattices (from 3 mg/mL to 9 mg/mL), PTEN was progressively down-regulated, in line with the increased levels of AKT phosphorylation for mTOR activation and attenuation of autophagy, marked by impediment of p62 turnover, ultimately resulting in YAP1 accumulation (Fig. 4D). To determine the causal role of PTEN in autophagy-mediated YAP1 degradation, we knocked down PTEN in KPC cells. Suppression of PTEN impeded autophagic flux, as indicated by the slowed turnover of p62 and LC3, resulting in YAP1 accumulation (Fig. 4E). Moreover, knockdown of PTEN impaired lysosome biogenesis and acidification, as indicated by a decreased number of Lyso-Tracker Red fluorescent puncta (Fig. 4F). Atg5, a critical component for phagophore initiation, along with others such as Atg12, was significantly up-regulated in KPC cells cultured in 3D collagen (Fig. 1F). Consequently, shRNA-mediated knockdown of ATG5 led to the restoration of YAP1, in agreement with the attenuation of autophagic flux, as indicated by accumulation of p62 and suppression of LC3 turnover (Fig. 4G). Likewise, ATG7 acts as an E-1 enzyme for the ubiquitin-like proteins, and its knockdown also impaired the autophagic flux, resulting in YAP1 accumulation (Fig. 4H).
Fig. 4.
Stromal softness up-regulates PTEN to promote autophagic-lysosomal flux for YAP1 degradation (A) RT-qPCR analysis of Pten mRNA levels in KPC cells cultured on 2D monolayers or embedded in 3D collagen. (B-C) Western blot analysis of KPC or BxPC-3 cells cultured on 2D monolayers or embedded in 3D collagen. (D) Western blot analysis of KPC cells embedded in 3D fibrin gel at different stiffnesses (0.6 kPa to 1.3 kPa). (E) Western blot analysis of control (sh-GFP) or PTEN knockdown (PTEN shRNA) KPC cells embedded in 3D collagen. (F) Confocal imaging of Lysotracker/Hoechst staining in control (sh-GFP) or PTEN knockdown (PTEN shRNA) KPC cells embedded in 3D collagen. (G and H) Western blot analysis of control (sh-GFP), ATG5 knockdown (ATG5 shRNA) or ATG7 knockdown (ATG7 shRNA) BxPC-3 cells embedded in 3D collagen. Representative images of experiments repeated 2–3 times are shown. The values are presented as mean ± SD; ***p < 0.001. Scale bar = 25 μm
In addition to the PTEN-mediated suppression of AKT for autophagic activation and increased autophagic flux in KPC cells, other cellular factors known for their ability to promote autophagy formation/flux are also up-regulated in cells cultured in soft 3D ECM. Approximately 24 factors known for autophagy promotion were increasingly expressed in cells with soft stroma. Conversely, 8 factors known for autophagy suppression were down-regulated in cells cultured in 3D gel. Taken together, these results suggest that multiple pathways for autophagy-lysosome biogenesis and flux are mobilized by the cancer cells in sense of stromal softness.
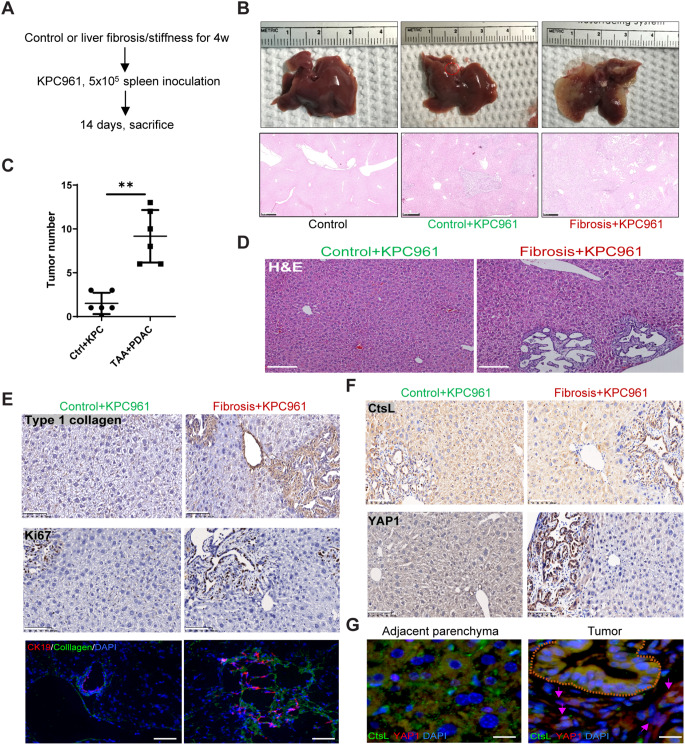
Liver fibrosis and stiffness promote PDAC tumorigenesis and metastasis
Given that the soft niche exerted cellular dormancy through YAP1 degradation, we predicted that tissue fibrotic stiffness may awaken the dormancy for tumorigenesis. To test this prediction, we generated liver fibrosis through repeated peritoneal injections of thioacetamide for 4 consecutive weeks, followed by inoculation with a small number KPC cells (Fig. 5A). Liver fibrosis was confirmed by Masson’s trichrome staining (Fig. S2D). On day 14 after tumor cell inoculation, the liver tissues injected with KPC cells were subjected to histological analysis. In this short period, tumorigenesis was barely detected in the livers of control mice, but strikingly appeared in the fibrotic liver (Fig. 5B-C). Histological examination revealed the formation of ductal tumors at the edge of liver lobules (Fig. 5D), indicating the origin from PDAC cells. Importantly, the tumorigenesis was intimately associated with type-I collagen deposition in the fibrotic liver (Fig. 5E). Immunofluorescence and IHC demonstrated the juxtaposition of tumor cells, marked by CK19 and Ki67, which was intimately associated with the fibrogenesis and stiffness, as indicated by the formation of fibrotic septa (Fig. 5E). CtsL was highly stained as lysosomal puncta in the parenchymal cells, but was reduced in the detached and less differentiated tumor cells (Fig. 5F). Intriguingly, the levels of CtsL in adjacent parenchyma of liver fibrosis was significantly lower than that in the control group (Fig. 5F), indicating the association between fibrosis/stiffness and lysosomal stress. In contrast, YAP1 was highly expressed in the tumor cells but was moderately expressed in the parenchymal cells (Fig. 5F). Again, using double immunofluorescence microscopy, we found that CtsL was mostly presented in the lysosomal puncta of the parenchyma and moderately present in the tumor epithelial cells in PanIN state, but was mostly absent in the less differentiated PDAC-like cells, beneath of the ductal cells, where YAP1 was heavily expressed (Fig. 5G). Thus, the stromal stiffness, generated by tissue fibrosis, can promote tumorigenesis by awakening cancer dormancy.
