Abstract
In two cases of parenteral transmission of human immunodeficiency virus type 1 (HIV-1) syncitium-inducing (SI) variants, we previously observed selection for macrophagetropic variants. Although infection of macrophages is generally mediated via CCR5, we found no selection for SI variants that could use CCR5 as coreceptor in addition to CXCR4, suggesting that features other than coreceptor usage account for the macrophagetropism of these transmitted SI HIV-1 variants.
Human immunodeficiency virus type 1 (HIV-1) isolates can display differences in biological properties, such as replication rate, syncytium-inducing (SI) capacity, and cytotropism (2, 6, 27, 29). In newly infected individuals, generally only non-syncytium-inducing (NSI) HIV-1 variants are present (27). These NSI variants persist during all stages of infection, while T-cell-line-tropic SI variants appear in the course of infection in about 50% of infected individuals (16, 29). The predominance of NSI HIV-1 variants in early stages of infection has been attributed to their macrophagetropism (38). Since mainly macrophagetropic HIV variants can be detected during primary infection, macrophages have long been considered to be the port of entry during virus transmission (30). Recently, however, in situ hybridization has identified CD4+ T cells as the only HIV-infected cells during primary infection (25, 37). How this fits with the tropism of early isolates remains to be established. The capacity to replicate in macrophages is generally determined by the capacity to use β-chemokine receptor CCR5 as coreceptor to infect CD4-positive cells (1, 10, 12, 19, 35). This CCR5 coreceptor usage is a characteristic of NSI HIV-1 variants, whereas SI HIV variants alternatively or additionally use the α-chemokine receptor CXCR4 as coreceptor (11, 13).
As might be expected since CCR5-restricted NSI variants generally initiate HIV-1 infection, individuals homozygous for a 32-bp deletion in the CCR5 gene (CCR5 Δ32/Δ32) are highly resistant to HIV-1 infection (7, 15, 20, 24). This is in concordance with the observation that exposed but uninfected individuals with a CCR5 wild-type (WT/WT) genotype have a low level of CCR5 expression and a high level of β-chemokine production (23). However, SI, CXCR4-using HIV-1 variants can be transmitted (32), as was also demonstrated by the rare cases of HIV-1-infected individuals with a CCR5 Δ32/Δ32 genotype (3, 4, 21).
Previously, we reported the transmission of SI HIV-1 variants in two parenteral transmission cases. In one case, a male recipient (Ams127) had been accidentally injected with a minute amount of blood from an HIV-1-infected male (ACH704) suffering from wasting syndrome CDC IVa (18). In the other case, a female recipient (ACH9012) was deliberately injected with a few milliliters of blood from an AIDS patient (Ams199) (31). The transmitted SI variants were highly macrophagetropic, more so than SI variants isolated from the donors (ACH704 and Ams199) at the time of transmission (30).
Since macrophagetropism is a feature generally attributed to CCR5-using NSI HIV-1 variants, we hypothesized that this selection for macrophagetropism of transmitted SI variants might be associated with a selection for SI variants that use CCR5 in addition to CXCR4. Therefore, we analyzed the coreceptor usage of biological HIV-1 clones obtained from the virus donors around the moment of transmission and from the recipients at one or two time points after seroconversion (27). These biological virus clones were previously obtained by coculture of cryopreserved patient peripheral blood mononuclear cells (PBMCs) with phytohemagglutinin (PHA)-stimulated healthy-donor PBMCs as previously described (27). Three clones from recipient Ams127 were isolated on monocyte-derived macrophages (MDM) (27). The envelope variable region 3 (V3) sequence of the biological virus clones was previously determined (30) (Table 1).
TABLE 1.
