Abstract
The Mediterranean fruit fly (Ceratitis capitata) is a globally invasive species and an economically significant pest of fruit crops. Understanding the evolutionary history and local climatic adaptation of this species is crucial for developing effective pest management strategies. We conducted a comprehensive investigation using whole genome sequencing to explore (i) the invasion history of C. capitata with an emphasis on historical admixture and (ii) local climatic adaptation across African, European, Central, and South American populations of C. capitata. Our results suggest a stepwise colonization of C. capitata in Europe and Latin America in which Mediterranean and Central American populations share an ancestral lineage. Conversely, South American invasion history is more complex, and our results partly suggest an old secondary invasion into South America from Europe or a colonization of South America directly from Africa, followed by admixture with an European lineage. Throughout its invasive range, C. capitata is challenged with diverse climatic regimes. A genome wide association study identified a relationship between allele frequency changes and specific bioclimatic variables. Notably, we observed a significant allele frequency shift related to adaptation to cold stress (BIO6), highlighting the species’ ability to rapidly adapt to seasonal variations in colder climates.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-76390-1.
Keywords: Phylogeography, Invasive species, Climate adaptation, Pest species
Subject terms: Genetic association study, Population genetics
Introduction
Biological invasions are a major driver of biodiversity loss and ecosystem disruption, often resulting in significant economic and ecological impacts. Understanding the mechanisms underlying the successful establishment and spread of invasive species is essential for mitigating their negative effects1–3. Central to this challenge is unraveling the complex interactions between genetic adaptation, environmental factors, and human-mediated dispersal4. In this context, studying the genetic and ecological dynamics of invasive species can provide critical insights into how they overcome geographic, climatic, and ecological barriers, enabling them to thrive in novel environments.
The phylogeography of a species is influenced by stochastic or neutral processes next to non-neutral processes like local adaptation. In the case of introduced, non-native species, genetic bottlenecks and elevated drift in concert with ecological adaptation act strongly on population genetic structure5,6. A comprehensive understanding of a species’ climatic niche and adaptive potential in the context of population genetic structure is often of paramount importance for developing effective management strategies for invasive organisms4,7–9.
One prime example of a damaging invasive species is Ceratitis capitata (Wiedemann) (Diptera: Tephritidae), also referred to as the Mediterranean fruit fly or “medfly”. Medfly poses a high threat to fruit production worldwide, inflicting substantial economic losses and impeding international trade10,11. Its ability to effectively colonize regions with diverse climatic conditions and exploit a wide range of host plants has contributed to its invasion success10,12,13. Originating in tropical Sub-Saharan Africa13–15, medfly has invaded numerous regions outside its native range during the last two centuries16–18. It is now established in northern Africa, all Mediterranean countries, Central and South America, Western Australia, and the Hawaiian islands17,19,20 and is annually detected in California2,21,22 and several Central European countries (see Egartner et al.25, for Austria, Chireceanu et al.23, for Romania and König et al.24, for Germany)23–25.
Current knowledge on C. capitata invasion history pinpoints East Africa as the region of origin, where genetic diversity is highest13,16,26. Numerous studies have underscored the important role of the Mediterranean region as a steppingstone for secondary invasions elsewhere in the world13,17,18,20,26. For instance, populations in Central and South America are likely the outcome of independent invasion events and exhibit divergent levels of similarity with European populations13. Several aspects of medfly invasion remain subject to ongoing debate. For example, recent findings by Arias et al.12 open the possibility for a completely new and independent invasion route into South America, specifically Brazil, directly from Africa. The latter example shows how fundamentals on medfly invasion are still a point of discussion and alludes to the complexity that often goes along with reconstructing the population phylogeography of highly mobile invasive species8,27,28. Currently, it largely remains unclear to which extent admixture has occurred throughout the invasion history of C. capitata12,13.
The number of markers utilized in phylogeographic studies of non-model species have increased dramatically in recent years. Estimating genetic relationships between closely related populations can be challenging and genetic data stemming from large genomic regions can increase accuracy29–31. Past studies have focused on pathways of invasion using mitochondrial sequences15,19, microsatellite loci13,18 or restriction site-associated DNA sequencing (RADseq)12. Here, we utilize single nucleotide polymorphis (SNP) data attained by whole genome re-sequencing and a high-quality reference genome. We investigated gene flow patterns among populations, with a specific interest in tracing back admixture events and invasion history. Moreover, by coupling genomic data with bioclimatic variables, we aimed to identify genetic markers associated with local climatic adaptation, shedding light on the molecular mechanisms underlying the species’ ability to thrive in contrasting environments across continents. To accomplish this, we employed a combination of statistical and modeling approaches, including Redundancy Analysis (RDA), Latent Factor Mixed Models (LFMM), PCadapt, and gradient forest analysis.
Overall, this study represents a significant advancement in our understanding of the geographic range expansion of the Mediterranean fruit fly, offering valuable insights into the evolutionary history and adaptive potential of one of the most damaging pest species worldwide. The integration of genomic and climatic data could enhance our understanding of the complex interplay between genetic variation and environmental factors leading to invasion success of a pest species in general. Ultimately, gene-environment association studies like this one can aid in the development of sustainable pest management strategies for economically or ecologically important pests.
Materials and methods
Sample collection, DNA extraction and whole genome resequencing
A total of 117 samples of C. capitata were collected from 25 locations in 20 countries between 2005 and 2021 (Table 1). Sampling locations are located in the following main geographical regions where C. capitata is well established: West Africa, East Africa, the Mediterranean, Central America, and South America. Samples with sufficient quality from Australia could not be attained and the invasion of C. capitata in Australia is not discussed in this study.
Table 1.
Region, location, year and number of samples considered in the current study.
