Abstract
Protein tyrosine kinase 6 (PTK6) is an intracellular tyrosine kinase that is distantly related to the SRC family of tyrosine kinases. It is expressed in epithelial linings and regulates regeneration and repair of the intestinal epithelium. Analysis of publicly available datasets showed Ptk6 is upregulated in tuft cells upon activation of type 2 immunity. We found that disruption of Ptk6 influences gene expression involved in intestinal immune responses. Administration of succinate, which mimics infection and activates tuft cells, revealed PTK6-dependent activation of innate immune responses in male but not female mice. In contrast to all wild type and Ptk6−/− female mice, Ptk6−/− male mice do not activate innate immunity or upregulate differentiation of the tuft and goblet secretory cell lineages following succinate treatment. Mechanistically, we found that PTK6 regulates Il25 and Irag2, genes that are required for tuft cell effector functions and activation of type 2 innate immunity, in organoids derived from intestines of male but not female mice. In patients with Crohn’s disease, PTK6 is upregulated in tuft cells in noninflamed regions of intestine. These data highlight roles for PTK6 in contributing to sex differences in intestinal innate immunity and provide new insights into the regulation of IL-25.
Subject terms: Biochemistry, Cell signalling, Innate immune cells
Introduction
The rapidly regenerating intestinal epithelium contains anchored stem cells that give rise to absorptive cells and different secretory cells, including enteroendocrine, goblet, Paneth, and tuft cells. Mucus-secreting goblet cells and anti-microbial peptide-secreting Paneth cells are well-recognized regulators of host-microbe interactions [1, 2]. More recently, tuft cells were identified as activators of innate immunity and part of the surveillance system for extracellular pathogens such as helminths, protists, and bacteria [3–6]. Emerging studies also suggest roles for tuft cells in repair processes, such as the attenuation of chronic inflammation [7].
Tuft cells are rare epithelial cells in the intestine under normal conditions. They specifically express interleukin (IL)-25, a cytokine that is necessary for the activation of innate immunity [5]. Tuft cells express G protein-coupled receptors such as taste receptors [8, 9], the succinate receptor 1 (SUCNR1) [10–12], or the vomeronasal receptor Vmn2r26 [6], which detect pathogen metabolites in their environment. In mice, administration of the metabolite succinate in drinking water is sufficient to induce an innate immune response in the intestine [10–12]. Following activation of these receptors, tuft cells secrete IL-25 to alarm immune cells in the lamina propria that pathogens are present. IL-25 signals to innate lymphoid cells type 2 (ILC2s) to produce IL-13 and activate the immune cascade [5]. IL-13 drives the type 2 immune response through the recruitment of other immune cells and upregulation of Gasdermin C proteins (GSDMCs), which have roles in pyroptosis, caspase-1-dependent programmed cell death associated with inflammation [6, 13]. In addition, IL-13 promotes secretory cell differentiation, including differentiation of tuft and goblet cells that increase in number [3–5]. Increases in mucus production, fluid secretion, and muscle contraction generate a “weep and sweep” response to eliminate infections [14, 15].
Protein tyrosine kinase 6 (PTK6, also known as BRK or Sik) is an intracellular non-receptor tyrosine kinase distantly related to the SRC family, which belongs to the PTK6 family of protein kinases together with FRK and SRMS (reviewed in [16]). PTK6 is expressed in regenerating epithelial linings of the skin and gastrointestinal tract [17–19] and has been shown to regulate intestinal growth and differentiation [18, 20]. PTK6 regulates the WNT/β-catenin signaling pathway [21], as well as signaling downstream of several growth factor receptors, including EGFR, IGF1R, and MET (reviewed in [16]). PTK6 expression can be induced by different stress signals, including DNA damage, from γ-irradiation [22, 23], ultraviolet radiation [24], azoxymethane [25], to hypoxia [26]. Damaging signals lead to the upregulation of PTK6 in the progenitor cell/stem cell compartment in the intestine, to promote regeneration and repair of intestinal epithelium [22, 25].
New bulk RNA sequencing (RNA-seq) data revealed differences in mucosal innate immune response gene expression in mice with germline disruption of Ptk6. In publicly available datasets [27, 28], we detected increased Ptk6 expression in the intestine following infections with the nematode parasite Heligmosomoides bakeri (Hb) or Salmonella, different activators of type 2 immunity. Here we demonstrate that PTK6 is essential for activation of type 2 immunity in male but not female mice. These data reveal important mechanistic differences in activation of type 2 immunity between sexes, which may have an impact on sexual disparities in disease pathogenesis.
Results
PTK6 regulates sex-specific gene expression in the intestine
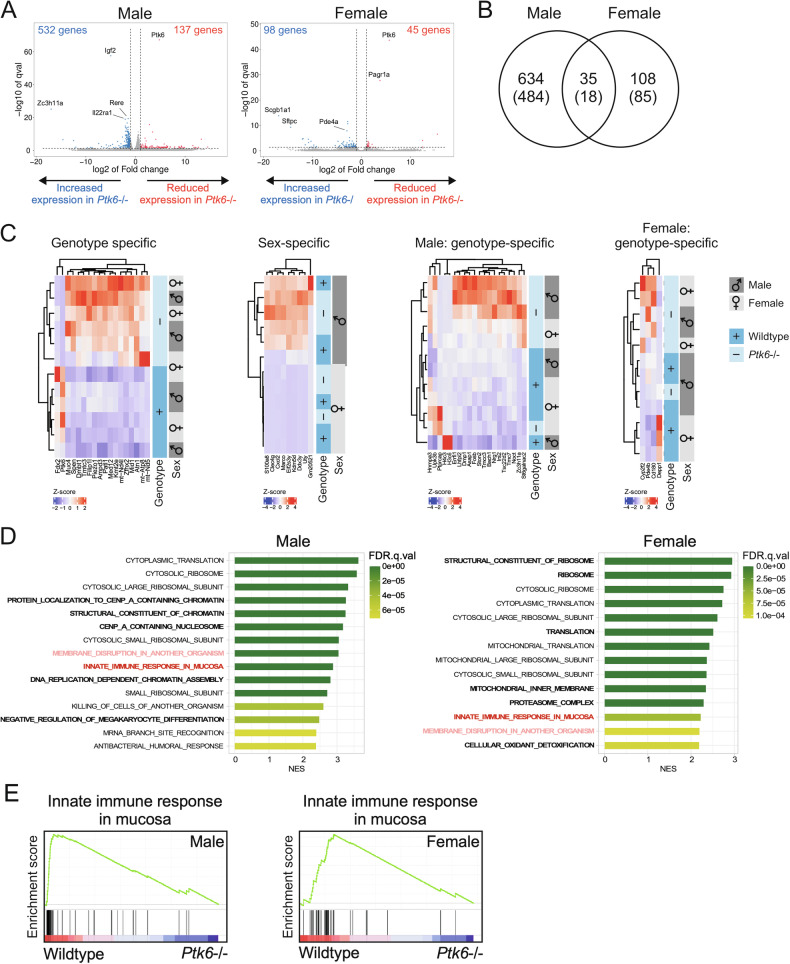
Previous in vivo studies of PTK6 in the intestine utilized only male mice [18, 22, 25]. To determine if there are sex-specific differences in PTK6 functions, we performed bulk RNA-seq using total RNA from distal jejunums of male and female wild type and Ptk6−/− mice. Disruption of Ptk6 led to more significant changes in gene expression in males than females. When compared with wild-type controls, increased expression of 532 genes was detected in male Ptk6−/− mice, compared with increased expression of only 98 genes in Ptk6−/− female mice (Fig. 1A). Similarly, loss of Ptk6 led to decreased expression of 137 genes in males and 45 genes in females when compared with controls. Only 35 differentially expressed genes (DEG), of which 18 are protein coding, were shared between males and females (Fig. 1B, Table S1). One of the most significantly upregulated genes in Ptk6−/− male mice is Il22ra1, which is expressed in all cell types, but is required for IL-22-induced differentiation of Paneth cells [29], as well as a specific subtype of goblet cells [30]. Heatmaps show the expression of top significant protein-coding DEGs between genotypes, and sexes, as well as significantly different expressed genes in male and female tissues dependent on genotype (Fig. 1C).
Fig. 1. PTK6 regulates sex-specific gene expression and innate immunity genes in the intestine.
A Analysis of jejunal RNA expression in male and female wild type and Ptk6−/− mice suggests unique roles for PTK6 in males and females. The volcano plots of differentially expressed genes (DEGs) show that male mice have more DEGs than female mice, with more genes changing in expression in male mice upon Ptk6 loss. (N = 3 per group). B Male and female wild type and Ptk6−/− mice share only 35 genes among significantly DEGs. A Venn diagram shows number of male DEGs only, female DEGs only and mutual DEGs in the overlap. The number of all genes is shown on top, while protein coding genes are shown below in parenthesis. C Heatmaps show expression of significant DEGs between genotypes, sexes, as well as significantly different genes in males and females that are genotype specific. D Gene set enrichment analysis of genes expressed in male and female wild type and Ptk6−/− mice. Innate immune response in mucosa (bold red) is one of the top different gene sets in both male and female mice, based on normalized enrichment score (NES). Another top gene pathway is “Membrane disruption in another organism” (bold light red), which shares genes with “Innate immune response in mucosa”. Male mice have some specific gene sets that do not display significantly different gene expression in females, and vice versa (bold black). The top 15 gene sets are shown. NES represents normalized enrichment score to account for the gene set size. E The enrichment plots of the innate immune response in mucosa gene set from GSEA for male and female mice. The leading-edge subset shows a cluster of genes that are responsible for high enrichment scores (ES) in wild-type mice. For male mice: ES = 0.74977636, NES = 2.876303, NOM p-value < 0.0001, FDR q-value < 0.0001. For female mice: ES = 0.7265350, NES = 2.2186077, NOM p-value < 0.0001, FDR q-value < 0.0001.
