Abstract
The beneficial effects of exercise are partly mediated via local or systemic functions of the insulin-like growth factor-1 (IGF-1) system. As IGF-1 increases local brain hemoglobin beta (Hbb) transcripts, we hypothesized that exercise could have similar effects. Mice were single-housed with free access to running wheels for seven days. After sacrifice and saline perfusion, the expression of 13 genes was quantified using real-time quantitative polymerase chain reaction (RT-qPCR) in three brain regions: the prefrontal cortex, motor cortex, and hippocampus. In addition, plasma insulin, glucose, homeostatic model assessment of IR (HOMA-IR), C-peptide, and IGF-1 were investigated. We show that hemoglobin-related transcripts (Hbb and 5’-aminolevulinate synthase 2 [Alas2]) increased 46–63% in the running group, while IGF-1-related genes [Igf1 / growth hormone receptor (Ghr)] decreased slightly (7%). There were also moderate to large correlations between Hbb- and IGF-1-related genes in the running group but not in the sedentary group. HOMA-IR, plasma glucose, and insulin changed marginally and non-significantly, but there was a trend toward an increase in plasma-IGF-1 in the running group. In conclusion, seven days of running increased Hbb-related transcripts in three brain regions. Hbb-related transcripts correlated with components of the brain IGF-1 system only in the running group.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-77489-1.
Keywords: Brain, Rodent, Exercise, Sedentary, Insulin resistance, Insulin-like growth factor-1
Subject terms: Neuroscience, Biomarkers, Endocrinology, Medical research
Authors:
Introduction
It is widely recognized that endurance exercise improves mental well-being1, increases cerebral blood flow2, alters neurotransmitter release3, and modulates the growth factors insulin-like growth factor-1 (IGF-1) and brain-derived neurotrophic factor (BDNF)4–6. Exercise also improves neuroprotective resilience to ischemia7 and recovery after stroke8. During exercise, growth hormone (GH) is released9, and exercise also has effects of a similar type as GH administration on the brain and neuroprotection in hypophysectomiced (Hx) rodents10,11. For example, exercise increases neural progenitor cell proliferation, neurogenesis, and cognitive function12,13, quite similar to the effects of GH and IGF-1 administration14,15.
Circulating GH crosses the blood-brain barrier16 and binds to GH receptors expressed in the brain17. GH stimulates IGF-1 expression in the liver18 and, to a lesser extent, in other organs, including the brain19,20. IGF-1 is actively transported over the blood-brain barrier via carrier-mediated uptake21,22. Some of the effects of GH in the brain are thought to be mediated by endocrine IGF-123–25. Indeed, the presence of both IGF-1 and IGF-1 receptors (IGF-1R) in neurons, glial, and endothelial cells26,27 enables endocrine and paracrine local effects of IGF-1 28–30.
Hemoglobin beta (Hbb) is expressed in neurons31,32, specifically in dopaminergic neurons, and to some extent in mature oligodendrocytes and cortical and hippocampal astrocytes33. Brain Hbb is thought to have biological functions similar to those in erythrocytes34, acting as a functional oxygen reservoir33. Several precursors, including Hbb and heme, form hemoglobin. The oxygen-binding heme group is synthesized through a pathway whereby Alas2 catalyzes the first step by converting glycine and succinyl-CoA into 5-aminolevulinic acid (ALA)35. ALA is then further processed in several steps, the final product being heme, which is united with two hemoglobin alpha and two hemoglobin beta to a functional tetrameric hemoglobin35,36. The enzyme Alox 15 is expressed in the mouse brain and is important for memory function37. Specifically, Alox15 acts via oxidizing polyunsaturated fatty acids to generate bioactive lipid metabolites such as eicosanoids and lipoxins and has a role in oxidative and inflammatory reactions38,39. Alox15 probably also has another functional link with hemoglobin, as suggested by results from an Alox15b knock-in mouse, which has lower erythrocyte counts, hematocrit, and hemoglobin, possibly mediated by cell membrane instability and increased hemolysis40. Altogether, Hbb, Alas2 and Alox15 can be considered as being functionally related to oxygen metabolism. Notably, Alox15 is increased in the hippocampus in response to GH treatment25. Previously, we have shown that GH administration significantly increased the production of Hbb, Alas2 and Alox15 transcripts in the rodent brain41,42. Also, in the GH/IGF-1 deficient Lewis dwarf (dw/dw) rat, GH administration increased hemoglobin in the brain25. Considering that GH or IGF-1 administration has many similarities to exercise-induced effects in the brain, we hypothesized that exercise would also increase Hbb-like transcripts in the brain.
The present study aimed to investigate whether seven days of running exercise affects Hbb-related transcripts previously shown to be affected by GH/IGF-1, and whether there is evidence that the IGF-1 system mediates the effects seen. In mice subjected to voluntary running for one week, we analyzed three different parts of the brain, the prefrontal cortex, the motor cortex, and the hippocampus, all of which are involved in exercise43–45. We assessed 13 gene transcripts, further grouped into four categories: Hbb-like, neuron-, glia-, and IGF-1-related. Finally, we analyzed plasma glucose, insulin, C-peptide, and IGF-1 as exercise improves these markers of glucose metabolism46.
Method
Animals and running procedures
We used 24 six-month-old male mice of the strain C57BL/6J (Charles River, Germany) kept under standard temperature conditions (24 °C) and 50–60% relative humidity. The sample size (n = 12 in each group) was determined for allowing detection of a transcript 40% difference provided a 30% coefficient of variation, with 90% power and 5% significance level. A 12-h light/dark cycle was maintained with light from 19:00 to 07:00 (reversed) and ad libitum access to food and water.
Running wheel experiments were performed as previously described47. Mice were housed individually in cages with free access to low-profile running wheels (ENV-047, Med Associates, Fairfax, VT, USA). After one week of acclimatization, half of the running wheels were unlocked (i.e., animals were randomized to voluntary running or sedentary controls), allowing simultaneous access for the animals in the running group. Running wheel activity was wirelessly recorded throughout the experiment.
Mice in the running group could run at their discretion, typically during their active dark phase, while the wheels for the sedentary group remained locked, preventing any running activity. Running activity was continuously monitored using a telemetry system integrated with the wheels, ensuring accurate tracking of distance and time spent running. This data was used to assess each animal’s level of voluntary activity. Since the running wheels for sedentary animals were locked and could not rotate, their activity was not recorded. Other confounders were not controlled for, and analysis was not blinded. Animals were sacrificed after one week of running during their active phase in the dark cycle. No specific criteria for inclusions or exclusions were used other than healthy appearance (other specific humane endpoints not used). After sacrifice some laboratory analysis was not successful, but the numbers of included animals are reported in the tables.