| Region | Location | Year | Latitude | Longitude | Altitude (m) | n | n per region |
|---|---|---|---|---|---|---|---|
| East Africa | Burundi - Kanyosha | 2013 | -3.415 | 29.408 | 1098 | 3 | 22 |
| Kenya - Ruiru | 2011 | -1.133 | 36.917 | 1571 | 2 | ||
| Mozambique - Cuamba | 2007 | -14.816 | 36.535 | 1133 | 5 | ||
| South Africa - Limpopo | 2010, 2011 | -23.926 | 29.463 | 1118 | 4 | ||
| Tanzania - Morogoro | 2005 | -6.817 | 37.667 | 1408 | 5 | ||
| Zambia - Kaoma | 2007 | -14.783 | 24.8 | 1244 | 3 | ||
| West Africa | Benin | 2006 | 9.5 | 2.25 | 364 | 5 | 9 |
| Senegal - Keur Mouseu | 2010 | 14.781 | -17.111 | 17 | 4 | ||
| Mediterranean | Croatia - Opuzen | 2021 | 43.011 | 17.472 | 23 | 6 | 57 |
| Croatia - Split | 2021 | 43.522 | 16.429 | 110 | 7 | ||
| Greece- Thessaloniki | 2020 | 40.609 | 22.771 | 45 | 7 | ||
| Greece - Volos | 2020 | 39.319 | 23.03 | 18 | 5 | ||
| Italy - Galatina | 2020 | 40.116 | 18.172 | 49 | 5 | ||
| Italy - Lazio | 2021 | 41.755 | 13.029 | 219 | 7 | ||
| Italy - Molise | 2021 | 41.896 | 15.097 | 11 | 7 | ||
| Spain - Ibiza | 2021 | 39.011 | 1.424 | 24 | 7 | ||
| Spain - La Pobla del Duc | 2020 | 39.011 | 1.424 | 24 | 6 | ||
| South America | Argentina | 2016 | -34.727 | -58.525 | 1125 | 5 | 13 |
| Bolivia | 2011 | -17.467 | -66.152 | 167 | 2 | ||
| Brazil | 2013 | -8.544 | -37.639 | 429 | 6 | ||
| Central America | El Salvador | 2013 | 13.723 | -89.27 | 13 | 4 | 16 |
| Guatemala | n.a. | 14.694 | -90.454 | 11 | 6 | ||
| Nicaragua | 2012 | 12.081 | -86.138 | 2 | 2 | ||
| Panama | 2013 | 8.77 | -82.439 | 3 | 4 |
DNA extraction was performed using the Qiagen Blood & Tissue extraction kit following the manufacturer’s recommendations and an elution volume of 120 µl. Subsequently, DNA extracts were submitted for whole genome sequencing (150 bp paired-end) on the NovaSeq 6000 platform with a minimum output of 6 Gb. Reads were quality trimmed with ‘fastp’ (https://github.com/OpenGene/fastp, Chen et al.32) using a phred-quality score of 20 while also removing adapter sequences. Trimmed reads were then aligned to the seven largest C. capitata reference scaffolds (87.7% of the total genome length) of the C. capitata reference available at the CoGe platform (genomevolution.org/coge/, genomeID:67550) using ‘bwa’ (https://github.com/lh3/bwa, Li and Durbin33). Next, variants were called using using GATK GenotypeGVCFs (https://gatk.broadinstitute.org/hc/en-us, McKenna et al.34) embedded in the elPrep pipeline (https://github.com/ExaScience/elprep, Herzeel et al.35). Following GATK hard-filters were applied to the variants: QD < 2, QUAL < 30, SOR > 3, FS > 60, MQ < 40, MQRankSum < -12.5 and ReadPosRankSum < -8. Variants were filtered to be biallelic SNPs, have a minor allele frequency of 0.05, a minimum depth of three and no missingness. Additionally, a second dataset was created to account for linkage disequilibrium (LD) adopting a correlation threshold of 0.1 and window size of 10,000 bp in the snpgdsLDpruning function from the package ‘SNPRelate’36. Lastly, a similar LD pruned dataset was created without missingness filter to be used as input for ADMIXTOOLS 237.
Phylogeography, historical admixture and invasion history of C. capitata
First, we inferred admixture between populations using the sNMF function within the ‘LEA’ package implemented in R38. The LD pruned dataset was used as input and K varied from 2 to 7 while performing 100 replicate runs for each K. Ancestral proportions (Q values) were then visualized for K = 2 up to the value of K with the lowest cross-entropy level (i.e. optimal K) + 1 to investigate further subdivision beyond the optimal K using ‘pong’ with sim_threshold = 0.9639. ‘Pong’ also analyses the ancestral proportions within values of K by grouping runs with similar result together as a mode, and reporting all modes and their frequencies for each K. In this manner, we do not only take into account the run with lowest cross-entropy, but we also consider alternative clustering solutions.
In addition to the sNMF analysis, we also modeled admixture events using the ADMIXTOOLS 2 software in R37. To choose the most plausible root for the admixturegraph, we have calculated the population-wise nucleotide diversity using pixy40. The population, and in extension, the region, with the highest diversity is assumed to be closest to the origin of C. capitata13–15,17. We employed PLINK 1.941 to prepare bed, bim and fam files from the vcf to calculate f2 statistics with extract_f2, followed by f2_from_precomp. The find_graphs function from ADMIXTOOLS 2 was then called using the precomputed f2 blocks to search for admixture graphs with parameters stop_gen = 200, stop_gen2 = 30, and East Africa as outgroup for 200 iterations and each count of admixture events (m) ranging from 0 to 6. Individuals were lumped per country in order to speed up the graph finding algorithm, whilst retaining sufficient resolution and power. We retained the top ten graphs with highest likelihood for each value of m. Out-of-sample scores were calculated for these models using the qpgraphs function with an f2_blocks_test argument, in order to make fair comparisons between graphs with different m. Final model selection was carried out by fitting competing models using bootstrap resampled SNP blocks (1000 bootstraps) with qpgraph_resample_multi, yielding score distributions for each graph. Two-sided bootstrap P-value testing was performed to assess significant differences in fit between graphs using the compare_fits function. When competing models with different m were not different (P-value < 0.05), the most parsimonious model was selected. When no significant difference could be found between graphs with an equal value for m, all unique topologies were retained. When the best fitting model did not differ from models with the same value of m, we visualize all graphs with unique topology and a representative for graphs that were present multiple times in the set of ten winning graphs. Common features of the competing models were then summarized. To test the robustness of the analysis, the above procedure was applied on two different SNP sets (MAF > 0.05 and MAF > 0.025).
Lastly, to explore genetic distance between populations, hierarchical clustering was performed by calculating the pairwise cosine type dissimilarity distances between sampling locations using the LD pruned dataset as input (https://github.com/gkanogiannis/fastreeR). A UPGMA clustering method was applied on the resulting distance matrix (‘stats’ package, R Core Team, 2023) and results were visualized using the iTOL webinterface (https://itol.embl.de/).
Association of SNPs with local climatic conditions
A dataframe of 0’s, 1’s, and 2’s was created for every SNP locus within the dataset containing no missing data, representing the number of reference alleles at each position. This data was then used as response variable for two gene-environment association (GEA) analyses and one outlier analysis described below.