To understand the differences between wildtype and Ptk6−/− mice, we used gene set enrichment analysis (GSEA). Transcriptome analysis identified the “innate immune response in mucosa” as one of the significantly different biological pathways between wild type and Ptk6−/− mice of both sexes (Fig. 1D, Table S1). Annotated genes included mostly Paneth cell genes (Defa) and some type 2 immunity genes (Il4), and the majority of these, including Defa35, Defa27 and Defa34, were among the leading-edge genes in both male and female mice (Fig. 1E). Even though tuft and goblet cell marker genes were not in this gene set, there is a crosstalk between tuft and Paneth cells [31, 32], as well as tuft and goblet cells [5]. This suggested that PTK6 could have a role in the regulation of intestinal innate mucosal immunity responsible for defense against different pathogens, including bacteria, protists, helminths, fungi, and some viruses [4, 6, 33, 34].
Ptk6 is upregulated in tuft cells by pathogens that activate intestinal type 2 immunity
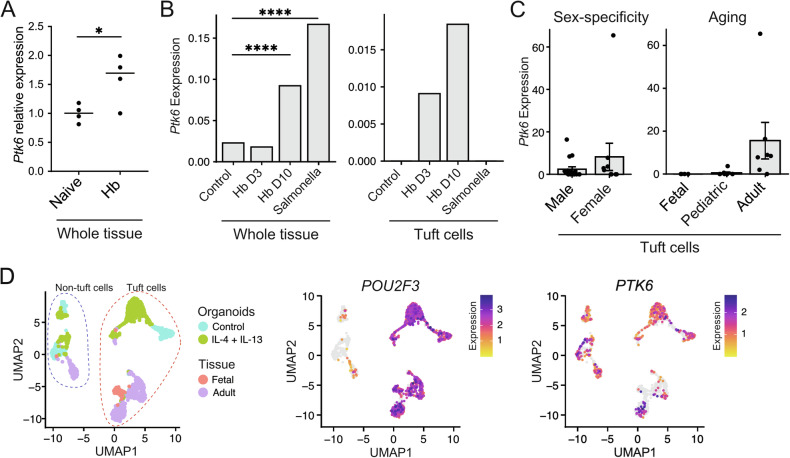
We examined publicly available datasets from different intestinal infection models for Ptk6 expression. Analysis of RNA-seq data from mice infected with Heligmosomoides bakeri (Hb), showed increased expression of Ptk6 at 24 h after infection (Fig. 2A) [28]. In a single cell (sc) RNA-seq study [27], we found Ptk6 is significantly upregulated in small intestinal tissue at 10 days post-Hb infection (Hb D10) and 2 days after Salmonella infection (Fig. 2B). At the single-cell level, Ptk6 expression exhibits an upward trend in tuft cells after Hb infection but was undetectable in tuft cells in uninfected controls and in Salmonella-infected intestines (Fig. 2B).
Fig. 2. Ptk6 is upregulated in tuft cells upon activation of type 2 immunity.
A Ptk6 mRNA expression increases in duodenum 24 h after infection with the helminth Heligmosomoides bakeri (Hb), shown as the change in relative expression from bulk-RNA-seq data [28]. B Ptk6 expression increases by 10 days (D10) in whole intestinal tissue from mice infected with Hb or by 2 days following Salmonella infection [27]. Increased Ptk6 expression was detected specifically in tuft cells in scRNA-seq data post Hb infection. Ptk6 expression is shown as average expression from all cell types per condition (left) or for tuft cells only per condition (right). Significant upregulation of Ptk6 at: day 10 upon H. bakeri infection p. adj = 2.120863e-15; day 2 upon Salmonella infection p.adj = 1.468545e-42. C In the human gastrointestinal tract, PTK6 mRNA expression is upregulated with age in tuft cells, while it appears similar between sexes [35]. D PTK6 expression is upregulated in tuft cells upon IL-13/IL-4 treatment of human intestinal organoids [36]. UMAP shows clustering of tuft and non-tuft cells, and their origins. POU2F3 is shown as a marker for tuft cells. PTK6 is predominantly found in organoid tuft cells, and to a lesser extent in tissue-derived tuft cells.
Exploring data from Elmentaite and colleagues, we detected PTK6 expression in tuft cells in both male and female samples [35]. Interestingly, the majority of PTK6 expression was present in adult tuft cells (Fig. 2C). Upregulation of PTK6 in human tuft cells upon activation of type 2 immunity is confirmed in single-cell RNA-seq data from organoids treated with IL-13 [36] (Fig. 2D). PTK6 expression is particularly high in the population that is upregulated upon IL-4/IL-13 treatment. This suggested that PTK6 may play roles in regulating tuft cell differentiation and function.
Male mice lacking PTK6 fail to upregulate differentiation of secretory cell lineages after succinate treatment
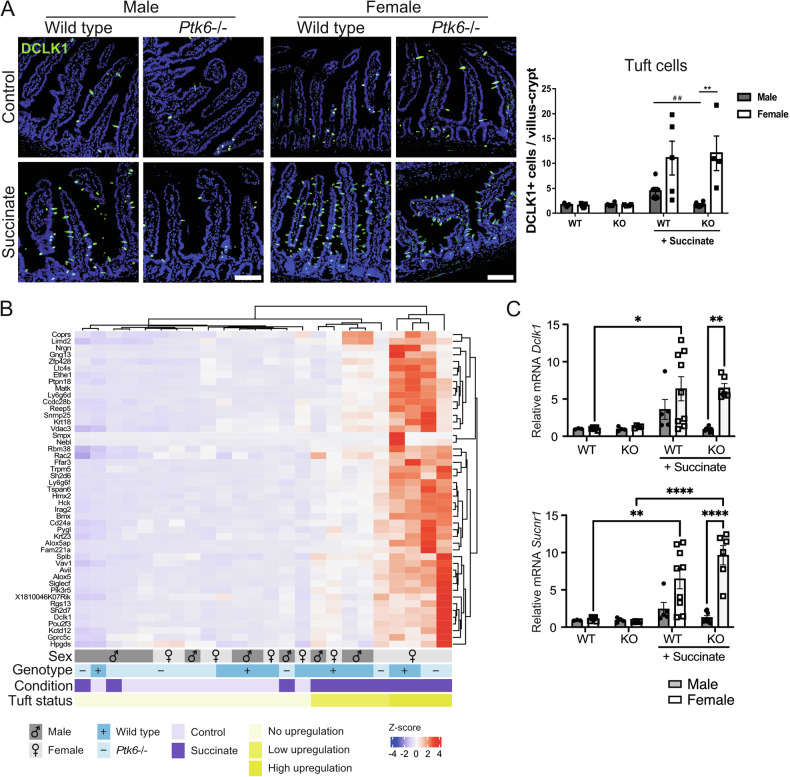
We used the succinate-based system to explore roles of PTK6 in type 2 immunity and tuft cell differentiation [10–12]. Wild type and Ptk6−/− mice were treated with succinate for 7 days and sections of jejunum were stained for expression of the tuft cell marker DCLK1, revealing sex-specific differences in induction of tuft cells. Male Ptk6−/− mice do not exhibit an increase in tuft cells following succinate treatment (Fig. 3A), while both female wild type and Ptk6−/− mice have more tuft cells, than male wild type mice (Fig. 3A). Likewise, tuft cell marker gene expression [27] is significantly upregulated in female jejunums following succinate treatment (Fig. 3B, Table S2). Ptk6−/− male mice treated with succinate express tuft cell marker genes similar to untreated males and females of both genotypes (Fig. 3B). We confirmed differences in expression of the tuft cell markers Dclk1 and Sucnr1 by qRT-PCR (Fig. 3C). Significantly upregulated genes in male and female wild type and Ptk6−/− female mice upon succinate treatment are predominantly part of the type 2 tuft cell gene signature [27] (Fig. S1A, B).
Fig. 3. Tuft cell expansion is impaired in Ptk6−/− male mice after activation of type 2 immunity with succinate.
A Induction of tuft cells is impaired in male Ptk6−/− (KO) mice following succinate treatment. Females exhibit an upward trend for stronger induction of tuft cells than males. Representative images of anti-DCLK1 staining in tuft cells of jejunal tissues from male and female wild type and Ptk6−/− mice. Scale bar, 50 μm. Tuft cell numbers were determined as the total number of DCLK1+ cells per villus-crypt unit. B Expression of tuft cell marker genes identified by Haber et al. [27] was analyzed in jejunal RNA-seq data from male and female wild type and Ptk6−/− mice. Female wild type and Ptk6−/− mice exhibit strong upregulation of tuft cell marker genes, while male Ptk6−/− mice have impaired upregulation of these genes following succinate treatment. C Relative mRNA expression of tuft cell markers Dclk1 and Sucnr1. qRT-PCR was performed with jejunal RNAs from wild type and Ptk6−/− male and female mice. Gene expression is normalized against Rps17 and compared with wild type control mice. For all experiments: RNA-seq n = 3; quantification and qRT-PCR n = 3 to 9 mice. Graphs depict mean ± SEM. Statistics: Two-way ANOVA, adjusted for multiple comparisons using Tukey test. *p-value < 0.05, **,##p-value < 0.01, ****p-value < 0.0001. # is used for comparison within the same sex when shown on the same graph with all comparisons.
