Abstract
Chronic obstructive pulmonary disease (COPD) is an etiologically complex disease characterized by acute exacerbations and stable phases. We aimed to identify biological functions modulated in specific COPD conditions, using whole blood samples collected in the AERIS clinical study (NCT01360398). Considered conditions were exacerbation onset, severity of airway obstruction, and presence of respiratory pathogens in sputum samples. With an integrative multi-network gene community detection (MNGCD) approach, we analyzed expression profiles to identify communities of correlated genes. The approach combined different layers of gene interactions for each explored condition/subset of samples: gene expression similarity, protein-protein interactions, transcription factors, and microRNAs validated regulons. Heme metabolism, interferon-alpha, and interferon-gamma pathways were modulated in patients at both exacerbation and stable-state visits, but with the involvement of distinct sets of genes. An important gene community was enriched with G2M checkpoint, E2F targets, and mitotic spindle pathways during exacerbation. Targets of TAL1 regulator and hsa−let−7b − 5p microRNA were modulated with increasing severity of airway obstruction. Bacterial infections with Moraxella catarrhalis and, particularly, Haemophilus influenzae triggered a specific cellular and inflammatory response in acute exacerbations, indicating an active reaction of the host to infections. In conclusion, COPD is a complex multifactorial disease that requires in-depth investigations of its causes and features during its evolution and whole blood transcriptome profiling can contribute to capturing some relevant regulatory mechanisms associated with this disease. In this work, we explored multi-network modeling that integrated diverse layers of regulatory gene networks and enhanced our comprehension of the biological functions implicated in the COPD pathogenesis.
Subject terms: Diseases, Systems analysis
Introduction
Chronic obstructive pulmonary disease (COPD) is a complex disease characterized by inflammation of the airways and pulmonary parenchymal disruption, with progressive and irreversible airflow obstruction over time1. The most common respiratory symptoms are dyspnea, cough, and sputum production, and patients generally experience periods of stable chronic inflammation alternating with acute exacerbations of COPD (AECOPD) or transient periods of aggravated symptoms requiring additional medical treatment. The disease and, particularly, exacerbations can be triggered by different agents, including bacterial and viral infections or inhaled particles2,3. Current treatment relies on bronchodilators, corticosteroids, and, when necessary, oxygen and antibiotics1. Moreover, influenza and pneumococcal vaccinations reduce the risk of exacerbations4, supporting the development of new vaccines against other respiratory pathogens that may trigger AECOPD.
While the primary site of illness is the lung, with different levels of airway obstruction determining COPD severity1, additional systemic inflammation, metabolic alterations, and skeletal muscle dysfunction make COPD a systemic disease5–7. Consequently, various studies have used blood transcriptome profiling to retrieve information about host response and disease evolution, such as the attempt to identify systemic biomarkers for AECOPD via the evaluation of specific genes, pathways, and gene modules8, or genes and pathways linked to inflammation, the immune system, and ceramide metabolism9, but no single biomarker has gained wide acceptance10. A meta-analysis of lung tissue gene expression data from different studies, which used Gene Set Enrichment Analysis (GSEA) to rank genes according to their differential expression11, showed the same biological functions linked to COPD despite largely non-overlapping lists of differentially-expressed genes (DEGs)12. Subsequently, Morrow et al. showed that the expression of individual genes in whole blood was not significantly associated with the frequency of exacerbations but, with network modeling methods, a statistically significant community of co-expressed genes was identified related to exacerbation frequency13. Similarly, in a meta-analysis of two observational studies of patients with COPD, COPDGene14, and ECLIPSE15, Reinhold et al. identified communities of co-expressed genes in blood associated with airflow obstruction, emphysema, immune response, and with genes specific for natural killer cells, dendritic cells, and neutrophils16.
Overall, these studies suggest that the complexity of COPD is reflected on mRNA transcripts modulated in whole blood of patients in a non-trivial way, and not described by simple single-gene signatures. Pathways and gene modules appear to better capture the disease features, playing a more relevant role in the description of COPD evolution. Due to these observations and similarities with other complex diseases and systemic biology networks17–21, we explored whole blood transcriptomic data from COPD patients, using multi-omic integrative methods based on network modeling to identify gene communities that interact in a complex and articulated way, while removing apparent noisy signals22. Specifically, we followed the rationale used by Cantini and colleagues for the identification of the driver gene in different cancer types21 and used multilayer network modeling of the gene interactions to describe the signaling and regulatory network of the immune system at a whole blood level during the evolution of COPD. As shown by Cantini et al. 21 and other comprehensive studies23,24, the approach has the advantage to more realistically represent the interactions exhibited in complex regulatory processes, for example in the evolution of cancer or other systemic diseases, compared to the simple observation of gene expression similarities and the consequent identification of clusters of co-expressed genes. In fact, these regulatory pathways are controlled at transcriptional and post-transcriptional levels and their alteration often involves modified expression levels of genes that are at the same time partners in a protein-protein interaction25 and targeted by the same set of transcription factors26 and microRNAs27. These interactions were modeled in our multilayer network-based analysis, in which genes are considered interactors in a network subdivided in multiple layers, with each layer representing a specific kind of gene-gene interaction, such as protein-protein interactions, transcription factor co-targeting networks, and microRNA co-targeting networks (Fig. 1). These layers are defined a priori since they model biological processes and interactions collected from public reference databases. To this stack of layers, two further layers are added, representing the actual co-expression network derived from whole blood transcriptomic data of COPD patients in each specific explored ‘condition’ or subset of samples, thus modeling different multi-layer networks for each explored condition (Fig. 1a). This enables the consideration of two aspects of gene expression, gene expression profiles correlation (co-expression) and gene expression level similarity (radial basis function (RBF) expression similarity), with results that are relatively simple to interpret. Because of this, and because multilayer integrative techniques have recently showed very informative results, particularly in cancer research21,22,28–30, multilayer clustering was selected over other forms of data integration.
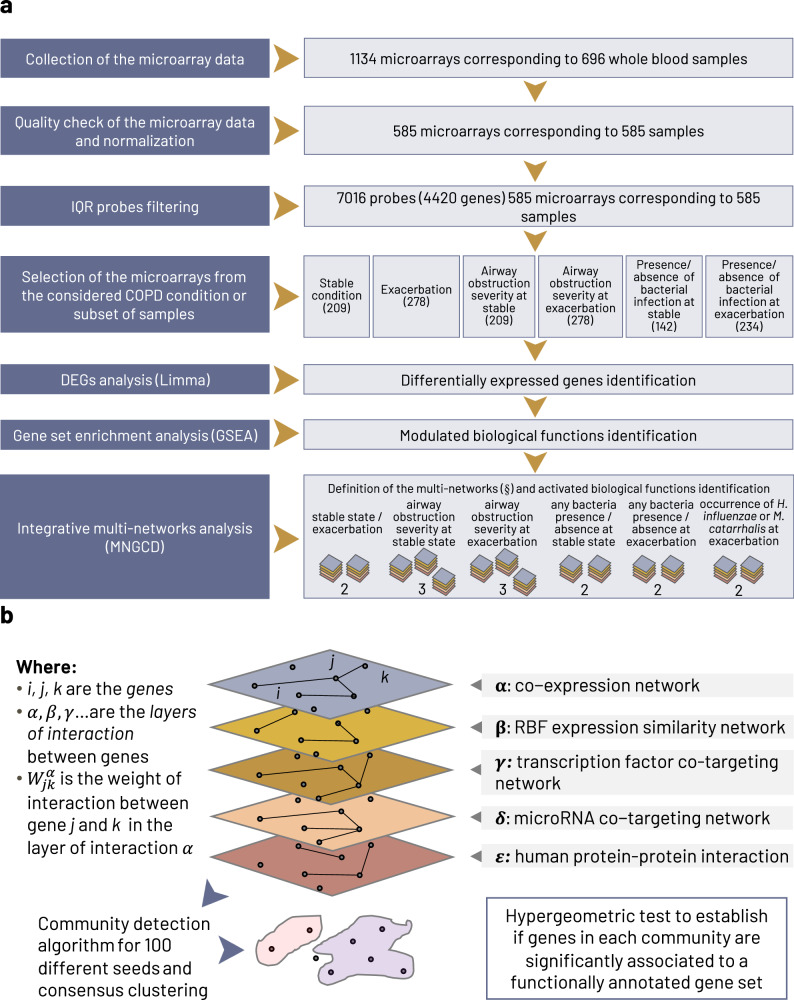
Fig. 1. Overview of the analysis pipeline.