Latent factor mixed modeling (LFMM)
With the aim of identifying SNPs that may be under the influence of local adaptation to climatic conditions, we applied latent factor mixed modelling (LFMM) as implemented in the ‘LEA’ package in R38. LFMM can be considered as a powerful method to survey for significant associations between allele frequency differences and features in the environment that exert selective pressure. Specifically, we have extracted bioclimatic data (average for the 1970–2000 time period) for all sampled locations from the WorldClim database (https://www.worldclim.org/data/bioclim) using the ‘raster’ package in R. Since gradient forest do not correct for collinearity, we followed authors’ instructions and have selected five uncorrelated climatic variables: BIO2 = mean diurnal range (°C*10), BIO5 = max temperature of warmest month (°C*10), BIO6 = min temperature of coldest month (°C*10), BIO18 = precipitation of warmest quarter (mm), BIO19 = precipitation of coldest quarter (mm) (See Fig. S1 for pairwise correlations and S2 for a manhattan plot of outlier loci). The number of latent factors for the LFMM analysis was inferred by the optimal number of clusters given by the SNMF analysis implemented in the LEA package. Significance values are calibrated by using the “gif” or “genome inflation factor”. This correction method uses a robust estimate of the variance of z-scores. Additionally, P-values were adjusted for multiple testing by applying the Benjamini & Hochberg procedure42. SNPs with a P-value > 0.01 were candidate SNPs under selection.
Redundancy analysis (RDA)
In contrast to LFMM, a redundancy analysis can be run simultaneously for every bioclimatic variable and corrects for underlying genetic structure similarly like LFMM. Multicollinearity among predictors was assessed by checking of the vif score for all variables using the ‘vif.cca’ function from the “vegan” package43. All values were well below 10, so all variables could be retained for further analysis. To factor out variance explained by geography, latitude and longitude were included as additional explanatory variables44. We applied a ± 3 SD cut-off on the SNP loadings to identify candidate SNPs.
PCadapt
Complementary to the association-based analyses mentioned above, we also applied a population clustering based method for outlier detection in the form of the ‘pcadapt’ algorithm as implemented in the R package “pcadapt”45. This approach uses a principal component analysis with a Mahalanobis distance-based approach to identify outlier loci. First, the number of clusters in the data was assessed using a thinned set of SNPs by setting the LD-clumping argument with default parameters. Secondly, the full set of SNPs was used as input and the optimal value for K was used as suggest by the screeplot. Adjusted P-values were calculated using the Benjamini & Hochberg procedure42.
Only outliers picked up in at least two methods independently were taken into further consideration as putative selective loci.
A gradient forest (GF) model was applied to map the allelic turnover (genomic composition) of putative selective SNPs to a geographic map spanning a large part of the distribution range where C. capitata is established. For this, the putative selective loci were used as predictors in the ‘gradientForest’ function as implemented in the R package “gradientForest”46. To account for spatial structure, we included Principal Coordinates of Neighbor Matrices (PCNMs) as calculated using the “vegan” R package43. Cumulative distributions of the standardized importance for each SNP was calculated separately and collectively and scaled by R2, for every examined bioclimatic variable. Next, using the ‘predict’ function implemented in the “raster” package, we simulated a value for each of the five bioclimatic variables for every pixel in the geographic raster image spanning the range of C. capitata. The ‘prcomp’ function from the “stats” package was then used to reduce the dimensionality and visualize results using an RGB color scheme.
Finally, a population level phylogeny was constructed using only the 49 outlier loci in order to explore discordances between the ‘neutral’ phylogeny of C. capitata and adaptive evolutionary processes. For this a UPGMA phylogenetic tree was constructed by computing the cosine dissimilarity between samples (https://github.com/gkanogiannis/fastreeR) and was compared with the phylogeny using the LD pruned SNP dataset.
Results
Population structure and invasion pathways
A total of 220,242 SNPs were retained after applying all filtering steps and 35,591 variants were retained after linkage pruning. Average individual depth was 12.15 (± 2.91). The highest nucleotide diversity (π) can be observed in populations within East Africa (regional π = 0.283; Mozambique, Tanzania, South Africa = 0.286; Kenya, Burundi = 0.280; Zambia = 0.275) closely followed by West Africa (regional π = 0.268; Benin = 0.272; Senegal = 0.264) (Tables S1 and S2).
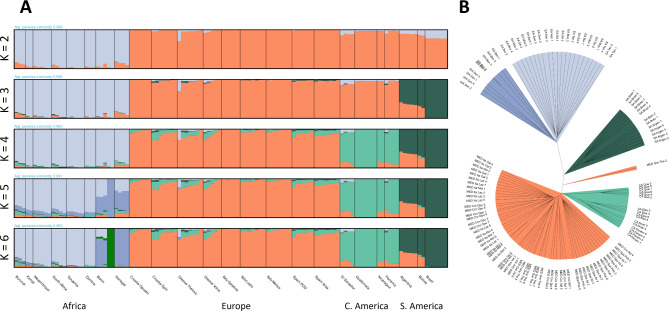
The admixture analysis pointed towards an optimal number of genetic clusters equal to five (Fig. 1A, Scree plot: Fig. S1). Cross-Entropy values for K equal to 2, 3, 4, 5 and 6 were 0.573, 0.566, 0.560, 0.558 and 0.559 respectively. African samples form a strong distinct cluster at K = 2 in the admixture plot with the invasive samples coinciding with the other cluster. At K = 3 and K = 4, the South American and Central American samples are separated from the rest respectively. The extra genetic cluster at K = 6 could be associated with the high genetic similarity between the samples from Benin (Figs. 1A and S2).
Fig. 1.
sNMF admixture coefficients as plotted with “pong” (A) and whole genome UPGMA clustering on a pairwise cosine type dissimilarity matrix (B).
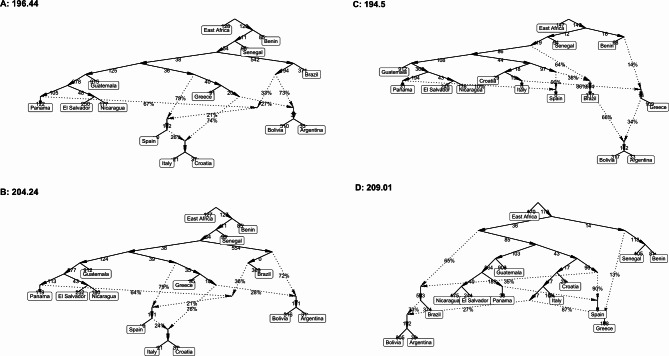
When using the SNP set excluding rare alleles (MAF > 0.05), the ADMIXTOOLS 2 model likelihood reached an overall optimum at four admixture events (m = 4) (Fig. 2). Hence, graphs modeling four admixture events were considered optimal. The most notable common features of the models can be summarized as follows: (i) The Bolivian and Argentinian populations share close ancestry with the Brazilian cluster and experienced admixture from Mediterranean genotypes. (ii) European and Central American populations share close ancestry through a common stepping stone. (iii) Guatemala is separated by strong drift from the other Central-American populations. (iv) An admixture event going from a basal branch to a European lineage suggests gene flow from a lineage from outside Europe or a basal, unsampled European population (Fig. 2).