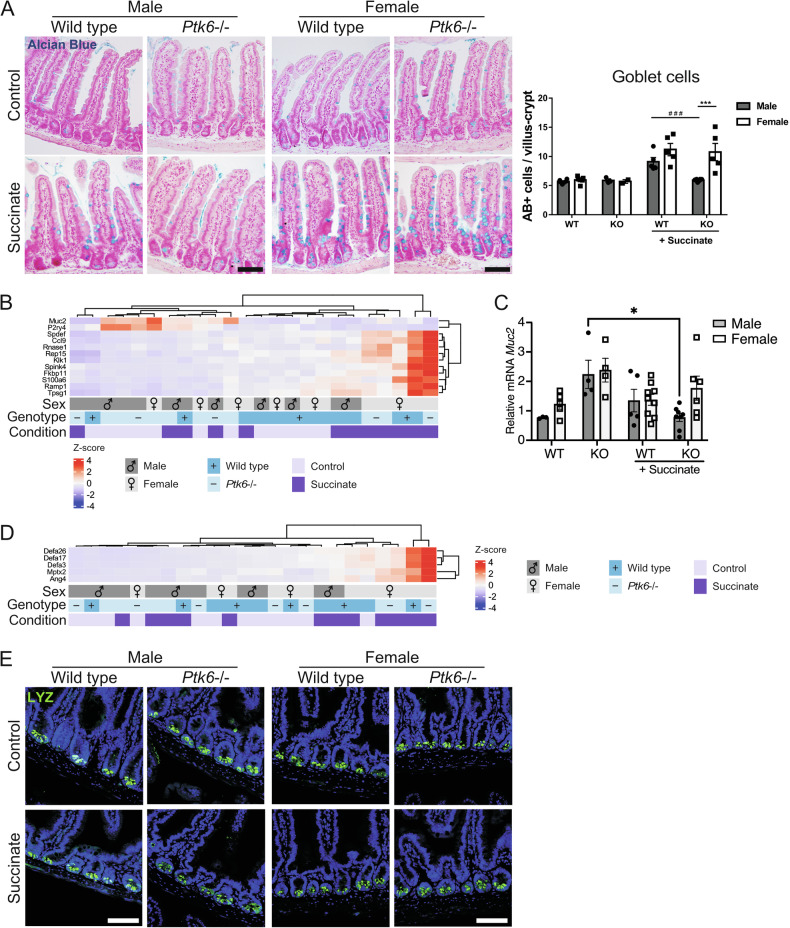
Since goblet cell expansion is characteristic for activation of type 2 immunity [3–5], we examined goblet cell differentiation following succinate treatment of wild type and Ptk6−/− mice. In human tissues, PTK6 is found in both male and female goblet cells and is upregulated with aging (Fig. S1C), consistent with upregulation of secretory cell lineages with aging [37]. In jejunal sections stained with Alcian Blue to detect mucins, we observed sex differences in goblet cell numbers upon activation of type 2 immunity with succinate (Fig. 4A). The numbers of goblet cells in wild type and Ptk6−/− female mice show a positive upward trend, in comparison with wild type males. However, male Ptk6−/− mice do not exhibit any expansion of goblet cells (Fig. 4A). Several goblet cell marker genes are upregulated like tuft cell-related genes after the succinate treatment, particularly in female mice of both genotypes (Fig. 4B). In contrast, Muc2 (mucin 2) and P2ry4 (pyrimidinergic receptor P2Y4), expressed by goblet cells, are more highly expressed in untreated male and female Ptk6−/− controls, and their expression is reduced upon succinate treatment (Fig. 4B). Higher Muc2 mRNA expression was validated by qPCR (Fig. 4C). Muc2 expression and goblet cells were upregulated in Ptk6−/−Frk−/−Srms−/− mice [38], suggesting that PTK6 may be a suppressor of Muc2 production, although loss of Ptk6 alone was not sufficient to upregulate basal goblet cell numbers.
Fig. 4. Male Ptk6−/− mice have impaired upregulation of goblet cells, as well as goblet and Paneth cell genes upon succinate treatment.
A Expansion of goblet cells is impaired in male Ptk6−/− mice upon treatment with succinate. Representative images of Alcian Blue (AB) stained jejunum from male and female wild type and Ptk6−/− mice. Scale bar, 50 μm. Quantification of goblet cells is shown as total number of AB+ cells per villus-crypt unit. ***,### p-value <0.001. B Heatmap of significantly expressed goblet cell genes in RNA-seq data. Ptk6−/− control mice express higher levels of Muc2 and P2ry4, goblet cell genes, compared with succinate treated mice. C qPCR was used to validate Muc2 expression; relative Muc2 mRNA expression is normalized against Rps17. *p-value < 0.5. D Expression of Paneth cell markers, identified by Haber et al. [27]. E There is no change in Paneth cell numbers upon succinate treatment for 7 days. Jejunums were stained for expression of lysozyme (LYZ), a Paneth cell marker. Scale bar, 50 μm.
Paneth cell marker genes are upregulated in wild type and Ptk6−/− female mice (Fig. 4D), but staining of jejunal sections with antibodies against lysozyme, a Paneth cell marker, did not show any changes in Paneth cell numbers following succinate administration (Fig. 4E), as has been previously described [5]. This may be due to the long life of Paneth cells, which have a turnover time of up to 2 months [39]. Additionally, we did not observe differences in cell proliferation, identified by Ki67 staining (Fig. S2), and we did not detect any significant changes in the expression of proliferation markers in RNA-seq analysis, including Mki67, Pcna, and Mcm2.
Type 2 immunity genes are significantly upregulated in female mice following succinate treatment
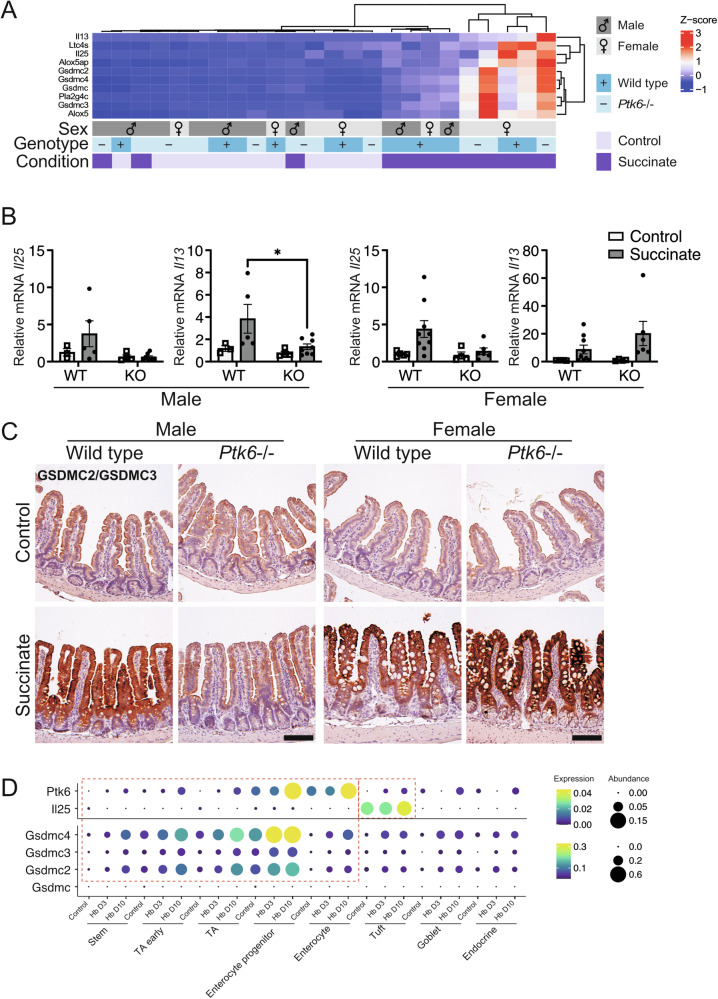
To further understand the effect of Ptk6 disruption on type 2 immunity, we examined the expression of genes that are upregulated following activation of the innate immune response [5, 13, 40, 41]. We focused on mediators of effector functions: enzymes for eicosanoid synthesis, cytokines, and pyroptotic genes. Several of these genes are significantly upregulated in succinate-treated wildtype and female Ptk6−/− mice in our dataset (Fig. 5A). We used qRT-PCR to validate RNA-seq data for cytokines Il25 and Il13 and observed an upward trend in the expression of these genes in wildtype males and females, as well as Ptk6−/− females, in response to succinate treatment (Fig. 5B).
Fig. 5. Tuft cell effector functions are impaired in male Ptk6−/− mice.
A A heatmap shows a significant increase in expression of tuft cell effector and type 2 immunity genes in succinate-treated female mice, and impaired upregulation in male succinate-treated Ptk6−/− mice. Gasdermin C genes (Gsdmc1-4) involved in type 2 immunity related pyroptosis, and Pla2g4c are some of the most significantly upregulated genes in the dataset. B Examination of gene expression using qRT-PCR shows impaired induction of type 2 immunity cytokine genes in male Ptk6−/− mice. Relative expression of Il25 from tuft cells, and Il13, an ILC2-produced cytokine needed for differentiation and expansion of tuft cells, in male and female mice is shown. Gene expression is normalized against Rps17. Graphs depict mean ± SEM. Statistics: Two-way ANOVA, adjusted for multiple comparisons using Tukey test. *p-value < 0.05. C GSDMC2 and GSDMC3 expression is upregulated upon succinate treatment in intestines of male wild type and female wild type and Ptk6−/− mice. Male Ptk6−/− do not display upregulation of pyroptotic genes upon succinate treatment. Intestinal sections were stained with an antibody that detects both GSDMC2 and GSDMC3. Scale bar, 50 μm. D Analysis of Haber et al. scRNA-seq data [27] shows that Ptk6 and Il25 are upregulated in tuft cell population upon helminth infection (Hb). Gsdmc1-4 are upregulated in stem, transient amplifying (TA) and enterocyte populations in a pattern like Ptk6. Expression of genes in specific populations are surrounded by red dashed lines.