Genotypes and phenotypes of biological HIV-1 clones obtained from two parenteral donor-recipient pairsa
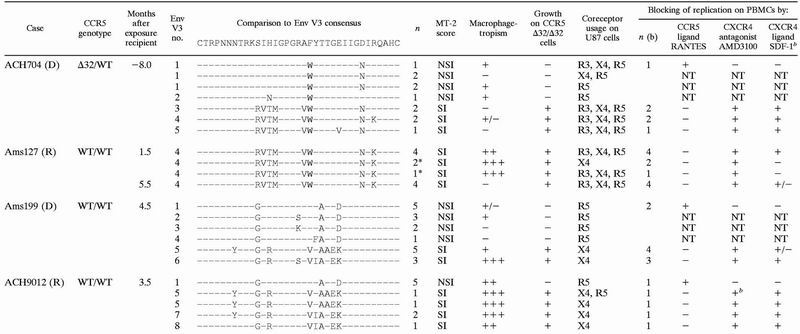 |
D, donor; R, recipient; Env V3 no., envelope V3 sequence number; n, number of clones with the designated genotype and phenotype; ∗, clones isolated on MDM. Macrophagetropism of the clones was previously determined by infection of MDM obtained from two different blood donors for each of the biological clones (see references 27 and 30) and is depicted as the percentage of infected MDM cultures (−, 0%; +/−, 1 to 25%; +, 26 to 50%; ++, 51 to 75%; +++, 76 to 100%). R3, CCR3; X4, CXCR4; R5, CCR5. Blocking of replication of the biological clones was determined by infection of PHA-stimulated PBMCs in the absence or presence of RANTES, AMD3100, or SDF-1α. n (b), number of clones with the designated genotype and phenotype tested for blocking of replication on PBMCs; +, replication was blocked; −, replication was not blocked; +/−, replication of half of these clones was blocked. NT, not tested.
Residual p24 production was observed for one biological virus clone with AMD3100 and for all biological virus clones inhibited by SDF-1α.
Biological virus clones were analyzed for coreceptor usage by using U87 astroglioma cells stably transfected with CD4 and coreceptor CCR3, CCR5, or CXCR4. Virus stocks were prepared on PHA-stimulated PBMCs. The U87 cells were inoculated with at least 100 50% tissue culture infective doses (TCID50) of the different biological HIV-1 clones. The emergence of HIV p24 antigen in the culture supernatant was indicative of productive infection and reflected the capacity of the biological HIV-1 clone to use the coreceptor that was expressed by the inoculated cells. The expression of CD4 and additional HIV-1 coreceptors by the cells used was routinely confirmed by fluorescence-activated cell sorter (FACS) analysis (data not shown). Furthermore, replication in PHA-stimulated PBMCs from a healthy donor homozygous for the 32-bp deletion in CCR5 was determined to investigate if the different biological virus clones were dependent on CCR5 expression for replication in primary cells (Table 1).
A total of 11 biological virus clones were available from a blood sample obtained from donor ACH704 8 months before the moment of transmission. Six of these biological virus clones had the NSI phenotype and five had the SI phenotype, as determined with the MT-2 cell line (16) (Table 1). The majority of the NSI clones was able to infect MDM, whereas only one of the SI variants (no. 4) from donor ACH704 was macrophagetropic (30) (Table 1). Interestingly, all biological virus clones isolated from recipient Ams127 had the SI phenotype and the same envelope V3 sequence as the macrophagetropic SI variant (no. 4) from donor ACH704, possibly due to the low inoculum concentration. The biological virus clones that were obtained early after seroconversion (n = 7) were all macrophagetropic, whereas the clones obtained 5.5 months after seroconversion (n = 4) lacked the capacity to replicate in macrophages. As concluded previously (30), this implies that sequence changes in the genomes of the different biological HIV-1 clones outside the V3 loop region may account for the difference in phenotypic properties of the clones.
Almost all biological virus clones from ACH704 and Ams127 were able to use CCR5 as a coreceptor on the U87 cells (Table 1). The majority of SI clones additionally used CCR3 and CXCR4 when expressed on U87 CD4-positive cells. However, the two highly macrophagetropic SI clones from recipient Ams127 isolated on MDM 1.5 months after exposure were restricted to CXCR4 usage on the U87 cells.