Fig. 5.
PDAC tumorigenesis and metastasis are promoted in the fibrotic liver (A) Schematic representation of the experimental design used to determine the impact of liver fibrosis/stiffness on tumorigenicity. The mice were treated with or without TAA for 4 weeks, followed by inoculation of UN-KPC961 cells for 2 weeks (n = 6). (B) Gross examination (upper row) and H&E staining (lower row) of control and tumor-bearing livers. Scale bar = 500 μm. (C) Quantitation of tumor numbers in normal or fibrotic liver sections. (D) H&E staining of normal or fibrotic liver sections, scale bar = 200 μm. (E) Immunohistochemical staining of type-I collagen, Ki67 and CK19 in normal or fibrotic liver sections, scale bar = 100 μm. (F) Immunohistochemical staining of YAP1 and CtsL in normal or fibrotic liver sections, scale bar = 100 μm. (G) Immunofluorescence staining of CtsL and YAP1 in PanIN and adjacent hepatic parenchyma. The PanIN ducts are indicated by brown dashed lines, and malignant cells are indicated by pink arrows, scale bar = 25 μm. The values are presented as mean ± SD; **p < 0.01
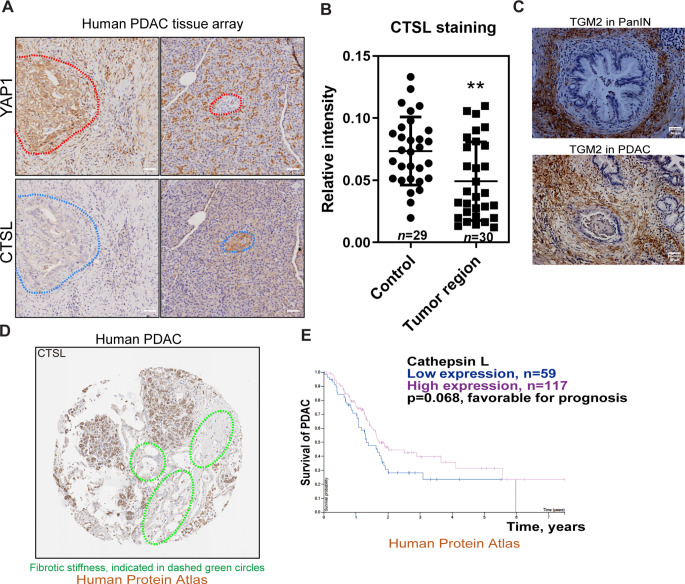
Lysosomal cathepsins are down-regulated in the human PDAC tissues, in association with fibrotic stiffness, YAP1 accumulation and poor prognosis in PDAC patients
Finally, we examined the potential inverse association between the cysteine cathepsins and YAP1 in the context of fibrotic stiffness in human PDAC tissues. We determined the protein expression levels of YAP1 and CTSL in human PDAC through tissue microarray analysis (n = 30 for PDAC, n = 29 as control). Immunohistochemical analysis showed that YAP1 was overwhelmingly stained in the cells of pancreatic ducts, and cancerous stromal cells as well (Fig. 6A-B). Conversely, CTSL was mostly absent in the corresponding cancer cells. Fibrotic septa and tensile stiffness in the PDAC tissues were indicated by the expression of tissue transglutaminase (TGM2) (Fig. 6C), which can crosslink ECM proteins to generate tensile force and stromal stiffness. We also retrieved the data from the Human Protein Atlas and examined the expression of CTSL and survival time of PDAC patients. We found that malignancy was negatively related to CTSL expression in adenocarcinomas. A typical CTSL-stained image of human PDAC tissues derived from the Human Protein Atlas was presented in Fig. 6D, which shows negligible staining of cathepsin L in the fibrotic ductal zooms. Cox proportional hazards regression was used to assess the relationship between CTSL expression levels and survival outcomes of PDAC patients. The hazard ratio (HR) was calculated to be 1.47, indicating that high expression levels of CTSL are associated with the favorable prognosis of PDAC (Fig. 6E). In contrast, high expression levels of cystatin A and cystatin B, the endogenous CTS inhibitors as an indication of lysosomal stress, are related to the unfavorable prognosis in PDAC patients (Fig. S2A, B). Taken together, these lines of clinical data, in agreement with our in vitro and in vivo results, support the hypothesis that stromal softness and its exerted autophagic-lysosomal flux can confine the oncogeneically transformed cancer cells to cellular dormancy.
Fig. 6.