At the time of euthanasia, mice were deeply anaesthetized with a peritoneal injection of 50 mg/kg sodium thiopental during the animals’ active phase. Blood was extracted by cardiac puncture with a 27-gauge needle and collected in an EDTA/heparin tube. Immediately after blood sampling, animals were transcardially perfused with a cold saline solution (0.9% NaCl). Perfusion was performed at a rate of 10 ml/min for 2–3 min per animal, corresponding to 20–30 ml. Perfusion was continued until 30 s after the perfusion fluid became completely transparent. Brain tissue from the prefrontal cortex, motor cortex and hippocampus were micro-dissected following the technique described by Chiu et al. (2007)48. The brain was removed from the skull and rinsed with ice-cold phosphate-buffered solution (PBS) to remove any surface blood. It was then placed on an ice-cold Petri dish and cut in half with a razor blade to separate the right and left hemispheres. Details of the dissection can be found in the Supplementary methods - dissection (item 1).
The dissected tissue was immediately treated with RNAlater™ according to the manufacturer’s instructions and subsequently stored at −20℃. The Gothenburg Ethical Committee for Animal Research approved all experiments (#181–2015), which were performed under relevant local and national guidelines and regulations. Additionally, the experiments were conducted following the ARRIVE guidelines 2.0, presented in the Supplemental file – ARRIVE 2.0 author checklist.
Quantitative reverse transcription polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the prefrontal cortex, motor cortex, and hippocampus according to Chomczynski and Sacchi with minor modifications described previously49,50. Optical density (OD) measurements 260/280 nm via NanoDrop 1000 (Thermo Scientific, USA) were used for RNA quantification. We also checked the RNA quality by running each sample in gel electrophoresis, where all samples showed intact 18 S and 28 S RNA bands, which is indicative of good quality. cDNA was prepared from 250 ng of total RNA using the HighCapacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster city, CA, USA). RT-qPCR with preformed TaqMan gene expression assays (Applied Biosystems, USA; Table 1) together with a QuantStudio™ 3 real-time PCR system (Applied Biosystems, Carlsbad, CA, USA), was used to quantify the gene transcripts, for further information see http://www.appliedbiosystems.com). For details about RT-qPCR quantification, see the previously published supplementary information42. All transcript levels were normalized to the expression of Gapdh and thus are arbitrary but linear for each transcript, and comparisons between treatment groups represent actual linear changes. We tested two housekeeping genes. For GAPDH, we observed 10.1% lower values (p = 0.0004) in the running group, while Cyclophilin A (assay number: Mm02342430_g1) had 13.0% (p = 0.0002) lower values in the running group. We therefore chose to use only GAPDH for normalization (results for Cyclophilin A are not shown any further). Transcripts were grouped into four categories Hbb-like (Hbb, Alas2, Alox15), neuron-related (Bdnf, Bax, Grin2a, Grin2b), glia-related (Hif1a, Gfap), and IGF-1-related (Igf1, Igf1r, Insr, Ghr), as found in Table 1.
Table 1.
Information on transcripts.
| Abbreviation in Ms. | Assay number | Fullname | Category | Main function |
|---|---|---|---|---|
| Hbb | Mm01611268_g1 | Hemoglobin, beta adult major chain | Hbb-like | Neuroprotection |
| Alas2 | Mm00802083_m1 | 5’-aminolevulinate synthase 2 | Hbb-like | Rate-controlling enzyme of heme biosynthesis (for functional hemoglobin assembly) |
| Alox15 | Mm00507789_m1 | Arachidonate 15-lipoxygenase | Hbb-like | Lipid peroxidating enzyme, cell membrane stability, memory, knock-in reduces hemoglobin |
| Bdnf | Mm04230607_s1 | Brain-derived neurotrophic factor | Neuron | Regulator of synaptic transmission and plasticity |
| Bax | Mm00443039_m1 | BCL2-associated X protein, apoptosis regulator | Neuron | Apoptosis regulator |
| Grin2a | Mm00433802_m1 | Glutamate receptor, ionotropic, 2a | Neuron | Brain plasticity |
| Grin2b | Mm00433820_m1 | Glutamate receptor, ionotropic, 2a | Neuron | Brain plasticity |
| Hif1a | Mm00468869_m1 | Hypoxia-inducible factor 1. alpha subunit | Glia | Hypoxia-induced signaling protein |
| Gfap | Mm01253033_m1 | Glial fibrillary acidic protein | Glia | Structural protein / morphogenesis |
| Igf1 | Mm00439560_m1 | Insulin-like growth factor 1 | IGF-I-related | Brain plasticity |
| Igf1r | Mm00802831_m1 | Insulin-like growth factor 1 receptor | IGF-I-related | Brain plasticity |
| Insr | Mm01211875_m1 | Insulin receptor | IGF-I-related | Glucose metabolism/brain plasticity |
| Ghr | Mm00439093_m1 | Growth hormone receptor | IGF-I-related | Brain plasticity |
| Gapdh | Mm99999915_g1 | Glyceraldehyde-3-phosphate dehydrogenase | Reference gene | N/A |
Main function for the transcripts, gene names and abbreviations of commercially available probes. The probes are assay-on-demand mixes of primers and TaqMan MGM probes (FAM dye labeled). References for the main function description are found in Supplementary Table S1 (ST1) (item 2). Further details can be obtained at http:/www.appliedbiosystems.com.
Plasma analysis
The biochemical analysis for glucose, collected during the saline perfusion, was quantified using a glucose colorimetric assay kit, ab65333 – Glucose Assay kit, (Abcam, Cambridge, UK), according to the manufacturer’s instruction. In our experiments, the inter-assay variability was 3.8%. C-peptide quantification was performed using a sandwich-type immunoassay ELISA kit, Cat# 80-CPTMS-E01, (Alpco, Salem, NH, U.S.A), according to the manufacturer’s instruction. In our experiments, the inter-assay variability is 1.8%, and according to the manufacturer, the intra-assay variability is 4.1%. Insulin quantification was performed using a mouse insulin ELISA kit, Cat# EMINS, (Thermo Fisher Scientific, Carlsbad, CA 92008, U.S.A), according to the manufacturer’s instruction. The inter-assay variability was 8.7%, and according to the manufacturer, the intra-assay variability was < 10%. IGF-1 quantification was performed using an immunoassay solid phase ELISA, Cat# MG100 (R&D Systems, Inc, Minneapolis, MN 55413, U.S.A), according to the manufacturer’s instruction. The inter-assay variability was 5.4%, and according to the manufacturer, the intra-assay variability was 4.1%.