Surprisingly, we found that three of the six NSI biological virus clones from donor ACH704 replicated in the CXCR4-expressing U87 cells and one even replicated in the CCR3-expressing cells (Table 1). Sequence analysis following infection of the CXCR4-expressing U87 cells confirmed the NSI type envelope V3 sequences (14) found previously (30) (data not shown), excluding contamination with other HIV-1 clones. However, the observation that these NSI virus clones did not replicate in PHA-stimulated PBMCs from a healthy CCR5 Δ32/Δ32 donor indicated their dependence on CCR5 expression for replication in primary cells, irrespective of their coreceptor repertoire in U87 cells.
In the other transmission case, both NSI and SI HIV-1 variants of donor Ams199 were transmitted to the recipient (ACH9012). We previously described a majority of highly macrophagetropic HIV-1 clones in the recipient and selective expansion of NSI variants (30). All NSI clones from donor Ams199 (n = 10) and recipient ACH9012 (n = 5) tested in this study were CCR5 restricted in the U87 astroglioma cells. The five donor-derived SI clones with V3 sequence no. 5 were CXCR4 restricted, whereas one of the two SI clones (no. 5) from the recipient could use both CXCR4 and CCR5. All SI variants of both donors and recipients could replicate in PHA-PBMCs with a CCR5 Δ32/Δ32 genotype (Table 1), indicating that these clones can use coreceptors other than CCR5, also on primary cells. None of the NSI variants could replicate in these CCR5-lacking PBMCs (Table 1), indicating that the NSI variants are indeed dependent on CCR5 expression for replication in primary cells.
Since some highly macrophagetropic SI variants from both recipients were found to be CXCR4 restricted on the U87 cells, we wanted to test whether these transmitted SI variants were also CXCR4 restricted on primary cells. Therefore, we cultured the biological virus clones on PHA-stimulated PBMCs in the presence of the CXCR4 antagonist AMD3100 (26) or CXCR4 ligand SDF-1α (5). In addition, RANTES was used to investigate if replication of the macrophagetropic SI variants would be inhibited by blocking CCR5. A mixture of PBMCs from seven healthy donors with a homozygous CCR5 WT genotype was prepared, cryopreserved, and used for all inhibition studies. PHA-PBMCs (106 cells/ml) were incubated for 3 h in the presence or absence of inhibitory concentrations of RANTES (1.25 μg/ml; National Institutes of Health AIDS reagents program), AMD3100 (1 μg/ml; synthesized by G. Bridger, AnorMed, Langley, Canada), or SDF-1α (2.5 μg/ml; Stratmann Biotech, Hannover, Germany). Then, 105 cells were inoculated with at least 50 TCID50 of the different biological HIV-1 clones. After overnight inoculation, the cells were washed once and fresh medium with or without RANTES, AMD3100, or SDF-1α was added. The emergence of HIV p24 antigen in the culture supernatant, harvested 7, 11, and 14 days after inoculation, was indicative of productive infection. In the absence of any blocking agent, all biological virus clones produced high levels of p24 antigen on PHA-PBMCs. The absence of p24 production was taken to be indicative of complete inhibition of replication. All measurements were performed in triplicate.
All NSI variants tested (n = 4) were completely inhibited by RANTES irrespective of coreceptor usage on the U87 cells, whereas addition of AMD3100 and SDF-1α had no effect on in vitro replication of these variants (Table 1). This indicated that these NSI variants are indeed dependent on CCR5 usage for replication in PBMCs, which is in agreement with their inability to replicate in CCR5 Δ32/Δ32 PBMCs.
The replication of all SI variants tested (n = 28) was blocked by AMD3100, except for the SI variant isolated from recipient ACH9012 that could use both CXCR4 and CCR5 on U87 cells. Some residual p24 production was observed when cells were inoculated with this variant in the presence of AMD3100, suggesting that it indeed has the ability to use CCR5 or another coreceptor apart from CXCR4 on PBMCs. Nevertheless, RANTES inhibited the p24 production of none of the SI HIV-1 variants (Table 1).
SDF-1α was not as efficient as AMD3100 in the blocking of CXCR4 usage. Replication of most of the SI variants was inhibited by SDF-1α, but residual p24 production could always be observed (Table 1).
These results indicate that, except for the dualtropic SI variant isolated from recipient ACH9012, all SI variants are dependent on CXCR4 usage on PBMCs irrespective of their coreceptor usage repertoire on the U87 cell line, since their replication is totally inhibited by addition of AMD3100.