High expression of lysosomal-cathepsins correlates with YAP1 down-regulation, less fibrosis and survival of PDAC patients (A) Representative immunohistochemistry images of YAP1 and CTSL staining in pancreatic tissues from PDAC patients (n = 30 for PDAC, n = 29 for control). (B) Semi quantitation of CTSL staining in PDAC cells vs. the control cells was determined by densitometry analysis. (C) Fibrotic stiffness in human PDAC tissues was determined as expression of tissue transglutaminase 2 (TGM2) in adjacent to the tumor ducts by immunohistochemistry. (D) A typical image of human pancreatic cancer slide stained for CTSL, from the Human Protein Atlas database. Fibrotic stiffness is indicated by dashed green circles. (E) Decreased expression of CTSL is associated with poor prognosis in PDAC patients (from the Human Protein Atlas database). The values are presented as mean ± SD; **p < 0.01. Scale bar = 50 μm
Discussion
The pancreas is a very soft organ with an elastic modulus of 1.2 kPa at 40 Hz, whereas the stiffness of late-stage pancreatic cancer can be tripled to 3.8 kPa [32]. In contrast, the liver is a relatively hard organ with an elastic modulus of 2–6 kPa, and the stiffness of the cirrhotic liver can reach to 8–12 kPa [5]. Such extreme softness of pancreas may partially explain the very late onset of PDAC that occurs at a median age of 71 years old, indicating a potential mechanism to confine the transformed cells to cellular dormancy. In contrast, the liver cancer has a relative early onset, which commonly occurs in middle-aged individuals. In this study, we found that soft ECM, including type-I collagen and fibrin lattices, restrains cancer cell dormancy. Importantly, we linked such cancer cell dormancy to the degradation of YAP1. We further demonstrated that the accelerated YAP1 degradation in cells cultured on soft matrix is independent of Hippo/Lats and the consequent proteosome pathway. Compelling evidence provided in this study demonstrated that cysteine cathepsins, the lysosomal acidic proteinases, mediate YAP1 degradation in cells in response to stromal softness. Lysosomal stress induced by pharmacologic inhibitors, hydrophobic amines such as E64 and Chloroquine, can rescue YAP1 in the cells embedded in soft 3D ECM. Cystatin A, an inhibitor of CtsB/H/L, can also restore YAP1. The lysosomal-localized CTSL is critical for YAP1 degradation, while the nucleus-located CTSL variants are incapable. Co-transfection experiments showed that initial cleavage occurs at the N-terminus of YAP1. In vitro, purified CTSL can directly digest YAP1 under acidic conditions, and such hydrolysis can be inhibited by the same hydrophobic amines. Importantly, the C-terminal fragments generated from in vitro degradation were closely matched with those found in the co-transfection experiments, demonstrating YAP1 as an authentic substrate for CTSL in lysosomal degradation.
Given the liver is one of the most common sites of pancreatic cancer metastasis [33], we evaluated the impacts of hepatic fibrosis and stiffness on PDAC tumorigenesis and metastasis. We showed that preexisting liver fibrosis could exacerbate cancer growth in the liver. The liver is a relatively hard organ, and inoculated pancreatic cancer cells can quickly colonize for growth. On the other hand, PDAC-derived exosomes can promote hepatic fibrosis and amply PDAC growth and metastasis [34]. Our clinical data and those from the Human Protein Atlas showed that CTSL expression is negatively associated with YAP1 expression in many types of carcinomas, while CTSH and CTSL are negatively or weakly expressed in PDAC cells. In contrast, high expression levels of cystatin A and cystatin B, the endogenous CTS inhibitors, are related to the poor prognosis in PDAC patients. It has been suggested that overexpression of certain cathepsins, including the cysteine-type cathepsins, is related to tumor progression and promotion [35]. These tumor-related cathepsins are often secreted from cancer cells, and the extracellular cathepsins may trigger inflammation and damage surrounding tissues.
Here, we propose a working model showing how soft stroma can maintain the oncogene-transformed cancer cells in cellular dormancy. In sensing stromal softness, the autophagic-lysosomal-cathepsin axis is activated to promote YAP1 degradation and tumor suppression. Lysosomal biogenesis is accelerated in cells in response to stromal softness, as indicated by up-regulation of a series of lysosomal hydrolases together with the increased expression of autophagy-related proteins such as Atg5 and Atg7 (graphical summary). Specifically, we found that in sensing of stromal softness, PTEN is accumulated and activated, leading to inactivation of Akt/mTORC and promotion of auto-lysosomal degradation of YAP1 involving p62/Sqstm1 and lysosomal cathepsins (CtsH and CtsL). While autophagy-lysosome system is mobilized and lysosomal flux is enhanced in cells with soft stroma, UPS is seemingly impeded, leading to the accumulation of p21 and PTEN for tumor suppression. Indeed, Skp2, a key protein kinase for p21 phosphorylation and UPS-based degradation [36], was down-regulated in cells with soft ECM. Thus, in the soft niche and in sensing of softness, the impediment of UPS for p21/PTEN accumulation thus can mobilize autophagic-lysosomal flux for YAP1 degradation, which ultimately may confine the dormancy of transformed cancer cells. Moreover, the softness-imposed cancer cell dormancy is niche dependent. The dormant cancer cells can be quickly awakened in sensing of stromal stiffness. Such notion is supported by our animal work, showing that preexisting tissue fibrosis can exacerbate tumor cell colonization and growth.
Most risk factors for cancer malignancy, including tobacco smoking, alcohol abuse, and chronic viral infection, are capable to activate myofibroblasts, leading to fibrotic stiffness, which can thus awaken the dormant cancer cells through integrin/FAK activation and its downstream pathways for YAP activation [37]. On the other hand, therapeutic strategies for alleviating tissue fibrosis may serve as effective ways to normalize the tumor microenvironment and prevent tumor progression [38, 39]. Given the critical roles of YAP in sensing extracellular matrix stiffness, YAP may serve as the promising therapeutic target for preventing the recurrence and metastasis of dormant cancer cells. Furthermore, suppressed expression of YAP1 and the downstream target gene of Ctgf in the cancer cells may likely impede myofibroblast activation and consequent fibrogenesis, maintaining tissue softness. In summary, the physiological functions of autophagic-lysosomal biogenesis and flux by the epithelial cells in soft 3D stroma may be essential for the cellular homeostasis and epithelial tight junctions for cellular quiescence and epithelial phenotypes. Such suppression mechanism may also occur in the oncogenetically transformed cells as long as the softness exists in adjacent niches.
