Abstract
Initial studies on the immunogenicity of SARS-CoV-2 (CoV-2) S glycoprotein (“spike”) as a protein subunit vaccine suggested sub-optimal efficacy in mammals. Although protein engineering efforts have produced CoV-2 spike protein sequences with greatly improved immunogenicity, additional strategies for improving the immunogenicity of CoV-2 protein subunit vaccines are scaffolding and the use of adjuvants. Comparisons of the effectiveness of engineered protein-only and engineered protein-nanoparticles vaccines have been rare. To explore this knowledge gap, we inoculated mice with two doses of either sequence-optimized trimeric spike protein or one of several sequence-optimized spike nanoparticles. We measured their immune response up to two months after the first dose. We also measured the immune response and protection against live virus in hamsters inoculated with either sequence-optimized trimeric spike protein or a liposome-based sequence-optimized spike nanoparticle. We found that in the presence of adjuvant, the antibody and neutralization titers elicited by spike-nanoparticles were not significantly greater than those elicited by spike-only in mice, even at doses as low as 0.1 µg/animal. Hamsters vaccinated with spike-only or spike-nanoparticles were equally protected from live virus one month after their first inoculation. These results suggest that sequence-optimized protein subunit vaccines in the form of individual prefusion-stabilized trimers can be as effective in improving immunogenicity as scaffolded forms.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-76377-y.
Keywords: Protein subunit vaccine, Protein engineering, Scaffold, Adjuvant, Nanoparticle, SARS-CoV-2 (CoV-2) spike protein
Subject terms: Viral infection, Preclinical research, Molecular medicine, Lipidomics, Viral proteins, Cryoelectron microscopy, Drug delivery, Protein design
Introduction
Following the release of the SARS-CoV-2 genome in February 20201, researchers and governments rushed to produce the first COVID-19 vaccines for widespread public dissemination. Through unprecedented efforts, just 10 months later, the first COVID-19 vaccine received Emergency Use Authorization from the Food and Drug Administration of the United States2. Thanks to decades of preparatory research and the development of synthetic lipid vesicle-based drug delivery systems, the first and second COVID-19 vaccines used during the COVID-19 pandemic were based on mRNA technology. A more “traditional” protein-based vaccine, developed by Novavax3, was only made available in the United States in July of 2022.
A key goal in designing new vaccine candidates is to maximize both immunogenicity and safety. First-generation vaccines of the 19th century, consisting of weakened or inactivated pathogens, were highly immunogenic but of questionable safety. Thus, in the 20th century, there was a move toward isolating specific protein antigens that could trigger a protective immune response without risk of causing disease4.
The determination of the structure of the influenza virus hemagglutinin glycoprotein (HA) by Don Wiley et al. opened the door to an understanding of the conformational transition of class-1 fusion proteins5. Like flu HA and HIV gp140, the coronavirus is an RNA virus that shares the same class of fusion proteins. These glycoproteins can be purified from a recombinant gene without a transmembrane domain and stabilized in trimeric form by introducing a trimerization tag such as the foldon-tag derived from bacteriophage T4 fibritin6. The lessons learned from the studies of influenza HA were implemented in all subsequent trimeric class-1 fusion protein antigen designs, including those meant for development of protein subunit vaccines against betacoronaviruses.
Although the immunogenicity of protein subunit vaccines can vary greatly from antigen to antigen, in some cases, lower immunogenicity can be overcome by sequence optimization of the target antigen. Similar to the other class-1 fusion proteins, the stabilization of its prefusion conformation by the introduction of a 2-proline substitution mutation into the MERS-CoV-S glycoprotein has been used to increase the neutralizing activity of sera in vaccinated mice7. This stabilization of the trimeric pre-fusion conformation not only increases the lifetime of the antigen but keeps it in the relevant conformation for eliciting neutralizing antibodies. This strategy has been applied and expanded upon in the case of the trimeric SARS-CoV-2 S glycoprotein (“spike”), where the homologous two proline mutations in combination with deletion of the furin cleavage site (S_ΔCS-PP8), cleavage of which is required to trigger the fusion transition, has been used to increase neutralizing activity of sera and protection from live virus in mice. Recently, further optimization of the prefusion-stabilized S2 domain of the spike protein has been shown to elicit broadly neutralizing protective antibodies, paving the way towards a next generation of pan-coronavirus vaccines9.
Among the common strategies to improve immunological response to vaccines in an immunogen-independent manner are the use of adjuvants and the presentation of antigens as protein nanoparticles. Adjuvants, which are regularly included in protein vaccine formulations to boost the immunogenic effect, are highly varied in composition and mechanism, but can generally consist of immunostimulatory molecules, formulations which aid in antigen delivery, or a combination of both10. Two recent examples are the solid lipid-nanoparticles (SLNs) developed to encase and effectively deliver the mRNA vaccines developed by Pfizer and Moderna into cells.
The presentation of antigens in protein nanoparticles can also take various forms and use different mechanisms to increase neutralization titers, amplify humoral response, and even allow for customizable retention in target tissues11,12. Here, we refer to nanoparticle vaccines as antigens assembled in clusters, antigens fused to a molecular scaffold, or a liposome acting as a carrier particle for antigens. This strategy is generally thought to increase immune response by increasing the “visibility” of the antigens to the immune system through substantial cross-linking of B cell receptors.
During the onset of the coronavirus pandemic leading up to the first emergency use authorization, initial vaccine design goals were safety and efficacy, with little time dedicated to optimizing ease of preparation and storage. Since the WHO officially declared the end of the COVID-19 pandemic on June 5, 202313 the need for yearly updates to the COVID-19 vaccine has become evident. Much money and effort have been expended on the utilization of viral display14–16, nanoparticles17–20, and various adjuvants21–23 to develop second-generation vaccines which can be periodically updated and be produced and stored at low cost. The emphasis on these techniques was partially due to initial studies demonstrating that the widely used 2-proline mutation of the SARS-CoV-2 (CoV-2) prefusion-stabilized trimerized spike protein was a poor inducer of neutralizing antibodies at low doses24,25 and unstable when stored at 4 °C for 1 month. Although sequence optimization has gradually improved the immunogenicity and stability of soluble trimeric spike protein over the past 4 years, side-by-side comparisons of these recent spike constructs and their nanoparticle variants are rare.
To directly compare the effects of nanoparticle presentation and addition of adjuvant on the immunogenicity of SARS-CoV-2 spike protein-based vaccines in animal models against protein-only formulations, we vaccinated mice with four engineered antigens in multiple trials performed in parallel in the presence and absence of adjuvant (Fig. 1). The first antigen (henceforth referred to as “spike”) was a prefusion-stabilized trimeric SARS-CoV-2 S glycoprotein based on a previously reported sequence containing six proline mutations and a loss-of-function mutation in the furin-cleavage site (HexaPro spike26) but with an added mono-streptavidin tag. Next, inspired by use of SLNs to encase and deliver mRNA vaccines, was a liposome-nanoparticle scaffold in which His-tagged spike proteins (spike-His) decorate the functionalized surface of a lipid nanoparticle (liposome: spike). Finally, we produced a self-assembled spike-His protein cluster (henceforth referred to as “spike rosettes”) and a spike: ferritin nanoparticle. Spike and liposome: spike were further tested for immunological response and protection in a challenge study with “live” SARS-CoV-2 in golden Syrian hamsters.
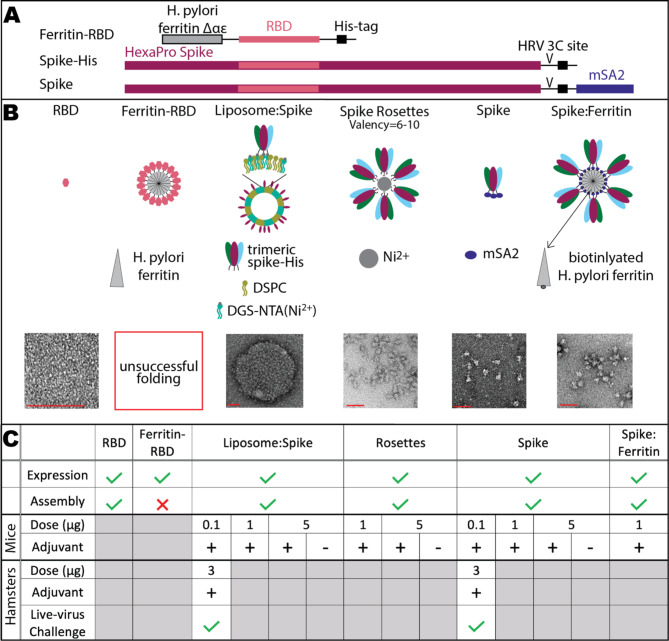
Fig. 1.