Homeostatic model of assessment of insulin resistance (HOMA-IR).
HOMA-IR was calculated by multiplying fasting insulin in IU/mL by fasting glucose in mmol/L, the sum divided by a factor of 22.5 51.
Statistical analysis
Values are presented as the mean ± 95% confidence interval (CI). Two-tailed t-test analyses were used for plasma and transcript-specific analyses for each brain region. The RT-qPCR gene expression values were logarithmically transformed to better approximate normal distribution, where appropriate. Apart from the individual t-tests for each brain region and transcripts, a mixed model was used to evaluate the aggregated effects of running concerning all brain regions and transcripts categories. Specifically, mixed models allow examining both fixed effects (as in the example of ordinary ANOVA) and random effects. Mouse was used as a random effect to account for the within-mouse correlation. We used maximum likelihood to deal with variations in data. Contrasts were constructed to compare the different brain regions and categories of transcripts rather than including category as a factor in the model. Spearman correlations were used to evaluate correlations, and rho (r), n, and p-values were presented. Correlation strengths (rho values; r) were reported according to Cohen52, being very small for r < 0.1, small for r < 0.1 to 0.3; moderate for r < 0.3 to 0.5, and large for r < 0.5. P values < 0.05 were considered statistically significant. SPSS (version 29.0.1.1) was used for statistical analyses.
For the running distance data, four out of twelve animals had partially missing data due to connectivity issues with the wireless running wheel, with recordings absent for two to five days. The missing values are presented in two ways (Supplementary Fig. 1 (item 3)). They were either set to 0 (without interpolation), or secondly, they were replaced with the average running distance for the dark or light cycle (with interpolation). Specifically, if a value was missing in the dark cycle, it was replaced by the average of all recorded dark cycle values for the same animal. Conversely, if a value was missing in the light cycle, it was replaced by the average of all recorded light cycle values for that animal.
Results
Running affects systemic and metabolic parameters
The running mice’s body weight tended to be slightly higher than that of the sedentary group (28.4 g vs. 27.1 g, p = 0.09) at the start of the experiment, which involved one week of non-running acclimatization with a locked running wheel. The average daily running distance (mean and 95% CI) was 3.07 ± 1.67 km/day; see Supplementary Fig. 1 (item 3); interpolated data, see Methods). The average percentage of time running during the day (mean and 95% CI) was 47.7 ± 12.3% and the average percentage of running activity during the active phase (mean and 95% CI) was 83.8 ± 4.3%. Running activity was not recorded in the sedentary group because the running wheel was locked. In Table 2, the measures reflecting systemic metabolism are presented. There were no differences between the groups regarding C-peptide, glucose, insulin plasma levels, or HOMA-IR index. For IGF-1, there was a tendency towards a slightly higher plasma level in the running group (+ 16%, p = 0.09; Table 2).
Table 2.
Metabolic parameters.
| Unit | Sedentary | Runner | ||
|---|---|---|---|---|
| Mean ± 95% CI | Mean ± 95% CI | p-value | ||
| C-peptide | pmol/L | 285 ± 32 | 262 ± 31 | 0.33 |
| Glucose | mmol/L | 5.71 ± 0.59 | 5.88 ± 0.41 | 0.63 |
| Insulin | mU/L | 1.65 ± 0.91 | 1.21 ± 0.32 | 0.39 |
| IGF-1 | ng/mL | 172 ± 23 | 200 ± 21 | 0.09 |
| HOMA - IR | Insulin x Glucose / 22.5 | 0.44 ± 0.26 | 0.33 ± 0.087 | 0.43 |
Abbreviations are homeostatic model of assessment of insulin resistance for HOMA-IR and insulin-like growth factor 1 for IGF-1. P-values are derived from T-tests. C-peptide (n = 11 for sedentary and n = 11 for runner), Glucose (n = 10 for sedentary and n = 11 for runner), Insulin (n = 9 for sedentary and n = 9 for runner) and IGF-1 (n = 11 for sedentary and n = 11 for runner). There was the same missing plasma sample in the running and sedentary group for all four parameters. For Glucose there was one hemolytic sample excluded whereas for Insulin, the additional missing values in the sedentary and running groups were due to levels in the range of blank (n = 2) or above the standard curve (n = 2).
Running increases Hbb-related transcripts and decreases IGF-1-related transcripts.
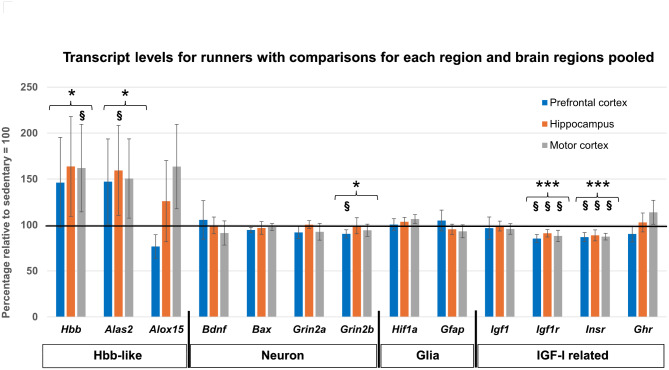
The transcripts for Hbb and Alas2 were upregulated after running in all three brain regions: prefrontal cortex, motor cortex, and hippocampus (Fig. 1; Table 3). The transcripts of the Igf1r and Insr were slightly downregulated (10–15%) by running in all three brain regions investigated. In addition, there was a slight decrease in the transcript Grin2b but only in the prefrontal cortex. The response to running was non-significant for the other transcripts investigated in the three brain regions examined.
Fig. 1.
This Figure visually shows the levels of each transcript also shown in Tables 3 to facilitate comprehension of the pattern of changes in the specific brain regions and aggregates of the three brain regions. Total transcript levels for all three brain regions (prefrontal cortex, hippocampus, and motor cortex) are shown for runners as percentage vs. sedentary animals. Transcript levels have been normalized to Gapdh. Sedentary levels = 100%. Data is presented as mean ± 95% Cl. Individual transcript comparisons in each brain region are made with t-tests, and aggregated effects (brackets) in the three brain regions are made with mixed models (see Methods). *P < 0.05, ***P < 0.001, or t-test, §P < 0.05, were used for statistical comparison as described in Methods.