In this work, we propose the hypothesis and potential mechanism by which oncogenically transformed cancer cells can be confined to a dormant state, which may partially explain the delayed onset of PDAC in association with the softness of pancreatic tissue. In particular, we noticed that autophagic biogenesis and flux are up-regulated for YAP1 degradation when the cancer cells are cultured under soft conditions. Such notion was initially reported in our previous works, showing that soft cultivation can up-regulate lysosomal cathepsin L for HDAC4 degradation, which confers the MMP loci into an epigenetic opening state for injury responsiveness [31, 40]. Limitations of this study appear in several ways. First, we performed 2D cell culture on collagen coated surface to mimic fibrotic stiffness, while embedded cells in 3D collagen lattices to simulate softness of pancreatic stroma. Although proper controls were used, 2D vs. 3D comparison is not perfect for the evaluation of matrix stiffness, since other factors, such as cell shape and cell–substrate interaction, may be affected [41]. Second, due to the limitation of collagen preparation, it’s unable to cast 3D collagen lattices with different crosslinking conditions and stiffness. Instead, fibrin 3D gel that can be casted in a wide range of crosslinking conditions and stiffness was chosen for our studies. Indeed, we have tested a line of polymeric ECM, including collagen gel, Matrigel, fibrin gel, and hydrogel, to evaluate tissue softness. Each of them has coherent limitations and failure to reflect the in vivo changes in tissue rigidity during pathogenesis. On the other hand, similar results were always obtained, showing the quiescent phenotype of cells in soft gel regardless of the chemical nature. Third, we still don’t know how autophagic biogenesis is promoted in the cells under soft conditions. Although we found the up-regulation of several key lysosomal cathepsins and accelerated autophagic flux in cells under soft conditions, we didn’t observe the up-regulation of transcription factor EB (TFEB), a master regulator of lysosomal biogenesis. Thus, a TFEB-independent pathway may be involved in lysosomal biogenesis. Indeed, we detected the activation of PTEN in cells cultured in soft gel, but how the key phosphatase is activated was unclear. Fourth and foremost, it is challenging for us to induce pancreatic softness in Kras/p53 mice, since oncogenes can quickly induce fibrosis and presumable stiffness, so that it is hard to observe the dormancy in vivo. Overall, this work may shed light on the mechanism underlying the late onset of pancreatic cancer, where the soft nature of the pancreas can confine the dormancy of the genetically transformed cells.
Methods
Key reagents
Type I collagen (#5005) was purchased from Advanced Matrix. Collagenase (#195109) was purchased from MP Biomedicals. Fibrinogen (F8630), MG132 (M7449), chloroquine (#1118000), E64 (E3132), DIOC6 (#318426) and purified human cathepsin L (C6854) were obtained from Sigma-Aldrich, and cathepsin inhibitor 1 (S2847) was obtained from Selleck. Dispase II (#1105759) was purchased from Gibco. Fetal bovine serum (#1822477) was obtained from Biological Industries, and Dulbecco’s Modified Eagle’s Medium (AD24926277) was obtained from Hyclone. Antibiotic cocktail was purchased from Solarbio. Antibodies against YAP1 (#1407S), HA (#3724S), HDAC4 (#2072), Histone H3 (4499 S), and LC3 (#306019) were purchased from Cell Signaling Technology. Antibodies against Cathepsin H (#6496), Cathepsin B (#13985) were purchased from Santa Cruz Biotechnology. Another antibody against Cathepsin H (70R-16662) was purchased from R&D. Antibodies against Cathepsin L (#58991), p21 (#109199) and CK19 (#52625) were purchased from Abcam. Antibodies against Lats1 (SAB2105023), and FLAG (F1804) were purchased from Sigma Aldrich. The antibody against tGM2 (#3222) was purchased from Epitomics. The antibody against Coll-I (#1310) was purchased from Southern Biotech. The antibodies against Lats2 (#614271), GAPDH (200306-7E4), and CDK4 (#31036) were purchased from Zen BioScience.
Embedding cells in 3D ECM and seeding on 2D monolayer culture
The 3D ECM culture model was constructed as previously described [31]. Generally, KPC (UN-KPC961, KrasG12D/Trp53R172H) or KC cells (KrasG12D) cells (2.0 × 105) were embedded in 0.5-cm3 of type-I collagen or fibrin lattices, and loaded into the 24-well plates. Briefly, type-I collagen cocktail was prepared in cold by mixing 0.3 mL of 10xMEM, 0.7 mL of DMEM, 2.4 mL of type I collagen (3.2 mg/mL), and 0.1 M NaOH was used to adjust the pH to 7.6. Cells grown in 10-cm dishes were harvested by trypsin treatment and FBS neutralization. The cells were suspended in DMEM at 1.6 × 106/ml. Finally, 3.0 ml type-I collagen cocktail was mixed with 1.0 ml cell suspension, and 0.5 ml gel (2 × 105 cells) was loaded into the wells of the 24-well plate. The plates were incubated at 37oC for 30–60 min. After gel polymerization, 0.5 ml of DMEM supplemented with 10% FBS was loaded on the top of gel lattices. For stiff 2D culture, cold collagen or fibrin cocktail was loaded into the wells and kept at 4oC without polymerization for 4 h. The gel liquid was removed and the wells were subsequently washed with DMEM. The cell suspension in 1.0 ml of DMEM supplemented with 10% FBS was loaded into the wells. The fibrin cocktail was prepared by dissolving fibrinogen powder in DMEM at concentrations ranging from 3.0 mg/mL to 24.0 mg/mL. To cast the fibrin lattices, 0.5 mL of KPC cell (2.0 × 105) suspension and 0.5 mL of fibrinogen solution were seeded into a 24 well plate, which contained 10 µL of thrombin (0.1 U/µL). The plates were incubated at 37oC for 10 min for fibrin polymerization. For the mitochondria membrane potential staining, the live cells were treated with 50 nM DIOC6 for 10 min, followed by imaging with a Nikon microscope.
Plasmid construction
Through PCR, the cDNAs of CTSL (#11249), CTSB (#11250), YAP1 (#66853) and YAP5SA (#33096) from Addgene, were amplified, and confirmed by sequencing analysis. The genes of interest were inserted into pLVX-Puro to establish stable cell lines or pcDNA3.1 for transient transfection. The shRNAs were subsequently cloned and inserted into pLKO.1. Epitope tags, including FLAG and HA (human influenza hemagglutinin), were added to the N-terminus of the cDNA of YAP1 or CTSL variants by PCR amplification. The plasmids of pcDNA3.1-CtsH and pcDNA3.1-CtsHC139S were described previously [42]. The variants of CTSL, including M1-CTSL (the first Met was mutated to Ala to skip the signal peptide) and ΔSP-CTSL (the signal peptide was deleted), were generated by PCR and the mutation of CTSLC138S was generated by QuikChange II (#200521), from Agilent. Then, an HA tag was added at the C-terminus through PCR amplification and the resulting sequence was insertd into pcDNA3.1. Tandem monomeric RFP-GFP-tagged LC3 was a kind gift from Dr. Ying Tong [43]. The plasmid FLAG-CSTA-PMV was generated by Wuxi Qinglan Biotech Co. Ltd. The sequence information of the primers used for sub-cloning is described in Table 1.
