Abstract
Neural crest cells are central to vertebrate development and evolution, endowing vertebrates with a “new head” that resulted in morphological, physiological, and behavioral features that allowed vertebrates to become active predators. One remarkable feature of neural crest cells is their multi-germ layer potential that allows for the formation of both ectodermal (pigmentation, peripheral glia, sensory neurons) and mesenchymal (connective tissue, cartilage/bone, dermis) cell types. Understanding the cellular and evolutionary origins of this broad cellular potential in the neural crest has been a long-standing focus for developmental biologists. Here, we review recent work that has demonstrated that neural crest cells share key features with pluripotent blastula stem cells, including expression of the Yamanaka stem cell factors (Oct3/4, Klf4, Sox2, c-Myc). These shared features suggest that pluripotency is either retained in the neural crest from blastula stages or subsequently reactivated as the neural crest forms. We highlight the cellular and molecular parallels between blastula stem cells and neural crest cells and discuss the work that has led to current models for the cellular origins of broad potential in the crest. Finally, we explore how these themes can provide new insights into how and when neural crest cells and pluripotency evolved in vertebrates and the evolutionary relationship between these populations.
Keywords: neural crest, stem cells, pluripotency, cellular potential, evolution
1.1. Introduction
Conrad Waddington famously depicted cellular potential in the form of a landscape where balls travel down a hill, segregating into valleys, each representing a distinct cell lineage [1]. This model suggests that once a cell initiates a trajectory towards lineage restriction, the fates it can contribute to become progressively restricted coinciding with a gradual loss of potential. A cell type that has historically challenged the paradigm portrayed by Waddington’s landscape is the vertebrate neural crest. Although these cells arise in the ectoderm between the neural plate and non-neural ectoderm, the neural crest gives rise to both ectodermal (pigmentation, peripheral glia, sensory neurons) and mesenchymal (connective tissue, cartilage/bone, dermis) cell types (Fig. 1) [2–4]. Recently, models have been put forth to explain the origins of this multi-germ layer potential within the crest. These include a model for (1) retention of blastula-like potential within the neural crest or (2) in vivo reprogramming of the crest that reverses the trajectory down Waddington’s landscape. [5–8]. In this review, we outline parallels between blastula stem cells and the neural crest, highlight the work that has led to each of these models, discuss the possible evolutionary origins of the neural crest’s multi-germ layer potential, and finally propose additional experimentation needed to gain a more comprehensive understanding of the nature and origin of cellular potential in the neural crest.