Immunogens used in this study. A Protein constructs used to produce immunogens. HexaPro spike is a prefusion-stabilized trimeric spike construct containing six proline mutations and a loss-of-function mutation in the furin-cleavage site26. RBD: CoV-2 S protein receptor binding domain. HRV 3 C site: HRV 3 C protease cleavage site. mSA2: mono-streptavidin sequence. B Conceptual design (cartoon) and representative electron micrographs of negative-stained prepared immunogens (bottom row). The red scale bar indicates 50 nm. Full micrographs of all immunogens, including that for poorly folded ferritin-RBD, can be found in Supplementary Figs. 1 and 2. Liposome: spike nanoparticle was prepared by mixture of 8xHis-tagged spike protein (A, spike-His) with DSPC (1,2-distearoyl-sn-glycero-3-phosphocholine) liposomes intercalated with DGS-NTA(Ni2+)(1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] nickel salt). C Summary of results of animal inoculation experiments for selected immunogens. Check marks (✓) and ×’s denote that an experiment was carried out successfully or unsuccessfully, respectively. Plus sign (+) and minus sign (-) indicate the inclusion or exclusion of adjuvant (AddaVax™), respectively
Results
Preparation of immunogens
We initially prepared one soluble spike construct and four scaffold-conjugated spike-based constructs (Fig. 1) as immunogens for animal testing. Of the spike-based scaffolds, although the CoV-2 spike receptor binding domain (RBD) alone expressed and folded as expected (Fig. 1B, Supplementary Fig. 1A), ferritin-RBD did not fold properly (Supplementary Fig. 1B) and elicited no immune response in mouse inoculation experiments (Supplementary Fig. 1C).
Spike protein was tethered to the surface of liposome nanoparticles (liposome: spike, Fig. 1B, Supplementary Fig. 2A) by exploiting the 8xHis-tag already present in the HexaPro spike construct (henceforth called spike-His). His-tagged proteins have been commonly purified from cell extracts through the use of Ni-coordinated nitrilo-tri-acetic acid (Ni: NTA) for three decades27,28. We reasoned that by including NTA-conjugated lipid in our liposome formulation, in the presence of Ni2+ and spike-His, spike-His-coated liposome nanoparticles would spontaneously assemble. Purified spike-His was mixed with Ni: NTA-conjugated liposome nanoparticles to form the liposome: spike particles, and the product was incubated with Ni: NTA beads for 1 h at room temperature to ensure removal of any untethered spike-His. Following removal of Ni: NTA beads by gravity filtration, to ensure complete removal of unbound spike-His, the liposome: spike particles present in the flow through were pelleted by high-speed centrifugation and resuspended in PBS. In a control experiment omitting Ni2+ from the liposome formulation, no liposome: spike was observed in the flow through.
While imaging a liposome: spike control mixture of spike-His and Ni2+ only, we observed that spike-His particles had clustered into groups of 6–10 with their His-tag-containing C-termini grouped together, presumably coordinated to a free Ni2+. This structure was reminiscent of the NovaVax rosette nanoparticle, in which the full-length, trimeric spike protein in pre-fusion conformation forms rosette-like structures as a result of the interaction of hydrophobic spike protein tails17. We reasoned that our Ni-induced clusters (spike rosettes, Fig. 1B, Supplementary Fig. 2B) could represent this type of nanoparticle well in our vaccine study.
The third immunogen was created to facilitate conjugation of spike protein to a nanoparticle scaffold using the highly specific mono-streptavidin tag (mSA2), a monomeric form of streptavidin capable of binding biotin (spike, Fig. 1B, Supplementary Fig. 2C). The tag mSA2 was inserted at the C-terminal end of the spike-His construct. Mono-streptavidin-tagged spike protein spike (henceforth referred to as “spike”) expressed well and could be purified with high yield (2–3 mg/200 mL of mammalian cell culture).
Ferritin nanoparticles have been utilized in several recent vaccine candidates involving trimeric antigens29–32. Thus, for our final nanoparticle, we designed a spike: ferritin nanoparticle consisting of spike protein linked to biotinylated ferritin nanoparticles (spike: ferritin, Fig. 1B, Supplementary Fig. 2D). Nanoparticles were prepared by mixing purified biotinylated ferritin with at least 100-fold excess of purified spike protein and incubating at room temperature for two hours. Unbound spike protein was removed by size exclusion chromatography.
After preparation, immunogens were sterile-filtrated to remove any bacterial contaminants larger than 0.22 μm using a sterile syringe filter and stored at 4 °C until animals were ready for vaccination. A few days prior to inoculation, inoculants were prepared by diluting the indicated dose of immunogen with PBS to a final volume of 25 µL. Just before inoculation, adjuvant (AddaVax™) or PBS was added 1:1 by volume, for a final inoculant volume of 50 µL. AddaVax™, a squalene-based oil-in-water nano-emulsion adjuvant, was chosen as adjuvant in these experiments due to the similarity of its formulation with that of MF59®, which is licensed for use in influenza vaccines for individuals 65 years of age and older in over 30 countries, including the United States33. MF59® has been shown to elicit both cellular and humoral immune responses, and the mechanism of this response has been linked to the recruitment of antigen-presenting cells to the injection site34.
Biophysical and biochemical characterization of immunogens
Formation of the spike rosettes, spike protein-only, spike: ferritin immunogens, and liposome: spike were verified by negative stain electron microscopy (Fig. 1B, cutouts). Most spike rosettes were observed to be composed of clusters of 6–10 trimeric spike-His proteins (entire micrograph, Supplementary Fig. 2B). Spike rosettes did not form in the absence of Ni ion (data not shown). Spike protein folded as expected (entire micrograph, Supplementary Fig. 2C). The spike: ferritin immunogens had an average occupancy of 78%, containing at least 6–8 spike trimers per ferritin nanoparticle (entire micrograph, Supplementary Fig. 2D). At the observed occupancies and expected geometries, we estimate that the antigen spacing of spike rosettes and spike: ferritin nanoparticles was 20–30 nm. Occupancies were determined by counting the occupancy of nanoparticles in a representative micrograph of the negatively stained sample.
Electron micrographs of negatively stained liposome: spike nanoparticles (Fig. 1B, Supplementary Fig. 2A) showed liposomes with a high surface density of spike-His, resulting in an antigen spacing similar to spike rosettes and spike: ferritin. Size exclusion chromatography (Fig. 2A) and dynamic light scattering (DLS) experiments (Supplementary Fig. 3) with liposome: spike revealed the formation of a homogeneous liposome whose hydrodynamic diameter increased slightly from 200 to 300 nm after the addition of spike-His.
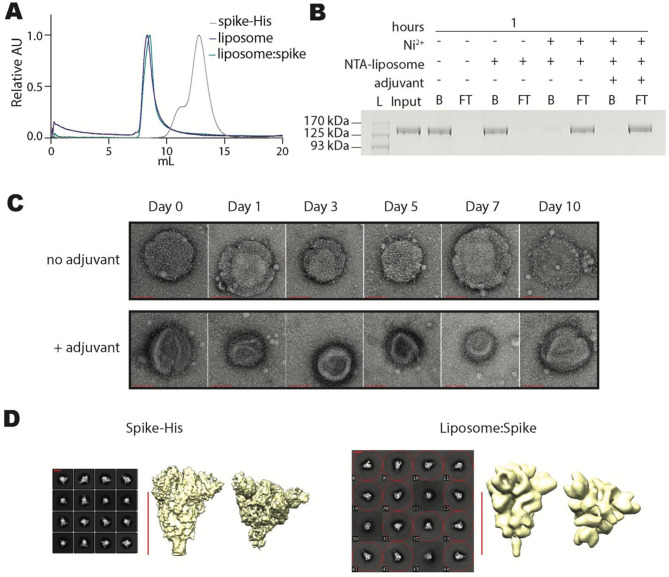
Fig. 2.