Table 3.
Quantitative reverse transcription polymerase chain reaction (RT-qPCR) values in runners and sedentary animals.
| Transcript | Group | Prefrontal Cortex | Hippocampus | Motor Cortex | Aggregated# |
|---|---|---|---|---|---|
| Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | ||
| Hbb | Runner | 146.2 (97.1–195.3) | 163.7 (109.3–218.0) | 161.9 (114.3–209.5) | 157.2 (117.3–197.2) |
| Sedentary | 100.0 (71.1–128.9) | 100.0 (69.8–130.2) | 100.0 (71.1–128.9) | 100.0 (60.0–140.0) | |
| Alas2 | Runner | 147.2 (100.8–193.6) | 159.4 (110.4–208.3) | 150.6 (107.4–193.7) | 152.4 (116.0–188.8) |
| Sedentary | 100.0 (72.0–128.0) | 100.0 (73.7–126.3) | 100.0 (73.0–127.0) | 100.0 (63.6–136.4) | |
| Alox15 | Runner | 76.6 (63.9–89.4) | 125.9 (81.8–170.1) | 163.5 (117.6–209.5) | 123.0 (95.8–150.2) |
| Sedentary | 100.0 (66.3–133.7) | 100.0 (60.3–139.7) | 100.0 (43.3–156.7) | 99.7 (72.6–126.8)£ | |
| BDNF | Runner | 105.5 (84.5–126.5) | 99.5 (90.4–108.6) | 91.3 (78.1–104.5) | 98.8 (87.9–109.7) |
| Sedentary | 100.0 (89.0–111.0) | 100.0 (89.6–110.4) | 100.0 (88.7–111.3) | 100.0 (89.1–110.9) | |
| Bax | Runner | 94.5 (91.6–97.4) | 96.8 (89.9–103.7) | 97.8 (94.1–101.6) | 96.4 (93.3–99.5) |
| Sedentary | 100.0 (95.0–105.0) | 100.0 (94.5–105.5) | 100.0 (96.4–103.6) | 100.0 (96.9–103.1) | |
| Grin2a | Runner | 91.9 (85.1–98.7) | 100.4 (96.1–104.7) | 92.6 (83.5–101.7) | 95.0 (91.0–98.9) |
| Sedentary | 100.0 (91.1–108.9) | 100.0 (95.4–104.6) | 100.0 (93.5–106.5) | 100.0 (96.1–103.9) | |
| Grin2b | Runner | 90.3 (85.9–94.8) | 99.2 (90.4–107.9) | 94.3 (87.6–100.9) | 94.6 (90.9–98.3) |
| Sedentary | 100.0 (93.6–106.4) | 100.0 (96.0–104.0) | 100.0 (94.9–105.1) | 100.0 (96.3–103.7) | |
| Hif1a | Runner | 100.5 (94.2–106.8) | 103.5 (98.7–108.3) | 106.4 (101.6–111.3) | 103.5 (99.4–107.5) |
| Sedentary | 100.0 (94.4–105.6) | 100.0 (94.8–105.2) | 100.0 (95.5–104.5) | 100.0 (96.0–104.0) | |
| Gfap | Runner | 104.8 (93.5–116.2) | 95.3 (89.8–100.9) | 93.2 (86.1–100.2) | 100.8 (93.2–108.5) |
| Sedentary | 100.0 (85.5–114.5) | 100.0 (91.0–109.1) | 100.0 (85.5–114.5) | 100.0 (92.4–107.6) | |
| Igf1 | Runner | 96.5 (84.4–108.6) | 98.8 (93.5–104.2) | 95.7 (89.9–101.5) | 97.0 (92.1–101.9) |
| Sedentary | 100.0 (94.1–105.9) | 100.0 (95.6–104.4) | 100.0 (94.4–105.6) | 100.0 (95.1–104.9) | |
| Igf1r | Runner | 85.2 (80.9–89.6) | 90.9 (86.9–94.9) | 88.1 (81.9–94.2) | 88.1 (84.8–91.3) |
| Sedentary | 100.0 (95.5–104.5) | 100.0 (95.1–104.9) | 100.0 (95.3–104.7) | 100.0 (96.8–103.2) | |
| Insr | Runner | 86.8 (81.8–91.7) | 88.7 (82.8–94.7) | 87.3 (83.7–91.0) | 87.6 (83.4–91.8) |
| Sedentary | 100.0 (94.9–105.1) | 100.0 (93.1–106.9) | 100.0 (94.9–105.1) | 100.0 (95.8–104.2) | |
| Ghr | Runner | 90.2 (81.6–98.8) | 102.8 (92.5–113.1) | 113.8 (100.8–126.8) | 102.3 (95.1–109.4) |
| Sedentary | 100.0 (92.9–107.1) | 100.0 (92.8–107.2) | 100.0 (90.1–109.9) | 100.0 (92.9–107.1) |
The values are normalized against sedentary = 100. Mean and variation is given as 95% confidence intervals (95% CI). Significant differences (p < 0.05) are in bold and designate t-test comparisons between runners and sedentary for each brain region. #For all three brain regions aggregated, the mixed model was used for statistical comparison between sedentary and runner (see Methods). The average sedentary values for each brain region were normalized to 100. £The reason for that sedentary was 99.7 for Alox15 in the three brain regions were some missing values (motor cortex; n = 2, prefrontal cortex; n = 1, hippocampus; n = 1). The mixed model algorithm considers this and balances missing values slightly, which slightly changed the calculated mean for this group. The other transcripts had no missing values, and adjusted sedentary means were all 100.0. There were however also missing values in the running group (prefrontal cortex; n = 3, hippocampus; n = 1), which also slightly changed the calculated means (but this is not as easily observed, as means were not expected to be 100). This is also graphically shown in Fig. 2. Transcript abbreviations are found in Table 1.
