Abstract
As the third most important temperate fruit, Pear (Pyrus spp.) exhibits a remarkable genetic diversity and is classified into two mainly categories known as Asian pear and European pear. Although several pear genomes are available, most of the released versions are fragmented and not chromosome-level high-quality. In this study, we report two high-quality genomes for Pyrus bretschneideri Rhed. cv. ‘Danshansuli’ (DS) and Pyrus communis L. cv. ‘Conference’ (KFL), which represent the predominant Asian and European cultivars, respectively, with nearly telomere-to-telomere (T2T) gap-free level. The finally assembled genome sizes for DS and KFL were 510.98 Mb and 510.71 Mb, respectively, with Contig N50 of 29.47 Mb and 30.47 Mb, where each chromosome was represented by a single contig. The DS and KFL genomes yielded a total of 46,394 and 44,702 protein-coding genes, respectively. Among these genes, the functional annotation accounted for 96.47% and 96.46% in the DS and KFL genomes. The two novels nearly T2T genomic information offers an invaluable resource for comparative genomics, genetic diversity analysis, molecular breeding strategies, and functional exploration.
Subject terms: Whole genome amplification, Comparative genomics, Agricultural genetics
Background & Summary
The pear (Pyrus spp.) is the third most widely cultivated fruit tree in temperate regions, following the apple (Malus pumila) and grape (Vitis vinifera)1, and it consisted of over 22 species, as well as more than 5000 accessions exhibiting diverse morphological, physiological, and adaptive characteristics2. Based on their morphology and original distribution, the genus Pyrus can be classified into two major native groups, Asian pears (Oriental pears, P. pyrifolia, P. bretschneideri, P. ussuriensis and Pyrus sinkiangensis) and European pears (Occidental pears, P. communis)3. ‘Dangshansuli’ (Pyrus bretschneideri Rehd.), a commercially significant cultivar of Asiatic pear, is cultivated worldwide with an annual production exceeding 4 million tons. With a cultivation history in China spanning over 500 years, it holds immense importance in the field. The European pear cultivar ‘Conference’ (P. communis L.) is widely recognized as an exceptional variety, serving as the predominant cultivated choice in countries including the United Kingdom, Germany, and France.
Since the first genome assembly of ‘Dangshansuli’ was published in 20134, subsequent genome sequences have been made available for ‘Bartlett’ European pear (P. communis)5, pear rootstock [(P. ussuriensis × P. communis) × spp.]1, Asian wild pear (P. betulifolia)6, Chinese sand pear (P. pyrifolia)7 and Japanese sand pear (P. pyrifolia)8. These genomes have facilitated the advancement of functional genomics and provided valuable insights for pear breeding; however, technological limitations have resulted in existing gaps within these genomes, leading to a loss of genetic information and impeding our comprehensive understanding of pear genome structure and evolution. High-accuracy gapless genomes are more informative and can greatly facilitate molecular breeding and gene characterization. In recent years, multiple telomere-to -telomere (T2T) sequence assemblies were reported for several plant species, including Arabidopsis thaliana9, rice10,11, barley12, banana13–15, maize16, tea tree17, tomato18, watermelon19, bitter melon20, kiwifruit21, Brassica rapa22, lemons23, strawberry24, Jujube25, apple26 and pear27. These genomes accurately represent high-complexity sequences in telomeric, centromeric, and high repeat regions, and provide an opportunity to explore genetic variations, repetitive sequences, and duplication events in these formerly ‘dark matter’ regions.
In this study, we generated the T2T gap-free genome sequence of ‘Dangshansuli’ and ‘Conference’, by incorporating PacBio HiFi reads, Nanopore ultra-long reads, and high throughput chromatin conformation capture (Hi-C) paired reads(Figs. 1 and 2). These genomic data can be valuable resources for comparative or functional genomic studies and pear breeding.
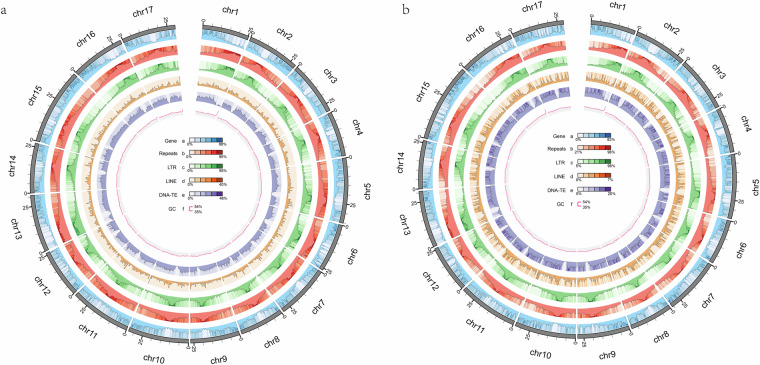
Fig. 1.