Characterization of liposome: spike. Biochemical experiments to observe formation and size distribution of liposome: spike immunogens. A FPLC size exclusion chromatograms, overlaying the elution curves of trimeric spike-His (grey), liposome (purple), and liposome: spike (green). Each chromatogram was scaled relative to its maximum value. B Anti-spike western blot of Ni: NTA bead capture assay 1 h after mixing Ni: NTA-liposome and spike-His. Image has been cropped and lanes have been rearranged for ease of comprehension. Original image can be found in Supplementary Fig. 5. (L, protein ladder; Input, spike-His in the absence of Ni: NTA-liposome; B, bead-captured fraction; S, supernatant). C Negative stain transmission electron microscopy images of liposome: spike demonstrating particle stability in 20% human serum at 37 °C for up to 10 days in the presence or absence of adjuvant. The red scale bar is equal to 100 nm. D TEM analysis of the spike-His-only and spike-His bound to the liposomes. Selected 2D class averages (left) and 3D reconstructions (right). The red scale bar is equal to 10 nm
A Ni: NTA bead-capture assay was used to confirm the binding of spike-His to Ni: NTA liposomes (Fig. 2B). Spike-His only, spike-His mixed with NTA-liposomes, spike-His mixed with NTA-liposomes and Ni ion, and spike-His mixed with NTA-liposomes, Ni ion, and adjuvant were incubated with Ni: NTA beads for 1 h before application to a gravity flow column and collection of flow-through. After washing and elution of the bound fraction from the Ni: NTA beads, western blot showed a high concentration of spike-His remaining in the flow through in the presence of NTA-liposomes and Ni ion and faint or undetectable amounts of spike-His in the eluted Ni-NTA bead-bound fraction, suggesting strong binding of spike-His to Ni: NTA-liposomes. In the absence of Ni ion, however, the spike-His was primarily found in the bound fraction.
To kinetically characterize the binding of spike-His to Ni: NTA-liposomes, a similar experiment was carried out in a time-dependent matter (Supplementary Fig. 4). Spike-His was mixed with Ni: NTA-liposomes and Ni: NTA beads in the presence and absence of adjuvant and incubated for 0.5, 1, 3, or 6 h before application to a gravity flow column and collection of flow-through. The Ni-NTA bead-bound fraction was then washed and eluted as described previously. Undetectable levels of spike-His in the bound fraction after 6 h indicated nearly complete binding of spike protein to Ni: NTA-liposome after 3–6 h.
Liposome: spike nanoparticles viewed by negative stain electron microscopy appeared stable in 20% human serum for up to 10 days (Fig. 2C), suggesting that liposome: spike nanoparticles will also be stable during in vivo experiments.
We further confirmed the correct folding and conformation of spike-His by cryo-electron microscopy. Free spike-His and spike-His present on the surface of liposome: spike particles were observed by cryo-electron microscopy and negative stain electron microscopy, respectively (Fig. 2D). Comparison of the 2D classes and 3D reconstructions of both samples revealed that liposome-bound spike-His particles maintain the trimeric state and overall conformation of unbound spike-His.
Antibody response to spike-based immunogens in a mouse model
For each immunogen and dose, 3–6 mice were inoculated via intramuscular injection with one dose of immunogen and received an additional dose 15 days later. Control mice were inoculated with PBS. Dose values indicate the mass of total protein present in the inoculant as measured by light absorbance at 280 nm (1 AU = 1 mg/ml). A previous study comparing spike-based nanoparticles to spike alone (2-proline mutant) had indicated that spike and spike-nanoparticle antibody and neutralization responses were similar when using a high dose, but the efficacy of spike-only dropped off significantly at lower doses25. With this in mind, we performed the experiment at a range of doses covering over an order of magnitude (0.1 µg, 1 µg, and 5 µg). Figure 3A summarizes the timetable of the 2-month experiment. The experiment was performed in four trials with different groupings of immunogen and dose, but always included a minimum of three biological replicates for each inoculant.
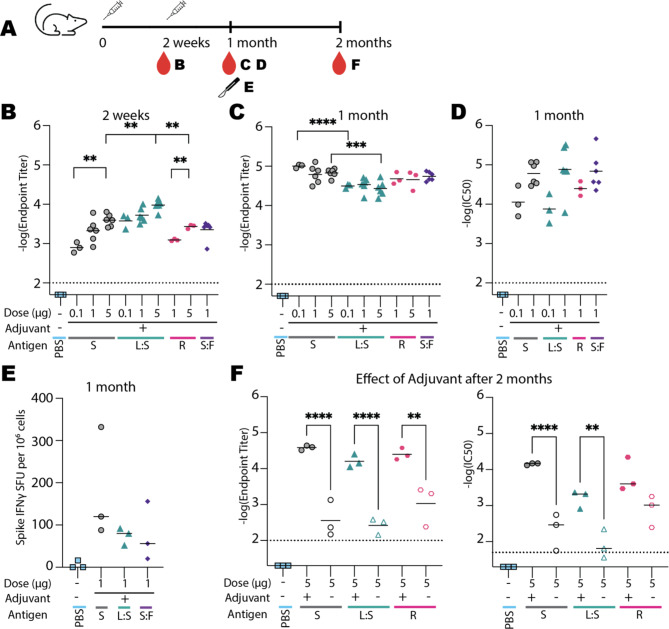
Fig. 3.
Antibody response to vaccination in the mouse model. A Timeline of inoculation (time points indicated by syringe cartoons), bleeding (depicted by red droplets), and harvesting of splenocytes (scalpel). Letter(s) next to each icon indicate(s) related panels. After one month, three mice from control (-), spike (S), liposome: spike (L: S), and spike: ferritin (S: F) groups were sacrificed to obtain fresh splenocytes for spike IFN quantification. The remaining mice were kept for an additional month. B, C Antibody titers of sera from mice inoculated with each immunogen for 2 weeks (B) and 1 month (C) after the first inoculation. D Pseudovirus neutralization titers from sera collected one month after the first dose. E Spike IFN levels one month after the first dose. F Effect of adjuvant on antibody titers (left) and on pseudovirus neutralization (right) 2 months after first inoculation for selected immunogens. Horizontal lines indicate mean values. Mean values of immunogens of the same dose, doses of the same immunogen, and immunogens with and without adjuvant were compared using the Bonferroni test. Asterisks indicate statistical significance. *p < 0.0332; **p < 0.0021; ***p < 0.0002; ****p < 0.0001. S, spike; L:S, liposome: spike; R, spike rosettes; S:F, spike: ferritin
All antibody titers were measured by detection of total anti-spike IgG. Initial antibody titers at two weeks post first dose (Fig. 3B) had increased roughly corresponding to increased doses. The antibody titers for each dose group were similar across the spike, spike rosette, and spike: ferritin groups. However, at the 5 µg dose, the mean antibody titer of the liposome: spike was statistically higher than that of spike and spike rosettes.
After the second inoculation, however, antibody titers within each immunogen group were no longer dose-dependent. At both 0.1 µg and 5 µg doses, the mean antibody titers of the spike group were statistically higher than those of liposome: spike (Fig. 3C). In contrast to the antibody titer results, the mean pseudovirus neutralization titers of serum samples from spike and liposome: spike groups increased with dose. Although these increases were not statistically significant, the average pseudovirus neutralization titer of the 1 µg dose was roughly one order of magnitude higher than the 0.5 µg dose for both spike and liposome: spike immunogen groups. 1 µg of spike trimer was as effective as the nanoparticle immunogens (Fig. 3D).
To confirm that the mSA2 tag was not a factor in the immunogenicity of spike protein, we also measured the antibody response of mice to spike protein which had been cleaved with HRV 3 C protease to remove the mSA2 sequence. No significant differences were observed between endpoint titers or IC50s of spike and spike lacking mSA2 (Supplementary Fig. 6A, 6B). Lastly, we tested liposome-only as an immunogen and found no detectable anti-spike antibody response (Supplementary Fig. 6C).
Moreover, ELISpot analysis revealed that induction of IFN-g-producing spike-specific T cells was comparable between mice immunized with spike alone, spike: ferritin, and liposome: spike (Fig. 3E).