Table 1.
The primers used for PCR amplification and subcloning
| Plasmid name | Sequence of forward strand (5’-3’) | Sequence of reverse strand (5’-3’) |
|---|---|---|
| pcDNA3.1-hCTSL | CGCGGATCCGCCACCATGAATCCTACACTCAT | CCGCTCGAGTCACACAGTGGGGTAGC |
| pcDNA3.1-hCTSB | CGCGGATCCGCCACCATGTGGCAGCTCTGGGC | CCGCTCGAGTTAGATCTTTTCCCAGTAC |
| pcDNA3.1-hCTSL-C138S | GGGTCAGTGTGGTTCTAGTTGGGCTTTTAG | GTAGCACTAAAAGCCCAACTAGAACCACAC |
| pcDNA3.1-hCTSL-HA | CGCGGATCCGCCACCATGAATCCTACACTCATCCTTGCTG | CCGCTCGAGTTAAGCGTAATCTGGAACATCGTATGGGTAAACAGTGGGGTAGCTGGCTG |
| pcDNA3.1-hCTSL-M1-HA | CGCGGATCCGCCACCTTCAATCCTACACTCATCCTTGCTG | CCGCTCGAGTTAAGCGTAATCTGGAACATCGTATGGGTAAACAGTGGGGTAGCTGGCTG |
| pcDNA3.1-hCtsH-HA | CCCAAGCTTGCCACCATGTGGACTGCGCTGCCCCTGC | CCGGAATTCTTAAGCGTAATCTGGAACATCGTATGGGTATACCTGAGGGATGGGGTAGG |
| pcDNA3.1-YAP1-FLAG-HIS | CCCAAGCTTGCCACCATGGACTACAAGGACGACGATGACAAGGATCCCGGGCAGCAGC | TTTTCCTTTTGCGGCCGCCTAATGGTGATGGTGATGATGTAACCATGTAAGAAAGC |
| pLVX-YAP1-FLAG-HIS | CCGGAATTCGCCACCATGGACTACAAG | CTAGTCTAGACTAATGGTGATGGTGATGATG |
| pLVX-hCSTA-FLAG | CCGGAATTCGCCACCATGATACCTGGAGGCTTATCTG | CCGGGATCCCTACTTATCGTCGTCATCCTTGTAATCAAAGCCCG |
| pcDNA3.1-hCTSL-ΔSP-HA | CGCGGATCCGCCACCATGCACAACAGATTATACGGCATG | CCGCTCGAGTTAAGCGTAATCTGGAACATCGTATGGGTAAACAGTGGGGTAGCTGGCTG |
Cell culture and transfection
KPC (UN-KPC961, KrasG12D/Trp53R172H) and KC (KrasG12D) cells were described previously (42). BxPC3 (P53Y220C, Ras WT) and HEK293T cell lines were cultured under an atmosphere of 5% CO2 at 37℃. KPC, MIAPaCa-2 and HEK293T cells were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum, with penicillin (100 U/ml), and streptomycin (100 mg/ml). BxPC-3 cells were cultured in RPMI-1640 Medium supplemented with 10% fetal bovine serum. DNA transfections were carried out with Lipofectamine 3000 (Life technologies) according to the manufacturer’s instructions.
Lentivirus-mediated shRNA knockdown
Lentiviral particles were prepared by transiently transfecting HEK293T cells with lentiviral vectors together with packaging vectors (pMD2.G and psPAX2). After 48 h, the lentiviral particles were harvested from the supernatant, and the virus in the supernatant was concentrated by filtration. Fluorescence titering assay was used to determine the titer of lentiviral particles. In brief, 75,000 of target cells were seeded into each well of a 6-well dish, followed by incubation with 1.5 mL of complete cell culture medium containing a series dilution of the lentiviral preparation for 48 h. Then, the percentage of GFP-positive target cells was determined using fluorescence microscope. The titer of lentiviral particles was calculated as follows: Titer (TU/mL) = number of cells transduced *fraction of GFP-positive cells* dilution factor/transduction volume (mL). Only the target cells with less than 40% of GFP-positive cells were considered for titer calculation, and the average of the titers obtained from different dilutions of lentiviral preparation was calculated. The targeting shRNAs were designed by Life Technologies, and synthetic shRNA segments were cloned and inserted into pLKO.1. The sequence information of the inserts for shRNA is provided in Table 2.
Table 2.