The telomere-to-telomere genomic characteristics of Pyrus bretschneideri ‘DS’ (a) and Pyrus communis. L ‘KFL’ (b).
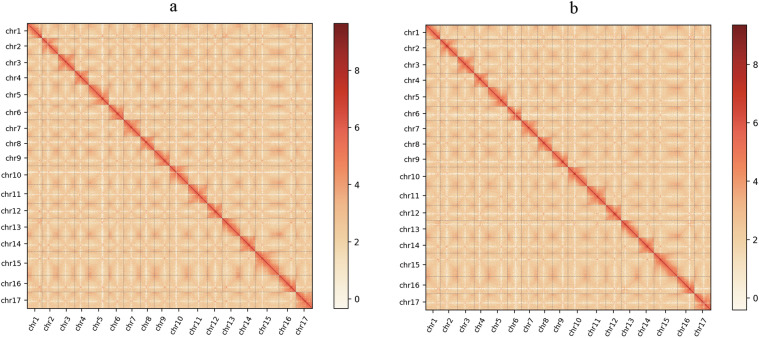
Fig. 2.
Hi-C intra-chromosomal interaction map in the genome P. bretschneideri ‘DS’ (a) and P. communis.L ‘KFL’ (b).
Methods
Sample collection
To obtain the representaive genetic resources, Pyrus bretschneideri Rhed. Asian cultivar ‘Danshansuli’ (DS) and Pyrus communis L. European cultivar ‘Conference’ (KFL) were sampled from Dangshan County, Anhui Province, China and National Germplasm Repository of Pear in the Research Institute of Pomology, Chinese Academy of Agricultural Sciences (CAAS), Xingcheng, China, respectively. Fresh young pear leaves were harvested from both cultivars, for DNA extraction. Additionally, RNA extraction and sequencing were performed using pooling samples from various tissues, including young leaves, mature leaves, and fruits at different developmental stages. The samples were rapidly frozen using liquid nitrogen and subsequently stored in freezers at a temperature of −80 °C.
Long-read library construction and sequencing
The high molecular weight (HMW) genomic DNA was extracted from fresh young leaves of both cultivars using DNeasy Plant Kit (Qiagen). Long-read sequencing libraries were prepared for both PacBio and Nanopore platforms. For PacBio sequencing, ~20 kb insert libraries were generated for each cultivar using the SMRTbell Express Template Prep Kit 2.0 (Pacific Biosciences, USA). The Nanopore ultra-long sequencing was performed with two libraries constructed according to the manufacturer’s instructions using the Ligation sequencing 1D kit (SQK-LSK109, Oxford Nanopore Technologies, Oxford, UK). The highly accurate long-read sequencing data were generated by the PacBio Revio SMRT cell equipped with HiFi model, while the sequencing of Nanopore cells were performed on the PromethION platform (Oxford Nanopore Technologies), resulting in a total of 28.6 Gb and 40 Gb for DS and KFL, respectively (Table S1). The N50 length of CCS reads was 18,248 bp and 16,031 bp for DS and KFL, respectively (Table S1 and Figure S1). A total of 182.77 Gb (6.76 million reads) and 149.43 Gb (4.76 million reads) ONT long reads were obtained, with the longest reads measuring 558,196 bp and 484,688 bp for DS and KFL, respectively (Table S2).
Hi-C library preparation, sequencing
Hi-C data utilization has proven indispensable and highly effective for achieving chromosome-level assembly. In this study, the fresh young leaves of two pear cultivars were fixed in 1% formaldehyde (Sigma) for cross-linking. The samples were subsequently resuspended in lysis buffer and chromatin was fragmented using MboI (NEB) restriction endonucleases. After biotin labelling, crosslinking using T4 DNA Ligase (ENZYMATICS), the DNA fragments were captured by employing Streptavidin-coated magnetic beads (Thermo Fisher SCIENTIFIC). Then, an “A” base at the 3′-end of each strand were added with KAPA HYPER PREP KIT (KAPA). Finally, two Hi-C libraries were obtained with DNBSEQ-T7 (PE150). A total of 114.8 Gb and 111.15 Gb clean data were sequenced for DS and KFL, respectively (Table S3), following the removal of low-quality reads and adapter contamination using SOAPnuke v2.11 (-n 0.01 -l 20 -q 0.1 -i -Q 2 -G 2 -M 2 -A 0.5)28.