Fig. 2.
Admixture graphs (ADMIXTOOLS 2) showing competing scenarios under m = 4.
For the SNP set of common and rare variants (MAC > 3), the graph finding algorithm converged for graphs with three admixture events. This final graph has virtually the same topology as scenario B of the graphs built with the common SNP set, except that Brazil is not admixed. By considering output generated with a different SNP set, we demonstrate that the common features shared between models are robust across different MAF cutoffs.
In the phylogeny, a clear distinction between samples from the native and invasive range can be observed with samples collected in East and West Africa forming two distinct sister groups (Fig. 1B). Within the invasive range, samples from South America are most distantly related to the rest of the samples, corroborating the pattern observed in the admixture graph (Fig. 1A). Samples from Central America and the Mediterranean share a recent origin. The Mediterranean clade can largely be divided in a Spanish clade and another clade comprising Italian, Croation, and Greek samples (Fig. 1B).
Local adaptation to climatic variables
LFMM revealed a total of 72 SNPs associated to five uncorrelated climatic variables: BIO2 (n = 12), BIO5 (n = 2), BIO6 (n = 23) and BIO19 (n = 35) (Figs. S3 and S4). The number of latent factors for the regression model was set to five, according to the number of clusters with the optimal BIC in the SNMF analysis (Fig. 1A).
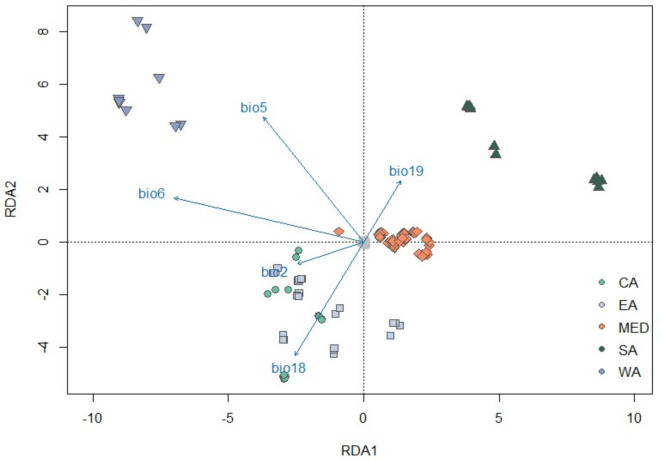
In the RDA, the first three components explained, 0.505, 0.191, and 0.127% of the variation, respectively and revealed a total of 945 putatively selective SNPs (Fig. 4; adjusted R2 = 0.074). All five RDA axes showed significant variation with genotypes (P-values < 0.001), but only the first three were taken into further consideration due to the low amount of variation explained by axes beyond this number. Variance inflation (vif) was low, with 4.00 as the highest value measured for BIO18. Values below 10 are considered to be acceptable47. Significant outliers were most associated with BIO5 (447 SNPs), followed by BIO18 (n = 173), BIO2 (n = 159), BIO6 (n = 152) and BIO19 (n = 13). The strongest correlation was found for SNP ‘Scaffold_4__1_contigs__length_86735756_34318211’ with BIO6 (R2 = 0.691). RDA1 and RDA2 largely split individuals in three groups; South America, West Africa, and a third group consisting of Mediterranean, East African, and Central American samples (Fig. 3). BIO5 and BIO6 suggest a breakup of East and West Africa with East African individuals grouping closer with Mediterranean and Central American individuals. The Mediterranean samples form a relatively tight cluster with association to BIO6.
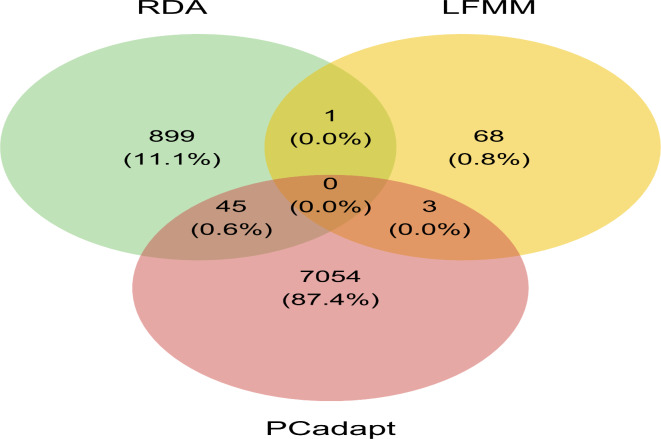
Fig. 4.
Venn-diagram showing the number of putative selective SNPs that were detected by each of the three analyses.
Fig. 3.
Redundancy analysis (RDA) using 220,242 SNP loci. This plot shows the first and second RDA axis with SNPs as grey filled circles. Individuals are colored and marked respective to their region of origin and the bioclimatic variables are represented by blue vectors. BIO2 = mean diurnal range (°C*10), BIO5 = max temperature of warmest month (°C*10), BIO6 = min temperature of coldest month (°C*10), BIO18 = precipitation of warmest quarter (mm), BIO19 = precipitation of coldest quarter (mm). The arrows represent the direction and strength of the relationship between the SNP variables and the bioclimatic variables while the angle between arrows indicates the correlation between variables.
PCadapt revealed a total of 7102 outlier loci using a total of five PC’s as suggested by the screeplot (Fig. S5).
Across approaches, one SNP was shared between LFMM and RDA (Fig. 4). PCadapt shared three and 45 SNPs with LFMM and RDA approach, respectively. No SNP could be found in the intersection of the three methods. A total of 49 SNPs were uncovered by a combination of two approaches and were retained for further analysis (Fig. 4, Table S3).