a The analysis pipeline of the microarray measurements from blood samples obtained from 127 patients enrolled in the AERIS study. After quality control filtering, the final dataset was composed of 585 microarray experiments corresponding to 112 patients. b A detail of the integrative multi-network approach to identify driving COPD genes and their associated biological functions. (§) The 14 defined multi-networks are: stable state and exacerbation (2 multi-networks); severity of airway obstruction at stable state (3 multi-networks) and at exacerbation (3 multi-networks); presence or absence of any bacteria at stable state (2 multi-networks) and at exacerbation (2 multi-networks); alternate occurrence of H. influenzae and M. catarrhalis infections at exacerbation (2 multi-networks). The 14 multi-networks used to perform the six comparisons of interest are described in detail in the Methods. COPD chronic obstructive pulmonary disease, DEG differentially-expressed genes, GSEA gene set enrichment analysis, IQR interquartile range, MNGCD multi-network gene community detection, RBF radial basis function.
Finally, we aimed to analyze these multi-layer networks using multi-network gene community detection (MNGCD) method21, based on the partition of maps of information algorithm31, to identify subnetworks of strongly interacting genes (‘gene communities’), which were then characterized for enrichment with annotated biological functions or sets of regulons (Fig. 1b).
The blood sample and clinical data considered in this work were collected in the Acute Exacerbation and Respiratory InfectionS in COPD (AERIS) observational cohort study of 127 patients with COPD32,33. Patients were followed for up to 2 years via monthly study visits, when patients had stable COPD symptoms, and additional visits when patients experienced exacerbation events. A complete description of the COPD evolution, together with exacerbation frequency and severity, have been reported32; the mean exacerbation rate was 3.04 per patient-year and most exacerbations (85.6%) were moderate in severity. Results from the AERIS cohort revealed the most common bacterial species associated with AECOPD were Haemophilus influenzae (Hi) and Moraxella catarrhalis (Mcat), and the most common virus was rhinovirus, with a higher rate of bacterial and viral coinfection at exacerbation than stable state, and a dependence with seasonality32. Analysis of lung microbiome diversity in the same patients also revealed that stability over time was more likely to decrease at exacerbation and in individuals with higher exacerbation frequencies3. These observations led us to conduct further assessments to better understand the association of bacterial and viral infections and worsening of COPD symptoms at a systemic level, in particular, in relation to the whole blood transcriptome of patients with COPD.
Results
Differentially-expressed genes across different conditions
Gene expression data were analyzed using blood samples collected during the AERIS study every six months from stable-state visits and at each exacerbation visit. Quality check rejection of microarrays and interquartile range (IQR) probes filtering across the samples returned an overall dataset of 585 arrays (corresponding to 112 patients with COPD) and 7016 probes (corresponding to 4420 genes).
Differential expression analysis was performed across numerous contrasts to identify DEGs between stable and AECOPD conditions in samples overall and also considering specific subsets of samples according to type of infection (bacterial, viral, or eosinophilic) or to the bacterial species present in sputum (see Supplementary Information Section 4 for a detailed list of the contrasts considered). None of the analyses identified DEGs, with the exception of the following contrasts:
Comparison of exacerbation samples with Hi present and Mcat absent versus Hi-negative and Mcat-positive exacerbation samples;
Comparison of patients with Hi and Mcat co-infections with patients without Hi or Mcat infections.
Both comparisons were done by defining three different sets of samples on the basis of the absence of other bacterial and viral infections, i.e., sets with: (1) any other pathogen, any virus; (2) no other pathogen, any virus; (3) no other pathogen, no virus. The DEGs identified are listed in Supplementary Information Section 4; these lists were input in a specific GSEA for Hallmark molecular signatures database (MSigDB) functional annotations, as reported in Supplementary information section 5. These analyses highlighted the modulation of heme metabolism and mitotic spindle for the first set (samples with any other pathogen, any virus), where there were concurrent Hi and Mcat infections at the gene-wise level. Heme metabolism was also modulated when samples with alternate occurrences of Hi and Mcat were compared. These cross signals suggested an important link between the presence of Hi and Mcat and the modulation of genes involved in the metabolism of heme iron-porphyrin complex and erythroblast differentiation. The roles of the two infections were then dissected by the multinetwork analysis that considered Hi- and Mcat-positive samples independently.
Communities of interacting genes identified in stable-state COPD and at exacerbation
Functions modulated during stable COPD state and exacerbation events were examined using GSEA and integrative MNGCD, considering the dataset of 4420 gene transcripts passing IQR filtering across the whole dataset.
The enriched gene communities identified by MNGCD and the enriched gene sets identified by GSEA from the Hallmark MSigDB functional annotations are summarized in Table 1. MNGCD identified gene communities associated to eight biological processes specifically modulated at exacerbation and four modulated at stable state. Three relevant processes, heme metabolism, interferon-alpha, and interferon gamma responses, were commonly identified by GSEA and MNGCD.
Table 1.
Comparison of enriched pathways and biological processes (annotated by Hallmark molecular signatures database) between COPD stable-state and exacerbation conditions
| Pathways and biological processes (Hallmark MSigDB) | Hallmark MSigDB systematic name | MNGCD analysis | GSEA | |
|---|---|---|---|---|
| Exacerbation, average p value (278 samples) | Stable, average p value 209 samples) | Exacerbation–stable contrast, corrected p value | ||
| E2F_TARGETS | M5925 | 10−5 | NS | NS |
| ESTROGEN_RESPONSE_LATE | M5907 | 5•10−4 | NS | NS |
| G2M_CHECKPOINT | M5901 | 10−7 | NS | NS |
| KRAS_SIGNALING_DN | M5956 | 3•10−3 | NS | NS |
| MITOTIC_SPINDLE | M5893 | 10−3 | NS | NS |
| HEME_METABOLISM | M5945 | 6•10−3 | 5•10−4 | 3•10−3 |
| INTERFERON_ALPHA_RESPONSE | M5911 | 8•10−6 | 2•10−4 | 3•10−3 |
| INTERFERON_GAMMA_RESPONSE | M5913 | 7•10−8* | 10-4* | 3•10−3 |
| ALLOGRAFT_REJECTION | M5950 | NS | 6•10−3 | NS |
Results of the MNGCD analysis are reported separately for stable and exacerbation conditions, and as direct contrast for the GSEA analysis (significance p < 0.01 after Benjamini-Hochberg correction). In the MNGCD analysis, for multiple communities enriched with the same Hallmark term, the minimum p value for that repetition was taken and p values of the 20 iterations were then log-averaged (details in Supplementary Information Section 3).
COPD chronic obstructive pulmonary disease, GSEA gene set enrichment analysis, MNGCD multi-network gene community detection, MSigDB molecular signatures database, NS not significant.
*p value was significant in more than 80% of the iterations but floating.