Effect of adjuvant on antibody response
We were surprised to see the similarity of antibody responses between spike-only and nanoparticle-spike antigens and wondered if the same trend would be observed in the absence of an adjuvant. To determine the role of adjuvant on the efficacy of vaccination, three mice were sequentially vaccinated with two 5-µg doses each of spike, liposome: spike, or spike rosette immunogens in the absence of adjuvant. Antibody and pseudovirus neutralization titers were measured in sera collected 2 months after the first dose and compared with sera from adjuvant-enhanced immunogen groups (Fig. 3F). Detectable antibody and pseudovirus neutralization titers were measured two months post first dose. Spike and liposome: spike groups in the presence of adjuvant had statistically higher antibody and neutralization titers than the groups of the same immunogens in the absence of adjuvant; however, the difference between the pseudovirus neutralization titers of the spike rosette groups with and without adjuvant was not statistically significant. After two months, neutralization titers of the 5 µg dose spike group were all higher than those of the 5 µg dose liposome: spike group; however, due to the low sample number, the mean neutralization titers are not statistically different.
Antibody response to spike or liposome: spike in a golden Syrian hamster model
Spike and liposome: spike immunogens were also tested in the golden Syrian hamster model. Four golden Syrian hamsters were inoculated via intramuscular injection with one 3 µg dose of immunogen or saline solution, followed by an additional dose two weeks later. Dose values indicate the mass of total protein present in the inoculant as measured by light absorbance at 280 nm and assuming 1 AU = 1 mg/ml. Figure 4A summarizes the timetable of the 6-week experiment.
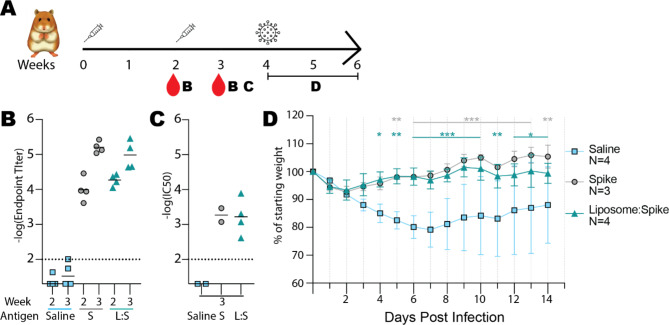
Fig. 4.
Antibody response to vaccination, and live virus challenge in hamster model. A Timeline of inoculation (syringes), bleeding (red droplets), and challenge (virus icon). Letter(s) next to each icon indicate(s) related panels. B Antibody titers of hamsters two weeks and three weeks after the first dose. The differences between immunogens at the same time point were not statistically different by the Bonferroni test. C Pseudovirus neutralization by each serum after two inoculations. Horizontal lines indicate mean values for (B) and (C). D Percent weight tracking of hamsters following infection with SARS CoV-2 virus four weeks after the first dose. Points indicate mean values of percent weight from saline-inoculated hamsters (blue), spike-inoculated hamsters (gray), and liposome: spike-inoculated hamsters (green). Bars show standard deviation. Two-way ANOVA was performed, followed by the Bonferroni test as a post hoc test. *p < 0.05 vs. saline, **p < 0.01 vs. saline and ***p < 0.001 vs. saline.
Total anti-S protein IgG antibody titer response to inoculation was comparably strong for spike and liposome: spike two weeks after the first dose and one week after the second dose, while the saline-injected negative control group showed minimal antibody response to inoculation (Fig. 4B). All tested serum samples from the spike and liposome: spike groups could neutralize CoV-2 pseudovirus, but no neutralization could be detected from the saline group serum samples (Fig. 4C). Serum volumes from two animals from the saline group and one animal from the spike group were insufficient for pseudovirus neutralization testing, and one animal from the spike group died during the 3-week blood draw due to anesthesia overdose.
Live virus challenge in golden Syrian hamster model
To observe the protection conferred by immunization, four weeks after the first inoculation, the hamsters above were infected with live SARS-CoV-2 virus, and the change in body weight was measured over time (Fig. 4D). Starting as early as three days post-infection, hamsters inoculated with spike or liposome: spike maintained their starting body weights significantly more effectively than those inoculated with saline only. There was no statistical difference between the body weight changes in the spike and liposome: spike groups.
Discussion
Our study compared prefusion-stabilized, soluble SARS-CoV-2 spike protein trimers (“spike”) with spike-decorated nanoparticles in the presence and absence of adjuvant and found that the adjuvanted nanoparticle assemblies had no significant advantage over the spike-only immunogen in antibody titers, neutralization titers, or spike-specific T cell responses in a mouse model. We also found that spike and liposome: spike vaccines were similarly immunogenic and protective against live virus in golden Syrian hamsters. This stands in contrast with previous reports comparing soluble spike protein with its derivative nanoparticles, which have typically used a 2-proline prefusion-stabilized spike sequence (S_2P)35, rather than the 6-proline mutant used in this study26. S_2P alone was significantly less effective than S_2P-based-spike-nanoparticles in eliciting neutralizing antibodies in mouse models and became ineffective at doses lower than 2 µg S_2P per mouse25.
The improvement in immunogenicity described above can likely be attributed to two factors related to the additional proline mutations and the removal of the furin cleavage site. First, there is the lower structural stability associated with S_2P. S_2P can only tolerate one freeze-thaw cycle before undergoing a sharp drop in antigenicity. The 6-proline spike mutant, on the other hand, is stable for up to three freeze-thaw cycles26. Furthermore, Olia et al. have documented a partial unfolding of S_2P after just eight days of storage at 4 °C36. Secondly, S_2P containing an intact furin site has been shown to undergo partial proteolysis during purification, and further in vivo cleavage after inoculation may result in a reduced immune response. However, Amanat et al. have shown that removal of the furin cleavage site in the two-proline mutant (∆CS_2P) prevents cleavage during purification8. Doses of ∆CS_2P spike protein trimer as low as 0.5 µg have shown robust immunological responses in the mouse model37. Our study utilizing the 6-proline mutant lacking the cleavage site reduces the reported required dose to an even lower level, demonstrating that in the presence of an adjuvant, 0.1 µg/mouse of sequence-optimized spike-alone can produce robust antibody and neutralization titers.
Our data suggest that with optimal antigen stability and effective adjuvants, monovalent spike protein trimers can elicit antibody and T-cell responses as efficiently as multivalent spike protein trimers. Antigen valency required for B cell responses can vary depending on antigen dose and characteristics, including the affinity to B cell receptors38. Antigen multivalency is generally considered desirable as it facilitates the cross-linking of B cell receptors (BCRs), promoting BCR signaling and subsequent B cell responses39. Indeed, correlations have been observed between antigen valency and B cell responses for specific antigens40. However, some monovalent or low-valent antigens can induce effective B-cell responses similar to multivalent antigens41–43. Given these, we speculate that stabilized monovalent spike protein trimers may crosslink BCRs through simultaneous recognition of epitopes by multiple BCRs, providing an optimal BCR signal that effectively induces the B cell response. Indeed, neutralizing mAbs-targeted epitopes have been characterized in a variety of locations on the spike protein, including the receptor-binding domain, the adjacent N-terminal domain, and the C-terminal S2 domain44.
All nanoparticles used in this study were approximately 20–400 nm in diameter. Nanoparticles within this size range can be effectively delivered to lymph nodes, although various factors affect their drainage and retention in lymph nodes45,46. Negative-stain micrographs indicate that epitope spacing in liposome: spike, spike rosettes, and spike: ferritin was approximately 20–30 nm, which is expected to support multivalency-dependent enhancement in activation of B cell receptor signaling47. Our nanoparticles do not likely have strong activity to change innate immunity induced by AddaVax adjuvant, but the liposomes may contribute to immunomodulatory effects48.
Throughout this study, we produced four novel spike-conjugated nanoparticles for testing as potential vaccine candidates. Although our version of the ferritin-RBD fusion construct was misfolded, other work has generated an alternatively designed RBD-ferritin fusion protein that folded properly and elicited an immune response in mice49. These different results highlight the effort and resources needed to generate an effective nanoparticle-immunogen fusion construct. Adaptable, modular nanoparticle formulations, such as the spike rosettes, liposome: spike, and spike: ferritin, allow quick generation of immunogen nanoparticles utilizing any protein that contains a poly-histidine (in the case of spike rosettes and liposome: spike) or streptavidin-based (for the spike: ferritin) purification tag. In the past several years, a few such modular antigen concepts have been described and generated25,50, but these systems involve binding partners that are not yet in common use. Our method has an advantage in that it makes use of two very common protein purification tags. Such broadly employable modules could considerably shorten the time required to generate and screen effective immunogens for use in nanoparticle-based vaccine studies.