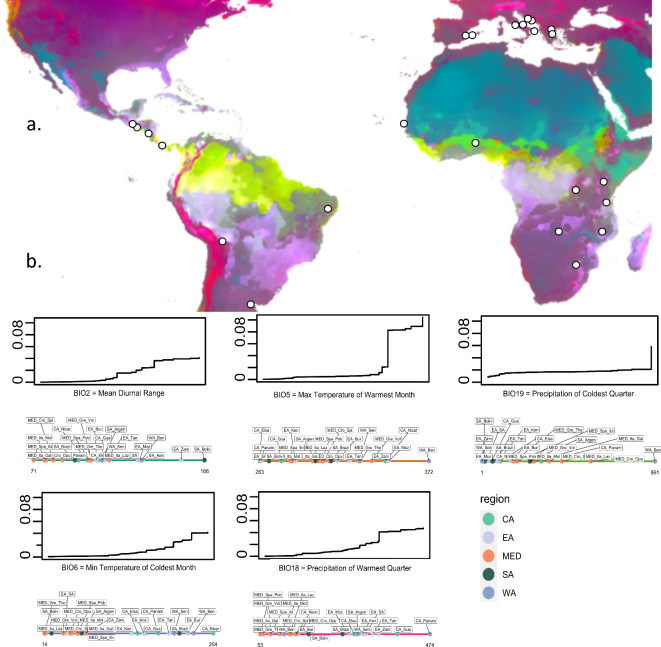
The gradient forest analysis reveals that BIO18 was the most important bioclimatic variable, explaining 6.66% of the variation within the 49 climate linked SNPs, closely followed by BIO5 (6.46%), BIO19 (6.36%) and BIO6 (5.91%). The spatial variables (PCNMs) and BIO2 (2.67%) account for a lower proportion of the variation (Fig. S6). A large allelic turnover at the 49 climate linked loci can be observed for BIO5 at around 345, which coincides with the position of the populations in Nicaragua and Mozambique (Fig. 5). For BIO2, BIO6, and BIO18 we can see a more gradual change in allelic composition (Fig. 5). Among climate linked SNPs, the SNP at position Scaffold_5__1_contigs__length_63380341_17630449 accounted for the largest variation in BIO5 (31.3%).
Fig. 5.
Gene-environment associations. (a) Spatial visualization of predicted genetic composition by the gradient forest algorithm for all 49 putatively selective loci. The projected rgb colors represent allelic-turnover across the map, including unsampled regions. Sampling locations are indicated with white circles. (b) Cumulative importance of allelic turnover functions of the 49 putatively selective loci with information on the value for each bioclimatic variable value at every sampling location below. Steeper slopes of the response curves indicate a more dramatic change in allelic composition along the bioclimatic gradient.
The 49 candidate SNP loci were dispersed across the genome (15 on scaffold 1; 11 on scaffold 2; seven on scaffold 3, scaffold 4, and scaffold 5; and two on scaffold 6). Twelve of them were located inside genes of which ten had a known homolog in another species (Table S4).
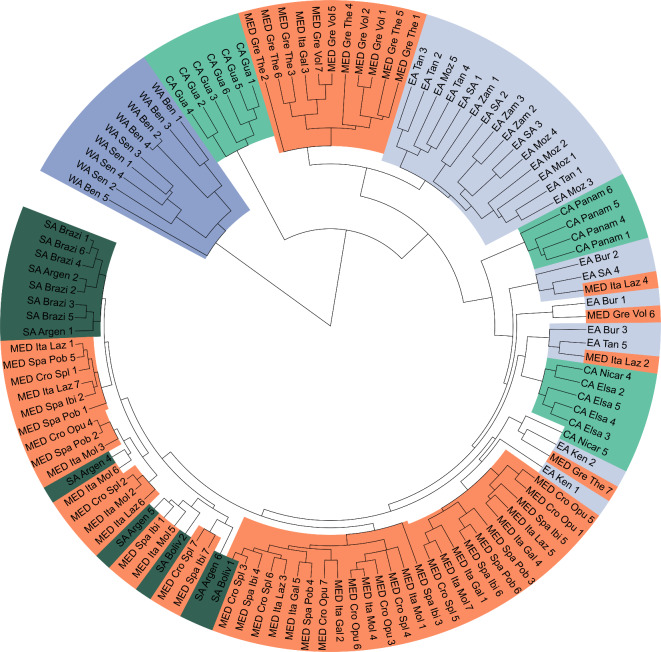
When investigating the neighbor joining tree constructed with only adaptive loci, all nine West African populations group together in one clade of which the substructuring corresponds to the population level (Fig. 6). Samples belonging to the invasive range of C. capitata share the clade with East African samples, which do not show strong geographical structuring. Central American samples are assigned to three clades (one consisting of Guatemalan samples, one consisting of Panamanian samples, and one containing a mixture of Nicaraguan and Salvadoran samples, Fig. 6).
Fig. 6.
Neighbor joining tree using the 49 candidate adaptive SNPs only. The position of the root corresponds with the deepest split in the tree, where West Africa splits from the rest of the samples.
Discussion
Invasion history and historical admixture
This study unravels and increases our understanding of the invasion history of the highly invasive and notorious pest C. capitata. Our findings largely corroborate earlier research but also provide new insights on the invasion history of C. capitata. We identified five main genetic groups within the extent of the medfly distribution sampled here. Mediterranean, Central American, and South American samples can be regarded as distinct genetic clusters and, as earlier studies have shown13,15,17,19,26, the Sub-Saharan region can be further divided in an Eastern and a Western part with a clear difference in genetic diversity (Table S1), ancestry proportions and branching in the hierarchical clustering analysis (Fig. 2). East Africa is most likely the ancestral native range of C. capitata in Africa as nucleotide diversity is highest in this region, confirming earlier findings13,15–17,26. The invasion of the Mediterranean countries likely took place in the 19th century, with the earliest record of medfly in Europe in 1842, during an age when Europe had strong ties with Africa via trade17. Colonization of Europe took place either through a Western (via the Iberian peninsula) or eastern (via Turkey) pathway or a combination of both13,16,17. Within Europe, Italy, Croatia, and Greece bear more similarities to each other than to Spain, which forms an almost country-specific clade, including samples from the Spanish mainland and from the island of Ibiza (Fig. 1B). This could be related to the more isolated position of the Iberian Peninsula within the European subcontinent. Sampling populations further east in the Mediterranean basin may help elucidate whether there is a genuine east-west divergence, potentially linked to the historical invasion pathways of C. capitata into Europe.
Additionally, the admixture graphs suggest consistent introgression of Central American genotypes in Spanish, Italian, and Croatian but not Greek populations, which could serve as an additional explanation for the observed sub-structuring in Europe. Although C. capitata has a relatively small range within the Mediterranean, the genetic relationships between Mediterranean populations and those from other regions are not fully resolved. This is likely due to Europe’s high level of connectivity with other parts of the world. (e.g. the introgression of Central American genotypes in the Mediterranean region, Fig. 2) and the stochastic nature of biological invasions where some genotypes establish by chance and others are filtered out. Additionally, divergent temporal shifts in genetic composition may be more pronounced in the Mediterranean due to stronger seasonality, potentially contributing to genetic differences even at fine scales. Further research is needed to clarify the interplay between population structure and seasonality in Mediterranean C. capitata.