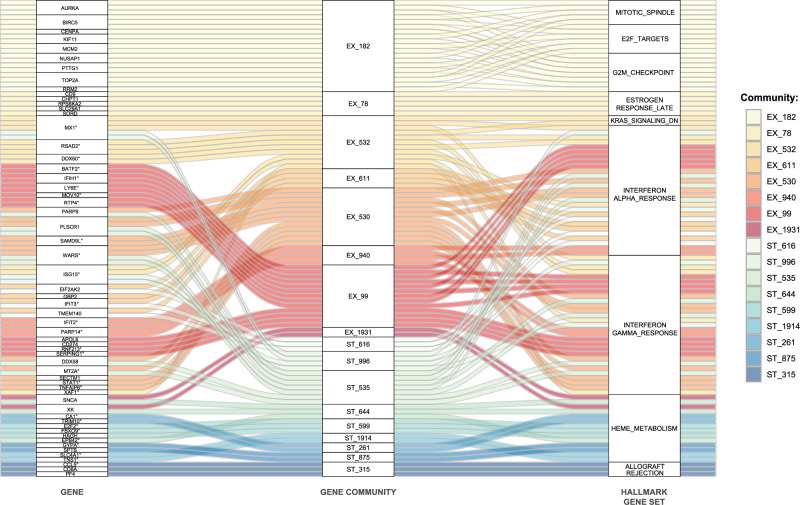
Genes identified by MNGCD in the 17 communities for exacerbation and stable conditions, together with enriched biological functions from the Hallmark collection, are shown as an alluvial plot in Fig. 2. The contents of the 17 gene communities are listed in Supplementary Information Section 6. G2M checkpoint, E2F targets, and mitotic spindle, which were enriched in the exacerbation dataset only, appeared in the same community of co-regulated genes. Estrogen response and KRAS signaling were co-regulated only in the exacerbation event, and involved a limited number of genes.
Fig. 2. Seventeen gene communities detected by multi-network gene community detection (MNGCD) in stable COPD and acute exacerbation visits from whole blood transcriptomic profiling.
The alluvial plot shows the relationships between gene communities detected by MNGCD (central column) and the genes belonging to these communities (left column) that contribute to significant enrichments of pathways and biological processes, as collected by Hallmark database (right column). The represented communities are those detected with a rate higher than 80% by iterating the analysis pipeline 20 times. Each color code is a different gene community (red/orange for exacerbation multiplex, blue/green for stable-state multiplex). ° Symbol in left column indicates genes in the core enrichment of gene set enrichment analysis (GSEA) with the Hallmark gene-set collection. COPD, chronic obstructive pulmonary disease.
hsa−let−7b − 5p targets, possibly in CD4+ T cells, might have a role in exacerbation
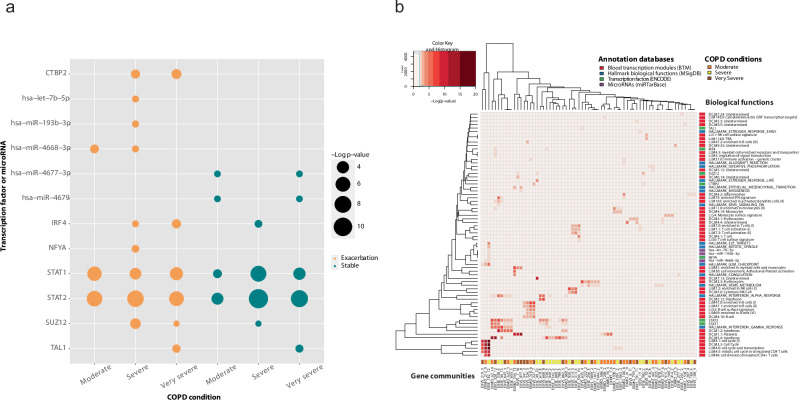
To further characterize stable-state and exacerbation conditions, we analyzed the identified communities with the enrichment of biological functions and processes annotated in three specific functional databases: the blood transcription modules (BTMs), the targets of transcription factors (ENCODE), and the targets of microRNAs (miRTarBase). As shown in Fig. 3a, the enrichment test for targets of transcription factors showed IRF4 was associated exclusively with one community in the stable condition (community id: ST_535), while STAT1 and STAT2 were significantly associated with gene communities for both the stable-state and exacerbation conditions. Moreover, STAT1 and STAT2 significantly targeted genes of the community EX_532 during exacerbation, also enriched with antiviral interferon signature and activated dendritic cells, which were not enriched in the stable-state multi-network.
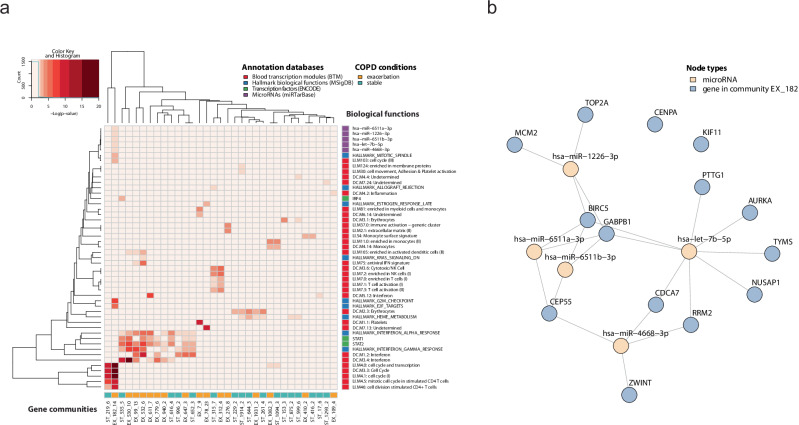
Fig. 3. Gene communities identified in COPD stable state and exacerbation involve multiple biological functions.
a A heatmap, where columns represent gene communities from stable visits and acute exacerbation events and the rows represent associated biological functions (annotated in blood transcription modules, microRNA targets, transcription factor targets, and Hallmark biological functions). Each cell of the heatmap is colored proportionally to the average Benjamini-Hochberg corrected log10 p value, testing the enrichment of the ith biological function in the jth gene community. b The microRNA regulatory network of the genes belonging to the community EX_182; in blue the genes of the community, yellow the significantly associated microRNAs. The links show regulatory interactions annotated in miRTarBase. COPD chronic obstructive pulmonary disease.
For microRNAs, there were no significant results in the stable state, while the exacerbation gene community, EX_182, was not only enriched with biological functions linked to cell cycle and mitotic cell cycle in stimulated CD4+ T cells, but also substantially over-represented targets of several microRNAs. Among these potential regulators, as shown in Fig. 3b, the most significant was hsa−let−7b − 5p; the Let-7 family is known to be associated with respiratory disease, including lung cancer and COPD34–38.
The targets of microRNA hsa−let−7b − 5p and of transcription factors NFYA, SUZ12, CTBP2, TAL1 modulate their expression with decreasing lung function
We examined if stable-state and exacerbation conditions were linked to a particular COPD severity stage and if limiting each analysis to a specific severity level could lead to the identification of other regulators, previously hidden in the stable-state and exacerbation groups overall.
For stable-state COPD in patients with moderate and very severe airway obstruction (Global Initiative for Chronic Obstructive Lung Disease (GOLD) grade classification, based on forced expiratory volume in 1 s (FEV1) test39), we identified targets of microRNAs hsa−miR−4677 − 3p and hsa−miR−4679, as shown in Fig. 4a. For exacerbations in patients with moderate COPD severity, we identified the enrichment of targets of hsa−miR−4668 − 3p, and for exacerbations in patients with severe COPD, we identified three microRNAs: hsa−miR−4668 − 3p, hsa−let−7b − 5p, and hsa−miR−193b − 3p (Fig. 4a).
Fig. 4. Gene communities identified in stable state and exacerbation with different levels of airway obstruction severity.
a The enriched transcription factors and microRNAs for each COPD severity grade in exacerbation and stable condition. b A heatmap, where columns represent genes communities from acute exacerbation events stratified by moderate (EXMO label), severe (EXSE), and very severe (EXVS) classification of the disease, and the rows represent associated biological functions (annotated in blood transcription modules, microRNA targets, transcription factor targets, and Hallmark biological functions). Each cell of the heatmap is colored proportionally to the average Benjamini-Hochberg corrected log10 p value, testing the enrichment of the ith biological function in the jth gene community. COPD, chronic obstructive pulmonary disease.