Genome assembly
The primary contigs of pear genome were initially generated by assembling PacBio HiFi data and nanopore data using Hifiasm (v0.19.6) with default parameters29. The initial assembled size of the DS genome was approximately 523.72 Mb, with a contig N50 value of 29.47 Mb, while the KFL genome had a size of 518.63 Mb and a contig N50 value of 29.42 Mb. The primary genome sequences of the two pear genomes underwent redundancy elimination due to their high heterozygosity rate. This was achieved using the Purge Haplotigs program with the parameters “-j 80 -s 80 -a 30”30. After filtering out redundant sequences, the assembled genome sizes for DS and KFL were 510.94 Mb and 514.99 Mb, respectively. Then, the Hi-C data were used for anchoring the assembled sequences to chromosome level. Firstly, the DS and KFL clean Hi-C reads were aligned to the contigs using BWA v 0.7.1231. The overall mapping rates of DS and KFL were 94.27% and 94.85%, respectively. After deduplication, the valid pairs reflecting contact interactions accounted for 28.49% and 31.49% of the total reads for DS and KFL, respectively. Then, the Hi-C contacts were calculated by juicer pipeline v 1.5 and the contigs were anchored onto chromosomes using the 3D-DNA pipeline v18092232 Subsequently, the verification and refinement processes were performed with JUICEBOX Assembly Tools (v 2.15.07)33. Finally, 98.36% (502.5 Mb) and 99.17% (506.4 Mb) of the assembled sequences for DS and KFL were successfully anchored and oriented onto all 17 chromosomes (Fig. 2). The ultra-long ONT reads were mapped to the chromosome-level genome using minimap2 to extend the telomere sequences34. The extension of telomeres is described in detail as follows: Firstly, all ONT ultra-long reads were aligned onto chromosomes using minimap2. Subsequently, all reads that exhibited a single alignment within 100 bp of the chromosome ends were collected. The read containing the artifacts sequence (the telomere regions are often misidentified as other types of repeats in a link-specific manner during nanopore sequencing, which is referred to as artifacts) is excluded from analysis. The read containing the artifacts sequence (the telomere regions are often misidentified as other types of repeats in a link-specific way during nanopore sequencing, which is called artifacts) is filtered out. Then, the comparison of the read is calculated, and the read of the extendible median length is defined as reference and the other is query. The reads of reference and query were assembled into consensus sequence. The consensus sequences were compared to both ends of each chromosome using Blastn, and the alignment sequences with coverage > = 90 were substituted based on their alignment position. Finally, telomere identification was conducted by searching the entire genome for characteristic sequences (CCCTAA/TTAGGG) in the telomere region and tallying the number of such sequences with at least four repeats within a 50 kb span at each end of every chromosome. To fill the remained gaps, the ultra-long ONT reads (>100 kb) were applied to generate gap-free genome using TGS-GapCloser (v 1.2.0) with the parameter “--min_match 2000 --min_nread 10”35. The comprehensive information provided through gap filling demonstrated strong support at a high level of depth (Table S4). The assembly pipeline is showed in Figure S2. The assembled genome sizes for DS and KFL were 510.98 Mb and 510.71 Mb, respectively, with Contig N50 values of 29.47 Mb and 30.47 Mb (Table 1), where each chromosome was composed by a single contig.
Table 1.
Comparison of the genome assemblies in pear (Pyrus bretschneideri ‘DS’, Pyrus communis.L ‘KFL’ and Pyrus pyrifolia ‘Yunhong No. 1’.
| Characteristics | P. bretschneideri DS (Wu. et al.4) | P. bretschneideri DS (this study) | P. communis KFL (this study) | P. pyrifolia Yunhong No. 1 (Sun et al.27) |
|---|---|---|---|---|
| Total assembly size (Mb) | 512.0 | 510.9 | 510.7 | 501.2 |
| Contig number | 25,312 | 71 | 57 | 20 |
| Contig N50 (kb) | 35.7 | 29,470.4 | 29,415.1 | 29,255.5 |
| Scaffold number | 2,103 | 71 | 57 | 20 |
| Scaffold N50 (kb) | 540.8 | 29,470.4 | 30,472.4 | 29,255.5 |
| Anchor ratio (%) | 75.5 | 98.36 | 99.17 | 99.81 |
| Number of gap-free chromosomes | 0 | 17 | 17 | 17 |
| Number of telomeres | 0 | 31 | 31 | 34 |
| Number of predicted centromeres | 0 | 17 | 17 | 17 |
| Genome BUSCOs (%) | 87.8 | 98.8 | 98.6 | 99.0 |
| Gene number | 42,812 | 46,394 | 44,702 | 41,969 |
| Gene BUSCOs (%) | 83.6 | 98.2 | 98.2 | 98 |
| Repeat sequence percentage (%) | 53.10 | 53.20 | 54.86 | 50.20 |
Genome annotation
After completing the assembly of the T2T genome, repeat and gene annotation processes were performed as the pipeline (Figure S3). For the repeat annotation, the Tandem repeats was performed using Tandem Repeats Finder (4.9) with default parameter36. Then, the homologous sequences of the two pear genomes were annotated using the software RepeatMasker (open-4.0.9)37 and RepeatProteinMask (v 4.0.7)38 based on the Repbase library (http://www.girinst.org/repbase)39. The databases of own repetitive sequence features were constructed using RepeatModeler open-1.0.1140 and LTR_FINDER_parallel 1.0.741, followed by repeat identification performed with RepeatMasker (open-4.0.9)37. Finally, 52.08% and 53.75% of assembled DS and KFL genomes were classified as transposable elements (TEs) (Table S5).