Central America and the Mediterranean shared a European steppingstone quite early in Medfly invasion, as suggested by the admixture graphs (Fig. 2). Central American populations form a distinct clade with Guatemala in a divergent position relative to El Salvador, Nicaragua and Panama. This divergence is most likely explained by the very low genetic diversity as a result of a (recent) bottleneck followed by strong drift in Guatemala (π = 0.106 in Guatemala while π = 0.126 in Central America as a region, Tables S1 and S2) rather than admixture between an ancestral Guatemalan population and the Mediterranean cluster. In admixture-type analyses, a genetic bottleneck and strong genetic drift can falsely resemble the same signal as observed with admixture with an extra ancestral population48. Most likely, the divergent position and low genetic diversity of Guatemala in the Central American clade can be attributed to efforts to suppress medfly numbers in Guatemala under the Moscamed program49. This program, initiated in 1975, is one of the largest collaborative medfly eradication efforts, aiming to achieve pest free areas in Guatemala and was later joined by Mexico and the USA50. The program effectively suppressed medfly numbers in northern parts of Guatemala, creating a buffer zone between Guatemala and Mexico and the USA50. Our study is the first that can possibly link the success of an eradication campaign, the Moscamed program, to changes in the genetic diversity of C. capitata. A more thorough sampling would be advisable to investigate further this matter.
There are two scenarios that can explain the South American population structure (Fig. 1) as defined by the admixture graphs (Fig. 2). First, South-American populations could be founded by flies with a European genotype, which then branched into a Brazilian clade on the one hand and a clade containing Bolivia and Argentina on the other hand. Second and perhaps more likely, Brazilian flies could be derived from African flies, which is in line with findings by Arias et al. (2022) who found support for a unique lineage in South America coinciding with a Brazilian population. Moreover, the same study highlights the unique, high diversity microbiome present in the Brazilian samples12. In light of our results and those of Arias et al.12, there is mounting evidence for an early invasion of a unique lineage in South America, via Europe or even directly from Africa. In this context it is noteworthy to mention that the first C. capitata in Brazil was detected in Sao Paolo in 1905, further confirming the species' most likely point of entry in South America and explaining the unique genetic composition of Brazilian flies51,52. Interestingly, in both scenarios, Bolivian and Argentinian populations are the result of admixture between South American and Mediterranean genotypes as supported by the admixture graphs (Fig. 2).
Adaptation to the local climate
In general, invasive species retain the same environmental niche in the invaded area compared to the native area and the majority of invasive species occupy similar climatic ranges53. However, there are examples of species that manage to quickly adapt to a new and highly different environment9,54–59.
We found proof for range-wide, fast adaptation over steep climatic barriers that cause strong changes in allelic composition at 49 outlier loci (Fig. 5). These climatic barriers could effectively impede gene exchange due to maladaptation and might underlie some of the phylogeographic patterns that we nowadays observe in C. capitata. For instance, the clear separation of the African populations into an eastern and a western clade could, at least partially, be attributed to a climatic barrier running along the equator (yellows and light greens in Fig. 5b). Furthermore, gene exchange between Central and South American populations could be limited to a certain extent13 because of abrupt changes in climatic conditions along a latitudinal gradient and could partially explain the limited genetic admixture between both regions (Fig. 1).
In European C. capitata populations, we could observe a significant allele frequency shift related to adaptation to cold winter temperatures (BIO6) (Fig. 5b). This response is shared with populations from temperate climates from countries such as South Africa and Argentina, where winter temperatures are also at the lower end. These results demonstrate a genetic basis for the high climatic adaptability of C. capitata to seasonality with cold (winter) temperatures. Recent experimental research focusing on cold-tolerance of C. capitata, exploring chill coma recovery time in several European populations found a modest yet significant link between locality and cold tolerance60. Working on another species, Noh et al.61 found that Drosophila melanogaster collected in colder years took less time to recover from chill coma. Furthermore, Bergland et al.62, could associate a rapid pattern of allele frequency fluctuation to seasonality in D. melanogaster, which suggests a mechanism for fast genetic adaptation to cold temperature. Further research could focus on how fast these changes in allele frequency can be manifested.
When comparing the phylogeny based on the linkage pruned SNP set (Fig. 1B) and the phylogeny where only the 49 climate associated SNPs (Fig. 6) were used, a clear uncoupling can be observed between the set of adaptative loci and genetic history. In other words, allele frequencies of loci associated with local adaptation can rapidly shift, independent of demographic processes or genetic history. Remarkably, East Africa and West Africa are more distantly related when looking at adaptive SNPs alone (Fig. 6), which is also demonstrated by the gradient forest prediction using adaptive SNPs (Fig. 6b). The genotypes within the invasive range bear more similarity with East African samples, which are nested within the invasive clade. This suggests that a genetic predisposition to some extent was already present in the native range and thus suggests selection on standing variation rather than the formation of denovo variants in Europe (Fig. 6). It is noteworthy to mention that the high genetic diversity of (East) African populations13,16 likely provides a head start for selection to act upon and low frequency variants might be upregulated in new environments with contrasting conditions (Table S1). No formal testing was performed to test this finding because a more elaborate sampling would be needed to better capture the genetic diversity within the native range.
Our findings highlight the potential for invasive fruit flies to rapidly expand their climatic niche by shifting allele frequencies in response to the climatic conditions encountered during invasion. For C. capitata, we uncovered evidence of adaptation to winter cold stress through changes in allele frequency, raising the question of whether other invasive tephritids might similarly be able to adapt to seasonal cold stress63,64. This opens new avenues for investigating how invasive species respond to climatic challenges in their new environments.
Conclusion
The colonization of C. capitata in Europe and Latin America occurred in a stepwise fashion in which Mediterranean and Central American populations share an ancestral lineage. Conversely, South American invasion history is more complex and results partly suggest an old secondary invasion into South America from Europe or a colonization of South America directly from Africa, followed by admixture with a European lineage. Throughout its range, C. capitata, is challenged with different climatic conditions and we uncovered a significant association between allele frequency at 49 SNP loci and five bioclimatic variables. Interestingly, we observed a strong allele frequency shift related to cold adaptation (BIO6) which could point towards the capacity of C. capitata to rapidly adapt to winter temperatures in temperate climates. To our knowledge this is the first study to focus on climatic adaptation in C. capitata on the genomic level, adding to former experimental studies on the matter. Our results could stimulate researchers to test more advanced hypotheses related to the adaptive potential regarding the invasiveness of C. capitata.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We explicitly thank the European Union’s Horizon 2020 program for funding this research.
Author contributions
P.D.: Designed research, analyzed data, writing, wet lab work. S.V.: analyzed data, wet lab work. M.V: designed research, reviewing, funding acquisition. A.S.: reviewing, sample collection. M.C.: reviewing, sample collection. V.R.: reviewing, sample collection. J.J.: reviewing, sample collection. M.B.: reviewing, sample collection. K.B.: reviewing, sample collection. N.P.: designed research, reviewing, funding acquisition. M.D.M.: designed research, reviewing, funding acquisition.
Funding
This work was supported by the European Union’s Horizon 2020 research and innovation program under Grant agreement No 818184.