Figure 1. In vivo cellular potential and shared gene regulatory circuitry of blastula and neural crest stem cells.
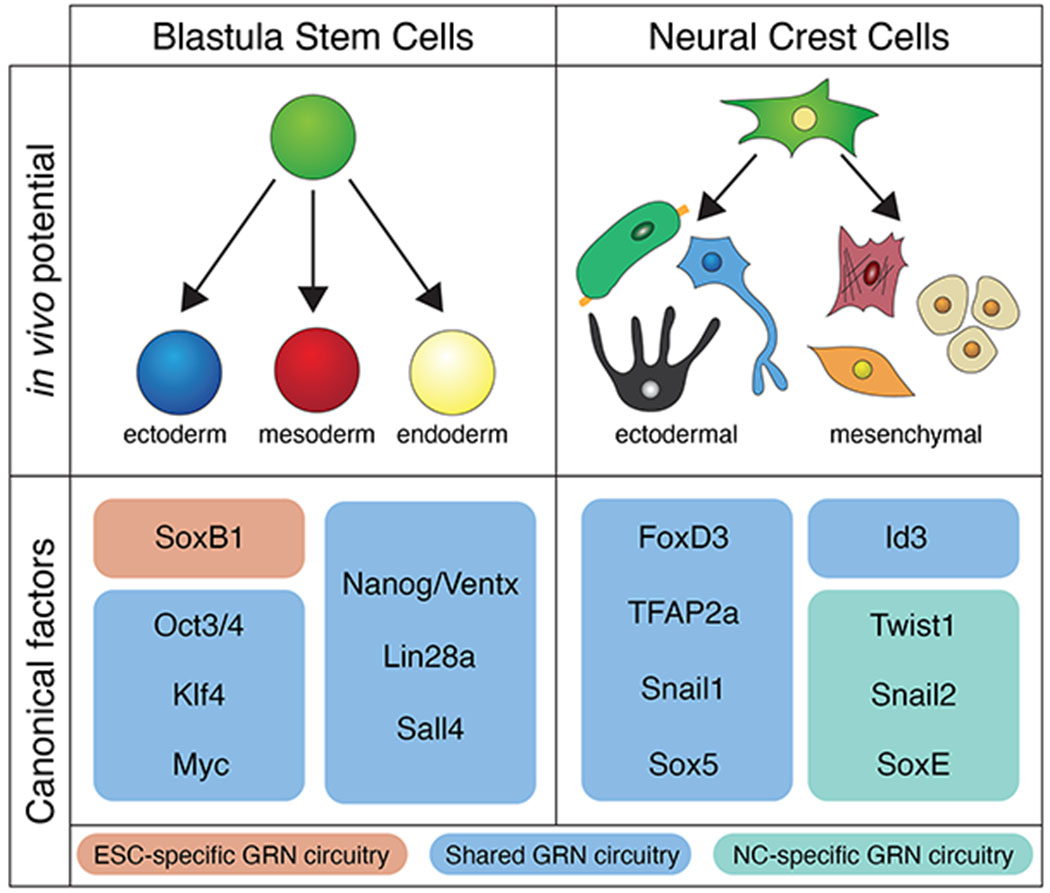
Top panels illustrate the in vivo potential of blastula stem cells (ectoderm, mesoderm, and endoderm) and neural crest cells (ectodermal: pigmentation, peripheral glia, sensory neurons and mesenchymal: connective tissue, cartilage/bone, dermis). Bottom panels show common gene regulatorys circuitry for blastula stem cells and neural crest factors. Circuity is grouped in boxes by shared (blue), ESC-specific (orange), and neural crest-specific (green) gene regulatory elements.
1.2. Cellular Potential in Stem Cells and the Neural Crest
During embryonic development initially totipotent cells progressively give rise to hundreds of different unipotent cell types. Embryonic stem cells, such as those found in the inner cell mass of blastula stage embryos, are defined as pluripotent as they self-renew and give rise to all embryonic cell types (Fig. 1). Pluripotency in vivo is a transient state governed by conserved gene regulatory networks [9]. Seminal work by Satoshi Yamanaka identified four transcription factors, Oct3/4, Sox2, Klf4, and c-Myc (referred to herein as the Yamanaka factors), whose combination could convert somatic cells into pluripotent stem cells. These reprogrammed somatic cells can self-renew, contribute to all three germ layers in teratoma assays, and maintain a pluripotent state in culture [10]. The reprogramming efficiency of the Yamanaka factors is generally less than 0.1%, however, occurs over weeks in vitro, and is dependent on significant alterations in epigenetic modifications [11]. Still, this hallmark discovery demonstrated that cell specialization is reversible all the way back to a pluripotent state and that a small number of factors are sufficient to initiate this process. Subsequent work has provided insights into how the Yamanaka factors orchestrate a gene regulatory network that not only can reprogram cells to a pluripotent state, but also maintain that state in vivo [12–14]. These mechanisms included driving feed forward transcriptional autoregulatory loops, promoting chromatin remodeling, and establishing bivalent epigenetic signatures at developmental gene promotors [9, 15, 16]. Importantly, a number of substitutes for the Yamanaka factors have been identified, including Lin28, Sall4, Esrrβ, Nr5a2, and Nanog, many of which are downstream targets of the Yamanaka factors in blastula stem cells [17–19].