Taken together, all our results indicate that the observed selection for macrophagetropism after transmission cannot be explained by selection for CCR5 coreceptor usage.
CCR5 expression may influence inter- and intrapatient selection of coreceptor usage of HIV-1 variants. The homozygous genotype of Ams127, the 32-bp deletion of CCR5, could have explained the absence of NSI variants in this recipient. Therefore, the CCR5 genes of all four individuals were analyzed for the 32-bp deletion by PCR analysis, as described previously (9). Donor ACH704 was found to be heterozygous for the 32-bp deletion in the CCR5 gene (Δ32/WT), and the other three were homozygous for the wild-type gene (WT/WT) (Table 1).
As coreceptor expression may be influenced by other polymorphisms, we additionally performed four-color flow cytometry on cryopreserved, unstimulated PBMCs from the recipients that had been collected around the time of transmission as well as from two HIV-negative, healthy donors. Staining was performed for 20 min at 4°C. For the analysis of coreceptor expression on lymphocytes, we used a combination of CD4 (-PerCP; Becton Dickinson [BD], San Jose, Calif.), CD45RO (-allophycocyanin; BD), CCR5 (2D7-fluorescein isothiocyanate; PharMingen, San Diego, Calif.), and CXCR4 (12G5-phycoerythrin; PharMingen). For the analysis of coreceptor expression on monocytes, we used a combination of CD4 (-PerCP; BD), CD14 (-allophycocyanin; Caltag Laboratories, Burlingame, Calif.), CCR5 (2D7-fluorescein isothiocyanate; PharMingen), and CXCR4 (12G5-phycoerythrin; PharMingen). Analysis was performed on a FACScalibur (BD).
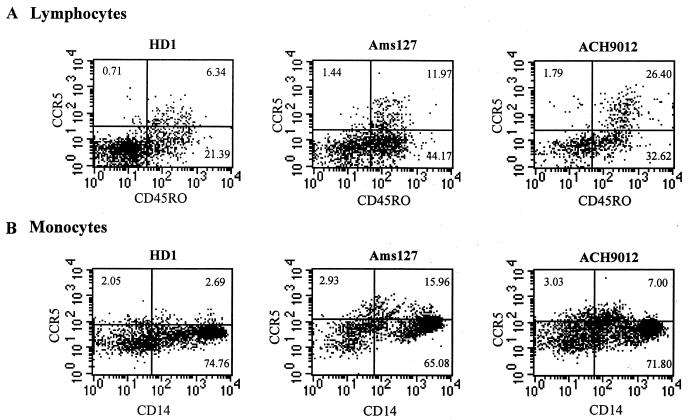
We observed high expression levels of CCR5 on CD4-positive lymphocytes isolated 1 day (Ams127) and 4 months (ACH9012) after the moment of transmission compared to the CCR5 expression level on cryopreserved PBMCs from an uninfected, healthy donor (Fig. 1a). CCR5 was mostly expressed on CD45RO+ (“memory”) lymphocytes. This high level of expression most likely reflects the activation of the immune system as a consequence of HIV-1 infection (8, 22). No difference was observed for the expression levels of CXCR4 in comparison to those in cells from healthy donors (data not shown). The level of coreceptor expression on the recipient's monocytes/macrophages might be particularly relevant with respect to transmission. We observed high expression levels of CCR5 on unstimulated, CD14+ monocytes isolated 1 day (Ams127) and four and a half months (ACH9012) after transmission (Fig. 1b). No difference was observed for the expression levels of CXCR4 on the monocytes in comparison to that of cells from healthy donors (data not shown). Therefore, the transmission of CXCR4 using SI HIV-1 variants cannot be explained by a low level of expression of CCR5 or a high level of expression of CXCR4 in the recipients.
FIG. 1.