Data availability
All paired-end illumina read data has been submitted to the NCBI Sequence Read Archive (SRA) under PRJNA1120743. The code to run admixtools and analyses associated to gene-climate association can be found within the GitHub repository: https://github.com/thesnakeguy/Ccap_Phylogeography_and_Climate_adaptation.
Declarations
Competing interests
The authors declare no competing interests.
Benefit-sharing
We included all the collaborators providing fly samples for this study as co-authors. The collection of samples in the field was performed in agreement with local regulation.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Alvarez, S., Evans, E. & Hodges, A. Estimated Costs and Regional Economic Impacts of the Oriental Fruit Fly (Bactrocera dorsalis) Outbreak in Miami-Dade County, Florida (2016).
- 2.Carey, J. R., Papadopoulos, N. & Plant, R. The 30-year debate on a multi-billion-dollar threat: Tephritid fruit fly establishment in California. Am. Entomol.63, 200–210 (2017). [Google Scholar]
- 3.Vandepitte, K. et al. Rapid genetic adaptation precedes the spread of an exotic plant species. Mol. Ecol.23, 2157–2164 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Matheson, P. & McGaughran, A. Genomic data is missing for many highly invasive species, restricting our preparedness for escalating incursion rates. Sci. Rep.12, 13987 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gatto-Almeida, F., Pichlmueller, F., Bodey, T. W., Samaniego, A. & Russell, J. C. The tails of two invasive species: Genetic responses to acute and chronic bottlenecks. Biol. Invasions. 24, 3263–3273 (2022). [Google Scholar]
- 6.Jaspers, C. et al. Invasion genomics uncover contrasting scenarios of genetic diversity in a widespread marine invader. Proc. Natl. Acad. Sci. USA118, e2116211118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferronato, P. et al. A phylogeographic approach to the Drosophila suzukii (Diptera: Drosophilidae) invasion in Brazil. J. Econ. Entomol.112, 425–433 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Garnas, J. R. et al. Complex patterns of global spread in invasive insects: Eco-evolutionary and management consequences. Biol. Invasions18, 935–952 (2016). [Google Scholar]
- 9.Sherpa, S. et al. Unravelling the invasion history of the Asian tiger mosquito in Europe. Mol. Ecol.28, 2360–2377 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Karsten, M. et al. Population genetics of Ceratitis capitata in South Africa: Implications for dispersal and pest management. PLoS ONE8, e54281 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magaña, C., Hernández-Crespo, P., Ortego, F. & Castañera, P. Resistance to malathion in field populations of Ceratitis capitata. J. Econ. Entomol.100, 1836–1843 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Arias, M. B. et al. Unveiling biogeographical patterns in the worldwide distributed Ceratitis capitata (medfly) using population genomics and microbiome composition. Mol. Ecol.31, 4866–4883 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Deschepper, P. et al. Looking at the big picture: Worldwide population structure and range expansion of the cosmopolitan pest Ceratitis capitata (Diptera, Tephritidae). Biol. Invasions23, 3529–3543 (2021). [Google Scholar]
- 14.De Meyer, M., Copeland, R. S., Wharton, R. A., McPheron, B. A. & Barnes, B. N. On the geographic origin of the medfly Ceratitis capitata (Wiedemann) (Diptera: Tephritidae). In Proceedings of the 6th International Symposium on Fruit Flies of Economic Importance (2004).
- 15.Ruiz-Arce, R. et al. Worldwide phylogeography of Ceratitis capitata (Diptera: Tephritidae) using mitochondrial DNA. J. Econ. Entomol.10.1093/jee/toaa024 (2020). [DOI] [PubMed]
- 16.Arias, M. B., Elfekih, S. & Vogler, A. P. Population genetics and migration pathways of the Mediterranean fruit fly Ceratitis capitata inferred with coalescent methods. PeerJ6, e5340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malacrida, A. R. et al. Globalization and fruitfly invasion and expansion: The medfly paradigm. Genetica. 131, 1–9 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Nikolouli, K. et al. Genetic structure and symbiotic profile of worldwide natural populations of the Mediterranean fruit fly, Ceratitis capitata. BMC Genet.21, 128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barr, N. B. Pathway analysis of Ceratitis capitata (Diptera: Tephritidae) using mitochondrial DNA. J. Econ. Entomol.102, 401–411 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Bonizzoni, M. et al. On the origins of medfly invasion and expansion in Australia. Mol. Ecol.13, 3845–3855 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Bonizzoni, M. et al. Microsatellite analysis of medfly bioinfestations in California. Mol. Ecol.10, 2515–2524 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Carey, J. R. Establishment of the Mediterranean fruit fly in California. Science253, 1369–1373 (1991). [DOI] [PubMed] [Google Scholar]
- 23.Chireceanu, C., Iamandei, M., Stanica, F. & Chiriloaie, A. The presence of the Mediterranean fruit fly Ceratitis capitata (Wied.) (Diptera: Tephritidae) in Romania. Rom J. Plant. Prot.6, 92–97 (2013). [Google Scholar]
- 24.König, S., Steinmöller, S. & Baufeld, P. Origin and potential for overwintering of Ceratitis capitata (Wiedemann) captured in an official survey in Germany. J. Plant. Dis. Prot.129, 1201–1215 (2022). [Google Scholar]
- 25.Egartner, A. & Lethmayer, C. Invasive fruit flies of economic importance in Austria: Monitoring activities 2016. J. Plant. Dis. Prot.123, 45–49 (2017). [Google Scholar]
- 26.Karsten, M., van Vuuren, J., Addison, B., Terblanche, J. S. & P. & Deconstructing intercontinental invasion pathway hypotheses of the Mediterranean fruit fly (Ceratitis capitata) using a bayesian inference approach: Are port interceptions and quarantine protocols successfully preventing new invasions? Divers. Distrib.21, 813–825 (2015). [Google Scholar]
- 27.Barr, N., Ruiz-Arce, R. & Armstrong, K. Using molecules to identify the source of fruit fly invasions. Trapp. Detect. Control Regul. Tephritid Fruit Flies 321–378. 10.1007/978-94-017-9193-9_10 (2014).