A major advance in our understanding of neural crest cell potential came from the realization that these cells share significant GRN architecture with the pluripotent cells of blastula-stage embryos [6] (Fig. 1). Indeed, early hints that the GRNs controlling pluripotent blastula and neural crest stem cells shared key factors came from studies showing that FoxD3 and Myc play key roles in both cell populations [20–22] and this was later shown to be true of another canonical neural crest gene TFAP2 [23–25]. Subsequently, it was shown that a broad range of neural plate border/neural crest factors were expressed in blastula stem cells [6]. Several of these neural crest/neural plate border factors (Snai1, Sox5, and ZIC3) are required to maintain pluripotency in blastula/embryonic stems cells [6, 26, 27]. Reciprocally, a combination of in situ hybridization, spatial genomic analysis, and single cell sequencing data demonstrate that core pluripotency factors are expressed in the neural plate border/neural crest progenitor population in multiple species [6–8, 28, 29]. Early foxd3 positive cells in zebrafish express multiple pluripotency factors [30]. In chick, expression of pluripotency factors is heterogenous with the KLF4/POUV/NANOG positive cells expressing markers for ectodermal and mesodermal cell types while MSI1/SOX2 positive cells predominately express markers for ectodermal cell types [31]. In addition to sharing transcriptional machinery, blastula stem cells and neural crest cells also share high levels of Erk activity and have similar epigenetic signatures – low levels of histone acetylation and bivalent promotors [32–36]. HDAC1 can enhance the generation of neural crest cells and also cooperates with NANOG to promote pluripotency in embryonic stem cells [32, 37]. Other chromatin remolding factors, such as Chd7 and Hmga1, help to maintain an undifferentiated state in embryonic stem cells and are necessary for neural crest formation with Chd7 being required for neural crest cell multipotency [38–43].
Functional studies have begun to shed light on the roles for some pluripotency factors for the multi-germ layer potential of neural crest cells. For example, in Xenopus, ventx2 (the functional ortholog of Nanog) and c-myc are essential for the formation of both ectomesenchymal and non-ectomesenchymal neural crest derivatives [7, 22]. Additionally, sall1 and sall4 morphants display reduced neural crest marker expression. A role for these factors in neural crest differentiation has yet to be explored, but SALL4 plays a crucial role in melanoma disease progression [44, 45]. In mouse, Oct4 is not required for the formation of non-ectomesenchymal cell types, but is important for ectomesenchymal derivatives [8]. LIN28A also promotes neural crest cell multipotency and is required for expression of early neural crest factors in chick in vivo [46]. Together, these data indicate that the broad cellular potential of blastula stem cells and neural crest cells are regulated by a shared set of transcription factors. The full extent to which these two cell populations overlap on a cellular, molecular, and epigenetic basis within an organism and across vertebrate species is an important area of ongoing investigation.
While there are significant similarities between GRNs of pluripotent blastula and neural crest stem cells, it is equally important to understand the differences as this can shed light on how GRNs evolve to support new cellular functions and behaviors. One of the most striking differences between these two GRNs is the utilization of SoxB1 (Sox1/2/3) factors in blastula stem cells but deployment of SoxE (Sox8/9/10) factors in the neural crest [47–52]. SoxE factor duplication coincided with the evolutionary emergence of the neural crest and its diversification [53]. Sox9 is required for differentiation of neural crest into cartilage, an ectomesenchymal derivative, while Sox10 is necessary for both melanocyte and sensory neuron/glia formation [47, 50, 54–60]. Given this, it is tempting to speculate that duplication and neofunctionalization of an ancestral SoxE factor may have directly contributed to the expanded cellular potential of the neural crest. This is an attractive hypothesis given the prominent role of SoxB1 factors in maintaining potential in stem cells. Importantly, it has been shown that other Sox factors, including Sox15 or Sox18, can substitute for SoxB1 factors in reprogramming [61]. Moreover, molecular replacement experiments conducted in Xenopus demonstrated that SoxE factors can rescue loss of cellular potential in SoxB1 morphants [62]. This suggests a model in which a molecular hand-off event in the pluripotency GRN between SoxB1 and SoxE factors is necessary to maintain cellular potential in the neural crest prior to lineage diversification. Subsequent to its role in promoting pluripotency SoxB1 expression becomes restricted to the neural plate and is essential for maintaining the neural progenitor pool, although a small subpopulation of Sox2+ cells at the neural plate border contribute to the migrating neural crest [63]. While the ultimate fates of those cells are unknown, those cells may have limited potential due to prolonged Sox2 expression and only contribute to the non-ectomesenchymal derivates. In the context of vertebrate evolution, the hand-off from SoxB1 to SoxE factors was likely not only important for maintaining the broad developmental potential of the neural crest, but also for positioning these cells to give rise to non-neural derivatives, a role that SoxB1 factors may be less suited for.