The predominant repetitive sequences identified in this study were long terminal repeats (LTRs), accounting for 36.03% (184,118,617 bp) and 37.31% (190,545,332 bp) of the genome in DS and KFL respectively (Table S6).
For gene annotation, protein-coding genes were predicted combining homology-based, de novo prediction, and RNA-Seq-based annotation. 1) The software of GlimmerHMM 3.0.442 with default parameters was used for de novo prediction. 2) The protein sequences of six representative plant species, namely Pyrus communis Bartlett DH Genome v2.05, Pyrus pyrifolia YunhongNO.127, Pyrus betuleafolia6, Prunus persica (GCF_000346465.2)43, Malus domestica (GCF_002114115.1)44, Arabidopsis thaliana (GCF_000001735.4)45 were retrieved from the National Center for Biotechnology Information (NCBI) or from available database for homology-based prediction. 3) The RNA-based prediction was carried out based on PacBio long-read transcriptome data. The ISO-Seq data was obtained using SMRT Analysis (v2.2). TransDecoder v 5.5.0 (https://github.com/TransDecoder/TransDecoder) with default parameters was used to predict the coding region. The integration of the three gene annotation strategies using Maker2 (2.31.10)46 yielded the final set of genes. Finally, a total of 46,394 and 44,702 protein-coding genes were obtained respectively for DS and KFL genomes (Table 1).
Functional annotation and genome evaluation
The gene set of the two pear genomes was functionally annotated based on the eight databases including NR version 2023-04-01 (NCBI nonredundant protein), TrEMBL version 2023-03-01 (http://www.uniprot.org), Swiss-Prot version 2023-03-01 (http://www.gpmaw.com/html/swiss-prot.html), KEGG version 105.0, 2023-01-01 (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg/), KOG version 2023-03-0147, PlantTFDB 5.0, InterPro 93.0 and GO Ontology (GO) version 2023-04-01. The functional annotation for DS and KFL genomes accounted for 96.47% and 96.46% of the annotated genes, respectively (Table S7).
Data Records
The sequencing dataset had been deposited in the Sequence Read Archive (SRA) under project number PRJNA1073018. The link of sequencing data is provided below: https://www.ncbi.nlm.nih.gov/sra?linkname=bioproject_sra_all&from_uid=1073018.
DNA sequencing data from Hi-C library of Pyrus bretschneideri Rehd. were deposited in the SRA at SRR27896858-SRR2789686548.
DNA sequencing data from PacBio HiFi library of Pyrus bretschneideri Rehd. were deposited in the SRA at SRR2789687749.
DNA sequencing data from ONT library of Pyrus bretschneideri Rehd. were deposited in the SRA at SRR2789687650.
DNA sequencing data from Hi-C library of Pyrus communis L. were deposited in the SRA at SRR27896866-SRR2789687351.
DNA sequencing data from PacBio HiFi library of Pyrus communis L. were deposited in the SRA at SRR2789687552.
DNA sequencing data from ONT library of Pyrus communis L. were deposited in the SRA at SRR2789687453.
The genome sequences and annotation are presented as follows: 10.6084/m9.figshare.25139555.
The genome assembly has also been deposited to NCBl under the accessionnumber of JBFSJW00000000054 and JBFSJV00000000055.