The His-tag: Ni: NTA-liposome system, which benefits from the cost-effectiveness, adaptability, and broad commercial availability of the His-tag: Ni: NTA purification system, has the added advantage of being highly accessible to researchers across the globe. Furthermore, although we did not observe any adjuvant-like advantage from our DSPC-cholesterol-NGS-NTA liposomes, other liposomes have been shown to increase immunological reactions, acting as a “self-adjuvant.” Combining the accessible and flexible His-tag: Ni: NTA system with a “self-adjuvanting” liposome may be helpful in vaccine development for other non-COVID diseases. We recognize that the presence of nickel in a vaccine meant for humans may cause complications due to toxicity or allergenic potential at high concentrations; however, no adverse reactions were observed in the mouse or hamster models.
We observed that liposome: spike induced higher antibody titers than spike-only antigens when used for an initial vaccination. Although this difference is not statistically significant when considering the number of repetitions, it could suggest that the larger size and multivalency of liposome: spike may be advantageous in activating naïve B cells. However, when used for prime-boost vaccination, liposome: spike was less effective to induce anti-spike antibodies than spike-only antigens, and neutralizing antibodies induced from liposome: spike were less persistent at high doses compared with those of spike, suggesting that spike-only antigens may be more effective to activate effector or memory B cells than liposome: spike in the booster dose of vaccination. Since effector or memory B cells can acquire immunoglobulins with high affinity to antigens during germinal center reactions51, unlike naïve B cells, they may not need antigen multivalency. We also speculate that the specific orientation of spike-His antigens tethered on liposomes may make certain epitopes less accessible to B cells compared to spike-only antigens.
Our experiment in mice comparing immunogens in the presence and absence of adjuvant demonstrated that the addition of adjuvant increased antibody titers by more than one order of magnitude for spike, liposome: spike, and spike rosettes (Fig. 3F). Interestingly, the adjuvant effect on pseudovirus neutralization titers was least pronounced for the spike rosettes vaccination group, in which the addition of adjuvant did not result in a neutralization titer that was statistically different from the immunogen in the absence of adjuvant. This may indicate that in the absence of an adjuvant, the scaffolding technique utilized in the spike rosettes offers a slight advantage over spike-alone. However, since the spike-alone and spike rosettes pseudovirus neutralization titers were not statistically different without adjuvant, more data would be required to make this assertion confidently.
The fact that low-dose spike-alone can be as effective as spike nanoparticles in the presence of adjuvant highlights the enduring potential of protein subunit vaccines as vaccine candidates. The inability of initial CoV-2 spike protein subunit vaccine candidates to elicit high titers of neutralizing antibodies led researchers to begin work on spike-nanoparticle engineering immediately. However, as our work has emphasized, spike-only antigenicity can rival or surpass that of spike-based nanoparticles after just a few protein engineering steps, such as the knock-out of the multibasic furin and TMPRSS2 protease cleavage sites between domains S1/S252 and the stabilization of the prefusion states53. The present study did not account for different variants of the coronavirus, and only a single strain (which was close the original Wuhan strain; see methods) was tested in hamsters. Although our study is limited by the number of biological replicates in comparing vaccine effectiveness, more extensive comparison of spike and spike nanoparticles in mice and in higher organisms could demonstrate that this engineering-based advantage holds true in the context of different nanoparticle types and in larger mammals. Issues faced during the production and distribution of COVID-19 vaccines and recent global supply chain issues have underscored the value of easy-to-produce vaccines that require few ingredients. Therefore, simply because of their ease of preparation, soluble subunit vaccines such as the sequence-optimized spike protein investigated here have a clear advantage over nanoparticle vaccines and may even prove to be more effective in conferring disease resistance.
Methods
Cloning
SARS-CoV-2 S HexaPro plasmid was a gift from Jason McLellan (Addgene plasmid # 154754)26. A mono-streptavidin sequence (mSA2, Kerafast, Accession ID: P22629) was inserted by restriction enzyme digest and ligation at the BstBI restriction site to make the spike construct.
The DNA sequence for AviTag-ferritin was ordered as a long single-stranded DNA oligo from Integrated DNA Technologies (IDT). The encoded protein, from N-terminal to C-terminal, consists of a His-tag, AviTag (GLNDIFEAQKIEWHE), a rigid linker (SLSTPPTPSTPPT), a bullfrog linker (Accession ID: P07797; residues 2–9 with N8Q mutation), followed by the Helicobacter pylori ferritin sequence (Accession ID: Q9ZLI1; residues 3-167 with I7E, N19Q, C31S mutations). Bullfrog linker and H. pylori ferritin sequence were adapted from a previous publication54.
Production, purification, and assembly of spike-based antigens
SARS-CoV-2 S HexaPro (spike-His) and mSA2-tagged-HexaPro (spike) plasmid was transfected, expressed, and purified as previously reported55. To prepare control spike protein which lacked mSA2, mSA2 from purified spike protein was removed by proteolytic cleavage at the internal HRV 3 C cleavage site.
Expression of AviTag-ferritin was carried out in E. coli strain CVB-T7 POL (Avidity, LLC), which overexpresses the E. coli biotin ligase BirA for in vivo biotinylation of Avi-tagged protein under the control of T7 promoter. The overnight culture of transformation colonies was diluted 200-fold into fresh Terrific Broth medium containing 0.1% MgSO4 and 0.04% glucose and incubated at 37° C with vigorous shaking. IPTG and biotin were added to the final concentration of 0.5 mM and 50 μm, respectively, when the OD600 reached 0.7. Expression and in vivo biotinylation were carried out at 20° C overnight. The harvested frozen pellet was resuspended in lysis buffer (50mM HEPES pH 7, 300mM NaCl, 30mM imidazole) with Roche complete protease inhibitor without EDTA. After lysis by sonication (Misonix S-4000), the centrifuge-cleared lysate was applied to Ni: NTA (Ni sepharose FF, Cytiva) pre-equilibrated with lysis buffer. Protein was eluted with 500 mM imidazole and desalted on a PD-10 desalting column.
To form the spike: ferritin nanoparticles, purified biotinylated AviTag-ferritin were mixed with at least a 100-fold excess of purified spike protein and incubated at room temperature for two hours. The sample was then fractionated on a Superose 6 increase 10/300 GL column, and the largest molecular weight peak was used for mouse inoculation experiments.
Liposomes used for liposome: spike were prepared in two steps. First, a 4:4:2 mixture of 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), cholesterol, and 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] nickel salt (DGS-NTA(Ni)) (Avanti Polar Lipids) were solubilized in chloroform and deposited as a thin film under nitrogen gas. Second, after rehydration in PBS, liposomes were extruded at a temperature above 60° C (the Tm of DSPC), with 30 passes through a 100 nm membrane with an Avanti Polar Lipids Extruder. The concentration of lipids was calculated according to established methods56.
To prepare liposome: spike immunogen, the liposome product was mixed with purified his-tagged spike protein in PBS with a final concentration of 4.7 nM and 1.1 µM, respectively. Ni: NTA beads were added to the sample, and the mixture was incubated for 1 h at room temperature to ensure no unbound spike-His remained in solution. Ni: NTA beads were removed by gravity filtration, and liposome: spike was further isolated by high-speed centrifugation at 4 ℃ and resuspended in PBS.
Spike rosettes were formed in a solution containing 20 µM NiCl2 and 0.55 µM of purified his-tagged spike protein incubated at room temperature overnight. The valency of spike rosette nanoparticles was estimated by visual inspection of negative-stain EM micrographs.
Characterization of liposome: spike
Size-exclusion chromatography of spike, liposome: spike, and liposome was carried out on a Superdex 200 column (GE Health Sciences) pre-equilibrated to PBS and run at 4 ℃ at 0.5 ml/min.
DLS measurements of liposome and liposome: spike were performed on the Zetasizer Nano. Samples of liposome, spike, or liposome: spike were filtered through a 0.22 μm Ultrafree-MC Centrifuge Filter (Durapore PVDF, Millipore) before loading to replicate preparation of vaccine samples. Although 0.22 μm is smaller than the peak diameter of liposome: spike as measured in this experiment, we reason that the liposome is flexible and can be deformed temporarily so as to fit through a narrow pore size. Buffer conditions (20 mM HEPES (pH 7.5) and 150 mM KCl) and cell holder temperature (25° C) were held constant during the measurement. Samples were placed at 25 °C for 15 min and then measured at the same temperature. All measurements were performed in triplicate, and an average value was calculated with standard error.