(A) FACS analysis of CD45RO and CCR5 expression on unstimulated, cryopreserved CD4-positive lymphocytes from a healthy donor (HD1) and two HIV-1 recipients, Ams127 and ACH9012. PBMC samples were obtained 1 day (Ams127) and 4 months (ACH9012) after the moment of transmission. In three quadrants of each plot, the percentage of total CD4-positive lymphocytes is depicted. (B) FACS analysis of CD14 and CCR5 expression on unstimulated, cryopreserved monocytes from a healthy donor (HD1) and HIV-1 recipients Ams127 and ACH9012. PBMC samples were obtained 1 day (Ams127) and four and a half months (ACH9012) after transmission. In three quadrants of each plot, the percentage of total monocytes is depicted. The quadrants are set according to the immunoglobulin G isotype control for each blood donor in each experiment.
In conclusion, in contradiction with our hypothesis, our results imply that the selection for macrophagetropic SI HIV-1 variants during transmission is not associated with a selection for CCR5 usage in addition to the CXCR4 usage of these SI variants on primary cells. On the contrary, in both recipients we found CXCR4-restricted SI clones that were able to efficiently infect macrophages, which confirms the finding that infection of macrophages can be mediated via coreceptors other than CCR5 (28, 34). These results suggest that features other than coreceptor usage must account for the macrophagetropism of the transmitted SI variants investigated in this study.
As both recipients had a wild-type CCR5 genotype and high expression levels of this β-chemokine receptor on CD4-positive lymphocytes and monocytes, a lack of CCR5 expression could not explain the transmission and persistence of CXCR4-restricted SI variants in these two cases.
The efficiency of transmission of different HIV-1 variants may depend on the route of transmission. This has also been indicated by the controversial results on possible protection of the CCR5 Δ32/Δ32 genotype against parenteral transmission of HIV-1 (17, 33, 36). Whether or not vertical or sexual transmission is established only by CCR5 using viruses remains to be established.
Acknowledgments
We thank Dan Littman for the U87 cell lines, G. Bridger for providing AMD3100, Ronald van Rij for helpful discussions, Jos Dekker for technical assistance, and Wendy van Noppen for critically reading the manuscript. RANTES was obtained through the AIDS Research and Reference Reagent Program, NIH (contributed by PreproTech, Inc.).
This study was performed as part of the Amsterdam Cohort Studies on HIV infection and AIDS, a collaboration between the Municipal Health Service, the Academic Medical Center, and the CLB, Amsterdam, The Netherlands, and was financially supported by The Netherlands Ministry of Public Health, by The Netherlands Foundation for Preventive Medicine (grant no. 1305), and by The Netherlands Organization for Scientific Research (NWO), project number 901-02-222.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Asjo B, Albert J, Karlsson A, Morfeldt-Månson L, Biberfeld G, Lidman K, Fenyö E M. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet. 1986;ii:660–662. [PubMed] [Google Scholar]
- 3.Balotta C, Bagnarelli P, Violin M, Ridolfo A L, Zhou D, Berlusconi A, Corvasce S, Corbellino M, Clementi M, Clerici M, Moroni M, Galli M. Homozygous Δ32 deletion of the CCR5 chemokine receptor gene in an HIV-1 infected patient. AIDS. 1997;11:F67–F71. doi: 10.1097/00002030-199710000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Biti R, French R, Young J, Bennetts B, Stewart G, Liang T. HIV-1 infection in an individual homozygous for the CCR5 deletion allele. Nat Med. 1997;3:252–253. doi: 10.1038/nm0397-252. [DOI] [PubMed] [Google Scholar]
- 5.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–832. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 6.Cheng-Mayer C, Seto D, Tateno M, Levy J A. Biologic features of HIV-1 that correlate with virulence in the host. Science. 1988;240:80–82. doi: 10.1126/science.2832945. [DOI] [PubMed] [Google Scholar]