Technical Validation
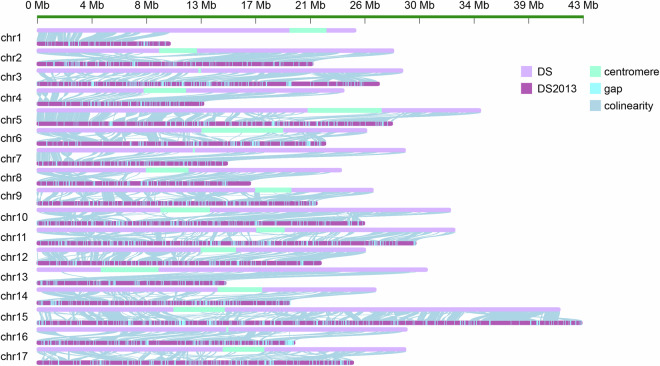
In comparison to the previously published genome, our novel T2T genome assembly for DS exhibits a substantial enhancement in contiguity metrics, with an impressive increase in contig N50 from 35.7 Kb to 29.47 Mb, representing an approximately 825-fold improvement (Table 1). The alignment results between the draft DS genome and T2T version demonstrated significant improvements in both anchoring rate and orientation compared to the previous genome (Fig. 3). The proportion of chromosome anchoring has been updated from 75.5% to 98.36%, and a total of 19,176 gaps have been filled. (Table 2).
Fig. 3.
The comparative analysis of draft version and novel T2T version of the DS genome. (Gap display with more than 500 bp).
Table 2.
The comparison statistics of the improved pear genome (DS) at chromosome-level.
| Chr ID | P. bretschneideri Danshansuli (Wu. et al.4) | P. bretschneideri Danshansuli (this study) | ||||
|---|---|---|---|---|---|---|
| Gap number | Gap length | Chr length | Gap number | Gap length | Chr length | |
| Chr1 | 560 | 127,399 | 10,691,755 | 0 | 0 | 25,508,628 |
| Chr2 | 1,049 | 282,340 | 22,098,781 | 0 | 0 | 28,547,938 |
| Chr3 | 1,308 | 535,648 | 27,392,285 | 0 | 0 | 29,270,603 |
| Chr4 | 676 | 202,901 | 13,384,095 | 0 | 0 | 24,572,644 |
| Chr5 | 1,432 | 393,963 | 28,442,882 | 0 | 0 | 35,484,103 |
| Chr6 | 1,131 | 480,795 | 23,112,003 | 0 | 0 | 26,394,572 |
| Chr7 | 800 | 220,735 | 15,267,112 | 0 | 0 | 29,470,448 |
| Chr8 | 878 | 214,883 | 17,110,699 | 0 | 0 | 24,371,048 |
| Chr9 | 1,158 | 330,362 | 22,428,363 | 0 | 0 | 26,902,160 |
| Chr10 | 1,241 | 514,530 | 26,220,497 | 0 | 0 | 33,080,020 |
| Chr11 | 1,499 | 780,917 | 30,316,187 | 0 | 0 | 33,429,468 |
| Chr12 | 1,070 | 349,691 | 22,757,174 | 0 | 0 | 26,290,821 |
| Chr13 | 756 | 168,848 | 15,147,870 | 0 | 0 | 31,223,542 |
| Chr14 | 1,087 | 383,605 | 20,263,496 | 0 | 0 | 27,130,315 |
| Chr15 | 2,163 | 878,535 | 43,574,056 | 0 | 0 | 41,821,467 |
| Chr16 | 1,083 | 424,041 | 20,649,150 | 0 | 0 | 29,614,284 |
| Chr17 | 1,285 | 403,377 | 25,332,008 | 0 | 0 | 29,508,640 |
| Total | 19,176 | 6,692,570 | 384,188,413 | 0 | 0 | 502,620,701 |
The two T2T genome assemblies were compared with the recently published T2T pear Yuhong No.1 (YH) genome. The HiFi long reads were mapped to the assembled genome using minimap234, and the results showed the mapping rate of 99.99% and 99.97% for DS and KFL, respectively (Table S8). The average sequencing depth in DS was 52.2, with a mapping rate of 99.99% and a coverage of 99.99%. Additionally, the coverage reached 96.75% at a minimum depth of 20×. (Table S8). In KFL, the average sequencing depth was 74.84, with a mapping rate of 99.97% and a coverage rate of 99.99% and the coverage rate reached 99.69% at a minimum depth of 20 × (Table S8). The GC distribution based on the HiFi reads was utilized to assess potential contamination. The distribution of GC content and sequencing depth revealed that nearly all GC points were concentrated around 37.5%, indicating an absence of exogenous species pollution (Figures S4, S5).
The completeness of the assembled genome sequences of the two pear cultivars was assessed using Benchmarking Universal Single-Copy Orthologs (BUSCO, v 5.3.1)56 with the embryophyta_odb10 database, which comprises 1614 genes. The BUSCO evaluation of the DS genome revealed that 99.19% of the BUSCOs were classified as complete, with 64.93% representing single-copy and complete BUSCOs, while 34.26% were identified as duplicated and complete BUSCOs (Table S9). The BUSCO evaluation of the KFL genome revealed that 99.38% of the BUSCOs were classified as complete, with 65.37% representing single-copy and complete BUSCOs, while 34.01% were identified as duplicated and complete BUSCOs (Table S9).