Sequences of primers used for shRNA
| shRNA name | Sequence of forward strand (5’-3’) | Sequence of reverse strand (5’-3’) |
|---|---|---|
| CtsB-1# | CCTTTGATGCACGGGAACAATCTCGAGATTGTTCCCGTGCATCAAAGGTTTTTG | AATTCAAAAACCTTTGATGCACGGGAACAATCTCGAGATTGTTCCCGTGCATCAAAGG |
| CtsB-2# | CCGGCCTGCTTACTTGCTGTGGTATCTCGAGATACCACAGCAAGTAAGCAGGTTTTTG | AATTCAAAAACCTGCTTACTTGCTGTGGTATCTCGAGATACCACAGCAAGTAAGCAGG |
| CtsL-1# | CCGGCCAGCTATCCTGTCGTGAATTCTCGAGAATTCACGACAGGATAGCTGGTTTTTG | AATTCAAAAACCAGCTATCCTGTCGTGAATTCTCGAGAATTCACGACAGGATAGCTGG |
| CtsL-2# | CCGGCAGAAGACTGTATGGCACGAACTCGAGTTCGTGCCATACAGTCTTCTGTTTTTG | AATTCAAAAACAGAAGACTGTATGGCACGAACTCGAGTTCGTGCCATACAGTCTTCTG |
| Atg5-1# | CCGGGCAGAACCATACTATTTGCTTCTCGAGAAGCAAATAGTATGGTTCTGCTTTTTG | AATTCAAAAAGCAGAACCATACTATTTGCTTCTCGAGAAGCAAATAGTATGGTTCTGC |
| Atg5-2# | CCGGCCTTGGAACATCACAGTACATCTCGAGATGTACTGTGATGTTCCAAGGTTTTTG | AATTCAAAAACCTTGGAACATCACAGTACATCTCGAGATGTACTGTGATGTTCCAAGG |
| Atg7-1# | CCGGGCCAACATCCCTGGATACAAGCTCGAGCTTGTATCCAGGGATGTTGGCTTTTTG | AATTCAAAAAGCCAACATCCCTGGATACAAGCTCGAGCTTGTATCCAGGGATGTTGGC |
| Atg7-2# | CCGGTTCTGTCACGGTTCGATAATGCTCGAGCATTATCGAACCGTGACAGAATTTTTG | AATTCAAAAATTCTGTCACGGTTCGATAATGCTCGAGCATTATCGAACCGTGACAGAA |
Animal experiments
The animals were housed at 25℃ by an IVC machine (SPF level). BALB/c nude mice at 4–5 week were purchased from GemPharmatech (Beijing, China). Liver fibrosis was induced by repeated intraperitoneal injections of thioacetamide at 200 mg/kg body weight twice a week for 4 weeks. Then, the KPC cell suspension (0.5 million/100 µL) prepared from monolayers was inoculated through splenic injection. The mice were sacrificed after 14 days for analysis, where the liver tissues injected with KPC cells were collected and subjected to H&E staining to determine the number of tumors. All animal experimental protocols were reviewed and approved by the Animal Ethics Committee of West China Hospital and College of Life Sciences, Sichuan University.
Immunohistochemistry and immunofluorescence staining
The liver tissue was fixed in 4% paraformaldehyde for 2–3 days at 4oC. After the standardized process the tissues were embedded in paraffin. The tissue slides were subjected to H&E staining (Beyotime, #C0105). The slides were subjected to antigen retrieval using sodium citrate-hydrochloric acid buffer solution or Tris EDTA buffer at pH 8.8. Endogenous peroxidases were quenched with 3% H2O2 for 10 min. The slides were treated with 0.3% Triton X-100 for 10 min and blocked with 5% BSA for 1 h at room temperature. Primary antibodies (anti-YAP1 at 1:200 from CST #1407S; anti-CtsL at 1:200 from R&D, #70R-16662) were diluted with 5% BSA and incubated with the slides at 4℃ overnight. HRP-conjugated secondary antibodies (ZSGB-Bio, Beijing, China, #PV9003) were applied, followed by DAB color development. Alternatively, rhodamine- or FITC-conjugated secondary antibodies (Sigma Aldrich, SAB3700860, SAB3700329) were applied for fluorescence imaging microscopy analysis, while nuclei were stained with DAPI (ZSGB-BIO, China, #ZLI-9557). The optical density of CtsH in the microscopic images was calculated with Image Proplus, and the CtsH-positive region of CtsH was semi-quantified.
Western blotting analysis
We applied two protocols for collecting the cellular proteins from 3D ECM culture for Western blot analysis. First, the gel lattices in the wells of the 24-well plates were transferred to 1.5-ml microtubes, and the collagen lattices were collapsed into small pellets by centrifugation at 5000 × g for 5 min at 4oC. The supernatant was aspirated from the pellets. Then, 0.1 mL of SDS-PAGE sample buffer with reducing agent (62.5 mmol/L pH 6.8 Tris-HCl, 2% SDS, 10% glycerol, 0.0025% bromophenol blue, and 0.1 mol/L dithiothreitol or 5% β-mercaptoethanol) was added to the emptied wells to collect the residual cellular proteins after removing the gel lattices. The lysates in the wells were transferred to the microtubes to combine with the gel pellets. The gel was completely melted by heating at 98℃ for 5 min. Second, 0.5 ml of 0.5 mg/mL collagenase in DMEM was added to the collagen lattices in 24-well plates, followed by incubation at 37oC for 20 min to release the cells from collagen. The released cells were collected by centrifugation at 5,000 g for 5 min. Fibrin gel lattices were digested with 500 µL of 0.5 U/mL Dispase II in DMEM at 37℃ for 10 min to release the cells from the fibrin clots. Collected cells were dissolved in RIPA buffer supplemented with protease inhibitor. Proteins in the lysates were resolved by 12% SDS-PAGE, followed by transferring to PVDF membranes (Bio-Rad, #1620177). After blocking with 5% nonfat milk, the membranes were hybridized to appropriate primary antibodies and horseradish peroxidase-conjugated secondary antibodies for subsequent detection with Clarity TM Western ECL Substrate (Bio-Rad, #1705060).
In Vitro YAP1 degradation assay
Recombinant human YAP1 expressing a His tag at the C-terminus was generated using lentiviral-transfected 293T cells. The recombinant protein was purified by cobalt-based IMAC resin (GE Healthcare, #10258736). Cell lysis in RIPA buffer was loaded on the cobalt-resins, followed by extensive washing with 20 mM imidazole, 150 mM NaCl and 20 mM Tris-HCl at pH 7.5. The bound protein was eluted with 300 mM imidazole, 150 mM NaCl, and 20 mM Tris-HCl at pH 7.5. Then through gel filtration the buffer system of the elute was exchanged to 150 mM NaCl, 20 mM Tris-HCl at pH 7.5. Digestion assay was set up in a 50 µl system containing purified YAP-6His, purified CtsL (50–400 ng), bovine serum albumin (20 µg), 50 mM dithiothreitol, and 33 µL of 150 mmol/L NaCl and 50 mmol/L HAc at pH 4.0. After incubation at 37oC for 30 min, the reactions were resolved by Western blotting for YAP1.