Similar to the transition from SoxB1 factors in the blastula to SoxE factors in the neural crest, the Snail family of transcription factors (Snai1, Snai2) also show comparable patterns of temporal variation in expression that may hold clues to the developmental origins of neural crest potential. In Xenopus, for example, Snai1 is expressed in the pluripotent blastula cells and is essential for expression of Yamanaka factors in vivo and lineage restriction to endomesoderm ex vivo. Shortly thereafter, Snai1 expression gradually resolves to the neural plate border, with low expression retained in the neural and non-neural ectoderm. By contrast, Snai2 expression is entirely absent from the blastula and becomes detectable only during late gastrulation at the neural plate border, overlapping with Snai1 and other early neural crest markers [6]. Given recent work in mouse showing a role for Snai1 in regulating the exit from naïve pluripotency to lineage restriction [64], it is tempting to speculate that the shift from a Snai1+/Snai2- state in the blastula to a Snai1+/Snai2+ state in the neural crest might be instrumental in mediating the emergence of the neural crest GRN from the pluripotency GRN in the early blastula. These examples provide further evidence for the role of paralogous substitutions [65] in shaping the developmental trajectory of the neural crest and the transition from pluripotency to lineage restriction. From an evolutionary perspective, it also highlights the importance of gene duplication—followed by neo- or sub-functionalization—as a potential mechanism by which neural crest and blastula stem cell GRNs originated and evolved in vertebrates.
1.2.1. Models for Neural Crest Potency
Historically, the multi-germ later potential of the neural crest had been viewed as a regaining of embryonic potential following an initial commitment to the ectodermal lineage. Following the discovery that neural crest cells share GRN features with pluripotent blastula cells, a new model was proposed in which neural crest cells retain blastula-like potential even as neighboring cells undergo lineage restriction [6]. From the perspective of Waddington’s landscape, this posits that neural crest cells remaining near the apex of landscape even as other cells descend toward lineage restriction (Fig. 2A). Support for this hypothesis comes from work in multiple model organisms, including frog, chick, mouse and fish, where neural crest factors are expressed in blastula stem cells and pluripotency factors in the neural crest, [6–8, 28–30]. In Xenopus, expression of many of these factors (soxB1, tfap2a, ventx2, snai1, zic1, ets1) appears to resolve temporally through gastrulation to position them at the neural plate border; however, this data from static in situs lacks the resolution to show continuous retention. Importantly, however, functional data using animal pole explants demonstrates that neural crest cells, when given appropriate queues, can form cells from all three germ layers, suggesting that the cellular potential of these cells is most akin to the pluripotent blastula stem cells [6]. Additional work is needed to probe the degree of overlap between the GRNs of blastula stem cells and neural crest cells. Moreover, a robust temporal analysis of the pluripotency factor expression from the blastula through neural crest formation is needed along with an investigation into the temporal nature of factor substitution (such as SoxB1 for SoxE) and its relevancy to GRN evolution. In addition, the mechanisms by which neural crest cells escape lineage restriction remain unknown. Data from chick suggests that the neural crest induction occurs at blastula stages [66], so these cells may possess an intrinsic capacity to avoid lineage restriction promoted by robust regulatory networks that drive early fate commitment. Finally, the extent to which this retention model applies to all other vertebrates, and in particular basal vertebrates, has not been fully investigated.