The targets of the microRNAs, hsa−miR−4668 − 3p and hsa−let−7b − 5p, were modulated significantly (p < 0.01, after Benjamini-Hochberg (BH) correction) in the pooled exacerbation condition and not in the stable state (Fig. 3a). Moreover, the targets of the microRNAs, hsa−miR−4668 − 3p and hsa−let−7b − 5p, were not modulated in stable state for each airway obstruction severity level (Fig. 4a), suggesting those microRNAs may play a role during exacerbation.
Focusing on the role of transcription factors, the targets of STAT1 and STAT2 were modulated for each airway obstruction severity level, in agreement with the results of the previous grouped analysis. The number of identified transcription factors that targeted the gene communities significantly increased with disease severity, both for stable state and exacerbation condition. The newly detected regulators were IRF4 (only for severe and very severe airway obstruction at exacerbation, and severe obstruction at stable state), NFYA (for severe airway obstruction at exacerbation), SUZ12 (for severe and very severe airway obstruction at exacerbation and severe at stable state), and CTBP2 (for severe and very severe at exacerbation). Moreover, the targets of the transcription factor TAL1 were modulated for very severe airway obstruction status, for both stable state and exacerbation.
As shown in Fig. 4b, the targets of IRF4, STAT2, and STAT1 were enriched in gene communities that were modulated by interferon. The gene community enriched with NFYA, the EXSE_182, was the same as the one enriched with targets of hsa−miR−4668 − 3p and hsa−let−7b − 5p, and was modulated with functions connected to cell cycle and mitosis in stimulated CD4+ T cells. SUZ12 was enriched together with a non-annotated BTM in the group with severe COPD at exacerbation. However, in the very severe condition, together with SUZ12, we identified the communities enriched in myeloid cells and monocytes, a non-annotated module and platelets. Concerning the transcription factor CTBP2, for the very severe condition at exacerbation we detected, together with its targets, the enrichment for B cell surface signature and B cells. Furthermore, for gene communities that were enriched with targets of TAL1, there was an enrichment with erythrocytes.
Bacterial infection in the lungs during AECOPD modulates specific functions in the blood transcriptome
After checking for differences between exacerbation and stable conditions, we examined the impact of bacterial infections and co-infections, as detected in sputum samples, on the blood transcriptome during those episodes.
As shown in Table 2, modulation of E2F targets, G2M checkpoints, mitotic spindle, estrogenic response, and KRAS signaling were found for exacerbation with bacterial infection only. The same functions were also enriched for the exacerbation event overall (Table 1). Moreover, as observed before, heme metabolism signal was activated in all groups. Interferon-alpha and interferon-gamma responses were activated in both bacteria-positive and -negative groups at exacerbation and stable state in the presence of bacterial infection only.
Table 2.
Comparison of enriched pathways and biological processes (annotated by Hallmark molecular signatures database) at COPD stable-state and exacerbation conditions in presence or absence of bacterial infection
| Pathways and biological processes (Hallmark MSigDB) | Hallmark MSigDB systematic name | Exacerbation, Bacteria-negative, average p value (88 samples) | Exacerbation, Bacteria-positive, average p value (146 samples) | Stable, Bacteria-negative, average p value (63 samples) | Stable, Bacteria-positive, average p value (79 samples) |
|---|---|---|---|---|---|
| E2F_TARGETS | M5925 | NS | 10-5 | NS | NS |
| ESTROGEN_RESPONSE_EARLY | M5906 | NS | 7•10-3 | NS | NS |
| ESTROGEN_RESPONSE_LATE | M5907 | NS | 6•10-4 | NS | NS |
| G2M_CHECKPOINT | M5901 | NS | 10-7 | NS | NS |
| HEME_METABOLISM | M5945 | 5•10-4 | 6•10-3 | 5•10-4 | 5•10-4 |
| INTERFERON_ALPHA_RESPONSE | M5911 | 2•10-6* | 10-6* | NS | 2•10-4* |
| INTERFERON_GAMMA_RESPONSE | M5913 | 5•10-10* | 8•10-7* | NS | 2•10-4* |
| KRAS_SIGNALING_DN | M5956 | NS | 4•10-3* | NS | NS |
| MITOTIC_SPINDLE | M5893 | NS | 10-3 | NS | NS |
| OXIDATIVE_PHOSPHORYLATION | M5936 | NS | 2•10-3 | NS | 2•10-3 |
Results of the multi-network gene community detection (MNGCD) analysis reported separately for stable-state and exacerbation conditions combined with presence (positive) and absence (negative) of bacterial infection. The p values of the hypergeometric enrichment are corrected by Benjamini-Hochberg method (significance p < 0.01). For multiple communities enriched with the same Hallmark term, the minimum p value for that repetition was taken and p values of the 20 iterations were then log-averaged (details in Supplementary information section 3).
COPD chronic obstructive pulmonary disease, MSigDB molecular signatures database, NS not significant.
*p value was significant in more than 80% of the iterations but floating.
Haemophilus influenzae in absence of Moraxella catarrhalis induces modulation of heme metabolism and interferon
To examine the specific response to two bacterial pathogens, Hi and Mcat, associated with exacerbation occurrences32, blood samples taken at exacerbation from patients with Hi-positive and Mcat-negative sputum samples (79 blood samples) were compared to those from patients with Mcat-positive and Hi-negative sputum samples (20 blood samples).
Several gene communities, which were enriched with biological functions that were detected at exacerbation but not in stable state, were also present in Hi-positive and Mcat-negative exacerbations. In particular, of the four gene communities that were enriched with terms present at exacerbation and not stable state, three were present with the same biological functions for Hi-positive and Mcat-negative exacerbations.
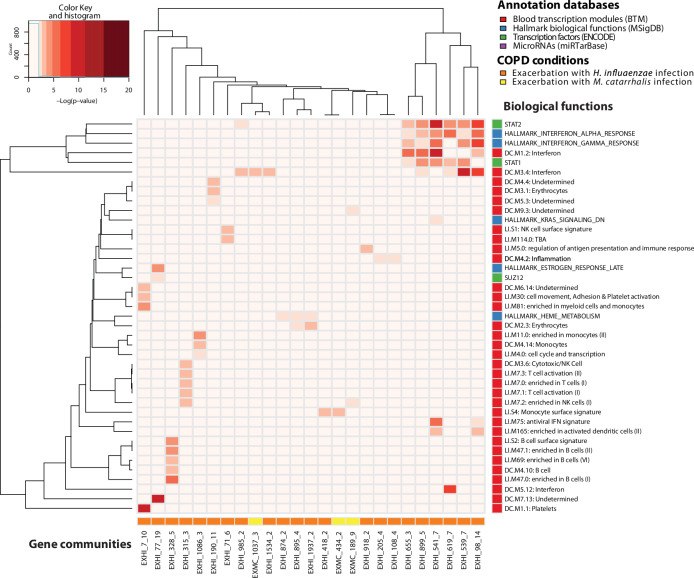
As shown in Fig. 5, EX_532 was enriched with the same biological functions as for gene community EXHI_541, which was also associated with antiviral interferon signature (LI.M75), activated dendritic cells (LI.M165), and interferon (DC.M1.2) from BTMs. For the gene set collection involving targets of transcription factors, both communities were over-represented by STAT1 and STAT2 targets. Also, the gene community EX_7, which was enriched for the exacerbation group with the terms platelets (DC.M1.1), myeloid cells, and monocytes (LI.M81), and one undetermined one (DC.M6.14), shared these biological functions with gene community EXHI_7, also enriched with cell movement, adhesion, and platelet activation (LI.M30). Moreover, EX_78, which was enriched in the exacerbation group with an undetermined biological function (DC.M7.13), had as parallel gene community, EXHI_77, enriched with the same term.
Fig. 5. Gene communities in H. influenzae and M. catarrhalis infections during exacerbation and their associated biological functions.