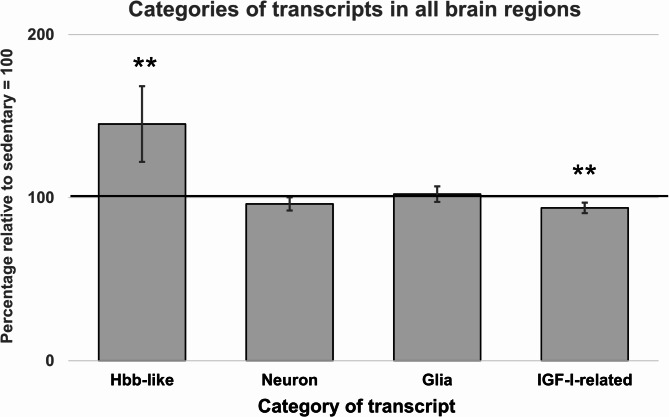
These patterns became even more apparent when the three brain regions were aggregated into one group (Fig. 2). Thus, these analyses confirmed that the running mice had significantly higher transcript levels in the Hbb-like category (45%) and significantly lower transcript levels in the IGF-1-related category (7%) (Fig. 2).
Fig. 2.
Levels of categories of transcripts in all brain regions (prefrontal cortex, hippocampus, and motor cortex). Aggregated comparisons are made with mixed model analysis, as described in Methods (statistical analysis). Sedentary levels = 100%. Data are presented as means ± 95% confidence intervals, **P < 0.01.
Hbb-related gene expression correlates with the IGF-1 system components in running animals
To further indicate changes in relationships between runners and sedentary animals, we performed correlation analyses between Hbb-related transcripts and systemic and local brain components of the IGF-1 system (Table 4). In the running group, Hbb and Alas2 moderately negatively correlated with both Igf1 and Ghr, while Alox15 had a moderate positive correlation with Ghr. In the sedentary group, there were no significant correlations between these transcripts (Table 4). For plasma glucose, there was a moderate negative correlation with Igf1 and Igf1r in the running group. The sedentary group had a moderate negative correlation between glucose and Insr. In the running group, HOMA-IR showed a large negative correlation with Igf1r and a positive moderate correlation with Insr and Ghr. Still, there were no significant correlations in the sedentary group.
Table 4.
Correlations between Hbb-related transcripts, plasma-IGF-I, glucose and HOMA-IR in running and sedentary mice.
| Correlations | Runner | Sedentary | |||||||
|---|---|---|---|---|---|---|---|---|---|
| (Spearman’s rho) | Igf1 | Igf1r | Insr | Ghr | Igf1 | Igf1r | Insr | Ghr | |
| Hbb | rho | −0.38 | −0.25 | −0.074 | −0.33 | 0.26 | 0.025 | −0.19 | 0.037 |
| p | 0.023 | 0.14 | 0.67 | 0.048 | 0.12 | 0.88 | 0.27 | 0.83 | |
| N | 36 | 36 | 36 | 36 | 36 | 36 | 36 | 36 | |
| Alas2 | rho | −0.35 | −0.27 | −0.016 | −0.34 | 0.33 | 0.047 | −0.18 | 0.015 |
| p | 0.038 | 0.11 | 0.93 | 0.043 | 0.051 | 0.79 | 0.29 | 0.93 | |
| N | 36 | 36 | 36 | 36 | 36 | 36 | 36 | 36 | |
| Alox15 | rho | 0.13 | 0.25 | −0.095 | 0.49 | −0.15 | −0.23 | −0.17 | −0.005 |
| p | 0.47 | 0.17 | 0.60 | 0.004 | 0.43 | 0.22 | 0.37 | 0.98 | |
| N | 33 | 33 | 33 | 33 | 32 | 32 | 32 | 32 | |
| Plasma-IGF1 | rho | −0.16 | −0.21 | −0.36 | −0.50 | −0.12 | −0.35 | -0.40 | 0.20 |
| p | 0.36 | 0.25 | 0.042 | 0.003 | 0.50 | 0.049 | 0.022 | 0.26 | |
| N | 33 | 33 | 33 | 33 | 33 | 33 | 33 | 33 | |
| Glucose | rho | -0.35 | -0.37 | 0.31 | −0.017 | 0.034 | -0.10 | -0.41 | -0.018 |
| p | 0.044 | 0.036 | 0.078 | 0.92 | 0.86 | 0.60 | 0.024 | 0.92 | |
| N | 33 | 33 | 33 | 33 | 30 | 30 | 30 | 30 | |
| HOMA-IR | rho | -0.33 | -0.53 | 0.46 | 0.36 | 0.39 | 0.15 | −0.14 | −0.14 |
| p | 0.094 | 0.005 | 0.016 | 0.067 | 0.060 | 0.48 | 0.50 | 0.52 | |
| N | 27 | 27 | 27 | 27 | 24 | 24 | 24 | 24 | |
The Spearman correlations are between the values for all values of all brain regions versus the levels of each transcript. Abbreviations are as in Table 1. and HOMA-IR is the homeostatic model of assessment of insulin resistance. N represents the pooled individual samples with successful analysis for all three brain regions. The reasons for not reaching N = 36 (i.e. missing numbers) are given in Tables 2, 3.
Discussion
The main finding of the present study is that exercise increased the expression of the Hbb and Alas2 transcripts in three regions of the mouse brain. Additionally, running slightly decreased IGF-1-related transcripts, whereas systemic components were unaffected. Interestingly, the correlation analyses only yielded significant results in the running group and for Hbb-like transcripts (Table 4). However, a negative correlation was found between local brain Insr and glucose in the sedentary group. We have previously observed increased Hbb-like transcripts when Hx rodents received replacement with GH and IGF-1 41. In the present study, the increase in these transcripts after running appears to be similar but of less magnitude to that seen after GH or IGF-1 replacement in Hx rodents. Furthermore, we found a tendency towards a 16% increase in peripheral plasma IGF-1 after running. This is in line with reports showing that effects of exercise on brain function are mediated by increased uptake of circulating IGF-1 4, which is necessary for the generation of new hippocampal neurons53.
Regarding Hbb and Alas2 in the brain, it could be questioned whether the measured transcript levels represent their expression in the brain or originate from the bloodstream. We have previously shown54, that Hbb and Alas2 detected in the brain at most have minor contributions (approximately 1–10%) from remaining blood cells. Therefore, the presently detected levels of Hbb and Alas2 are indeed from the brain, although some minor contamination from the bloodstream cannot be ruled out.
We also detected that running decreased the expression of the Insr and Igf1r transcripts. Previously, it has been shown that expression of Insr decreases in running vs. sedentary rats55. Furthermore, another study reports that insulin transport across the blood-brain barrier is augmented following exercise, although serum insulin levels are unchanged56. Moreover, higher IGF-1 in plasma may have produced a lowering effect on the expression of Igf-1r potentially induced by negative feedback. In contrast, our study could not detect any difference in plasma insulin levels, glucose levels, or HOMA-IR index between the running and sedentary mice. Body weight was similar in the two groups, which suggests that metabolism was minimally affected by the 7-day running paradigm.