Figure 2. Proposed models for cellular potential in the neural crest.
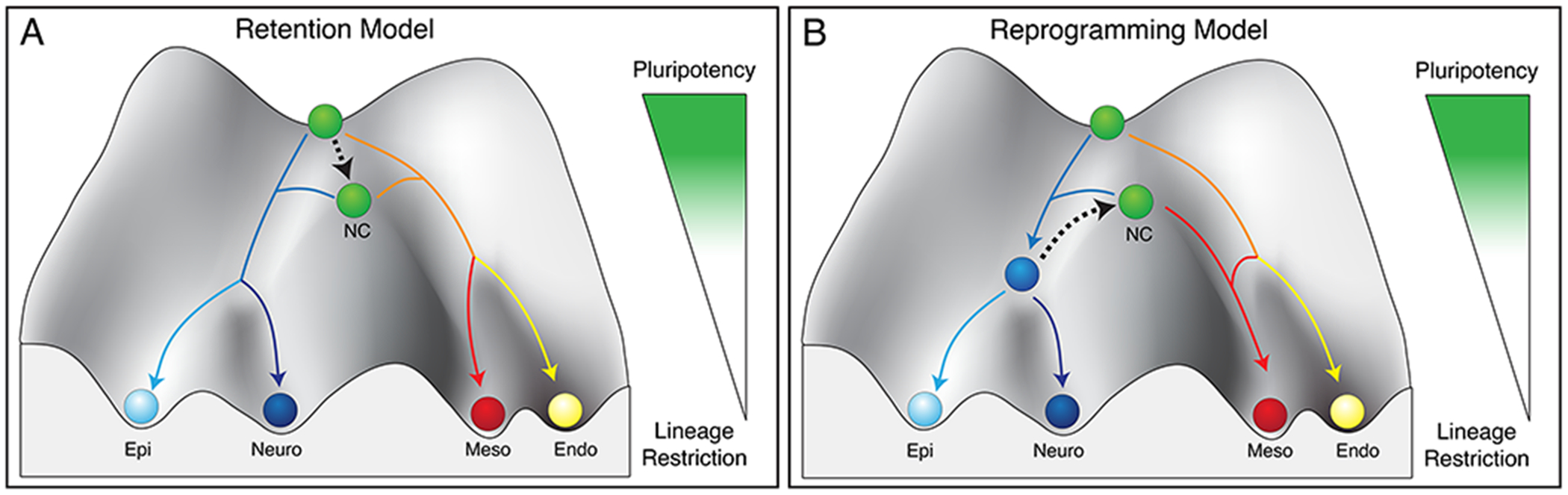
(A) Neural crest cells avoid lineage restriction and instead retain blastula-like cellular potential (black arrow). (B) Neural crest cells follow the trajectory of ectodermal cells along Waddington’s landscape and then undergo a reprogramming event (black arrow), conferring them with enhanced cellular potential.
Neural crest (NC), epidermis (Epi), neural (Neuro), mesoderm (Meso), endoderm (Endo).
A second model based on recent work in mouse embryos puts a new spin on the historic model (ball pushed back up hill; Fig. 2B) by providing evidence for an in vivo reprogramming event that occurs in the neural crest progenitor population [8]. This work postulates that Yamanaka (or Yamanaka-like) factors are transiently re-expressed in prospective murine neural crest cells resulting in a cellular reprogramming back to a state with increased cellular potential. The work that led to the genesis of this hypothesis showed that mouse Oct4, after being down-regulated in the rostral neuroectoderm, is re-expressed in the prospective neural crest. Ablation of the Oct4+ progenitor pool leads to dramatic defects in the craniofacial complex while genetic knockout of Oct4 from this pool leads to milder facial phenotypes, but significant alterations to the ectomesenchymal progenitor population. This work is the first to show evidence of a potential in vivo reprogramming event in the neural crest. While cell reprogramming efficiency is quite low in somatic cells in culture, embryonic cells may be more amenable to reprogramming. Further work needed to validate this model include functional assays testing the cellular potential of these Oct4+ cells and assessing the requirement for other pluripotency factors in this reprogramming event. Finally, given the documented differences between neural crest formation in mice and other vertebrates [67], the extent to which this reprogramming model applies to all other vertebrates, and in particular basal vertebrates, needs to be more fully examined.