In this heatmap, columns represent gene communities from acute exacerbation events presenting H. influenzae (EXHI label) and M. catarrhalis (EXMC) infections, and the rows represent associated biological functions (annotated in blood transcription modules, microRNA targets, transcription factor targets and Hallmark biological functions). Each cell of the heatmap is colored proportionally to the average Benjamini-Hochberg corrected log10 p value, testing the enrichment of the ith biological function in the jth gene community. AECOPD acute exacerbations of COPD; COPD chronic obstructive pulmonary disease.
Biological functions modulated in exacerbation but not at stable state were not modulated in exacerbations with Mcat-positive and Hi-negative sputum samples. Furthermore, Hi-positive and Mcat-negative exacerbations showed modulation of heme metabolism, erythrocytes, and of targets of STAT1, STAT2, and SUZ12. All these enriched communities, except SUZ12, were enriched for each condition in the previous analyses, reflecting possibly the higher prevalence of Hi in sputum samples. Moreover, the GSEA on both the genes passing IQR filtering and on DEGs determined the enrichment of heme metabolism. With MNGCD, no significant result was obtained in terms of microRNAs enrichment when comparing the two types of infections.
Discussion
We aimed to characterize, through blood gene expression profiling, acute exacerbations, the severity of airway obstruction, and specific respiratory tract infections in COPD, to better understand the disease and possibly develop a signature for stratifying COPD patients taking part in future clinical trials. We analyzed blood gene expression data collected every six months from stable-state visits and at all exacerbation visits over a two-year period from 127 patients with COPD taking part in the AERIS study32,33. Although COPD is a mainly pulmonary disease, it is also associated with prolonged inflammation and exacerbations with systemic presentations. An impact on the transcriptomic modulations of whole blood cells was therefore expected. Despite this, our search for significant DEGs was, for the great majority of compared conditions, unsuccessful. This might be due to the heterogeneity that characterizes COPD40 and to the number of tested patients in our study, which may have been too low to overcome the false discovery rate control linked to fold-change in gene expression. Another reason could be that whole blood transcriptome results from the activity of many cell types, some in low abundance, and modulations of relevant genes of specific cell types might be diluted in the total peripheral blood transcripts. Also, our findings are consistent with other reports of difficulties in clearly identifying DEG signatures from whole blood transcripts profiling associated with other disease conditions, contributing to the recent shift in focus to identifying networks of interacting genes associated with particular COPD conditions13,16,40–44. Consequently, we used MNGCD, a multilayer gene community detection approach that integrates transcriptomic data with other kinds of omic information21, and compared results using a well-established functional analysis method, GSEA. This enabled us to describe the diverse regulatory gene communities and the associated functional modules in the different conditions of the study, rather than examining the dependence of the results on the different layers of interaction, which was beyond the scope of the analysis. Both approaches are based on the coherent (correlated or anticorrelated) behavior of genes participating in the same biological process, even if none is regulated in a statistically significant way. Moreover, this modeling technique has various advantages, including the simplicity to implement layers in a unique network framework without the need to treat different kinds of information with a different modeling formalism, and the scalability of network modeling algorithms for community detection of large numbers of samples and genes. Also, there was a need in this study to compare transcriptomes of tens of different samples groups/conditions and a method that allows the interchange of transcriptome layers, leaving the other layers unchanged (that were capturing other omics interactions), was particularly useful for the interpretation. The transcriptomic modulation was thereby captured in two layers (gene co-expression and RBF expression similarity), which were changed (or recomputed) every time the group of selected samples was changed to answer a given question. When this is applied to multiple multilayers, the result shows which gene communities have reorganized their expression across the different conditions. Another important advantage was the interpretability of the identified communities that allowed their functional role and the kind of interactions behind their structure to be identified. Finally, the network modeling formalism allowed us to consider two aspects of the gene expression, gene co-expression and gene RBF expression similarity, in a unique framework, using two layers based on two different metric definitions that captured the two aspects.
The first key comparison performed was identification of transcriptomic differences between patients with stable COPD and patients with acute exacerbations, where no significant DEGs could be identified. Through MNGCD and GSEA methods, we identified interferon-alpha and interferon-gamma responses and heme metabolism at both exacerbation and stable-state conditions. GSEA identified a statistically significant difference for these functions in the contrast between samples at stable visits and exacerbation. MNGCD revealed these functions were activated in both stable and exacerbation conditions, but involved a different, diverse set of genes, suggesting an important role for these functions in COPD. In particular, the gene community enriched for the interferon response showed a higher number of modulated genes at exacerbation compared to stable state. Conversely, heme metabolism was modulated in both groups but with a higher number of genes in the stable-state dataset, as identified by both MNGCD and GSEA. Moreover, these functions were identified in other specific COPD conditions suggesting a physiological circumstance for patients with COPD. Also, with MNGCD, we identified different hallmark biological processes specific for the stable-state or exacerbation condition, with a prevalence of inflammatory and cell-cycle related pathways (mitotic spindle, G2M checkpoint, and E2F targets) during exacerbations. With the MNGCD approach, it was possible to identify multiple biological functions associated with one gene community, suggesting a possible interplay of functions via the modulation of these genes.
With more specific functional annotations by transcriptional regulator targets and BTMs, an interesting gene community was identified during exacerbations that linked the microRNA hsa−let−7b − 5p targets, mitotic cell cycle of CD4+ T cells, and G2M checkpoint and E2F target genes. One in particular was the interferon response in exacerbation, linked to STAT1 and STAT2 and dendritic cells activation.
The analysis of more specific conditions related to patients with severe obstruction at exacerbation identified significant modulation of targets of hsa−let−7b − 5p microRNA. This finding is in agreement with the literature for another member of let-7 microRNA family targets, although that analysis was performed on sputum samples from patients with COPD45. The Let-7 family is known to be involved in cell development and differentiation and has relationships with oncogenesis46, T cell immunity47, and the development of lung cancer from COPD34. Moreover, we found several transcription factors, NFYA, SUZ12, CTBP2, and TAL1, modulated the expression of their targets with decreasing lung function in COPD patients, as assessed by FEV1 (GOLD grade). These were identified in addition to STAT1 and STAT2, which were detected in all multi-network analyses stratified by FEV1, were in the same communities that were enriched with interferon, and have already been associated with COPD in previous studies26,48,49. Of these transcription factors, TAL1 is of particular interest as it was identified in our study for very severe airway obstruction only. In both exacerbation and stable state, TAL1 was enriched together with heme metabolism and erythrocytes. TAL1 was previously identified as the top upstream regulator of peripheral blood mononuclear cell gene expression in asthma and was detected together with miR-486, which has been identified previously as a potential marker of childhood asthma in plasma50. In particular, our analysis relates the activation of a community of genes enriched with targets of hsa−let−7b − 5p microRNA in CD4 T cell activity in exacerbation with worsening conditions of the airways. This finding improves our knowledge of the activity of genes specifically associated with CD4+, CD8+ T cells in COPD patients with more severe airway obstruction, as observed in other studies41.
Finally, we analyzed whole blood gene expression samples from exacerbations that were positive for bacterial infection, comparing samples positive for Hi and negative for Mcat with those that were Mcat-positive and Hi-negative. GSEA functional analysis of DEGs in samples containing concurrent or alternate infections of Hi and Mcat identified a key set of modulated genes involved in heme metabolism and erythroblast differentiation. No microRNA targets were modulated in either condition, while targets of STAT2, STAT1, and SUZ12 were modulated only with Hi presence and Mcat absence. Targets of STAT2 and STAT1 were modulated in the same communities enriched with interferon. Moreover, biological functions connected to heme were enriched only for Hi-positive and Mcat-negative exacerbations, suggesting specific modulation of heme metabolism linked to Hi infection in COPD51. The similarity between the AECOPD condition overall and its subset of Hi-positive and Mcat-negative exacerbations might be due to the fact that, in the AERIS dataset, a large proportion of exacerbation sputum samples were positive for Hi32. Of 278 exacerbation samples, 107 contained Hi and 48 contained Mcat, with or without the presence of other pathogens. Alternatively, since heme metabolism was modulated in almost all analyzed COPD conditions, this might reflect an increase in oxygen carrying molecules, as COPD is linked to breathing and oxygenation issues, but also the inflammation status, which may be linked to variations in specific enzymes like heme oxygenase-1, known to play a role in the immune system and viral infection52.