When selecting 6-month-old mice for this study, we aimed to achieve a high capacity for voluntary running to capture an exercise-induced upregulation of neuronal Hbb. Total voluntary running distance has been reported to decline by approximately 45% in mice around 10 months old compared to younger mice aged 5 to 14 weeks57. However, these animals are relatively healthy in terms of metabolism. The effects of endurance exercise on metabolic profile would likely have been more apparent in older or obese animals with insulin resistance. Also, other breeds of mice with a genetic risk of insulin resistance, such as GLUT4 +/- mice58,59 and db/db mice60, could have been used to find more robust effects of running on insulin resistance. The generalizability is probably high to other rodent strains, but somewhat less known to humans. However, in general, exercise and insulin resistance have rather similar relations in humans and rodents, with larger variations for use of different ages and comorbidities.
We identified that both Hbb and Alas2 correlated with Igf1 and Ghr in the running group, which further strengthens our hypothesis that there is an association between Hbb-like and IGF-1-related transcripts. While Alox15 was not significantly increased in any of the specific brain regions, there was a numerical increase (Table 3), which in the aggregated mixed model analysis for the three brain regions and Hbb, Alas2, and Alox15 became significant (Fig. 2). Although Alox15 has some relation with Hbb and Alas2 in terms of affecting hemoglobin levels40, and in terms of being increased by GH treatment in parallel with the equally increased Hbb and Alas241, its direct function in synthesizing lipid metabolites is nevertheless unrelated to hemoglobin synthesis. Therefore, it may not be surprising that Alox15 is less affected by running than Hbb and Alas2. Also, running in the present experiment increased plasma IGF-1 (16%, p = 0.09) less than in the GH treatment mentioned (≈ 2.5-fold41).
Further on, neither neuro-related nor glia-related category transcripts were affected by running exercise. Apart from direct effects on the levels of transcripts by running, correlation patterns may indicate the presence of actively functioning pathways, for example, through IGF1 stimulation of the IGF-1R. Interestingly, significant moderate to large correlations were found almost exclusively in the running group (Table 4), which further supports the notion that there is a relation between Hbb-like transcripts and the local IGF-1 system in the brain, which is activated in runners. In the literature, we could not find any evidence that an association between Hbb and IGF-1 in the brain has previously been shown. Also, it appears that some of the small (r = 0.1–0.3) non-significant correlations may have some biological significance but would have required higher statistical power, i.e., more animals, to have been detected significant. Also, it deserves to be mentioned that correlation patterns should be interpreted with caution and should not be regarded as causal.
Our results suggest that running may indeed increase brain Hbb-like transcripts. This presents a new mechanism of action for the previously known neuroprotective resilience of running61–63. Therefore, Hbb-like transcripts and proteins may be a target of intervention to convey neuroprotection in brain ischemia. This could be investigated in experimental stroke studies, perhaps using running intervention. Further, it would be interesting to check the single-cell expression of Hbb in different brain cell types and to do a knockout model of Hbb, specifically in neurons.
Summary
Running for seven days substantially increased Hbb-related transcripts in three brain regions. This may be due to an increase in peripheral IGF-1 and an increased transport of insulin into the brain after exercise, which in turn could contribute to a lowering of the local Insr and Igf1r transcripts. The correlations between Hbb-like and IGF-1 system transcripts in the running group but not in the sedentary group further support a link between IGF-1-related and Hbb-like transcripts in response to exercise.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
M.W., L.K., R.M., J.I., H-G.K. and N.D.Å. designed the study. L.K. and R.M. planned and carried out the experiments. M.W. performed the analysis and drafted the manuscript. N.D.Å. processed the experimental data and supervised the project. M.W., L.K., R.M., J.I., H-G.K., J.S. and N.D.Å. contributed to the interpretation of the results and critical revision of the final manuscript.
Funding
Open access funding provided by University of Gothenburg. Nils David Åberg: Stiftelsen Peter Erikssons Minnesfond, Grants from the Swedish Government and the county councils, the ALF-agreement (ALFGBG-965762, ALFGBG-719761), Swedish stroke association, Stiftelsen Hjalmar Svenssons forskningsfond (HJSV20200229). Hans Georg Kuhn: Grants from the Swedish Government and the county councils, the ALF-agreement (ALFGBG-726541), Swedish medical research council (VR-MH 2019 − 01637).