Genes encoding GSDMCs, regulators of pyroptosis, are one of the most significant classes of DEGs in our dataset (Table S2). Succinate-treated females and wild type males exhibited strong upregulation of GSDMC2 and GSDMC3 in the jejunum, while male Ptk6−/− mice failed to upregulate these proteins (Fig. 5C). This pattern of protein expression mimics the RNA expression. Analysis of the Haber et al. scRNA-seq dataset [27] revealed upward trend in Il25 and Ptk6 expression in tuft cells upon activation of type 2 immunity with Hb (Fig. 5D). Furthermore, Gsdmc1-4 are upregulated in the same populations as Ptk6 (Fig. 5D).
PTK6 regulates Irag2 and Il25 expression required for tuft cell mediated activation of type 2 immunity in organoids developed from male mice
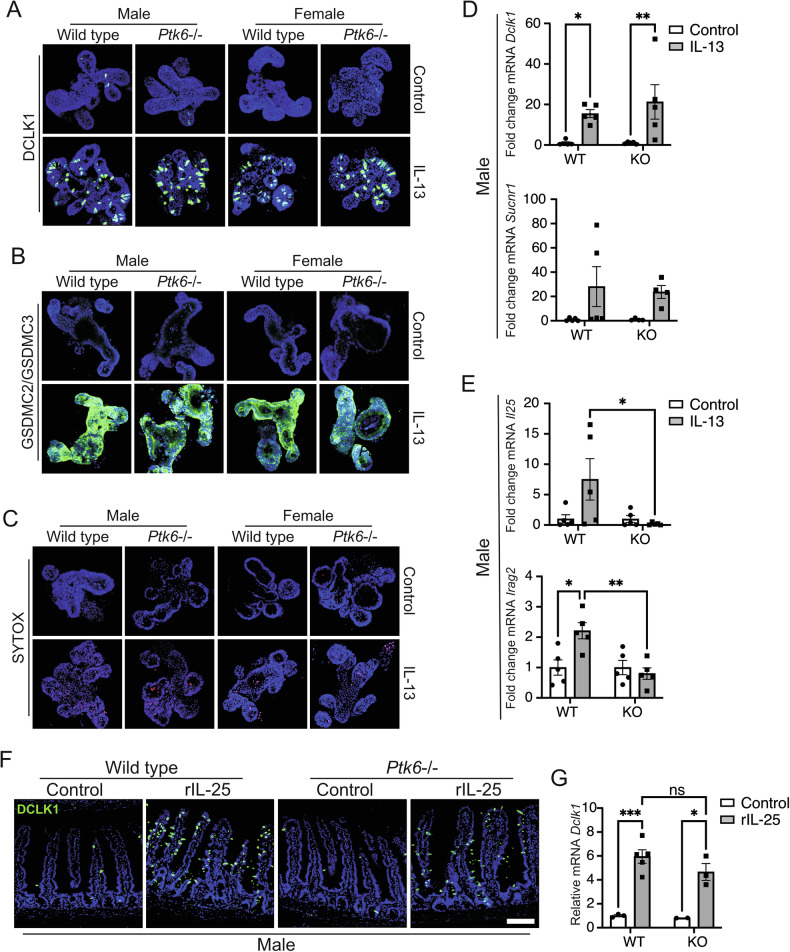
IL-13 treatment of intestinal organoids provides effective in vitro model for studying activation of type 2 immunity [30, 36], eliminating contributions of other immune cells and the microbiome to intestinal epithelium. IL-13 is normally produced by activated ILC2s, and organoids treated with IL-13 exhibit upregulation of tuft, goblet cell, and Paneth gene signatures [30]. We treated organoids developed from jejunal epithelium, with IL-13, and observed similar induction of tuft cells in organoids from male and female wild type and Ptk6−/− mice (Fig. 6A). No differences in GSDMC2 and GSDMC3 expression (Fig. 6B), or cell death, visualized by staining with SYTOX-red, were observed between the sexes or genotypes (Fig. 6C). The tuft cell marker genes Dclk1 and Sucnr1 were induced to a similar extent in organoids derived from the male and female mice of both genotypes (Fig. 6D, S3A). These data suggest that regulation of differentiation by IL-13 is intact in male and female Ptk6−/− mice and indicate that the defect in Ptk6−/− male mice is upstream of IL-13.
Fig. 6. Impaired induction of Il25 and Irag2 in organoids developed from male Ptk6−/− mice following treatment with IL-13.
A Ptk6 disruption does not affect tuft cell differentiation. Organoids were treated for 7 days with 30 ng/ml IL-13 and visualized using anti-DCLK1 antibody. IL-13, a cytokine secreted in vivo by ILC2s upon activation of tuft cells, induced expansion of tuft cells in organoids developed from intestines of male and female wild type and Ptk6−/− mice. B Gasdermins GSDMC2 and GSDMC3 are upregulated following IL-13 treatment in organoids derived from all mice. C Cell death is induced with IL-13 in all organoids, visualized with SYTOX-red. Organoids were treated with SYTOX-red 30 min prior fixation and staining with DAPI. D Tuft cell marker genes Dclk1 and Sucnr1 are induced after IL-13 treatment in organoids derived from wild type and Ptk6−/− male mice. Gene expression is normalized against Rps17 and shown as the fold change per genotype. E Upregulation of Il25 and Irag2 mRNA expression is impaired in organoids produced from Ptk6−/− male mice following IL-13 treatment. Gene expression is normalized against Rps17 and is shown as the fold change per genotype. F Exogenous IL-25 rescued activation of type 2 immunity in Ptk6−/− male mice. Wild type and Ptk6−/− have similar induction of tuft cells after injection for 7 days with rIL-25. Tuft cells are visualized with anti-DCLK1 antibody. Scale bar, 50 μm. G Relative expression of Dclk1 confirms upregulation of tuft cells upon rIL-25 administration to male Ptk6−/− mice. Gene expression is normalized against Rps17. For all experiments: Graphs depict mean ± SEM. Statistics: Two-way ANOVA, adjusted for multiple comparisons using Tukey test. *p-value < 0.05, **p-value < 0.01, ***p-value < 0.001.
Using qRT-PCR, we examined the expression of Il25, the tuft cell-specific cytokine that activates type 2 immunity in vivo. Interestingly, upregulation of Il25 was impaired in organoids developed from male Ptk6−/− intestinal cells, showing an expression pattern like intact animals (Fig. 6E). Expression of Irag2 is also impaired in male Ptk6−/− organoids (Fig. 6E). The product of Irag2, LRMP, mediates regulation of Ca2+ influx through interaction with IP3Rs, which is required for opening of transient receptor potential cation channel subfamily M member 5 (TRPM5) to induce membrane depolarization and IL-25 secretion [3, 42]. In organoids derived from female mice, expression of Il25 and Irag2 follows an upward trend after IL-13 treatment, but does not reach statistical significance (Fig. S3B). These data indicate that loss of PTK6 leads to a defect in effector functions of tuft cells in epithelial organoids developed only from males. Females may have higher basal expression of these genes, or there may be intermediate factors that more robustly activate their expression. Using scRNA-seq data from organoids [36], we confirmed that PTK6 and IRAG2 are expressed in the same tuft cell types upregulated upon IL-13 treatment and localized to the same cells (Fig. S3C, D).
To determine if impaired activation of innate immunity is dependent on lack of IL-25 in Ptk6−/− male mice, we asked if ectopic administration of IL-25 could rescue the secretory cell defects. We administered rIL-25 intraperitoneally to wild type and Ptk6−/− male mice. Exogenous IL-25 rescued activation of type 2 immunity in Ptk6−/− mice, detected by upregulation of tuft cells and Dclk1 in jejunums from male Ptk6−/− mice (Fig. 6F, G). Goblet cell differentiation was also rescued in male Ptk6−/− mice as visualized with Alcian blue staining (Fig. S4A). Exploring the expression of two genes impacted by Ptk6 loss, we show that Irag2 had upward trend in wild type male mice upon IL-25 treatment, while in Ptk6−/− males it remained unchanged. On the other hand, Il25 expression was rescued in male Ptk6−/− mice (Fig. S4B), suggesting that the administration of exogenous IL-25 can bypass the requirement for PTK6. These data further suggest that there are no immune cell defects in male Ptk6−/− mice, and that restoration of IL-25, expressed specifically by tuft cells, is sufficient to rescue ILC2 activation in Ptk6−/− male mice.