To perform Ni: NTA bead capture, sufficient beads were added to ensure full binding of the free spike-His in the sample. Liposome: spike was incubated with the beads for 1 h at room temperature before application to a gravity flow column and collection of flow-through. After washing with PBS, the bead-captured spike-His was collected with imidazole elution. The flow-through and elution were then analyzed by PAGE using a Novex 4–12% Bis-Tris acrylamide gel (Invitrogen). Bands were visualized with Coomassie staining.
Liposome: spike particle stability in 20% human serum in PBS was observed in the presence and absence of AddaVax adjuvant (InVivoGen). Temperature was kept at 37 °C for the duration of the experiment.
Electron microscopy of spike constructs
Samples for imaging by negative-stain electron microscopy were applied to glow-discharged carbon-coated copper EM grids and stained with uranyl acetate according to standard procedures. Grids were examined and imaged on a Talos L120C TEM operating at 120 kV acceleration voltage using a Ceta camera (Thermo Fisher Scientific).
Samples prepared for inspection by cryo-electron microscopy were spike or liposome: spike. 3 µL of each were applied to holey carbon grids (Quantifoil 1.2/1.3 Cu 300 mesh, Quantifoil Micro Tools) plasma-cleaned in a mixture of H2 and O2, blotted from 3 s, and plunge-frozen into liquid ethane using an FEI Vitrobot Mark IV at 7 °C with 98% humidity. Data were collected on the FEI Talos Arctica (200 kV) microscope equipped with a Falcon III camera (Thermo Fisher). Movies were recorded at a nominal 96,000x magnification (1.1 Å/px) over 40 frames in integrating mode with a total accumulated dose of 40 e−/Å2 and a defocus range of -1.0 to -2.5 μm. Image stacks were aligned and summed using motioncor257 with dose weighting, and CTF estimation was performed with Gctf58. Templates from the 2D classification of manually picked particles were used for autopicking in RELION 3.0 or cisTEM59. After template-based autopicking and removal of poor picks by 2D classification, an initial 3D model with C3 symmetry was generated. Particles were then aligned with 3D auto-refinement. Multiple rounds of 2D and 3D classification were used to remove poor particle picks further. The final 3D auto-refinement was performed using a soft mask, followed by postprocessing with the same mask.
Inoculation of mice and serum collection
Immunizations were carried out by intramuscular injection of female C57BL/6J mice (SLC) with 50 µL of inoculant containing a mix of 25 µL of antigen in PBS and 25 µL of AddaVax adjuvant. Each mouse was inoculated once on Day 0 and once on Day 15. Blood draws were carried out on Day 0 and Day 14 through the tail veil. After 1 or 2 months, the mice were euthanized by inhalation of carbon dioxide using a displacement rate from 30 to 70% of the chamber volume/min, and a final blood draw was carried out through the inferior vena cava. Serum was extracted from whole blood by centrifugation after room temperature incubation with a separating agent. The adjuvant used in all experiments was AddaVax (InVivoGen).
ELIspot
Peptide pools of SARS-CoV-2 S (JPT; PM-WCPV-S-1) were dissolved in DMSO (500 µg/mL). Freshly isolated splenocytes (2.5 × 105 cells per well) were stimulated with 1 mg/mL peptide solution or equimolar amounts of DMSO for 18 h. IFN-γ ELISpot assays were performed using Mouse IFN-γ Single-Color Enzymatic ELISpot kits (CTL; 2CT1B-mIFNGp-2M2), according to the manufacturer’s protocol. CTL ImmunoSpot S6 Analyzer was used for spot counts and determined by subtracting background spot counts in a DMSO-treated well from a peptide-treated well for antigen-specific spot counts.
Animals
Female 6–8-week-old, specific pathogen-free C57BL/6J mice were obtained from Japan SLC. Protocols of all mouse experiments were performed in accordance with the relevant guidelines and regulations as approved by the Animal Care and Use Committee at the Okinawa Institute of Science and Technology Graduate University (approval number 2020-295). Golden Syrian hamsters, specifically pathogen-free, female, 3 weeks of age, were purchased from Japan SLC, Inc. (Hamamatsu, Japan). Food and water were given ad libitum. All hamster experiments were performed in accordance with the relevant guidelines and regulations as approved by the Tottori University Animal Care Committee (approval number 21-Y-46). This study was performed according to the ARRIVE guidelines.
Vaccine administration to hamsters
After one week of acclimation, the animals received the first vaccine dose via the intramuscular (i.m.) route. Animals were anesthetized by isoflurane inhalation before vaccination. Each hamster was held in one hand while the vaccine was injected into the deltoid muscle with a 23G needle. Either spike protein (N = 4) or liposome: spike (N = 4) were inoculated at 3 µg/ 50 µL volume (1:1 of antigen: adjuvant AddaVax volume) per animal. Control animals were inoculated with 50 µL saline (i.m.) (N = 4). Animals received the second vaccine dose via the same route two weeks after the first dose.
Serum isolation from hamsters
Pre-immune sera were isolated from the retro-orbital blood before the first vaccination. Post-immune sera were separated before the second vaccination and 1 week after the second vaccination. All sera were kept at -80 °C until analysis.
Virus infection
Two weeks after the second dose of vaccine, animals were anesthetized first with isoflurane inhalation, followed by injection of 1 mL fluid anesthesia (0.03 mg/mL medetomidine, 0.4 mg/mL midazolam and 0.5 mg/mL butorphanol) via subcutaneous (s.c.) route. Upon confirming the loss of the pedal withdrawal reflex, animals were inoculated with SARS-CoV-2 (0.2 mL, 105.5 TCID50/0.1 mL; strain SARS-CoV-2 WK-521, GISAID ID: EPI_ISL_40866760) via the intranasal (i.n.) route. SARS-CoV-2 was provided by the National Institute of Infectious Disease (NIID, Tokyo, Japan), propagated with VeroE6/TMPRSS2 at the BSL3 lab of Tottori University, and used at passage 9. Weight was measured and recorded daily until 14 days post-infection (DPI).
ELISA
50 µL/well of 2–4 µg/mL of spike was applied to Immulon 4 HBX plates and adsorbed at 4° C overnight. The next day, plates were washed with 200–300 µL of PBS + 0.1% Tween-20 (PBS-T) and blocked with 200 µL PBS-T + 3% milk solution for 1 h at 20 °C. After blocking, block solution was thrown off, 3x or 4x serial dilutions of serum samples in PBS-T + 1% milk were added at 100 µl /well, and plates were incubated for 2 h at 20 °C. After incubation, plates were washed with 200–300 µL of PBS-T and 100 µL of 1:3000 diluted goat anti-mouse IgG(Fab specific)-peroxidase antibody (Sigma) or goat anti-hamster IgG(H + L)-HRP antibody (Southern Biotech) in PBS-T + 1% milk was applied and incubated on the plates for an additional hour. Finally, plates were washed with 200–300 µL of PBS-T, and the bound antibody was detected with SigmaFast OPD solution according to manufacturer instructions. Each sample was tested in duplicate. Mean intensities were plotted against the log of reciprocal serum dilution factor in GraphPad Prism v8 and fitted with a sigmoidal, four-parameter logistic nonlinear regression. Endpoint titers are defined as the dilutions for which the calculated curves were equal to 0.2 absorbance units.
Production and titration of CoV-2 spike protein-pseudo-typed recombinant VSV
HEK-293T/17 (NIBSC CFAR catalog: #5016) cells grown in DMEM + GlutaMAX + 10% FCS + 1% v/v penicillin-streptomycin and at ~ 80% confluency in 10-cm culture dishes were transfected with 15 µg pCAGGS SARS-CoV-2 spike plasmid (NIBSC CFAR catalog: #100976) mixed with 60 µg of PEI transfection reagent using typical methods and incubated overnight at 37° C with 5% CO2 for protein expression. The morning of the next day, cell media was carefully removed, and cells were infected with G*ΔG-luc rVSV (KeraFast) at MOI of 0.1 in 5 mL of serum-free DMEM for 2 h at 37° C. After incubation the plate was washed 3 times with 3 ml PBS and left in 8 mL of DMEM + GlutaMAX + 10% FCS + 1% v/v penicillin-streptomycin for 24 h at 37 °C, 5% CO2. Cell culture supernatant was collected the next day, passed through a 0.45 μm filter, and frozen in 200 µL aliquots on dry ice.