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sutoo, D. & Akiyama, K. Regulation of brain function by exercise. Neurobiol. Dis. 13, 1–14. 10.1016/s0969-9961(03)00030-5 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Lucas, S. J., Cotter, J. D., Brassard, P. & Bailey, D. M. High-intensity interval exercise and cerebrovascular health: curiosity, cause, and consequence. J. Cereb. Blood Flow. Metab. 35, 902–911. 10.1038/jcbfm.2015.49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deslandes, A. et al. Exercise and mental health: many reasons to move. Neuropsychobiology 59, 191–198. 10.1159/000223730 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Carro, E., Nuñez, A., Busiguina, S. & Torres-Aleman, I. Circulating insulin-like growth factor I mediates effects of exercise on the brain. J. Neurosci. 20, 2926–2933 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaynman, S., Ying, Z. & Gomez-Pinilla, F. Hippocampal BDNF mediates the efficacy of exercise on synaptic plasticity and cognition. Eur. J. Neurosci. 20, 2580–2590. 10.1111/j.1460-9568.2004.03720.x (2004). [DOI] [PubMed] [Google Scholar]
- 6.Llorens-Martín, M., Torres-Alemán, I. & Trejo, J. L. Exercise modulates insulin-like growth factor 1-dependent and -independent effects on adult hippocampal neurogenesis and behaviour. Mol. Cell. Neurosci. 44, 109–117. 10.1016/j.mcn.2010.02.006 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Zhu, Y., Sun, Y., Hu, J. & Pan, Z. Insight into the mechanism of exercise preconditioning in ischemic stroke. Front. Pharmacol. 1310.3389/fphar.2022.866360 (2022). [DOI] [PMC free article] [PubMed]
- 8.Geng, H. et al. Early rehabilitation exercise after stroke improves neurological recovery through enhancing angiogenesis in patients and cerebral ischemia rat model. Int. J. Mol. Sci. 2310.3390/ijms231810508 (2022). [DOI] [PMC free article] [PubMed]
- 9.Wideman, L., Weltman, J. Y., Hartman, M. L., Veldhuis, J. D. & Weltman, A. Growth hormone release during acute and chronic aerobic and resistance exercise: recent findings. Sports Med. 32, 987–1004. 10.2165/00007256-200232150-00003 (2002). [DOI] [PubMed] [Google Scholar]
- 10.van Praag, H., Christie, B. R., Sejnowski, T. J. & Gage, F. H. Running enhances neurogenesis, learning, and long-term potentiation in mice. Proc. Natl. Acad. Sci. U. S. A. 96, 13427–13431. 10.1073/pnas.96.23.13427 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Praag, H., Kempermann, G. & Gage, F. H. Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat. Neurosci. 2, 266–270. 10.1038/6368 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Blackmore, D. G., Golmohammadi, M. G., Large, B., Waters, M. J. & Rietze, R. L. Exercise increases neural stem cell number in a growth hormone-dependent manner, augmenting the regenerative response in aged mice. Stem Cells 27, 2044–2052 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Blackmore, D. G. & Waters, M. J. The multiple roles of GH in neural ageing and injury. Front. Neurosci. 17, 1082449. 10.3389/fnins.2023.1082449 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bianchi, V. E., Locatelli, V. & Rizzi, L. Neurotrophic and neuroregenerative effects of GH/IGF1. Int. J. Mol. Sci. 1810.3390/ijms18112441 (2017). [DOI] [PMC free article] [PubMed]
- 15.Baltazar-Lara, R. et al. Neuroprotective effects of growth hormone (GH) and insulin-like growth factor type 1 (IGF-1) after hypoxic-ischemic injury in chicken cerebellar cell cultures. Int. J. Mol. Sci. 2210.3390/ijms22010256 (2020). [DOI] [PMC free article] [PubMed]
- 16.Pan, W. et al. Permeation of growth hormone across the blood-brain barrier. Endocrinology 146, 4898–4904 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Brooks, A. J. & Waters, M. J. The growth hormone receptor: mechanism of activation and clinical implications. Nat. Rev. Endocrinol. 6, 515–525. 10.1038/nrendo.2010.123 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Chia, D. J. Minireview: mechanisms of growth hormone-mediated gene regulation. Mol. Endocrinol. 28, 1012–1025. 10.1210/me.2014-1099 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’Ercole, A. J., Stiles, A. D. & Underwood, L. E. Tissue concentrations of somatomedin C: further evidence for multiple sites of synthesis and paracrine or autocrine mechanisms of action. Proc. Natl. Acad. Sci. U. S. A. 81, 935–939. 10.1073/pnas.81.3.935 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quik, E. H. The Somatotropic Axis: Effects on Brain and Cognitive Functions (Utrecht University, 2012).
- 21.Armstrong, C. S., Wuarin, L. & Ishii, D. N. Uptake of circulating insulin-like growth factor-I into the cerebrospinal fluid of normal and diabetic rats and normalization of IGF-II mRNA content in diabetic rat brain. J. Neurosci. Res. 59, 649–660 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Nishijima, T. et al. Neuronal activity drives localized blood-brain-barrier transport of serum insulin-like growth factor-I into the CNS. Neuron 67, 834–846. 10.1016/j.neuron.2010.08.007 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Aberg, N. D., Brywe, K. G. & Isgaard, J. Aspects of growth hormone and insulin-like growth factor-I related to neuroprotection, regeneration, and functional plasticity in the adult brain. Sci. World J. 6, 53–80. 10.1100/tsw.2006.22 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trejo, J. L. et al. Central actions of liver-derived insulin-like growth factor I underlying its pro-cognitive effects. Mol. Psychiatry 12, 1118–1128. 10.1038/sj.mp.4002076 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Yan, H. et al. Circulating IGF1 regulates hippocampal IGF1 levels and brain gene expression during adolescence. J. Endocrinol. 211, 27–37. 10.1530/joe-11-0200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D’Ercole, A. J., Ye, P., Calikoglu, A. S. & Gutierrez-Ospina, G. The role of the insulin-like growth factors in the central nervous system. Mol. Neurobiol. 13, 227–255. 10.1007/BF02740625 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Folli, F., Ghidella, S., Bonfanti, L., Kahn, C. R. & Merighi, A. The early intracellular signaling pathway for the insulin/insulin-like growth factor receptor family in the mammalian central nervous system. Mol. Neurobiol. 13, 155–183. 10.1007/BF02740639 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Andersson, I. K. et al. Differing expression of insulin-like growth factor I in the developing and in the adult rat cerebellum. Acta Physiol. Scand. 132, 167–173. 10.1111/j.1748-1716.1988.tb08314.x (1988). [DOI] [PubMed] [Google Scholar]