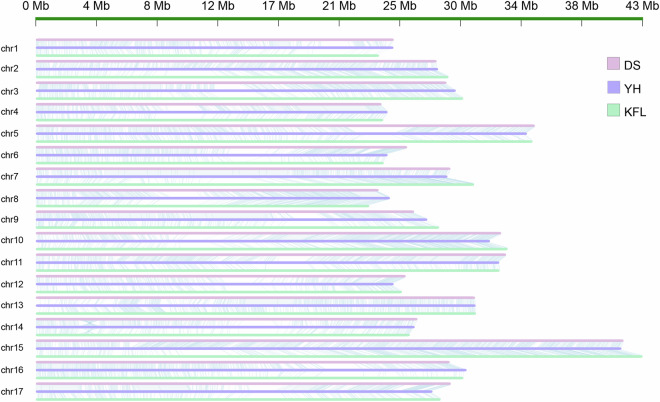
The three T2T genomes including ‘DS’ (T2T version), ‘KFL’ and Pyrus pyrifolia Yuhong No.1 were aligned with each other by MUMmer4 with nucmer model (-c 1000 --maxgap = 500, identity >95 and length >15 kb). The collinearity analysis of genome revealed high homology (>80%), and the chromosome anchoring was accurate (Fig. 4). A total of 5.46 M (YH vs DS) and 9.18 M SNPs (YH vs KFL) were detected (Table S10).
Fig. 4.
Collinearity analysis of P. bretschneideri ‘DS’ (T2T version), P. communis. L ‘KFL’ and Pyrus pyrifolia Yuhong No. 1 genomes.
A total of 31 telomeres were assembled and identified for both DS and KFL, except for each one of chromosome 1, 13, 16, suggesting that both genomes were assembled nearly telomere-to-telomere (Table S11). Using T2T genome sequences, we successfully predicted all 17 centromeric regions exhibiting the characteristic of centromere-specific monomers in both DS and KFL. Notably, these centromeric regions were predominantly composed of repetitive sequences, particularly LTR elements, with a significantly lower gene density compared to other genomic regions (Table 1 and Fig. 1).
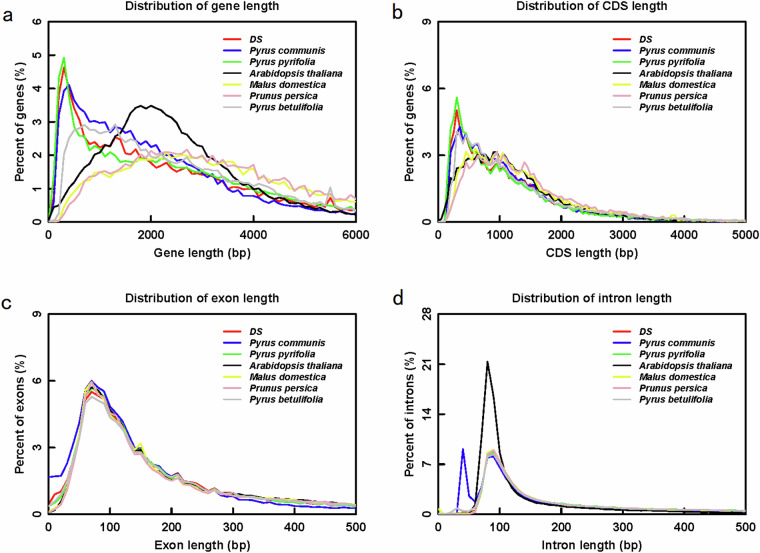
The comparison of gene features revealed that the same Pyrus genus exhibited a similar distribution pattern in terms of gene length, exon number, intron length, and exon length. (Fig. 5 and S6).
Fig. 5.
The composition of gene elements in P. bretschneideri ‘DS’ and other related species. (a) Distribution of gene length. (b) Distribution of CDS length. (c) Distribution of exon length. (d) Distribution of intron length.
The gene model, which is similarly structured, also indicates that the gene annotation is comparable. For the gene evaluation, 98.2% of completely gene BUSCOs in both DS and KFL, which is slightly higher than Yuhong No1 (98%) and significantly higher that that of draft DS genome (Table 1).
Supplementary information
Acknowledgements
This work was financially supported by the earmarked fund for Young Talents Program of Anhui Academy of Agricultural Sciences (QNYC- 202112), Earmarked Fund for China Agriculture Research System (CARS-28).