PTK6 is expressed in tuft cells of noninflamed ileum from Crohn’s disease patients
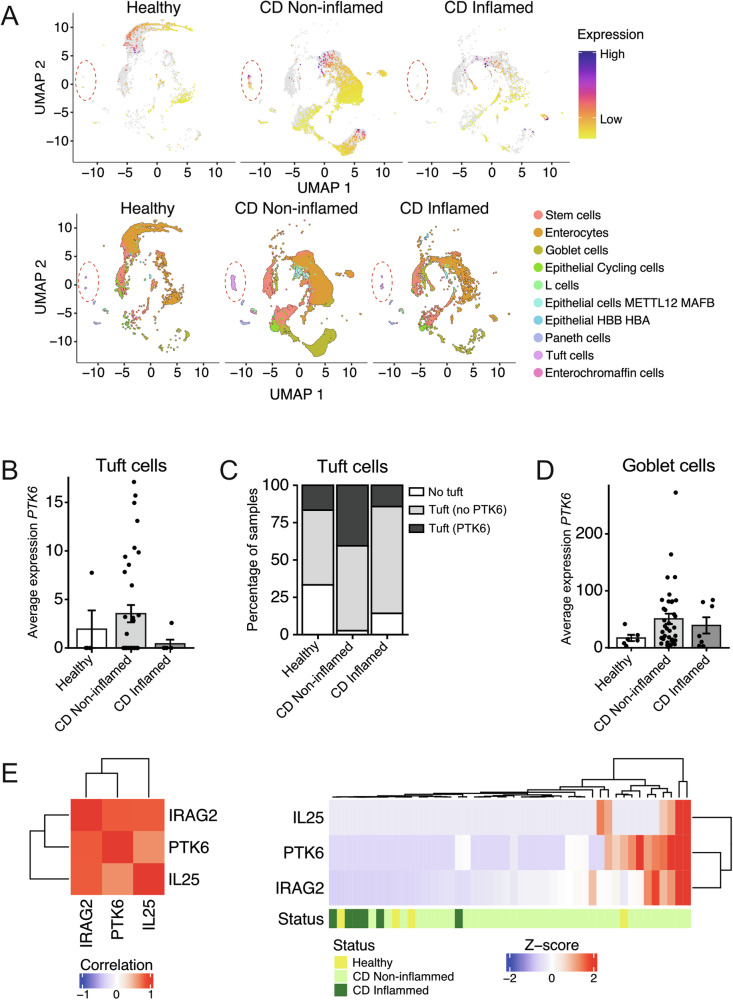
Alterations in tuft cell production have been reported in patients with inflammatory bowel disease (IBD). Banerjee and colleagues showed roles for tuft cells and the activation of type 2 immunity in suppression of the IL-23 - IL-17 axis of type 3 immunity characteristic for Crohn’s disease (CD), followed by reconstitution of the epithelium [7]. Other studies report alterations in tuft cell production in CD [43, 44], as well as in ulcerative colitis (UC) [45]. To further understand how our findings might relate to human disease, we examined scRNA-seq data from ileal tissues of patients with CD or healthy controls [44]. Greater numbers of tuft cells were present in noninflamed ileal tissues of patients with CD [44] (Fig. 7A), and PTK6 was higher in these samples (Fig. 7B, C). Like findings of Banerjee and colleagues [7], inflamed intestines had reduced numbers of tuft cells [44] when compared with non-inflamed intestine (Fig. 7C). PTK6 mRNA is expressed in tuft cells in ~15% of samples from healthy and inflamed tissues, while it is found in double the number of patients with tuft cells localized in non-inflamed regions (40%) (Fig. 7C). We did not observe significant differences in PTK6 expression in tuft cells between sexes (Fig. S5), as already observed (Fig. 2C). Correlation analysis shows high correlation between PTK6, IL25 and IRAG2 (Fig. 7E). IL25 was expressed only in tuft cells from noninflamed samples, and PTK6 was co-expressed with IL25 in 5/6 of samples (Fig. 7E), supporting potential crosstalk in tuft cells.
Fig. 7. PTK6 is upregulated in tuft cells from noninflamed intestine in Crohn’s disease patients.
A PTK6 is highly expressed in enterocytes and goblet cells in the ileum in patients with Crohn’s disease (CD) and healthy controls. Analysis of scRNA-seq data from Kong, et al. [44] revealed expression of PTK6 in other populations, including tuft cells (surrounded by red dashed circles). The bottom panel shows different cell populations in each sample group. All stem populations are grouped as “Stem cells”; all enterocyte populations are grouped as “Enterocytes”; all goblet cell populations are grouped as “Goblet cells”, following authors’ annotations [44]. B Average expression of PTK6 in tuft cells per patient from Kong et al. scRNA-seq data [44]. C PTK6 is expressed in tuft cells from a subset of samples. It is mostly expressed in tuft cells from non-inflamed CD tissues, and to a lesser extent in healthy controls and inflamed CD tissues. D PTK6 is expressed in goblet cells from human patients. E Correlation matrix and expression heatmap of PTK6, IL25 and IRAG2 from Kong, et al. dataset [44]. Expression of PTK6 per sample (sum of cell expression, per cell type) positively correlates with expression of IL25 and IRAG2 (PTK6-IRAG2: r = 0.81, p-value < 0.0001; PTK6-IL25: r = 0.64, p-value < 0.0001). Heatmap shows Z-scores of summed expression for each of the genes per cell type. IL25 is upregulated in tuft cells in a subset of non-inflamed tissues only. It is co-expressed with PTK6 in 5/6 samples.
Using the same dataset [44], we show that PTK6 is highly expressed in goblet cells in all groups of human samples (Fig. 7D). Together these data indicate that PTK6 may function in epithelial cells with roles in mucosal immunity, and it may regulate gene expression and/or function directly, or indirectly through regulating IL-25 expression and/or secretion.
Discussion
Research in the last decade has revealed important differences in biology between sexes, and here we report significant differences between male and female mice dependent on PTK6 (Fig. 1). Gene set enrichment pathway analysis identified the “innate immune response in mucosa” as one of the top differences between wildtype and Ptk6−/− animals in both sexes (Fig. 1D) and prompted us to further explore the role of PTK6 in innate immunity. We detected upregulation of Ptk6 upon infection with helminths and bacteria (Fig. 2A, B). Looking closely at the pathways, Ptk6 is upregulated in tuft cells upon activation of innate type 2 immunity in vivo and in vitro (Fig. 2B, D).
We show that PTK6 regulates tuft cell-mediated activation of type 2 immunity and secretory cell expansion in the intestines of male mice. Sex differences in innate type 2 immunity in the intestine are not well understood. Activation of innate type 2 immunity in various organs shows a strong sex bias, with females having stronger and more robust immune responses [46]. Sex hormones may regulate sex-bias in innate immunity. Androgens negatively regulate ILC2s, and ILC2s express higher levels of the androgen receptor [47, 48].
Female mice produce more tuft and goblet cells than males following succinate treatment (Figs. 3A, 4A), exhibit higher expression of tuft cell markers and stronger activation of type 2 immunity (Figs. 3–5). Female wild type and Ptk6−/− mice exhibit some differences in gene expression (Fig. 1), but they have similar upregulation of genes involved in tuft cell-mediated type 2 immunity. Expression of FRK and SRMS, two PTK6 family members, did not significantly change in female Ptk6−/− mice after succinate treatment (Fig. S6A, B). FRK is expressed in male and female tuft cells, and in tuft cells induced by IL-13 treatment (Fig. S6C, D).
Ectopic administration of the ILC2 cytokine IL-13 induced differentiation of tuft cells in wild type and Ptk6−/− organoid cultures (Fig. 6A), indicating stem cell responses are intact in the absence of PTK6. While IL-13-treated wild type and Ptk6−/− organoids produced equivalent numbers of tuft cells, we detected reduced expression of Il25 and Irag2 in male Ptk6−/− organoids compared with wild type organoids (Fig. 6E). Defects in IL-25-dependent effector functions in vivo would lead to impaired succinate-induced activation of ILC2s, secretion of IL-13 and secretory cell differentiation. Restoring IL-25 by administration of rIL-25 to Ptk6−/− male mice stimulated tuft cell differentiation to wild type levels (Fig. 6F).
Regulation of IL-25 in tuft cells at transcriptional, translational, and post-translational levels is not well understood. Epigenetic mechanisms may be involved, since sex-differences in PTK6 regulated Il25 expression are maintained in organoid cultures. Rescue of Il25 expression following administration of exogenous IL-25 (Fig. S4B) suggests that IL-25 may auto-regulate itself [49–51] independently of PTK6, and we cannot rule out induction of IL-25 transcription in rIL-25 treated mice by PTK6-independent mechanisms. IL-25 has been shown to be part of an autoregulatory loop that activates STAT3 [50, 51]. Interestingly, PTK6 regulates activation of STAT3 through direct tyrosine phosphorylation [25, 52], and STAT3 activation was impaired in Ptk6−/− male mice [25]. It is possible that impaired STAT3 activation could be corrected through activation of STAT3 by rIL25.
In humans, the IL25 gene maps to an inflammatory bowel susceptibility locus (14q11-12, IBD4). However, no mutations or polymorphisms were identified in the IL25 gene, leading investigators to conclude it had no association with IBD [53]. However, they did not consider contributions of transcriptional and posttranscriptional regulation. IL-25-mediated activation of type 2 immunity may have beneficial effects for patients with CD. Banerjee and colleagues reported a reduced number of tuft cells in inflamed ileums from patients and in a mouse model of CD [7]. PTK6 is expressed in tuft cells in almost half of patients in non-inflamed CD (Fig. 7C), and its expression shows an upward trend in tuft cells from noninflamed regions of CD ileums (Fig. 7B). Co-expression of IL25 and IRAG2 with PTK6 in a subset of patients with noninflammatory CD further suggests crosstalk and the potential for PTK6 to regulate tuft cells and inflammation in CD.
Understanding contributions of sex to disease is critical for precision medicine. For example, sex differences impact the prevalence, risk, and outcomes of autoimmune and cardiovascular diseases and cancer [54]. In the intestine, type 2 immune responses are important for clearing certain infections, such as helminths, which can lead to malnutrition, vitamin deficiencies and intestinal obstructions [55]. Resistance to existing anthelmintics is a public health concern and understanding mechanisms of helminth clearance will be important for developing novel treatments [56]. Understanding the sex-biased functions of PTK6 in regulation of innate immunity may lead to improved strategies for treatment of a variety of sex-related diseases.