- 28.Popa-Báez, Á. D. et al. Tracing the origins of recent Queensland fruit fly incursions into South Australia, Tasmania and New Zealand. Biol. Invasions23, 1117–1130 (2021). [Google Scholar]
- 29.Burley, J. T., Orzechowski, S. C. M., Sin, S. Y. W. & Edwards, S. V. Whole-genome phylogeography of the blue-faced honeyeater (Entomyzon cyanotis) and discovery and characterization of a neo-Z chromosome. Mol. Ecol.32, 1248–1270 (2023). [DOI] [PubMed] [Google Scholar]
- 30.Edwards, S. V., Tonini, J. F. R., Mcinerney, N., Welch, C. & Beerli, P. Multilocus phylogeography, population genetics and niche evolution of Australian brown and black-tailed treecreepers (Aves: Climacteris). Biol. J. Linn. Soc.138, 249–273 (2023). [Google Scholar]
- 31.McCormack, J. E., Hird, S. M., Zellmer, A. J., Carstens, B. C. & Brumfield, R. T. Applications of next-generation sequencing to phylogeography and phylogenetics. Mol. Phylogenet. Evol.66, 526–538 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Chen, S., Zhou, Y., Chen, Y. & Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenna, A. et al. The genome analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res.20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herzeel, C. et al. Multithreaded variant calling in elPrep 5. PLoS ONE16, e0244471 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng, X. A tutorial for the R package SNPRelate. (2022).
- 37.Maier, R. et al. On the limits of fitting complex models of population history to f-statistics. eLife12, e85492 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frichot, E. & François, O. L. E. A. An R package for landscape and ecological association studies. Methods Ecol. Evol.6, 925–929 (2015). [Google Scholar]
- 39.Behr, A. A., Liu, K. Z., Liu-Fang, G., Nakka, P. & Ramachandran, S. Pong: Fast analysis and visualization of latent clusters in population genetic data. Bioinformatics32, 2817–2823 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korunes, K. L. & Samuk, K. pixy: Unbiased estimation of nucleotide diversity and divergence in the presence of missing data. Mol. Ecol. Resour.21, 1359–1367 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang, C. C. et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience4, s13742-015-0047–s13742-015-0048 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjamini, Y. & Hochberg, Y. Controlling the false Discovery rate: A practical and powerful Approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol.57, 289–300 (1995). [Google Scholar]
- 43.Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci.14, 927–930 (2003). [Google Scholar]
- 44.Capblancq, T. & Forester, B. R. Redundancy analysis: A Swiss Army Knife for landscape genomics. Methods Ecol. Evol.12, 2298–2309 (2021). [Google Scholar]
- 45.Privé, F., Luu, K., Vilhjálmsson, B. J. & Blum, M. G. B. Performing highly efficient genome scans for local adaptation with R package pcadapt version 4. Mol. Biol. Evol.37, 2153–2154 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Ellis, N., Smith, S. J. & Pitcher, C. R. Gradient forests: Calculating importance gradients on physical predictors. Ecology. 93, 156–168 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Miles, J. Tolerance and variance inflation factor. In Wiley StatsRef: Statistics Reference Online (John Wiley & Sons, Ltd, 2014). 10.1002/9781118445112.stat06593.
- 48.Lawson, D. J., van Dorp, L. & Falush, D. A tutorial on how not to over-interpret STRUCTURE and ADMIXTURE bar plots. Nat. Commun.9, 3258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gorden Tween. MOSCAMED-Guatemala - An evolution of ideas (2002).
- 50.Enkerlin, W. et al. Area Freedom in Mexico from Mediterranean Fruit fly (Diptera: Tephritidae): A review of over 30 years of a successful Containment Program using an Integrated area-wide SIT Approach. Fla. Entomol.98, 665–673 (2015). [Google Scholar]
- 51.Malacrida, A. R. et al. Genetic aspects of the worldwide colonization process of Ceratitis capitata. J. Hered. 89, 501–507 (1998). [DOI] [PubMed] [Google Scholar]
- 52.Acioli, A. N. S., Silva, N. M., Adaime, R., Costa-Silva, F. C. & Zucchi, R. A. March to the North: Ceratitis capitata (Wiedemann, 1824) (Diptera, Tephritidae) reaches Manaus, state of Amazonas, Brazil. Entomol. Commun.6, ec06015 (2024). [Google Scholar]
- 53.Liu, C., Wolter, C., Xian, W. & Jeschke, J. M. Most invasive species largely conserve their climatic niche. Proc. Natl. Acad. Sci. USA. 117, 23643–23651 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo, W. Y., Lambertini, C., Li, X. Z., Meyerson, L. A. & Brix, H. Invasion of old world Phragmites australis in the new world: Precipitation and temperature patterns combined with human influences redesign the invasive niche. Glob Change Biol.19, 3406–3422 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Krehenwinkel, H., Rödder, D. & Tautz, D. Eco-genomic analysis of the poleward range expansion of the wasp spider Argiope bruennichi shows rapid adaptation and genomic admixture. Glob Change Biol.21, 4320–4332 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Medley, K. A., Westby, K. M. & Jenkins, D. G. Rapid local adaptation to northern winters in the invasive Asian tiger mosquito Aedes albopictus: A moving target. J. Appl. Ecol.56, 2518–2527 (2019). [Google Scholar]
- 57.Rollins, L. A., Richardson, M. F. & Shine, R. A genetic perspective on rapid evolution in cane toads (Rhinella marina). Mol. Ecol.10.1111/mec.13184 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Rosenthal, W. C. et al. Invasion and rapid adaptation of guppies (Poecilia reticulata) across the Hawaiian Archipelago. Evol. Appl.14, 1747–1761 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zenni, R. D., Bailey, J. K. & Simberloff, D. Rapid evolution and range expansion of an invasive plant are driven by provenance–environment interactions. Ecol. Lett.17, 727–735 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Moraiti, C. A., Verykouki, E. & Papadopoulos, N. T. Chill coma recovery of Ceratitis capitata adults across the Northern Hemisphere. Sci. Rep.12, 17555 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noh, S., Everman, E. R., Berger, C. M. & Morgan, T. J. Seasonal variation in basal and plastic cold tolerance: Adaptation is influenced by both long- and short-term phenotypic plasticity. Ecol. Evol.7, 5248–5257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bergland, A. O., Behrman, E. L., O’Brien, K. R., Schmidt, P. S. & Petrov, D. A. Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLOS Genet.10, e1004775 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ben-Yosef, M., Altman, Y., Nemni-Lavi, E., Papadopoulos, N. T. & Nestel, D. Effect of thermal acclimation on the tolerance of the peach fruit fly (Bactrocera zonata: Tephritidae) to heat and cold stress. J. Therm. Biol.117, 103677 (2023). [DOI] [PubMed] [Google Scholar]
- 64.Wang, J., Zeng, L. & Han, Z. An assessment of cold hardiness and biochemical adaptations for cold tolerance among different geographic populations of the Bactrocera dorsalis (Diptera: Tephritidae) in China. J. Insect Sci.14, 292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All paired-end illumina read data has been submitted to the NCBI Sequence Read Archive (SRA) under PRJNA1120743. The code to run admixtools and analyses associated to gene-climate association can be found within the GitHub repository: https://github.com/thesnakeguy/Ccap_Phylogeography_and_Climate_adaptation.