- 7.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O'Brien S J Hemophilia Growth and Development Study; Multicenter AIDS Cohort Study; Multicenter Hemophilia Cohort Study; San Francisco City Cohort; ALIVE study. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 8.De Roda Husman A M, Blaak H, Brouwer M, Schuitemaker H. CCR5 cell-surface expression in relation to CCR5 genotype and the clinical course of HIV-1 infection. J Immunol. 1999;163:4597–4603. [PubMed] [Google Scholar]
- 9.De Roda Husman A M, Koot M, Cornelissen M, Brouwer M, Broersen S M, Bakker M, Roos M T L, Prins M, De Wolf F, Coutinho R A, Miedema F, Goudsmit J, Schuitemaker H. Association between CCR5 genotype and the clinical course of HIV-1 infection. Ann Intern Med. 1997;127:882–890. doi: 10.7326/0003-4819-127-10-199711150-00004. [DOI] [PubMed] [Google Scholar]
- 10.Deng H K, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Suttons R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of the major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 11.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3 and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 12.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 13.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 14.Fouchier R A M, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau N R, Phair J, Ho D D, Koup R A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 16.Koot M, Keet I P M, Vos A H V, De Goede R E Y, Roos M T L, Coutinho R A, Miedema F, Schellekens P T A, Tersmette M. Prognostic value of human immunodeficiency virus type 1 biological phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 17.Kupfer B, Kaiser R, Brackmann H-H, Effenberger W, Rockstroh J K, Matz B, Schneweis K E. Protection against parenteral HIV-1 infection by homozygous deletion in the C-C chemokine receptor 5 gene. AIDS. 1999;13:1025–1028. doi: 10.1097/00002030-199906180-00004. [DOI] [PubMed] [Google Scholar]
- 18.Lange J M A, Boucher C A B, Hollak C E M, Wiltink E H H, Reiss P, Van Royen E A, Roos M T L, Danner S A, Goudsmit J. Failure of zidovudine prophylaxis after accidental exposure to HIV-1. N Engl J Med. 1990;322:1375–1377. doi: 10.1056/NEJM199005103221907. [DOI] [PubMed] [Google Scholar]
- 19.Lee B, Doranz B J, Rana S, Yi Y, Mellado M, Frade J M R, Martinez-A C, O'Brien S J, Dean M, Collman R G, Doms R W. Influence of the CCR2–V64I polymorphism on human immunodeficiency virus type 1 coreceptor activity and on chemokine receptor function of CCR2b, CCR3, CCR5 and CXCR4. J Virol. 1998;72:7450–7458. doi: 10.1128/jvi.72.9.7450-7458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lui R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiple exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 21.Michael N L, Nelson J A E, KewalRamani V N, Chang G, O'Brien S J, Mascola J R, Volsky B, Louder M, White II G C, Littman D R, Swanstrom R, O'Brien T R. Exclusive and persistent use of the entry coreceptor CXCR4 by human immunodeficiency virus type 1 from a subject homozygous for CCR5 Δ32. J Virol. 1998;72:6040–6047. doi: 10.1128/jvi.72.7.6040-6047.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ostrowski M A, Justement S J, Cantanzaro A, Hallahan C A, Ehler L A, Mizell S B, Kumar P N, Mican J, Chun T-W, Fauci A S. Expression of chemokine receptors CXCR4 and CCR5 in HIV-1-infected and uninfected individuals. J Immunol. 1998;161:3195–3201. [PubMed] [Google Scholar]
- 23.Paxton W A, Liu R, Kang S, Wu L, Gingeras T, Landau N R, Mackay C R, Koup R A. Reduced HIV-1 infectability of CD4+ lymphocytes from exposed-uninfected individuals: association with low expression of CCR5 and high production of β-chemokines. Virology. 1998;244:66–73. doi: 10.1006/viro.1998.9082. [DOI] [PubMed] [Google Scholar]
- 24.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C-M, Saragosti S, Lapouméroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 25.Schacker T, Little S, Connick E, Gebhard K, Zhang Z Q, Krieger J, Pryor J, Havlir D, Wong J K, Schooley R T, Richman D, Corey L, Haase A T. Productive infection of T cells in lymphoid tissues during primary and early human immunodeficiency virus infection. J Infect Dis. 2001;183:555–562. doi: 10.1086/318524. [DOI] [PubMed] [Google Scholar]