Author contributions
Y. Qi, Y. Cao, L. Tian and F. Zhan collected the samples, conducted experiments, N. NA, L. Lu, D. Shan and F. Ke performed bioinformatics analysis. Y. Qi, Z. Gao, J. Jian and Y. Xu conceived the study and wrote the manuscript. All authors have read and approved the final manuscript.
Code availability
No specific code was developed in this study. The data analyses were performed following the protocols described in the Methods section.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yongjie Qi, Dai Shan, Yufen Cao
Contributor Information
Yongjie Qi, Email: anhuiqyj@163.com.
Jianbo Jian, Email: jianjianbo@bgi.com.
Zhenghui Gao, Email: gzh96gao@163.com.
Yiliu Xu, Email: yiliuxu@163.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-024-04015-3.
References
- 1.Ou, S. et al. A de novo genome assembly of the dwarfing pear rootstock Zhongai 1. Scientific Data.6, 281 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li, J. et al. Pear genetics: recent advances, new prospects, and a road map for the future. HorticRes. 9 (2022). [DOI] [PMC free article] [PubMed]
- 3.Wu, J. et al. Diversification and independent domestication of Asian and European pears. Genome Biol.19, 77 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu, J. et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res.23, 396–408 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linsmith, G. et al. Pseudo-chromosome–length genome assembly of a double haploid “Bartlett” pear (Pyrus communis L.). GigaScience8 (2019). [DOI] [PMC free article] [PubMed]
- 6.Dong, X. et al. De novo assembly of a wild pear (Pyrus betuleafolia) genome. Plant Biotechnology Journal18, 581–595 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao, Y. et al. High-quality genome assembly of ‘Cuiguan’ pear (Pyrus pyrifolia) as a reference genome for identifying regulatory genes and epigenetic modifications responsible for bud dormancy. HorticRes.8, 197 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shirasawa, K. et al. Chromosome-scale genome assembly of Japanese pear (Pyrus pyrifolia) variety ‘Nijisseiki’. DNA Res.28, dsab001 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, B. et al. High-quality Arabidopsis thaliana Genome Assembly with Nanopore and HiFi Long Reads. Genomics, proteomics & bioinformatics10.1016/j.gpb.2021.08.003 (2021). [DOI] [PMC free article] [PubMed]
- 10.Li, K. et al. Gapless indica rice genome reveals synergistic contributions of active transposable elements and segmental duplications to rice genome evolution. Molecular plant14, 1745–1756, 10.1016/j.molp.2021.06.017 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Song, J. M. et al. Two gap-free reference genomes and a global view of the centromere architecture in rice. Molecular plant14, 1757–1767, 10.1016/j.molp.2021.06.018 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Navratilova, P. et al. Prospects of telomere-to-telomere assembly in barley: Analysis of sequence gaps in the MorexV3 reference genome. Plant biotechnology journal10.1111/pbi.13816 (2022). [DOI] [PMC free article] [PubMed]
- 13.Belser, C. et al. Telomere-to-telomere gapless chromosomes of banana using nanopore sequencing. Communications biology4, 1047, 10.1038/s42003-021-02559-3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, H. et al. Telomere-to-telomere haplotype-resolved reference genome reveals subgenome divergence and disease resistance in triploid Cavendish banana. Horticulture research10, 10.1093/hr/uhad153 (2023). [DOI] [PMC free article] [PubMed]
- 15.Liu, X. et al. The phased telomere-to-telomere reference genome of Musa acuminata, a main contributor to banana cultivars. Scientific Data10, 631 10.1038/s41597-023-02546-9 (2023). [DOI] [PMC free article] [PubMed]
- 16.Liu, J. et al. Gapless assembly of maize chromosomes using long-read technologies. Genome 854 biology21, 121, 10.1186/s13059-020-02029-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang, W. et al. Genome assembly of wild tea tree DASZ reveals pedigree and selection history of tea varieties. Nature communications11, 3719, 10.1038/s41467-020-17498-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Rengs, W. et al. A chromosome scale tomato genome built from complementary PacBio and Nanopore sequences alone reveals extensive linkage drag during breeding. The Plant journal: for cell and molecular biology110, 572–588, 10.1111/tpj.15690 (2022). [DOI] [PubMed] [Google Scholar]
- 19.Deng, Y. et al. A telomere-to-telomere gap-free reference genome of watermelon and its mutation library provide important resources for gene discovery and breeding. Molecular plant10.1016/j.molp.2022.06.010 (2022). [DOI] [PubMed]
- 20.Fu, A. et al. Telomere-to-telomere genome assembly of bitter melon (Momordica charantia L. var. abbreviata Ser.) reveals fruit development, composition and ripening genetic characteristics. Horticulture research10, uhac228, 10.1093/hr/uhac228 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yue, J. et al. Telomere-to-telomere and gap-free reference genome assembly of the kiwifruit Actinidia chinensis. Horticulture research10, uhac264, 10.1093/hr/uhac264 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang, L. et al. A near-complete genome assembly of Brassica rapa provides new insights into the evolution of centromeres. Plant biotechnology journal21, 1022–1032, 10.1111/pbi.14015 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bao, Y. et al. A gap-free and haplotype-resolved lemon genome provides insights into flavor synthesis and huanglongbing (HLB) tolerance. Horticulture research10, uhad020, 10.1093/hr/uhad020 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou, Y. et al. The Telomere to Telomere genome of Fragaria vesca reveals the genomic evolution of Fragaria and the origin of cultivated octoploid strawberry. Horticulture research10.1093/hr/uhad027 (2023). [DOI] [PMC free article] [PubMed]