RNA isolation and RT-qPCR analysis
UN-KPC961 cells were embedded in 3D collagen or seeded on monolayers for 24 h. The cells in 3D collagen were collected by centrifugation. Total RNA was extracted using Trizol (Transgen, #ER501), and the integrity of the total RNA was assessed by agarose gel electrophoresis. Reverse transcription was carried out with a PrimeScriptTM RT reagent Kit (TaKaRa, #RR047A). Quantitative real time qPCR was used for quantifying mRNA concentrations using the SYBR® Premix Ex Taq™ (TaKaRa, #RR4401A) and Bio-Rad cfx96 platform according to manufacturer’s protocol. Gene expression levels were normalized to that of GAPDH. The sequences of primers used for qPCR analysis are listed in Table 3.
Table 3.
Sequences of primers used for RT-qPCR analysis
| Primer Name | Sequence of forward strand (5’-3’) | Sequence of reverse strand (5’-3’) |
|---|---|---|
| mYap1 | AGGAGAGACTGCGGTTGAAA | CCCAGGAGAAGACACTGCAT |
| mCtgf | CAAGGACCGCACAGCAGTT | AGAACAGGCGCTCCACTCTG |
| mCtsB | GAAGAAGCTGTGTGGCACTG | GTTCGGTCAGAAATGGCTTC |
| mCtsL | TCTCACGCTCAAGGCAATCA | AAGCAAAATCCATCAGGCCTC |
| mCtsH | TCATGGCTGCAAAGGAGGT | TGTCTTCTTCCATGATGCCC |
| mPten | GGCTTCCGTCTGGAGGATTAT | AACCTGCCGAGATATTCCACA |
| mGapdh | GCACAGTCAAGGCCGAGAAT | GCCTTCTCCATGGTGGTGAA |
RNA-Seq and heatmap analysis
RNA-Seq was conducted at Novogene (Beijing, China). UN-KPC961 cells were cultured on 2D monolayers and in 3D Type I collagen for 24 h. Total RNA was extracted with Trizol. Gene expression levels were used as readouts to generate heatmaps using Cluster 3.0 and TreeView software.
Tissue microarray and the human protein atlas data retrieval
Pancreatic cancer tissue microarray was purchased from Shanghai Outdo Biotech (China). Images of CTSL-stained PDAC tissue and survival analysis of PDAC patients were obtained from the Human Protein Atlas.
Statistical analysis
Data recording, processing and calculating were processed with Microsoft Excel 2016 (Microsoft Corp.) and GraphPad Prism 6.0 (GraphPad Software Inc.). Data are presented as means ± SD. Differences in means were assessed using a t-test or one-way ANOVA, followed by Tukey’s multiple comparison test. Survival analysis was conducted using Cox proportional hazards regression, implemented in R software (version 4.4.1) with the ‘survival’ package. Statistical significance is indicated as *: p < 0.05, **: p < 0.01, ***: p < 0.001.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors gratefully acknowledge the Human Protein Atlas for sharing the data of PDAC patients.
Author contributions
T.Z., J.C., Y-P.H. and X.Z. conceived the project, designed the experiments and wrote the manuscript. T.Z., J.C., H.Y., X.S. Y.O., Q.W., M.E., S.Z., F.R., Y.T., C.H. performed the experiments and analyzed the data. R.H., J.L., Y.G. and S.J.P. critically revised the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (31571165, 82070846, 31771288), Sichuan Science and Technology Program (2021ZYCD016, 2022YFS0308), and Chengdu Science and Technology Program (2023-GH02-00083-HZ).
Data availability
Data will be made available on reasonable request.
Declarations
Consent for publication
Not applicable.
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tianci Zhang and Jingjing Chen authors shared co-first authorship.
Yuan-Ping Han and Xiaofeng Zheng authors shared co-senior authorship.
References
- 1.Almog N (2010) Molecular mechanisms underlying tumor dormancy. Cancer Lett 294:139–146. 10.1016/j.canlet.2010.03.004 [DOI] [PubMed] [Google Scholar]
- 2.Bissell MJ, Hines WC (2011) Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med 17:320–329. 10.1038/nm.2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Risson E, Nobre AR, Maguer-Satta V, Aguirre-Ghiso JA (2020) The current paradigm and challenges ahead for the dormancy of disseminated tumor cells. Nat cancer 1:672–680. 10.1038/s43018-020-0088-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aguirre-Ghiso JA (2007) Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer 7:834–846. 10.1038/nrc2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller S, Sandrin L (2010) Liver stiffness: a novel parameter for the diagnosis of liver disease. Hepat Med 2:49–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apte M, Pirola RC, Wilson JS (2015) Pancreatic stellate cell: physiologic role, role in fibrosis and cancer. Curr Opin Gastroenterol 31:416–423. 10.1097/MOG.0000000000000196 [DOI] [PubMed] [Google Scholar]
- 7.Anlas AA, Nelson CM (2020) Soft microenvironments induce Chemoresistance by increasing autophagy downstream of integrin-linked kinase. Cancer Res 80:4103–4113. 10.1158/0008-5472.CAN-19-4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson SM, Vander Heiden MG (2017) Critical functions of the lysosome in Cancer Biology. Annu Rev Pharmacol Toxicol 57:481–507. 10.1146/annurev-pharmtox-010715-103101 [DOI] [PubMed] [Google Scholar]
- 9.Chi C et al (2010) Disruption of lysosome function promotes tumor growth and metastasis in Drosophila. J Biol Chem 285:21817–21823. 10.1074/jbc.M110.131714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dennemarker J et al (2010) Deficiency for the cysteine protease cathepsin L promotes tumor progression in mouse epidermis. Oncogene 29:1611–1621. 10.1038/onc.2009.466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebert E et al (1997) Expression of cysteine protease inhibitors stefin A, stefin B, and cystatin C in human lung tumor tissue. Adv Exp Med Biol 421:259–265. 10.1007/978-1-4757-9613-1_34 [DOI] [PubMed] [Google Scholar]
- 12.Kuopio T et al (1998) Cysteine proteinase inhibitor cystatin A in breast cancer. Cancer Res 58:432–436 [PubMed] [Google Scholar]
- 13.Liang XH et al (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402:672–676. 10.1038/45257 [DOI] [PubMed] [Google Scholar]
- 14.Gorgulu K et al (2019) Levels of the Autophagy-Related 5 Protein Affect Progression and Metastasis of Pancreatic Tumors in Mice. Gastroenterology 156, 203–217 e220, 10.1053/j.gastro.2018.09.053 [DOI] [PubMed]