1.2.2. Lessons in cellular potential from the trunk neural crest
The multi-germ layer potential of the neural crest is greater at the cranial axial level as trunk neural crest do not give rise to cartilage and other ectomesenchymal derivatives in vivo [4]. The more restricted in vivo cellular potential of trunk neural crest allows for an interesting comparative analysis of the broader potential exhibited by the cranial neural crest. Under appropriate cellular contexts trunk neural crest cells are capable of contributing to ectomesenchymal derivatives. When cultured ex vivo under pro-chondrogenic conditions, trunk crest successfully formed bone/cartilage [68], and although premigratory trunk crest transplanted to the cranial regions failed to form cartilage in classic transplantation experiments, they do contribute to connective tissues, dermis, and muscle [69, 70]. Recent work revisiting this classic experiment demonstrated that trunk crest could contribute to cartilage in vivo if Sox8, Ets1, and Tfap2b were expressed in posterior epiblast cells prior to transplantation [71]. Similarly, overexpression of Twist1 in the trunk crest in vivo pushed cells towards ectomesenchymal lineages, albeit not cartilage, at the expense of neuronal sensory, autonomic, and glial fates [72]. Thus, while trunk neural crest cells seem to largely lack an intrinsic ability to contribute to cartilage in vivo, such potential can be unveiled given appropriate developmental cues. Conversely, cranial neural crest cells, when transplanted to a trunk axial level, still form cartilage indicating an intrinsic persistence of cellular potential despite changes in the environment [73].
Single cell sequencing data suggests that the process of delamination largely erases axial transcriptional signatures and primes cells for fate decisions [72]. But how do these findings relate to cellular potential? In the context of trunk to cranial transplantation, we can infer that intrinsic differences in cellular potential arise during development that cannot be fully erased or rewritten even in a new environmental context. However, a small number of transcription factors (Sox8/Tfap2b/Ets1) are sufficient to confer greater potential [71]. It would be interesting to test if expression of the Yamanaka factors in the trunk neural crest could lead to in vivo cartilage formation and assess if SOX8, TFAP2B, and ETS1 are direct transcriptional targets of these factors. The expression of core pluripotency factors in the trunk neural crest has not been fully characterized, so it is possible the absence of a single factor could limit potential. Notably, the dermal denticles of skates are derived from the trunk neural crest [73, 74], and it would also be interesting to examine the expression of the core pluripotency factors and Sox8/Tfap2b/Ets1 in these embryos. Finally, classic transplantation experiments paired with transcriptome/epigenome analyses should provide critical insights into differences between the cellular potential of cranial and trunk crest cells.
1.3. Evolutionary Origins of Neural Crest Potential
Given the central importance of neural crest cells to vertebrate development and evolution, one of the long-standing goals in evolutionary-developmental biology (“evo-devo”) has been to reconstruct the ancestral vertebrate neural crest GRN by comparing neural crest development between the two major lineages of vertebrates, the jawed (gnathostome) and jawless (cyclostome) clades [75, 76]. There have also been attempts to trace the pre-vertebrate origins of the neural crest and its cellular potential by studying the development of the closest extant relatives of the vertebrates—the invertebrate chordates [77–79]. In this section we discuss the evolutionary origins of the neural crest and neural crest cellular potential. We review briefly how recent work has shown the presence of “neural crest-like” cells with restricted potential in invertebrate chordates and speculate on how GRN evolution might have led to the broad cellular potential characteristic of the neural crest in vertebrates.