Methods and materials
Mice
Animal experiments were approved by the University of Illinois at Chicago Institutional Animal Care and Use Committee. All mice were maintained under specific pathogen-free conditions. The generation of the germline knockout of Ptk6−/− has been described [18]. This strain has been backcrossed more than 20 generations into the C57BL/6 background and has been made available at The Jackson Laboratory Repository with the JAX Stock No. 038346. All experiments utilized aged-matched animals from different litters, from the same breeding colony. All experiments ended at 9–11 weeks of age. For experiments on activation of type 2 immunity, male and female wild type and Ptk6−/− mice were given 100 mM succinate (S9637; Sigma Aldrich, St. Louis, MO) in drinking water for 7 days [11]. For rescue experiments with IL-25, male wildtype and Ptk6−/− mice were injected intraperitoneally 0.5 μg of rIL-25 (1399-IL; R&D systems, Minneapolis, MN) diluted in PBS, or PBS only for controls. Tissues were harvested and paraffin-embedded for histology or processed for RNA isolation. Jejunums were divided for immunohistochemistry (proximal and middle) and RNA preparation (distal).
Organoid cultures
Organoids were generated from small intestines of 5 male wild type, 5 female wild type, 5 male Ptk6−/− and 5 female Ptk6−/− mice according to Sato et al. [57] following the media manufacturer’s protocol (STEMCell Technologies, Vancouver, Canada). Briefly, proximal jejunums from male and female 8–9 weeks old mice were harvested, 8 cm of the proximal small intestine was washed, villi scraped off, and the intestine was cut into 2 mm pieces. The pieces were washed with PBS 10 times and incubated with 3 mM EDTA in PBS for 30 min at 4 °C with gentle rocking. The crypts removal was facilitated by vigorous shaking, and crypts were passed through a 70 μm cell strainer (352350; Corning, Corning, New York). Crypts were washed in DMEM/F-12, counted, and resuspended in Matrigel (356231; Corning, Corning, New York) in a ratio of 2:3 (60 μl) with a final count ~700 crypts/well. After establishing the culture, organoids were passaged every 6–7 days using Gentle Cell Dissociation Reagent (100–0485, STEMCell Technologies). Experiments were performed between the 2nd and 4th passage.
To mimic the activation of type 2 immunity and promote tuft cell differentiation in culture, organoids were treated with 30 ng/ml IL-13 and/or 10 mM sodium-succinate, and at days 1, 3, and 5 after plating. Organoids were plated in 24-well plates for RNA analysis (6 wells per treatment), or ibidi 8-well chambers with glass bottoms (80827; Ibidi, Fitchburg, WI) for immunofluorescence (at least 2 wells per treatment per antibody). The addition of sodium-succinate alone or in combination with IL-13 had no impact on tuft cell differentiation in the organoid models.
Pyroptosis analysis
Organoids were plated in each well of ibidi 8-well chambers, in 30 μl of 2:3 ratio of organoids/Matrigel mix per well. SYTOX-red (S34859; Invitrogen, Waltham, MA) was added to the medium at the concertation 0.5 μM, 30 min before the end of the experiment. After treatment, the organoids were washed, fixed in 4% paraformaldehyde for 30 min, and further analyzed.
Immunohistochemistry and immunofluorescence
Tissues containing proximal/middle jejunums were rehydrated and antigen retrieval was performed using sodium-citrate buffer pH 6, at 95 °C for 20 min. For DAB staining, tissues were incubated with H2O2 for 10 min. Tissues were blocked in 5% BSA supplemented with 2% of appropriate serum based on the source of the secondary antibody, for 1 h at room temperature. Primary antibodies were incubated overnight at 4 °C. Secondary antibodies were incubated for 1 h following the appropriate detection. DAB-stained tissues were counterstained with Hematoxylin dye, while immunofluorescent tissues were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen, Waltham, MA). The number of tuft cells is quantified as the number of DCLK1+ cells per crypt-villus. For the quantification, 4–5 of random areas from each tissue were taken with Leica DM8 fluorescent microscope at 10x magnification.
For organoid experiments, fixed organoids were permeabilized using 0.2% Triton-X in PBS for 30 min, washed in 0.1% Tween in PBS (PBST), and blocked in 5% BSA in PBST overnight at 4 °C. Primary antibodies were incubated overnight at 4 °C, secondary antibodies for 1.5 h at room temperature, and streptavidin-Alexa Fluor for 30 min at room temperature. Nuclei were stained with DAPI.
Antibodies
Primary antibodies used in the study: anti-DCLK dilution 1:2000 (ab31704; Abcam, Cambridge, UK), anti-GSDMC2/GSDMC3 dilution 1:2500 (Abcam), anti-Lysozyme dilution 1:200 (A0099, DakoCytoman, Denmark), anti-Ki-67 1:200 (ab16667, Abcam).
Alcian blue staining of goblet cells
Deparaffinized and rehydrated tissues were incubated in 3% glacial acetic acid solution prior to incubation with 1% w/v Alcian Blue solution (A3157; Sigma Aldrich, St. Louis, MO). Tissues were counterstained with Nuclear Fast Red (H3403; Vector Laboratories, Newark, California). The number of goblet cells is quantified as the number of Alcian Blue+ cells per crypt-villus. For quantification of goblet cells, 4–5 of random areas from each tissue were taken with Leica DM8 light microscope at 10x magnification.
Quantitative RT–PCR
Total RNA was isolated from distal jejunums using TRIzol (15596026, Invitrogen), following the manufacturer’s protocol. cDNA was prepared from total RNA using iScript (1708891; Bio-Rad, Hercules, CA), and qPCR was performed using SensiFAST SYBR No-ROX Kit (BIO-98005; Meridian Life sciences, Cincinnati, OH) following the manufacturer’s protocols. Primers used in the study:
Dclk1: Fwd: 5′-CAGCCTGGACGAGCTGGTGG-3′, and Rev: 5′-TGACCAGTTGGGGTTCACAT-3′; Il13: Fwd: 5′-TGAGCAACATCACACAAGACC-3′, and Rev: 5′-GGCCTTGCGGTTACAGAGG-3′; Il25: Fwd: 5′-ACAGGGACTTGAATCGGGTC-3′, and Rev: 5′-TGGTAAAGTGGGACGGAGTTG-3′; Irag2: Fwd: 5′-TTTGGACGTGACAAGAGGGT-3′, and Rev: 5′-ACTCTGGTCCTCGTTCATTCTG-3′; Muc2: Fwd: 5′-ATGCCCACCTCCTCAAAGAC-3′, and Rev: 5′-GTAGTTTCCGTTGGAACAGTGAA-3′; Rps17: Fwd: 5′-CGCCATTATCCCCAGCAAG-3′, and Rev: 5′-TGTCGGGATCCACCTCAATG-3′; Sucnr1: Fwd: 5′-GGCAGAGTTTTCTGTCGAGAC-3′, and Rev: 5′-ACATTCCCAAGCAGTCCAA-3′.
For all experiments, gene expression was normalized to Rps17 using the 2-ΔΔCT method. When analyzing gene expression in tissues, expression is shown relative to control wild type mice, for which average expression is set at 1. For organoids, gene expression is shown as the fold change in gene expression following each treatment, averaged per mouse genotype.
Bulk RNA-seq
Total jejunal RNA samples were processed by LC Sciences (Houston, TX). RNA quantity and purity were analyzed using Bioanalyzer 2100 and RNA 6000 Nano LabChip Kit (Agilent, CA, USA, 5067-1511), and high-quality RNA samples with RIN number > 7.0 were used to construct a sequencing library. mRNA was purified from total RNA (5ug) using Dynabeads Oligo (dT) (Thermo Fisher, CA, USA) with two rounds of purification, and fragmented into short fragments using divalent cations under elevated temperature (Magnesium RNA Fragmentation Module (NEB, cat. e6150, USA) under 94 °C 5–7 min). cDNA was generated using SuperScriptTM II Reverse Transcriptase (Invitrogen, cat. 1896649, USA). cDNA was then used to synthesize U-labeled second-stranded DNAs with E. coli DNA polymerase I (NEB, cat.m0209, USA), RNase H (NEB, cat.m0297, USA) and dUTP Solution (Thermo Fisher, cat. R0133, USA). An A-base was added to the blunt ends of each strand, preparing them for ligation to the indexed adapters. Each adapter contained a T-base overhang for ligating the adapter to the A-tailed fragmented DNA. Dual-index adapters were ligated to the fragments, and size selection was performed with AMPureXP beads. After the heat-labile UDG enzyme (NEB, cat.m0280, USA) treatment of the U-labeled second-stranded DNAs, the ligated products were amplified with PCR by the following conditions: initial denaturation at 95 °C for 3 min; 8 cycles of denaturation at 98 °C for 15 sec, annealing at 60 °C for 15 sec, and extension at 72 °C for 30 sec; and then final extension at 72 °C for 5 min. The average insert size for the final cDNA libraries was 300 ± 50 bp. At last, we performed the 2×150 bp paired-end sequencing (PE150) on an Illumina NovaseqTM 6000 following the vendor’s recommended protocol.
RNA-seq bioinformatics analysis
Using Illumina paired-end RNA-seq approach, we sequenced the transcriptome, generating a total of million 2 ×150 bp paired-end reads. Reads were further filtered by Cutadapt (https://cutadapt.readthedocs.io/en/stable/,version:cutadapt-1.9). The parameters were as follows: 1) removing reads containing adapters; 2) removing reads containing polyA and polyG; 3) removing reads containing more than 5% of unknown nucleotides (N); 4) removing low-quality reads containing more than 20% of low quality (Q-value ≤ 20) bases. Sequence quality was verified using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/, 0.11.9). including the Q20, Q30, and GC content of the clean data. Reads were aligned with the reference genome using the HISAT2 package (https://daehwankimlab.github.io/hisat2/,version:hisat2-2.0.4). The mapped reads of each sample were assembled using StringTie (http://ccb.jhu.edu/software/stringtie/,version:stringtie-1.3.4d) with default parameters. Then, all transcriptomes from all samples were merged to reconstruct a comprehensive transcriptome using gffcompare software (http://ccb.jhu.edu/software/stringtie/gffcompare.shtml,version:gffcompare-0.9.8). After the final transcriptome was generated, StringTie and ballgown (http://www.bioconductor.org/packages/release/bioc/html/ballgown.html) were used to estimate the expression levels of all transcripts and perform expression abundance for mRNAs by calculating FPKM (fragment per kilobase of transcript per million mapped reads) value.