Infection of VeroE6/TMPRSS2 cells (JCRB1819) by titrations of CoV-2 spike protein-pseudo-typed recombinant VSV was performed in 96-well plates. VeroE6/TMPRSS2 cells grown in a 10-cm culture plate to 90–100% confluency were detached with Trypsin-EDTA, and the cell suspension was diluted 6-8x to prepare sufficient volume for transfer of 100 µL/well into a 96-well culture plate. Cells were allowed to adhere to the plate for at least 2 h at 37° C, 5% CO2. After confirming cell adhesion, a 4-fold dilution series of CoV-2 spike protein-pseudo-typed recombinant VSV starting with 4x diluted virus stock in media was used to infect the cells at 100 µL/well in quadruplicate. Pseudovirus was incubated with cells for 24 h at 37° C, 5% CO2. The next day, infection was assessed using the Bright-Glo Luciferase Assay system (Promega) and measured on a DTX800 multimode plate reader (Beckman Coulter, Indianapolis, IN, USA). A viral dilution achieving, on average, 10,000–30,000 RLU was selected for use in pseudovirus neutralization assays.
Pseudovirus neutralization assays
Pseudovirus neutralization assays were carried out similarly to the virus titration experiment described above, but prior to infection, serial dilutions of sera samples in media were mixed with pseudovirus at 2× the selected viral dilution in 96-well plates. In parallel, one column of 2× diluted virus was diluted with an equal volume of media, and one column was filled with 120 µL/well of media only. The sera-pseudovirus plate was incubated for 1 h at 37° C, 5% CO2 before adding 100 µL of each well into the corresponding position of a 96-well plate containing adherent VeroE6/TMPRSS2 cells (prepared as described above). After 24 h at 37 °C, 5% CO2 infection was detected using the BrightGlo Luciferase Assay system, and %neutralization was calculated using the formula below, where RLU is the relative luciferase units of a given well, RLUcell, max is the maximum RLU of the cells mixed with media only, and RLUvirus, min is the minimum RLU of cell infection with pseudovirus in the absence of serum.
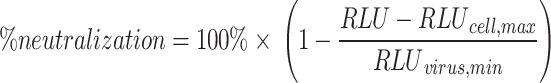 |
Each dilution series of serum was tested in duplicate or triplicate. The mean %neutralization for each dilution was plotted against the log of reciprocal serum dilution factor in GraphPad Prism v8 and fitted with a sigmoidal, four-parameter logistic nonlinear regression. The calculated equation was constrained to have a top value of 100% and a bottom value of 0%.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Giada Mattiuzzo and Emma Bentley provided initial protocols for Production and Titration of VSV Pseudotyped Virus Expressing SARS-CoV-2 S protein. We are grateful for the financial support of the hamster experiments by the OIST “Shinka” grant. M.W. was supported by the Platform Project for Supporting Drug Discovery and Life Science Research (BINDS) from AMED, under Grant Number JP18am0101076. We are grateful for use of the core facilities maintained by the OIST Scientific Computing and Data Analysis Section, Scientific Imaging Section, and Instrumental Analysis Section.
Author contributions
MMM designed, performed, analyzed, and interpreted ELISA and pseudovirus neutralization experiments. TGK designed, prepared, and characterized spike-based immunogens. Collected and analyzed TEM data. KYK designed and prepared liposome: spike and ferritin-RBD. FO and SS designed, performed, analyzed, and interpreted hamster immunization and live-virus challenge experiments. TI, HI, animal BSL3 experiments. HI planned and supervised mouse vaccination experiments. NS performed ELISA and maintained cell culture. HAI performed and analyzed pseudovirus neutralization experiments. VM prepared spike immunogens. TYH designed and prepared biotinylated ferritin. MC gave advice on neutralization assays and Syrian hamster experiments. MT designed, performed, and analyzed ELIspot assays. DS inoculated and performed blood draws on mice. JF supervised hamster immunization and helped obtain funding. PL, STP antigen design. MW study design, funding, organization, supervision, coordination. MMM wrote initial manuscript. All authors contributed to editing and writing of the manuscript.
Data availability
Requests for information or raw data should be directed to the corresponding author.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wu, F. et al. A new coronavirus associated with human respiratory disease in China. Nature . 579, 265–269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DA Takes Additional Action in Fight Against COVID-19 By Issuing Emergency Use Authorization for Second COVID-19 Vaccine. (2020). https://www.fda.gov/news-events/press-announcements/fda-takes-additional-action-fight-against-covid-19-issuing-emergency-use-authorization-second-covid
- 3.Heath, P. T. et al. Safety and Efficacy of NVX-CoV2373 Covid-19 vaccine. N. Engl. J. Med. 385, 1172–1183 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pollard, A. J. & Bijker, E. M. A guide to vaccinology: from basic principles to new developments. Nat. Rev. Immunol. 21, 83–100 (2021). 10.1038/s41577-020-00479-7 [DOI] [PMC free article] [PubMed]
- 5.Wiley, D. C. & Skehel, J. J. THE STRUCTURE AND FUNCTION OF TIlE HEMAGGLUTININ MEMBRANE GLYCOPROTEIN OF INFL1JENZA VIRUS. (1987). http://www.annualreviews.org [DOI] [PubMed]
- 6.Meier, S., Güthe, S., Kiefhaber, T. & Grzesiek, S. Foldon, the natural trimerization domain of T4 fibritin, dissociates into a monomeric A-state form containing a stable β-hairpin: atomic details of trimer dissociation and local β-hairpin stability from residual dipolar couplings. J. Mol. Biol. 344, 1051–1069 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Pallesen, J. et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. U S A. 114, E7348–E7357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amanat, F. et al. Introduction of two prolines and removal of the polybasic cleavage site lead to higher efficacy of a recombinant spike-based sars-cov-2 vaccine in the mouse model. mBio. 12, 1–10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsieh, C. L. et al. Prefusion-stabilized SARS-CoV-2 S2-only antigen provides protection against SARS-CoV-2 challenge. Nat. Commun. 15, 1553 (2024). [DOI] [PMC free article] [PubMed]
- 10.Reed, S. G., Orr, M. T. & Fox, C. B. Key roles of adjuvants in modern vaccines. Nat. Med. 19, 1597–1608 (2013). 10.1038/nm.3409 [DOI] [PubMed]
- 11.Ingale, J. et al. High-density array of Well-ordered HIV-1 spikes on synthetic liposomal nanoparticles efficiently activate B cells. Cell. Rep. 15, 1986–1999 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bo, Y. & Wang, H. Materials-based vaccines for infectious diseases. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 14 (2022). 10.1002/wnan.1824 [DOI] [PMC free article] [PubMed]
- 13.WHO Director-General’s opening remarks at the media briefing – 5. May (2023). https://www.who.int/news-room/speeches/item/who-director-general-s-opening-remarks-at-the-media-briefing---5-may-2023 (2023).