This study was limited, as mentioned previously, by its sample size and also by the use of a whole blood readout to investigate the transcriptome changes in a chronic pulmonary disease. Additionally, since the AERIS dataset was derived from samples from COPD patients only, with no healthy controls, it cannot be confirmed if some biological functions identified in the analyzes were specifically related to COPD. Moreover, in principle, it would have been interesting to analyze many other conditions or sample subsets, but not enough samples were available from the AERIS study to conduct these investigations.
In conclusion, the shift in focus from classical detection of DEGs to the identification of modulated gene communities helped us to navigate the high complexity of COPD. This approach has enhanced our comprehension of the biological functions implicated in the pathogenesis of this disease. With an innovative integrative network modeling method for analyzing the transcriptome, we identified biological functions and regulators that have the potential to characterize particular COPD conditions, especially severe obstruction of the airway during exacerbation. It would be of interest to test if microRNAs of the let-7 family can be used for the classification of patients with severe disease or to further investigate molecular targets identified in different pathways (such as heme) in the discovery of new drugs and therapies. Finally, we observed that the presence of bacterial infections with Mcat and, in particular, Hi, triggered a specific cellular and inflammatory response in AECOPD, indicating an active reaction of the host to infections. This supports the development of a vaccine against these specific pathogens as a strategy for preventing exacerbations in COPD.
Methods
An overview of the procedures for analyzing gene expression in blood samples from the AERIS study (gene expression profiling, GSEA, and MNGCD) is shown in Fig. 1.
AERIS clinical study and ethical approval
The AERIS study was a prospective, observational cohort study based at University Hospital Southampton, UK (ClinicalTrials.gov: NCT01360398)32,33. In this study, 127 patients with moderate, severe, or very severe COPD aged 40–85 years were followed for up to 2 years, collecting various data, including FEV1 at monthly visits and at exacerbation to classify airway obstruction level according to GOLD grade39. Anonymized individual participant data and study documents can be requested for further research from www.clinicalstudydatarequest.com.
Sputum samples collected at monthly visits and at exacerbation were analyzed for the presence of bacterial pathogens (H. influenzae, M. catarrhalis, Streptococcus pneumoniae, Staphylococcus aureus, and Pseudomonas aeruginosa) by DNA PCR test. Blood for gene transcript profile analysis was collected every 6 months during the study and within 72 h of the onset of exacerbation, resulting in 723 samples with no duplicates. The exact criteria used for the inclusion of stable-state and exacerbation visit samples are described in Supplementary information section 1.
The AERIS study was conducted in accordance with the Declaration of Helsinki and Good Clinical Practice, and was approved by the Southampton and South West Hampshire Research Ethics Committee. All participants provided written informed consent.
RNA extraction, microarray hybridization, and data pre-processing
Total RNA was extracted from the 723 PaxGene tubes and purified, as previously described in refs. 53,54. RNA quality was assessed using a Bioanalyzer-2100 (Agilent Technologies, CA, USA); 27 samples did not meet the quality control (QC) criteria (RNA integrity number (RIN) > 6). Total RNA 50 ng was used for complementary DNA (cDNA) amplification, fragmentation, and labeling using the Ovation whole blood kit (NuGEN, CA, USA). Fragmented cDNA was hybridized using hgu133 Plus2.0 GeneChip (Affymetrix, CA, USA; 54675 probe-sets, including long non-coding RNA and microRNAs, as shown in several works55–58). Microarrays failing initial pre-defined QC criteria (glyceraldehyde 3-phosphate dehydrogenase ratio < 2.5 and scale factor < 8) were repeated in two follow-up batches. Therefore, 1134 microarrays were generated, corresponding to 696 whole blood samples with technical, but not biological, replicates. The screening of arrays by standard QC metrics was applied to reject slides not passing QC metrics thresholds (described in Supplementary Information Section 2). Raw data were then normalized with Robust Multi-array Average (RMA) with the help of probe sequence and with GC-content background correction method (GCRMA R package v2.42.0), log2-transformed and IQR-filtered, taking those probes with IQR > 0.75. We then averaged the expression of probe sets referring to the same gene.
Whole blood transcriptome analysis
The IQR-filtered and probe-averaged transcript profiles were the input for the three parallel analysis pipelines written in R, namely the computation of significant DEGs in specific COPD conditions (Limma R package v3.34), the GSEA11 (clusterProfiler R package v3.6) and the MNGCD pipeline21 (based on an ad hoc R script to wrap the Infomap software v0.x execution), as depicted in Fig. 1. In the MNGCD pipeline, we integrated the community detection by Infomap with a consensus robustness two-level step assessment by averaging the results of the stochastic algorithm across 100 iterations repeated 20 times. Details of the MNGCD pipeline are described in Supplementary information section 3.
The aim of the GSEA and MNGCD analyses was to identify biological functions that were modulated in certain COPD conditions/subsets of samples.
GSEA, which requires a contrast between two diverse conditions, was extensively applied in our analysis (as detailed in Supplementary Information Section 5). Here we report the results of the following comparisons of interest, that can also be compared to MNGCD results:
Exacerbation versus stable state;
Presence versus absence of any bacteria in sputum during exacerbation and at stable state;
Exacerbations with presence in sputum of Hi and absence of Mcat versus exacerbations with Mcat presence and Hi absence.
MNGCD was applied if a minimum of 20 samples per condition were available21. For this reason, we considered 14 COPD conditions/subsets with enough samples for six comparisons of interest defined as follows:
AECOPD: visits at (1) exacerbation and (2) stable state;
Severity of airway obstruction at exacerbation: (3) moderate, (4) severe, and (5) very severe, according to FEV1 GOLD grade;
Severity of airway obstruction at stable state: (6) moderate, (7) severe, and (8) very severe, according to FEV1 GOLD grade;
(9) Presence or (10) absence of any bacterial infection in sputum sample at exacerbation;
(11) Presence or (12) absence of any bacterial infection in sputum sample at stable state;
(13) Exacerbations with presence of Hi and absence of Mcat, or (14) exacerbations with presence of Mcat and absence of Hi.
Therefore, as shown in Fig. 1a, we built 14 multi-networks, one for each COPD condition/subset of samples. We integrated gene expression data from the AERIS study for each selected condition with different layers of omics information, embodying experimentally validated interactions, as described in detail in Supplementary Information Section 3. In brief, the network layers in each multi-network were:
A gene co-expression network and an RBF expression similarity network, both based on the gene expression microarray data of this study;
A transcription factor co-targeting network, from ENCODE experimentally validated interactions59;
A microRNA co-targeting network from experimentally validated interactions by different assays and collected in miRTarBase database release 7.060;
A binary human protein-protein interaction network based only on experimentally validated interactions taken from the public database APID (Agile Proteins Interactomes DataServer)61.
Once the COPD conditions to be considered were defined and the corresponding samples were identified, only the first two network layers (gene co-expression network and RBF expression similarity network) captured information about gene expression from the AERIS study samples. The remaining three layers were defined a priori and not dependent on the considered samples/condition, acting as a filter for the interactions in the expression networks by excluding mere correlations21.
The identified communities can group together genes cooperating in relevant biological pathways/functions for the analyzed COPD condition. Therefore, we used the hypergeometric test (p < 0.01 after BH correction) to assess the over-representation in each community of biological functions defined by the Hallmark biological functions gene-set collection (MSigDB)62, the BTMs63,64 and the databases used to build the transcription factors and microRNAs networks59,60. Moreover, for greater confidence in the detected communities, the only enrichments kept were those detected in the consensus clustering with a rate higher than 80%, iterating the entire algorithm 20 times.