Differential gene expression analysis was performed using DESeq2 software [58] between two different groups (and by edgeR [59] between two samples). The genes with the parameter of false discovery rate (FDR) below 0.05 and absolute fold change ≥ 2 were considered differentially expressed genes.
Gene set enrichment analysis was performed using software GSEA (v4.1.0) and MSigDB to identify whether a set of genes in specific GO terms, KEGG pathways, DO terms, and Reactome shows significant difference between groups [60]. Briefly, we input gene expression matrix and rank genes by Signal2Noise normalization method. Enrichment scores and p-value was calculated in default parameters. GO terms, KEGG pathways (DO terms, Reactome) meeting this condition with |NES| > 1, NOM p-val<0.05, FDR q-val < 0.25 were considered to be different in two groups.
Whole raw and processed data files have been deposited in NCBI’s Gene Expression Omnibus [61] and are accessible through GEO Series accession number GSE243229. Other RNA-seq and scRNA-seq are publicly available from the referenced studies [27, 28, 44]
Public data analysis and statistics
RNA-seq data were explored for significant gene expression, and compared with previously identified marker genes of tuft cells [27], and type 2 immunity-related pyroptotic genes [13].
Publicly available RNA-seq data was obtained directly from El-Naccache et al. [28], and expression normalized to control values (naïve mice).
Publicly available single cell (sc) RNA-seq data were obtained from GEO (Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/geo/): GSE92332 [27] and GSE233451 [36]; from CZ CELL×GENE Discover [62]: Gut cell atlas [35]; from Broad Institute Single Cell Portal: SCP1884 [44]. Tuft cells from Haber et al. [27] originate from mouse small intestine, Kong et al. [44] from human ileum, while tuft cells from other studies [35, 36] were not separated based on tissue origin. For Elmentaite et al. study [35] we used only expression from healthy samples. The pseudo-bulk analysis was performed from data analyzed following authors’ preprocessing steps and annotated cell types based on the author’s information for all samples together or already available normalized data grouped per sample. Statistical analysis of Ptk6 upregulation upon infection [27] was performed using monocle3 [63–65]. Other single-cell analyses were performed based on the authors’ guidance and available markers for cell type annotations: Elmentaite et al. [35], and Kong et al. [44]: cell annotations originated from metadata, expression values from normalized matrix and visualized using with monocle3 [63–65] without normalization; Haber et al. [27]: cell annotations originated from metadata, SalmHelm UMI counts matrix was normalized using monocle3 default settings. Huang et al. [36]: gene expression normalized by following authors’ guidelines, cell populations grouped based on metadata.
Data were analyzed using R software. For the analysis of scRNA-seq data, we used the Seurat package (version 5) [66] and monocle3 [63–65]. For visualization, we used scCustomize [67], ggplot2 [68] and ComplexHeatmap packages [69].
Statistical analyses for quantification and qPCR data were performed using GraphPad Prism software version 9 (La Jolla, CA); two-way ANOVA and Mann-Whitney tests were used to determine significant differences between groups. *p-value < 0.05, **,##p-value < 0.01, ***,###p-value < 0.001, ****p-value < 0.0001. # is used for comparison within the same sex when shown on the same graph with all comparisons.
Supplementary information
Acknowledgements
The authors thank Dr. Wanian Alwanian for helpful comments, and Dr. Jalees Rehman for providing resources.
Author contributions
ALT, KV: generated the idea and hypothesis, designed experiments, wrote, and revised the manuscript. ALT: supervised the study, secured the funding and resources. KV, WB, MBG: performed animal experiments, analyzed tissues. KV: performed gene expression studies and bioinformatics analysis. KV, WB, MBG, ALT: read, revised, and approved the manuscript.
Funding
The work was supported in part by NIH grants R01DK44525 and R01CA188427 (ALT).
Data availability
Processed RNA-seq data from jejunums of wild type and Ptk6−/− mice are available in the article and supplementary materials. Additionally, the whole raw and processed data files have been deposited in NCBI’s Gene Expression Omnibus [61] and are accessible through GEO Series accession number GSE243229. Other datasets used in this study are listed in sections “Methods and Materials” and “Supplemental Material and Methods.” This study did not generate any new code.
Competing interests
The authors declare no competing interests.
Ethics statement
Although publicly available human datasets were analyzed, no human subjects were used in these studies. All mouse experiments were approved by the University of Illinois at Chicago Institutional Animal Care and Use Committee. The authors confirm that all methods were performed in accordance with relevant guidelines and regulations.
Footnotes
Edited by Hans-Uwe Simon
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-024-07149-9.
References
- 1.Gustafsson JK, Johansson MEV. The role of goblet cells and mucus in intestinal homeostasis. Nat Rev Gastroenterol Hepatol. 2022;19:785–803. [DOI] [PubMed] [Google Scholar]
- 2.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356–68. [DOI] [PubMed] [Google Scholar]
- 3.Howitt MR, Lavoie S, Michaud M, Blum AM, Tran SV, Weinstock JV, et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science (N. Y, NY). 2016;351:1329–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerbe F, Sidot E, Smyth DJ, Ohmoto M, Matsumoto I, Dardalhon V, et al. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature. 2016;529:226–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Moltke J, Ji M, Liang H-E, Locksley RM. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature. 2016;529:221–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong Z, Zhu X, Geng J, Xu Y, Wu R, Li C, et al. Intestinal Tuft-2 cells exert antimicrobial immunity via sensing bacterial metabolite N-undecanoylglycine. Immunity. 2022;55:686–700. e7 [DOI] [PubMed] [Google Scholar]
- 7.Banerjee A, Herring CA, Chen B, Kim H, Simmons AJ, Southard-Smith AN, et al. Succinate produced by intestinal microbes promotes specification of tuft cells to suppress ileal inflammation. Gastroenterology. 2020;159:2101–15. e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo X-C, Chen Z-H, Xue J-B, Zhao D-X, Lu C, Li Y-H, et al. Infection by the parasitic helminth Trichinella spiralis activates a Tas2r-mediated signaling pathway in intestinal tuft cells. Proc Natl Acad Sci. 2019;116:5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howitt MR, Cao YG, Gologorsky MB, Li JA, Haber AL, Biton M, et al. The taste receptor TAS1R3 regulates small intestinal tuft cell homeostasis. ImmunoHorizons. 2020;4:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nadjsombati MS, McGinty JW, Lyons-Cohen MR, Jaffe JB, DiPeso L, Schneider C, et al. Detection of succinate by intestinal tuft cells triggers a type 2 innate immune circuit. Immunity. 2018;49:33–41.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lei W, Ren W, Ohmoto M, Urban JF, Matsumoto I, Margolskee RF, et al. Activation of intestinal tuft cell-expressed Sucnr1 triggers type 2 immunity in the mouse small intestine. Proc Natl Acad Sci. 2018;115:5552–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider C, O’Leary CE, von Moltke J, Liang H-E, Ang QY, Turnbaugh PJ, et al. A metabolite-triggered tuft cell-ILC2 circuit drives small intestinal remodeling. Cell. 2018;174:271–84. e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xi R, Montague J, Lin X, Lu C, Lei W, Tanaka K, et al. Up-regulation of gasdermin C in mouse small intestine is associated with lytic cell death in enterocytes in worm-induced type 2 immunity. Proc Natl Acad Sci. 2021;118:e2026307118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grencis RK. Immunity to helminths: resistance, regulation, and susceptibility to gastrointestinal nematodes. Annu Rev Immunol. 2015;33:201–25. [DOI] [PubMed] [Google Scholar]
- 15.Anthony RM, Rutitzky LI, Urban JF Jr., Stadecker MJ, Gause WC. Protective immune mechanisms in helminth infection. Nat Rev Immunol. 2007;7:975–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilic MB, Tyner AL. Targeting protein tyrosine kinase 6 in cancer. Biochim Biophys Acta Rev Cancer. 2020;1874:188432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasioukhin V, Serfas MS, Siyanova EY, Polonskaia M, Costigan VJ, Liu B, et al. A novel intracellular epithelial cell tyrosine kinase is expressed in the skin and gastrointestinal tract. Oncogene. 1995;10:349–57. [PubMed] [Google Scholar]
- 18.Haegebarth A, Bie W, Yang R, Crawford SE, Vasioukhin V, Fuchs E, et al. Protein tyrosine kinase 6 negatively regulates growth and promotes enterocyte differentiation in the small intestine. Mol Cell Biol. 2006;26:4949–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haegebarth A, Heap D, Bie W, Derry JJ, Richard S, Tyner AL. The nuclear tyrosine kinase BRK/Sik phosphorylates and inhibits the RNA-binding activities of the Sam68-like mammalian proteins SLM-1 and SLM-2. J Biol Chem. 2004;279:54398–404. [DOI] [PubMed] [Google Scholar]