- 26.Schols D, Struyf S, Van Damme J, Este J A, Henson G, De Clercq E. Inhibition of T-tropic HIV strains by selective antagonization of the chemokine receptor CXCR4. J Exp Med. 1997;186:1383–1388. doi: 10.1084/jem.186.8.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, De Goede R E Y, Van Steenwijk R P, Lange J M A, Eeftink Schattenkerk J K M, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simmons G, Reeves J D, McKnight A, Dejucq N, Hibbits S, Power C A, Aarons E, Schols D, De Clercq E, Proudfoot A E I, Clapham P R. CXCR4 as a functional coreceptor for human immunodeficiency virus type 1 infection of primary macrophages. J Virol. 1998;72:8453–8457. doi: 10.1128/jvi.72.10.8453-8457.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tersmette M, Gruters R A, De Wolf F, De Goede R E Y, Lange J M A, Schellekens P T A, Goudsmit J, Huisman J G, Miedema F. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van 't A. B. Wout, Kootstra N A, Mulder-Kampinga G A, Albrecht-van Lent N, Scherpbier H J, Veenstra J, Boer K, Coutinho R A, Miedema F, Schuitemaker H. Macrophage-tropic variants initiate human immunodeficiency virus type1. infection after sexual, parenteral and vertical transmission. J Clin Investig. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veenstra J, Schuurman R, Cornelissen M, Van 't Wout A B, Boucher C A B, Schuitemaker H, Goudsmit J, Coutinho R A. Transmission of zidovudine-resistant human immunodeficiency virus type 1 variants following deliberate injection with blood of a patient with AIDS: characteristics and natural history of the virus. Clin Infect Dis. 1995;21:556–560. doi: 10.1093/clinids/21.3.556. [DOI] [PubMed] [Google Scholar]
- 32.Weiss S H, Goedert J J, Gartner S, Popovic M, Waters D, Markham P, Di Marzo Veronese F, Gail M H, Barkley W E, Gibbons J, Gill F A, Leuther M, Shaw G M, Gallo R C, Blattner W A. Risk of human immunodeficiency virus (HIV-1) infection among laboratory workers. Science. 1988;239:68–71. doi: 10.1126/science.3336776. [DOI] [PubMed] [Google Scholar]
- 33.Wilkinson D A, Operskalski E A, Busch M, Mosley J W, Koup R A. A 32-bp deletion within the CCR5 locus protects against transmission of parenterally acquired human immunodeficiency virus but does not affect progression to AIDS defining illness. J Infect Dis. 1998;178:1163–1166. doi: 10.1086/515675. [DOI] [PubMed] [Google Scholar]
- 34.Yi Y, Rana S, Turner J D, Gaddis N, Collman R G. CXCR-4 is expressed by primary macrophages and supports CCR5-independent infection by dual-tropic but not T-tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:772–777. doi: 10.1128/jvi.72.1.772-777.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaitseva M, Blauvelt A, Lee S, Lapham C K, Klaus-Kovtun V, Mostowski H, Manischewitz J, Golding H. Expression and function of CCR5 and CXCR4 on human Langerhans cells and macrophages: implications for HIV primary infection. Nat Med. 1997;3:1369–1375. doi: 10.1038/nm1297-1369. [DOI] [PubMed] [Google Scholar]
- 36.Zamarchi R, Indraccolo S, Minuzzo S, Coppola V, Gringeri A, Santagostino E, Vicenzi E, De Silvestro G, Biagiotti R, Baldassarre C, Chieco-Bianchi L, Amadori A. Frequency of mutated CCR-5 allele (D32) among Italian healthy donors and individuals at risk of parenteral HIV infection. AIDS Res Hum Retrovir. 1999;15:337–344. doi: 10.1089/088922299311303. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Z Q, Schuler T, Zupancic M, Wietgrefe S, Staskus K A, Reimann K A, Reinhart T A, Rogan M, Cavert W, Miller C J, Veazey R S, Notermans D, Little S, Danner S A, Richman D D, Havlir D, Wong J, Jordan H L, Schacker T W, Racz P, Tenner-Racz K, Letvin N L, Wolinsky S, Haase A T. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science. 2001;286:1353–1357. doi: 10.1126/science.286.5443.1353. [DOI] [PubMed] [Google Scholar]
- 38.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]