- 25.Yang, M. et al. Insights into the evolution and spatial chromosome architecture of jujube from an updated gapless genome assembly. Plant Communications, 10.1016/j.xplc.2023.100662 (2023) [DOI] [PMC free article] [PubMed]
- 26.Li, W. et al. Near-gapless and haplotype-resolved apple genomes provide insights into the genetic basis of rootstock-induced dwarfing. Nat Genet56, 505–516 (2024). [DOI] [PubMed] [Google Scholar]
- 27.Sun, M. et al. Telomere-to telomere pear (Pyrus pyrifolia) reference genome reveals segmental and whole genome duplication driving genome evolution. Horticulture Research10, uhad201, 10.1093/hr/uhad201 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen, Y. et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience7, 1–6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng, H. et al. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods18, 170–175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roach, M. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics19, 460 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dudchenko, O. et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science356, 92–95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durand, N. et al. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell systems3, 95–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu, G. et al. LR_Gapcloser: a tiling path-based gap closer that uses long reads to complete genome assembly. Gigascience8 (2019). [DOI] [PMC free article] [PubMed]
- 36.Benson, G. Tandem repeats finder:a program to analyze DNA sequences. Nucleic Acids Research27(2), 573–580, 10.1093/nar/27.2.573 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price, A. et al. De novo identification of repeat families in large genomes. Bioinformatics21(Suppl 1), i351–358 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Bao, W. et al. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mobile DNA6, 11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jurka, J. et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenetic and genome research110, 462–467 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Tarailo-Graovac, M. & Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Current protocols in bioinformatics Chapter 4, 4.10.11–14.10.14 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Ou, S. & Jiang, N. LTR_FINDER_parallel: parallelization of LTR_FINDER enabling rapid identification of long terminal repeat retrotransposons. Mobile DNA10, 48 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Majoros, W. et al. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics20, 2878–2879 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Verde, I. et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet45, 487–494 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Daccord, N. et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet49, 1099–1106 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Tabata, S. et al. Sequence and analysis of chromosome 5 of the plant Arabidopsis thaliana. Nature408, 823–826 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Holt, C. & Yandell, M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics12, 491 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korf, I. Gene finding in novel genomes. BMC Bioinformatics5, 59 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.NCBI Sequence Read Archivehttps://identifiers.org/insdc.sra:SRR27896858 (2024).
- 49.NCBI Sequence Read Archivehttps://identifiers.org/insdc.sra:SRR27896877 (2024).
- 50.NCBI Sequence Read Archivehttps://identifiers.org/insdc.sra:SRR27896876 (2024).
- 51.NCBI Sequence Read Archivehttps://identifiers.org/insdc.sra:SRR27896873 (2024).
- 52.NCBI Sequence Read Archivehttps://identifiers.org/insdc.sra:SRR27896875 (2024).
- 53.NCBI Sequence Read Archivehttps://identifiers.org/insdc.sra:SRR27896874 (2024).
- 54.NCBI GenBankhttps://identifiers.org/ncbi/insdc:JBFSJW010000000 (2024).
- 55.NCBI GenBankhttps://identifiers.org/ncbi/insdc:JBFSJV010000000 (2024).
- 56.Simão, F. et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- NCBI Sequence Read Archivehttps://identifiers.org/insdc.sra:SRR27896858 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/insdc.sra:SRR27896877 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/insdc.sra:SRR27896876 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/insdc.sra:SRR27896873 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/insdc.sra:SRR27896875 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/insdc.sra:SRR27896874 (2024).
- NCBI GenBankhttps://identifiers.org/ncbi/insdc:JBFSJW010000000 (2024).
- NCBI GenBankhttps://identifiers.org/ncbi/insdc:JBFSJV010000000 (2024).
Supplementary Materials
Data Availability Statement
No specific code was developed in this study. The data analyses were performed following the protocols described in the Methods section.