Finally, we also searched for significant DEGs in whole blood, comparing exacerbation and stable state, frequency of exacerbation episodes, different severities of airway obstruction, and presence versus absence of viruses and bacteria in sputum samples. Different Limma regression models were applied depending on the biological question (see Supplementary information section 4). Each regression model applied a false discovery rate threshold of 0.05.
Supplementary information
Acknowledgements
GlaxoSmithKline Biologicals SA funded this study and was involved in all stages of study conduct, including analysis of the data. GlaxoSmithKline Biologicals SA also took in charge all costs associated with the development and publication of this manuscript. The authors thank members of the AERIS Study Group: J. Alnajar (University of Southampton, Southampton, UK), R. Anderson (University of Southampton, Southampton, UK), E. Aris (GSK, Wavre, Belgium), W.R. Ballou (GSK, Rockville, MD, USA), A. Barton (University Hospital Southampton NHS Foundation Trust, Southampton, UK), S. Bourne (University of Southampton, Southampton, and Portsmouth Hospitals NHS Trust, Portsmouth, UK), M. Caubet (GSK, Wavre, Belgium), S.C. Clarke (University of Southampton, Southampton, UK), D. Cleary (University of Southampton, Southampton, UK), C. Cohet (GSK, Wavre, Belgium), N.A. Coombs (University of Southampton, Southampton, UK), J-M. Devaster (GSK, Wavre, Belgium), V. Devine (University of Southampton, Southampton, UK), N. Devos (GSK, Wavre, Belgium), E. Dineen (University Hospital Southampton NHS Foundation Trust, Southampton, UK), T. Elliott (University of Southampton, Southampton, UK), R. Gladstone (Wellcome Sanger Institute, Hinxton, UK), S. Harden (University Hospital Southampton NHS Foundation Trust, Southampton, UK), J. Jefferies (University of Southampton, Southampton, UK), V.L. Kim (University of Southampton and University Hospital Southampton NHS Foundation Trust, Southampton, UK), P. Moris (GSK, Wavre, Belgium), K. Osman (University of Southampton, Southampton, UK), K.K. Ostridge (University of Southampton and University Hospital Southampton NHS Foundation Trust, Southampton, UK), T.G. Pascal (GSK, Wavre, Belgium), M. Peeters (GSK, Wavre, Belgium), B. Sente (GSK, Wavre, Belgium), S. Schoonbroodt (GSK, Wavre, Belgium), K.J. Staples (University of Southampton and University Hospital Southampton NHS Foundation Trust, Southampton, UK), A.C. Tuck (University of Southampton, Southampton, UK), L. Welch (Solent NHS trust, Southampton, UK), V. Weynants (GSK, Wavre, Belgium), T.M.A. Wilkinson (University of Southampton and University Hospital Southampton NHS Foundation Trust, Southampton, UK), A.P. Williams (University of Southampton, Southampton, UK), N.P. Williams (University of Southampton and University Hospital Southampton NHS Foundation Trust, Southampton, UK), C. Woelk (University of Southampton, Southampton, UK), M. Wojtas (University of Southampton, Southampton, UK) and S.A. Wootton (University Hospital Southampton NHS Foundation Trust, Southampton, UK). The authors also thank Viviane Bechtold (GSK), Simona Rondini (GSK), Laura Taddei (GSK), and Vincent Weynants (GSK) for their contribution to the study and/or review of the manuscript. The authors also thank Business & Decision Life Sciences Medical Communication Service Center for editorial assistance, manuscript coordination and writing support, on behalf of GSK. Joanne Knowles (independent medical writer, on behalf of GSK) provided medical writing support. PaxGene is a trademark of PreAnalytiX GmbH. Ovation is a trademark of NuGEN Technologies, Inc. GeneChip is a trademark of Affymetrix, Inc.
Author contributions
R.M., V.B., M.C., G.D.G., C.L., D.M., T.M.A.W., and A.M. did the conceptualization of the work. R.M. developed the methodology. R.M., Y.E.A., V.B., M.C., G.D.G., C.L., D.M., T.M.A.W., and A.M. performed the formal analysis and interpreted the results. R.M. and A.M. developed the content and the structure of the initial draft of the manuscript. All authors worked on the revision of the manuscript and approved the final version. R.M. and Y.E.A. contributed equally.
Data availability
Anonymized individual participant data and study documents can be requested for further research from www.clinicalstudydatarequest.com.
Code availability
Codes can be requested for further research from www.clinicalstudydatarequest.com. An example of the scripts executed to perform the random walk clustering by Infomap and the enrichment analyses is available at https://github.com/muzzial1/MNGCD_for_COPD.
Competing interests
R.M. is a PhD student at the University of Turin, with a scholarship from GSK. Y.E.A. was a consultant of XPE Pharma & Science for GSK when the analyses were conducted. V.B., G.D.G., C.L., and A.M. are employees of GSK. D.M. was employed by GSK when the analyses were conducted. G.D.G holds financial equities in GSK. M.C. reports no financial conflict of interest. T.M.A.W. has received grants from AstraZeneca, Synairgen and MyMHealth and fees, reimbursement for travel and meeting attendance from Boehringer Ingelheim, Chiesi, and AstraZeneca, outside of the submitted work, grants from GSK during conduct of the study. In addition, T.M.A.W. has a patent 2018 US Patent 62/479550 – Immunogenic Composition, Use and Methods of Treatment – A novel vaccine to prevent exacerbations of COPD pending to GSK, and a patent 2018 US Patent 62/479562 – Immunogenic Composition, Use and Methods of Treatment – A novel vaccine to prevent exacerbations of COPD pending to GSK and is Founder and Director, MyMHealth Ltd. All authors declare no other financial and non-financial relationships and activities.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Roberta Marino, Yousef El Aalamat.
Supplementary information
The online version contains supplementary material available at 10.1038/s41540-024-00425-6.
References
- 1.Global Initiative for Chronic Obstructive Lung Disease. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. 2022 report. https://goldcopd.org/wp-content/uploads/2021/11/GOLD-REPORT-2022-v1.0-12Nov2021_WMV.pdf (2021).