- 14.Lu, M. et al. SARS-CoV-2 prefusion spike protein stabilized by six rather than two prolines is more potent for inducing antibodies that neutralize viral variants of concern. (2022). 10.1073/pnas [DOI] [PMC free article] [PubMed]
- 15.O’Kennedy, M. M. et al. Immunogenicity of adjuvanted plant-produced SARS-CoV-2 Beta spike VLP vaccine in New Zealand white rabbits. Vaccine. 10.1016/j.vaccine.2023.02.050 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang, Y. et al. A highly efficacious live attenuated mumps virus-based SARS-CoV-2 vaccine candidate expressing a six-proline stabilized prefusion spike. (2022). 10.1073/pnas [DOI] [PMC free article] [PubMed]
- 17.Bangaru, S. et al. Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate. Sci. (1979). 370, 1089–1094 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen, A. A. et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice (2021). [DOI] [PMC free article] [PubMed]
- 19.Park, K. S. et al. Lipid-based vaccine nanoparticles for induction of humoral immune responses against HIV-1 and SARS-CoV-2. J. Controll. Rel. 330, 529–539 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, J. et al. Self-adjuvanting lipoprotein conjugate αgalCer-RBD induces potent immunity against SARS-CoV-2 and its variants of concern. J. Med. Chem. 65, 2558–2570 (2022). [DOI] [PubMed] [Google Scholar]
- 21.Johnston, S. C. et al. A SARS-CoV-2 Spike Ferritin Nanoparticle Vaccine Is Protective and Promotes a Strong Immunological Response in the Cynomolgus Macaque Coronavirus Disease 2019 (COVID-19) Model. Vaccines (Basel). 10, (2022). [DOI] [PMC free article] [PubMed]
- 22.Kimoto, T. et al. Induction of systemic, mucosal, and cellular immunity against SARS-CoV‐2 in mice vaccinated by trans‐airway with a S1 protein combined with a pulmonary surfactant‐derived adjuvant SF‐10. Influenza Other Respir Virus. 17, e13119 (2023). [DOI] [PMC free article] [PubMed]
- 23.Liu, Z. et al. A novel STING agonist-adjuvanted pan-sarbecovirus vaccine elicits potent and durable neutralizing antibody and T cell responses in mice, rabbits and NHPs. Cell. Res. 32, 269–287 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walls, A. C. et al. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell. 183, 1367–1382e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, B. et al. A platform incorporating trimeric antigens into self-assembling nanoparticles reveals SARS-CoV-2-spike nanoparticles to elicit substantially higher neutralizing responses than spike alone. Sci. Rep. 10, 18149 (2020). [DOI] [PMC free article] [PubMed]
- 26.Hsieh, C. L. et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Sci. (1979). 369, 1501–1505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leonhardt, F. et al. A systematic review about affinity tags for one-step purification and immobilization of recombinant proteins: integrated bioprocesses aiming both economic and environmental sustainability. 3 Biotech. 13, Preprintathttpsdoiorg101007s13205–Preprintathttpsdoiorg101007s13023 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crowe, J. et al. 6xHis-Ni-NTA chromatography as a superior technique in recombinant protein expression/purification (ed Harwood, A. J.) 371–387 (Humana, Totowa, NJ, 10.1385/0-89603-258-2:371. (1994). [DOI] [PubMed] [Google Scholar]
- 29.Xu, D. et al. Design of universal Ebola virus vaccine candidates via immunofocusing. Proc. Natl. Acad. Sci. U S A. 121 (7), e2316960121 (2024). [DOI] [PMC free article] [PubMed]
- 30.Wang, W. et al. Self-assembled ferritin-based nanoparticles elicit a robust broad-spectrum protective immune response against SARS-CoV-2 variants. Int. J. Biol. Macromol. 264, 130820 (2024). [DOI] [PubMed]
- 31.Tapia, D., Reyes-Sandoval, A. & Sanchez-Villamil, J. I. Protein-based Nanoparticle Vaccine Approaches Against Infectious Diseases. Arch. Med.Res. 54, 168–175 (2023). 10.1016/j.arcmed.2023.02.003 [DOI] [PubMed]
- 32.Qiao, Y. et al. Hemagglutinin-based DNA vaccines containing trimeric self-assembling nanoparticles confer protection against influenza. J. Leukoc. Biol. 112, 547–556 (2022). [DOI] [PubMed] [Google Scholar]
- 33.Adjuvanted Flu Vaccine. (2022). https://www.cdc.gov/flu/prevent/adjuvant.htm
- 34.Ott, G., Barchfeldj-, G. L. & Van Nest, G. Enhancement of Humoral Response against Human Influenza Vaccine with the Simple Submicron Oil/Water Emulsion Adjuvant MF59. Vaccine. 13 (1995). [DOI] [PubMed]
- 35.Wrapp, D. et al. Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. (2019). https://www.gisaid [DOI] [PMC free article] [PubMed]
- 36.Olia, A. S. et al. SARS-CoV-2 S2P spike ages through distinct states with altered immunogenicity. J. Biol. Chem. 297, 101127 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao, H. C. et al. Low-dose SARS-CoV-2 S-Trimer with an Emulsion Adjuvant Induced Th1-Biased protective immunity. Int. J. Mol. Sci. 23 (9), 4902 (2022). [DOI] [PMC free article] [PubMed]
- 38.Avalos, A. M. & Ploegh, H. L. Early BCR events and antigen capture, processing, and loading on MHC class II on B cells. Front. Immunol. 5 (2014). 10.3389/fimmu.2014.00092 [DOI] [PMC free article] [PubMed]
- 39.Woodruff, M. F. A., Reid, B. & James, K. Effect of antilymphocytic antibody and antibody fragments on human lymphocytes in vitro. Nature. 215, 591–594 (1967). [DOI] [PubMed] [Google Scholar]
- 40.Goodnow, C. C. et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. (1988). [DOI] [PubMed]
- 41.Avalos, A. M. et al. Monovalent engagement of the BCR activates ovalbumin-specific transnuclear B cells. J. Exp. Med. 211, 365–379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minguet, S., Dopfer, E. P. & Schamel, W. W. A. Low-valency, but not monovalent, antigens trigger the B-cell antigen receptor (BCR). Int. Immunol. 22, 205–212 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Volkmann, C. et al. Molecular requirements of the B-cell antigen receptor for sensing monovalent antigens. EMBO J. 35, 2371–2381 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shafqat, A. et al. SARS-CoV-2 epitopes inform future vaccination strategies. Frontiers in Immunology vol. 13 Preprint at (2022). 10.3389/fimmu.2022.1041185 [DOI] [PMC free article] [PubMed]
- 45.Trevaskis, N. L., Kaminskas, L. M. & Porter, C. J. H. From sewer to saviour-targeting the lymphatic system to promote drug exposure and activity. Nature Reviews Drug Discovery vol. 14 781–803 Preprint at (2015). 10.1038/nrd4608 [DOI] [PubMed]
- 46.Oussoren, C., Zuidema, J., Crommelin, D. J. A. & Storm, G. Lymphatic uptake and Biodistribution of liposomes after Subcutaneous Injection. II. Influence of liposomal size, lipid composition and lipid dose. Biochim. Biophys. Acta. 1328 (2), 261–272 (1997). [DOI] [PubMed]
- 47.Veneziano, R. et al. Role of nanoscale antigen organization on B-cell activation probed using DNA origami. Nat. Nanotechnol. 15, 716–723 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.La-Beck, N. M. & Gabizon, A. A. Nanoparticle interactions with the immune system: clinical implications for liposome-based cancer chemotherapy. Front. Immunol. 8, 1-6 (2017). [DOI] [PMC free article] [PubMed]
- 49.Joyce, M. G. et al. SARS-CoV-2 ferritin nanoparticle vaccines elicit broad SARS coronavirus immunogenicity. Cell. Rep. 37 (12), 110143 (2021). [DOI] [PMC free article] [PubMed]
- 50.Ueda, G. et al. Tailored design of protein nanoparticle scaffolds for multivalent presentation of viral glycoprotein antigens. Elife. 9, 1–30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akkaya, M., Kwak, K. & Pierce, S. K. B cell memory: building two walls of protection against pathogens. Nat. Rev. Immunol. vol. 20 229–238 Preprint at (2020). 10.1038/s41577-019-0244-2 [DOI] [PMC free article] [PubMed]
- 52.Gobeil, S. M. C. et al. D614G mutation alters SARS-CoV-2 Spike conformation and enhances protease cleavage at the S1/S2 Junction. Cell. Rep. 34 (2), 108630 (2021). [DOI] [PMC free article] [PubMed]
- 53.Costello, S. M. et al. The SARS-CoV-2 spike reversibly samples an open-trimer conformation exposing novel epitopes. Nat. Struct. Mol. Biol. 29, 229–238 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanekiyo, M. et al. Rational design of an Epstein-Barr Virus Vaccine Targeting the receptor-binding site. Cell. 162, 1090–1100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Amanat, F. et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 26, 1033–1036 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mozafari, M. R., Mazaheri, E. & Dormiani, K. Simple equations pertaining to the particle number and surface area of metallic, polymeric, lipidic and vesicular nanocarriers. Sci. Pharm. 89 (2), 15 (2021).
- 57.Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, K. & Gctf Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsuyama, S. et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2- expressing cells. Proc. Natl. Acad. Sci. U S A. 117, 7001–7003 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Requests for information or raw data should be directed to the corresponding author.