- 15.Huang J, Wu S, Barrera J, Matthews K, Pan D (2005) The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122:421–434. 10.1016/j.cell.2005.06.007 [DOI] [PubMed] [Google Scholar]
- 16.Morvaridi S, Dhall D, Greene MI, Pandol SJ, Wang Q (2015) Role of YAP and TAZ in pancreatic ductal adenocarcinoma and in stellate cells associated with cancer and chronic pancreatitis. Sci Rep 5:16759. 10.1038/srep16759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dupont S et al (2011) Role of YAP/TAZ in mechanotransduction. Nature 474:179–183. 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- 18.Hingorani SR et al (2005) Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7:469–483. 10.1016/j.ccr.2005.04.023 [DOI] [PubMed] [Google Scholar]
- 19.Li Q, Rycaj K, Chen X, Tang DG (2015) Cancer stem cells and cell size: a causal link? Semin Cancer Biol 35:191–199. 10.1016/j.semcancer.2015.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francescangeli F et al (2023) Dormancy, stemness, and therapy resistance: interconnected players in cancer evolution. Cancer Metastasis Rev 42:197–215. 10.1007/s10555-023-10092-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han YP, Tuan TL, Wu H, Hughes M, Garner WL (2001) TNF-alpha stimulates activation of pro-MMP2 in human skin through NF- (kappa)B mediated induction of MT1-MMP. J Cell Sci 114:131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao B et al (2007) Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21:2747–2761. 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pohl C, Dikic I (2019) Cellular quality control by the ubiquitin-proteasome system and autophagy. Sci (New York N Y) 366:818–822. 10.1126/science.aax3769 [DOI] [PubMed] [Google Scholar]
- 24.Komura T et al (2017) Clinical features of cystatin A expression in patients with pancreatic ductal adenocarcinoma. Cancer Sci 108:2122–2129. 10.1111/cas.13396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duncan EM et al (2008) Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell 135:284–294. 10.1016/j.cell.2008.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soond SM et al (2019) Lost or forgotten: the nuclear cathepsin protein isoforms in cancer. Cancer Lett 462:43–50. 10.1016/j.canlet.2019.07.020 [DOI] [PubMed] [Google Scholar]
- 27.Hill R, Wu H (2009) PTEN, stem cells, and cancer stem cells. J Biol Chem 284:11755–11759. 10.1074/jbc.R800071200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ying H et al (2011) PTEN is a major tumor suppressor in pancreatic ductal adenocarcinoma and regulates an NF-kappaB-cytokine network. Cancer Discov 1:158–169. 10.1158/2159-8290.CD-11-0031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ueno T et al (2008) Loss of Pten, a tumor suppressor, causes the strong inhibition of autophagy without affecting LC3 lipidation. Autophagy 4:692–700. 10.4161/auto.6085 [DOI] [PubMed] [Google Scholar]
- 30.Hill R et al (2010) PTEN loss accelerates KrasG12D-induced pancreatic cancer development. Cancer Res 70:7114–7124. 10.1158/0008-5472.CAN-10-1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Z et al (2017) Cathepsin H-Mediated degradation of HDAC4 for Matrix Metalloproteinase expression in hepatic stellate cells: implications of Epigenetic Suppression of Matrix Metalloproteinases in Fibrosis through stabilization of class IIa Histone Deacetylases. Am J Pathol 187:781–797. 10.1016/j.ajpath.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi Y et al (2018) The use of magnetic resonance elastography in differentiating autoimmune pancreatitis from pancreatic ductal adenocarcinoma: a preliminary study. Eur J Radiol 108:13–20. 10.1016/j.ejrad.2018.09.001 [DOI] [PubMed] [Google Scholar]
- 33.Liu Z, Gou A, Wu X (2023) Liver metastasis of pancreatic cancer: the new choice at the crossroads. Hepatobiliary Surg Nutr 12:88–91. 10.21037/hbsn-22-489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie Z et al (2022) Exosome-delivered CD44v6/C1QBP complex drives pancreatic cancer liver metastasis by promoting fibrotic liver microenvironment. Gut 71:568–579. 10.1136/gutjnl-2020-323014 [DOI] [PubMed] [Google Scholar]
- 35.Turk V et al (2012) Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta 1824:68–88. 10.1016/j.bbapap.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, Wang C (2006) F-box protein Skp2: a novel transcriptional target of E2F. Oncogene 25:2615–2627. 10.1038/sj.onc.1209286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elbediwy A et al (2016) Integrin signalling regulates YAP and TAZ to control skin homeostasis. Development 143:1674–1687. 10.1242/dev.133728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao M et al (2022) Targeting fibrosis, mechanisms and cilinical trials. Signal Transduct Target Therapy 7:206. 10.1038/s41392-022-01070-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas D, Radhakrishnan P (2019) Tumor-stromal crosstalk in pancreatic cancer and tissue fibrosis. Mol Cancer 18:14. 10.1186/s12943-018-0927-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han YP, Yan C, Zhou L, Qin L, Tsukamoto H (2007) A matrix metalloproteinase-9 activation cascade by hepatic stellate cells in trans-differentiation in the three-dimensional extracellular matrix. J Biol Chem 282:12928–12939. 10.1074/jbc.M700554200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott KE, Fraley SI, Rangamani P (2021) A spatial model of YAP/TAZ signaling reveals how stiffness, dimensionality, and shape contribute to emergent outcomes. Proc Natl Acad Sci USA 118. 10.1073/pnas.2021571118 [DOI] [PMC free article] [PubMed]
- 42.Yang ZM et al (2017) Cathepsin H-Mediated degradation of HDAC4 for Matrix Metalloproteinase expression in hepatic stellate cells. Am J Pathol 187:781–797. 10.1016/j.ajpath.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimura S, Noda T, Yoshimori T (2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3:452–460. 10.4161/auto.4451 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on reasonable request.