- 2.Sethi, S. & Murphy, T. F. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N. Engl. J. Med.359, 2355–2365 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Mayhew, D. et al. Longitudinal profiling of the lung microbiome in the AERIS study demonstrates repeatability of bacterial and eosinophilic COPD exacerbations. Thorax73, 422–430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varkey, J. B., Varkey, A. B. & Varkey, B. Prophylactic vaccinations in chronic obstructive pulmonary disease: current status. Curr. Opin. Pulm. Med.15, 90–99 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Agustí, A. G. et al. Systemic effects of chronic obstructive pulmonary disease. Eur. Respir. J.21, 347–360 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Oudijk, E. J., Lammers, J. W. & Koenderman, L. Systemic inflammation in chronic obstructive pulmonary disease. Eur. Respir. J. Suppl.46, 5s–13s (2003). [DOI] [PubMed] [Google Scholar]
- 7.Barnes, P. J. & Celli, B. R. Systemic manifestations and comorbidities of COPD. Eur. Respir. J.33, 1165–1185 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Koutsokera, A., Stolz, D., Loukides, S. & Kostikas, K. Systemic biomarkers in exacerbations of COPD: the evolving clinical challenge. Chest141, 396–405 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Bahr, T. M. et al. Peripheral blood mononuclear cell gene expression in chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol.49, 316–323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stockley, R. A., Halpin, D. M. G., Celli, B. R. & Singh, D. Chronic obstructive pulmonary disease biomarkers and their interpretation. Am. J. Respir. Crit. Care Med.199, 1195–1204 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA.102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeskind, J. E., Lenburg, M. E. & Spira, A. Translating the COPD transcriptome: insights into pathogenesis and tools for clinical management. Proc. Am. Thorac. Soc.5, 834–841 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrow, J. D. et al. Identifying a gene expression signature of frequent COPD exacerbations in peripheral blood using network methods. BMC Med. Genomics8, 1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Regan, E. A. et al. Genetic epidemiology of COPD (COPDGene) study design. COPD7, 32–43 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vestbo, J. et al. Evaluation of COPD longitudinally to identify predictive surrogate end-points (ECLIPSE). Eur. Respir. J.31, 869–873 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Reinhold, D. et al. Meta-analysis of peripheral blood gene expression modules for COPD phenotypes. PLoS One12, e0185682 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Domenico, M. Multilayer modeling and analysis of human brain networks. Gigascience6, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett, L., Kittas, A., Muirhead, G., Papageorgiou, L. G. & Tsoka, S. Detection of composite communities in multiplex biological networks. Sci. Rep.5, 10345 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pournoor, E., Mousavian, Z., Dalini, A. N. & Masoudi-Nejad, A. Identification of key components in colon adenocarcinoma using transcriptome to interactome multilayer framework. Sci. Rep.10, 4991 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang, L. et al. Integrating data and knowledge to identify functional modules of genes: a multilayer approach. BMC Bioinforma.20, 225 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cantini, L., Medico, E., Fortunato, S. & Caselle, M. Detection of gene communities in multi-networks reveals cancer drivers. Sci. Rep.5, 17386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haas, R. et al. Designing and interpreting ‘multi-omic’ experiments that may change our understanding of biology. Curr. Opin. Syst. Biol.6, 37–45 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Didier, G., Brun, C. & Baudot, A. Identifying communities from multiplex biological networks. PeerJ3, e1525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choobdar, S. et al. Assessment of network module identification across complex diseases. Nat. Methods16, 843–852 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan, Y. P., Shi, Y. H. & Gu, W. C. Analysis of protein-protein interaction network in chronic obstructive pulmonary disease. Genet Mol. Res.13, 8862–8869 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Caramori, G., Casolari, P. & Adcock, I. Role of transcription factors in the pathogenesis of asthma and COPD. Cell Commun. Adhes.20, 21–40 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Hobbs, B. D. & Tantisira, K. G. MicroRNAs in COPD: small molecules with big potential. Eur. Respir. J.53, 1900515 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Duan, R. et al. Evaluation and comparison of multi-omics data integration methods for cancer subtyping. PLoS Comput. Biol.17, e1009224 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozturk, K., Dow, M., Carlin, D. E., Bejar, R. & Carter, H. The emerging potential for network analysis to inform precision cancer medicine. J. Mol. Biol.430, 2875–2899 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alcalá-Corona, S. A., Sandoval-Motta, S., Espinal-Enríquez, J. & Hernández-Lemus, E. Modularity in biological networks. Front Genet12, 701331 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosvall, M. & Bergstrom, C. T. Maps of random walks on complex networks reveal community structure. Proc. Natl Acad. Sci. USA.105, 1118–1123 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkinson, T. M. A. et al. A prospective, observational cohort study of the seasonal dynamics of airway pathogens in the aetiology of exacerbations in COPD. Thorax72, 919–927 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilkinson, T. M. A. et al. Drivers of year-to-year variation in exacerbation frequency of COPD: Analysis of the AERIS cohort. ERJ Open Res.5, 00248–02018 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin, J. I. & Brusselle, G. G. Mechanistic links between COPD and lung cancer: a role of microRNA let‑7? Nat. Rev. Cancer14, 70 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Tasena, H. et al. microRNA-mRNA regulatory networks underlying chronic mucus hypersecretion in COPD. Eur. Respir. J.52, 1701556 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Qian, Y. et al. Comprehensive analysis of miRNA-mRNA-lncRNA networks in non-smoking and smoking patients with chronic obstructive pulmonary disease. Cell. Physiol. Biochem.50, 1140–1153 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Xie, P. et al. Sequential serum Let-7 is a novel biomarker to predict accelerated reproliferation during fractional radiotherapy in lung cancer. Clin. Lung Cancer17, e95–e101 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Mironova, N. et al. MicroRNA drop in the bloodstream and microRNA boost in the tumour caused by treatment with ribonuclease A leads to an attenuation of tumour malignancy. PLoS One8, e83482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vestbo, J. et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am. J. Respir. Crit. Care Med.187, 347–365 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Chang, Y. et al. COPD subtypes identified by network-based clustering of blood gene expression. Genomics107, 51–58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Obeidat, M. et al. Network-based analysis reveals novel gene signatures in peripheral blood of patients with chronic obstructive pulmonary disease. Respir. Res.18, 72 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma, A. et al. Integration of molecular interactome and targeted interaction analysis to identify a COPD disease network module. Sci. Rep.8, 14439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, B. et al. Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods11, 333–337 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Li, C. X., Wheelock, C. E., Sköld, C. M. & Wheelock, Å. M. Integration of multi-omics datasets enables molecular classification of COPD. Eur. Respir. J.51, 1701930 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Van Pottelberge, G. R. et al. MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med.183, 898–906 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Gilles, M. E. & Slack, F. J. Let-7 microRNA as a potential therapeutic target with implications for immunotherapy. Expert Opin. Ther. Targets22, 929–939 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swaminathan, S. et al. Differential regulation of the Let-7 family of microRNAs in CD4+ T cells alters IL-10 expression. J. Immunol.188, 6238–6246 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Yew-Booth, L. et al. JAK-STAT pathway activation in COPD. Eur. Respir. J.46, 843–845 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Bakke, P. S. et al. Candidate genes for COPD in two large data sets. Eur. Respir. J.37, 255–263 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Schofield, J. P. R. et al. Morse-clustering of a topological data analysis network identifies phenotypes of asthma based on blood gene expression profiles. bioRxiv, 516328 (2020).
- 51.Ahearn, C. P., Gallo, M. C. & Murphy, T. F. Insights on persistent airway infection by non-typeable Haemophilus influenzae in chronic obstructive pulmonary disease. Pathog. Dis.75, ftx042 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cummins, N. W. et al. Heme oxygenase-1 regulates the immune response to influenza virus infection and vaccination in aged mice. FASEB J.26, 2911–2918 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burny, W. et al. Different adjuvants induce common innate pathways that are associated with enhanced adaptive responses against a model antigen in humans. Front. Immunol.8, 943 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Mot, L. et al. Transcriptional profiles of adjuvanted hepatitis B vaccines display variable interindividual homogeneity but a shared core signature. Sci. Transl. Med.12, eaay8618 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Sun, J. et al. A potential panel of six-long non-coding RNA signature to improve survival prediction of diffuse large-B-cell lymphoma. Sci. Rep.6, 27842 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu, H. et al. Identification and validation of long noncoding RNA biomarkers in human non-small-cell lung carcinomas. J. Thorac. Oncol.10, 645–654 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Liu, J. et al. Identification of LINC01279 as a cell cycle‑associated long non‑coding RNA in endometriosis with GBA analysis. Mol. Med. Rep.18, 3850–3858 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiménez-Ortega, R. F. et al. Identification of microRNAs in human circulating monocytes of postmenopausal osteoporotic Mexican-Mestizo women: A pilot study. Exp. Ther. Med.14, 5464–5472 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gerstein, M. B. et al. Architecture of the human regulatory network derived from ENCODE data. Nature489, 91–100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chou, C. H. et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res.46, D296–D302 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alonso-López, D. et al. APID interactomes: providing proteome-based interactomes with controlled quality for multiple species and derived networks. Nucleic Acids Res.44, W529–W535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst.1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chaussabel, D. et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity29, 150–164 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li, S. et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat. Immunol.15, 195–204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Anonymized individual participant data and study documents can be requested for further research from www.clinicalstudydatarequest.com.
Codes can be requested for further research from www.clinicalstudydatarequest.com. An example of the scripts executed to perform the random walk clustering by Infomap and the enrichment analyses is available at https://github.com/muzzial1/MNGCD_for_COPD.