- 20.Mathur PS, Gierut JJ, Guzman G, Xie H, Xicola RM, Llor X, et al. Kinase-dependent and -independent roles for PTK6 in colon cancer. Mol Cancer Res. 2016;14:563–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palka-Hamblin HL, Gierut JJ, Bie W, Brauer PM, Zheng Y, Asara JM, et al. Identification of beta-catenin as a target of the intracellular tyrosine kinase PTK6. J Cell Sci. 2010;123:236–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haegebarth A, Perekatt AO, Bie W, Gierut JJ, Tyner AL. Induction of protein tyrosine kinase 6 in mouse intestinal crypt epithelial cells promotes DNA damage-induced apoptosis. Gastroenterology. 2009;137:945–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gierut JJ, Mathur PS, Bie W, Han J, Tyner AL. Targeting protein tyrosine kinase 6 enhances apoptosis of colon cancer cells following DNA damage. Mol Cancer Ther. 2012;11:2311–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chastkofsky MI, Bie W, Ball-Kell SM, He YY, Tyner AL. Protein tyrosine kinase 6 regulates UVB-induced signaling and tumorigenesis in mouse skin. J Invest Dermatol. 2015;135:2492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gierut J, Zheng Y, Bie W, Carroll RE, Ball-Kell S, Haegebarth A, et al. Disruption of the mouse protein tyrosine kinase 6 gene prevents STAT3 activation and confers resistance to azoxymethane. Gastroenterology. 2011;141:1371–80. 80.e1-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Regan Anderson TM, Peacock DL, Daniel AR, Hubbard GK, Lofgren KA, Girard BJ, et al. Breast tumor kinase (Brk/PTK6) is a mediator of hypoxia-associated breast cancer progression. Cancer Res. 2013;73:5810–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, et al. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El-Naccache DW, Chen F, Palma MJ, Lemenze A, Fischer MA, Wu W, et al. Adenosine metabolized from extracellular ATP promotes type 2 immunity through triggering A(2B)AR signaling in intestinal epithelial cells. Cell Rep. 2022;40:111150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He G-W, Lin L, DeMartino J, Zheng X, Staliarova N, Dayton T, et al. Optimized human intestinal organoid model reveals interleukin-22-dependency of paneth cell formation. Cell Stem Cell. 2022;29:1333–45. e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindholm HT, Parmar N, Drurey C, Campillo Poveda M, Vornewald PM, Ostrop J, et al. BMP signaling in the intestinal epithelium drives a critical feedback loop to restrain IL-13–driven tuft cell hyperplasia. Sci Immunol. 2022;7:eabl6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu S, Balasubramanian I, Laubitz D, Tong K, Bandyopadhyay S, Lin X, et al. Paneth cell-derived lysozyme defines the composition of mucolytic microbiota and the inflammatory tone of the intestine. Immunity. 2020;53:398–416. e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coutry N, Nguyen J, Soualhi S, Gerbe F, Meslier V, Dardalhon V, et al. Cross talk between Paneth and tuft cells drives dysbiosis and inflammation in the gut mucosa. Proc Natl Acad Sci. 2023;120:e2219431120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ouellette AJ. Paneth cell α-defensins in enteric innate immunity. Cell Mol Life Sci. 2011;68:2215–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khatun A, Sakurai M, Okada K, Sakai Y, Morimoto M. Detection of α-defensin in eosinophils in helminth-infected mouse model. J Vet Med Sci. 2018;80:1887–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elmentaite R, Kumasaka N, Roberts K, Fleming A, Dann E, King HW, et al. Cells of the human intestinal tract mapped across space and time. Nature. 2021;597:250–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang L, Bernink JH, Giladi A, Krueger D, van Son GJF, Geurts MH, et al. Tuft cells act as regenerative stem cells in the human intestine. Nature. 2024. 10.1038/s41586-024-07952-6. [DOI] [PMC free article] [PubMed]
- 37.Nalapareddy K, Nattamai KJ, Kumar RS, Karns R, Wikenheiser-Brokamp KA, Sampson LL, et al. Canonical wnt signaling ameliorates aging of intestinal stem cells. Cell Rep. 2017;18:2608–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kikuchi I, Iwashita Y, Takahashi-Kanemitsu A, Koebis M, Aiba A, Hatakeyama M. Coevolution of the ileum with Brk/Ptk6 family kinases confers robustness to ileal homeostasis. Biochem Biophys Res Commun. 2023;676:190–7. [DOI] [PubMed] [Google Scholar]
- 39.Ireland H, Houghton C, Howard L, Winton DJ. Cellular inheritance of a Cre-activated reporter gene to determine Paneth cell longevity in the murine small intestine. Dev Dyn. 2005;233:1332–6. [DOI] [PubMed] [Google Scholar]
- 40.Nussbaum JC, Van Dyken SJ, von Moltke J, Cheng LE, Mohapatra A, Molofsky AB, et al. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature. 2013;502:245–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGinty JW, Ting HA, Billipp TE, Nadjsombati MS, Khan DM, Barrett NA, et al. Tuft-cell-derived leukotrienes drive rapid anti-helminth immunity in the small intestine but are dispensable for anti-protist immunity. Immunity. 2020;52:528–41. e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang C-Y, Wang J, Zhao Y, Liu J, Yang X, Yue X, et al. Tumor suppressor p53 regulates intestinal type 2 immunity. Nat Commun. 2021;12:3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maddipatla SC, Kolachala VL, Venkateswaran S, Dodd AF, Pelia RS, Geem D, et al. Assessing cellular and transcriptional diversity of ileal mucosa among treatment-naive and treated Crohn’s disease. Inflamm Bowel Dis. 2023;29:274–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kong L, Pokatayev V, Lefkovith A, Carter GT, Creasey EA, Krishna C, et al. The landscape of immune dysregulation in Crohn’s disease revealed through single-cell transcriptomic profiling in the ileum and colon. Immunity. 2023;56:444–58. e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kjaergaard S, Jensen TSR, Feddersen UR, Bindslev N, Grunddal KV, Poulsen SS, et al. Decreased number of colonic tuft cells in quiescent ulcerative colitis patients. Eur J Gastroenterol Hepatol. 2021;33:817–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol. 2016;16:626–38. [DOI] [PubMed] [Google Scholar]
- 47.Laffont S, Blanquart E, Savignac M, Cénac C, Laverny G, Metzger D, et al. Androgen signaling negatively controls group 2 innate lymphoid cells. J Exp Med. 2017;214:1581–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li F, Xing X, Jin Q, Wang X-M, Dai P, Han M, et al. Sex differences orchestrated by androgens at single-cell resolution. Nature. 2024;629:193–200. [DOI] [PubMed] [Google Scholar]
- 49.Zheng XL, Wu JP, Gong Y, Hong JB, Xiao HY, Zhong JW, et al. IL-25 protects against high-fat diet-induced hepatic steatosis in mice by inducing IL-25 and M2a macrophage production. Immunol Cell Biol. 2019;97:165–77. [DOI] [PubMed] [Google Scholar]
- 50.Xu M, Lu H, Lee YH, Wu Y, Liu K, Shi Y, et al. An interleukin-25-mediated autoregulatory circuit in keratinocytes plays a pivotal role in psoriatic skin inflammation. Immunity. 2018;48:787–98. e4 [DOI] [PubMed] [Google Scholar]
- 51.Wang WB, Yen ML, Liu KJ, Hsu PJ, Lin MH, Chen PM, et al. Interleukin-25 mediates transcriptional control of PD-L1 via STAT3 in multipotent human mesenchymal stromal cells (hMSCs) to suppress Th17 responses. Stem Cell Rep. 2015;5:392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu L, Gao Y, Qiu H, Miller WT, Poli V, Reich NC. Identification of STAT3 as a specific substrate of breast tumor kinase. Oncogene. 2006;25:4904–12. [DOI] [PubMed] [Google Scholar]
- 53.Büning C, Genschel J, Weltrich R, Lochs H, Schmidt H. The interleukin-25 gene located in the inflammatory bowel disease (IBD) 4 region: no association with inflammatory bowel disease. Eur J Immunogenetics. 2003;30:329–33. [DOI] [PubMed] [Google Scholar]
- 54.Shi Y, Ma J, Li S, Liu C, Liu Y, Chen J, et al. Sex difference in human diseases: mechanistic insights and clinical implications. Signal Transduct Target Ther. 2024;9:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.WHO. Soil-transmitted helminth infections: World Health Organization; 2023 [September 07 2024]. Available from: https://www.who.int/news-room/fact-sheets/detail/soil-transmitted-helminth-infections.
- 56.Reed EK, Smith KA. Using our understanding of interactions between helminth metabolism and host immunity to target worm survival. Trends Parasitol. 2024;40:549–61. [DOI] [PubMed] [Google Scholar]
- 57.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5. [DOI] [PubMed] [Google Scholar]
- 58.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abdulla S, Aevermann B, Assis P, Badajoz S, Bell SM, Bezzi E, et al. CZ CELL×GENE Discover: a single-cell data platform for scalable exploration, analysis and modeling of aggregated data. bioRxiv. 10.1101/2023.10.30.563174.
- 63.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14:979–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hao Y, Stuart T, Kowalski MH, Choudhary S, Hoffman P, Hartman A, et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat Biotechnol. 2024;42:293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marsh S, Salmon M, Hoffman P samuel-marsh/scCustomize: Version 1.1.1. Zenodo; 2023.
- 68.Wickham H. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York; 2016.
- 69.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Processed RNA-seq data from jejunums of wild type and Ptk6−/− mice are available in the article and supplementary materials. Additionally, the whole raw and processed data files have been deposited in NCBI’s Gene Expression Omnibus [61] and are accessible through GEO Series accession number GSE243229. Other datasets used in this study are listed in sections “Methods and Materials” and “Supplemental Material and Methods.” This study did not generate any new code.