- 29.Shinar, Y. & McMorris, F. A. Developing oligodendroglia express mRNA for insulin-like growth factor-I, a regulator of oligodendrocyte development. J. Neurosci. Res. 42, 516–527. 10.1002/jnr.490420410 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Popken, G. J., Dechert-Zeger, M., Ye, P. & D’Ercole, A. J. Brain development. Adv. Exp. Med. Biol. 567, 187–220. 10.1007/0-387-26274-1_8 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Richter, F., Meurers, B. H., Zhu, C., Medvedeva, V. P. & Chesselet, M. F. Neurons express hemoglobin α- and β-chains in rat and human brains. J. Comp. Neurol. 515, 538–547. 10.1002/cne.22062 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He, Y. et al. Brain alpha- and beta-globin expression after intracerebral hemorrhage. Transl. Stroke Res. 1, 48–56. 10.1007/s12975-009-0004-x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biagioli, M. et al. Unexpected expression of alpha- and beta-globin in mesencephalic dopaminergic neurons and glial cells. Proc. Natl. Acad. Sci. U. S. A. 106, 15454–15459. (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russo, R. et al. Hemoglobin is present as a canonical α2β2 tetramer in dopaminergic neurons. Biochim. Biophys. Acta Proteins Proteom. 1834, 1939–1943. 10.1016/j.bbapap.2013.05.005 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Sadlon, T. J., Dell’Oso, T., Surinya, K. H. & May, B. K. Regulation of erythroid 5-aminolevulinate synthase expression during erythropoiesis. Int. J. Biochem. Cell Biol. 31, 1153–1167 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Baron, M. H. Transcriptional control of globin gene switching during vertebrate development. Biochim. Biophys. Acta 1351, 51–72. 10.1016/s0167-4781(96)00195-9 (1997). [DOI] [PubMed] [Google Scholar]
- 37.Shalini, S. M. et al. Distribution of Alox15 in the rat brain and its role in prefrontal cortical resolvin D1 formation and spatial working memory. Mol. Neurobiol. 55, 1537–1550. 10.1007/s12035-017-0413-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivanov, I., Kuhn, H. & Heydeck, D. Structural and functional biology of arachidonic acid 15-lipoxygenase-1 (ALOX15). Gene 573, 1–32 10.1016/j.gene.2015.07.073 (2015). [DOI] [PMC free article] [PubMed]
- 39.Singh, N. K. & Rao, G. N. Emerging role of 12/15-Lipoxygenase (ALOX15) in human pathologies. Prog. Lipid Res. 73, 28–45. 10.1016/j.plipres.2018.11.001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schäfer, M. et al. Male knock-in mice expressing an arachidonic acid lipoxygenase 15B (Alox15B) with humanized reaction specificity are prematurely growth arrested when aging. Biomedicines 1010.3390/biomedicines10061379 (2022). [DOI] [PMC free article] [PubMed]
- 41.Walser, M. et al. Effects of peripheral administration of GH and IGF-I on gene expression in the hippocampus of hypophysectomised rats. Neuroendocrinol. Lett. 39, 525–531 (2019). [PubMed] [Google Scholar]
- 42.Walser, M. et al. Different modes of GH administration influence gene expression in the male rat brain. J. Endocrinol. 222, 181–190. 10.1530/JOE-14-0223 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Behrmann, M., Geng, J. J. & Shomstein, S. Parietal cortex and attention. Curr. Opin. Neurobiol. 14, 212–217. 10.1016/j.conb.2004.03.012 (2004). https://doi.org:. [DOI] [PubMed] [Google Scholar]
- 44.Yip, D. W. & Lui, F. in StatPearls, Physiology, Motor Cortical (StatPearls Publishing Copyright © 2023, StatPearls Publishing LLC., 2023). [PubMed]
- 45.Anand, K. S. & Dhikav, V. Hippocampus in health and disease: an overview. Ann. Indian Acad. Neurol. 15, 239–246. 10.4103/0972-2327.104323 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thyfault, J. P. & Bergouignan, A. Exercise and metabolic health: beyond skeletal muscle. Diabetologia 63, 1464–1474. 10.1007/s00125-020-05177-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karlsson, L. et al. Constitutive PGC-1α overexpression in skeletal muscle does not contribute to Exercise-Induced Neurogenesis. Mol. Neurobiol. 58, 1465–1481. 10.1007/s12035-020-02189-6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiu, K., Lau, W. M., Lau, H. T., So, K. F. & Chang, R. C. Micro-dissection of rat brain for RNA or protein extraction from specific brain region. J. Visualized Exp. JoVE 269. 10.3791/269 (2007). [DOI] [PMC free article] [PubMed]
- 49.Chomczynski, P. & Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159. (1987). [DOI] [PubMed] [Google Scholar]
- 50.Blomstrand, F., Åberg, N. D., Eriksson, P. S., Hansson, E. & Ronnback, L. Extent of intercellular calcium wave propagation is related to gap junction permeability and level of connexin-43 expression in astrocytes in primary cultures from four brain regions. Neuroscience 92, 255–265 (1999). [DOI] [PubMed] [Google Scholar]
- 51.Matthews, D. R. et al. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28, 412–419. 10.1007/bf00280883 (1985). [DOI] [PubMed] [Google Scholar]
- 52.Cohen, J. Statistical Power Analysis for the Behavioral Sciences 2nd edn (Routledge, 1988). 10.4324/9780203771587
- 53.Trejo, J. L., Carro, E. & Torres-Aleman, I. Circulating insulin-like growth factor I mediates exercise-induced increases in the number of new neurons in the adult hippocampus. J. Neurosci. 21, 1628–1634. 10.1523/jneurosci.21-05-01628.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walser, M. et al. Brain tissue haemoglobin expression in saline-perfused vs non-perfused rodents. Heliyon 1010.1016/j.heliyon.2023.e23343 (2024). [DOI] [PMC free article] [PubMed]
- 55.Machrina, Y., Purba, A., Lindarto, D. & Maskoen, A. M. Exercise intensity alter insulin receptor gene expression in diabetic type – 2 rat model. Open Access Maced J. Med. Sci. 7, 3370–3375. 10.3889/oamjms.2019.425 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown, C. et al. Insulin blood-brain barrier transport and interactions are greater following exercise in mice. J. Appl. Physiol. (Bethesda, Md. : 1985) 132 824–834. 10.1152/japplphysiol.00866.2021 (2022). [DOI] [PMC free article] [PubMed]
- 57.Manzanares, G., Brito-da-Silva, G. & Gandra, P. G. Voluntary wheel running: patterns and physiological effects in mice. Braz. J. Med. Biol. Res. 52, e7830. 10.1590/1414-431x20187830 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stenbit, A. E. et al. GLUT4 heterozygous knockout mice develop muscle insulin resistance and diabetes. Nat. Med. 3, 1096–1101. 10.1038/nm1097-1096 (1997). [DOI] [PubMed] [Google Scholar]
- 59.Nandi, A., Kitamura, Y., Kahn, C. R. & Accili, D. Mouse models of insulin resistance. Physiol. Rev. 84, 623–647. 10.1152/physrev.00032.2003 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Guest, C. P. & RahmouneH. Characterization of the db/db mouse model of type 2 diabetes. Methods Mol. Biol. (Clifton N. J.) 1916, 195–201. 10.1007/978-1-4939-8994-2_18 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Saraulli, D., Costanzi, M., Mastrorilli, V. & Farioli-Vecchioli, S. The long run: neuroprotective effects of physical exercise on adult neurogenesis from youth to old age. Curr. Neuropharmacol. 15, 519–533. 10.2174/1570159x14666160412150223 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vecchio, L. M. et al. The neuroprotective effects of exercise: maintaining a healthy brain throughout aging. Brain Plast. 4, 17–52. 10.3233/bpl-180069 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu, Y. et al. The beneficial effects of physical exercise in the brain and related pathophysiological mechanisms in neurodegenerative diseases. Lab. Invest. 99, 943–957. 10.1038/s41374-019-0232-y (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.