1.3.1. Invertebrate chordates—cells at the neural plate border with restricted developmental potential
Extant invertebrate chordates are represented by two main lineages: cephalochordates (amphioxus) and tunicates (sea squirts). Although all chordates have a common set of shared-derived characters (e.g., notochord, post-anal tail, pharyngeal slits), the invertebrate chordate body plan notably lacks many of the defining characters of the vertebrate clade, many of which are derived from the neural crest [80]. The most conspicuously absent of these features are ectomesenchymal derivatives, such as a robust craniofacial and jaw skeleton of cartilage and bone and smooth muscle, as well as parts of the teeth and heart [80]. Many of these features culminated in the formation of the vertebrate “new head” and are hypothesized to have enabled the first vertebrates to distinguish themselves morphologically, physiologically and behaviorally from their invertebrate relatives, thereby making the transition from small, passive filter feeders to large, active predators. Thus, many of the fundamental differences between the vertebrate and invertebrate body plans can be framed as differences in cellular potential. Understanding the molecular and genetic basis for this difference in cellular potential between invertebrate chordates and vertebrates is essential to understanding not just how neural crest cells evolved, but also how these cells acquired the ability to layer a broad range of new specialized cell types onto the ancestral chordate body plan.
1.3.2. Amphioxus
Historically, cephalochordates were recognized as the sister group to vertebrates, with tunicates as outgroup. This was supported by comparisons of morphological characters in both living forms and fossils, as well as phylogenetic analysis of single genes or small groups of genes [81–83]. However, with the advent of large-scale, genome-wide sequencing it was revealed that tunicates—rather than cephalochordates—are the closest living relatives of the vertebrates, forming a new clade (olfactores) with cephalochordates as outgroup [84, 85].
Regardless of these changes to the chordate phylogeny, there has never been strong evidence to suggest that amphioxus has neural crest cells or a cell type that might be homologized with the neural crest. While many of the transcription factors and signaling molecules that drive the formation of neural crest cells in vertebrates are encoded in the amphioxus genome they are also present in other invertebrates such as arthropods. Nevertheless, expression of a handful of these factors overlap in the neural plate and neural plate border, including Snail, Pax3/7, Tfap2a, Dlx, Msx, and Zic, and some are responsive to Bmp signaling, just as in vertebrates [86]. Other homologs of neural crest genes such as FoxD3, Twist, and SoxE are not expressed in the neural plate or neural plate border and are instead expressed in mesoderm or endomesoderm, consistent with co-option of these factors to the neural crest domain in early vertebrate history [86, 87].
There are also important similarities and differences between amphioxus and vertebrates in the expression of key genes known for regulating pluripotency. For example, only members of the SoxB1 family are known to be expressed in the animal pole of blastula stage amphioxus embryos [88]. SoxB1 expression in the neurula gradually resolves to the neural plate and neural plate border, overlapping with expression of genes such as Snail, Msx, Zic and others [86]. The amphioxus Myc homolog is not expressed in the neural plate or neural plate border, with transcripts instead localizing to the endomesoderm [86], and it is unknown if pluripotent blastula cells express Myc alongside SoxB1 genes. Strict orthologs of other core members of the vertebrate pluripotency GRN (e.g., Ventx/Nanog, Oct4) appear to be absent from the amphioxus genome [89–91], a feature likely shared with tunicates (described below).
1.3.3. Tunicates
In contrast to amphioxus, there is significant evidence that tunicates possess at least one or two cell types that might be homologized as a precursor cell type to the neural crest in vertebrates. In the mangrove tunicate, E. turbinata, vital dye labeling revealed migratory cells emanating from the neural tube in a manner reminiscent of neural crest [92]. These cells express the HNK1 epitope and a Zic ortholog and give rise to pigment cells that colonized the larval body wall and siphons. In another tunicate species (C. intestinalis) a neural crest-like population was traced to the a9.49 lineage in the tadpole head [78]. Cells derived from this lineage express Msx, Pax3/7, Zic, Id, Snail, Ets, FoxD and migrate before differentiating into pigmented sensory organs—the otolith and ocellus [78]. Normally, these cells migrate only a short distance before differentiating, however ectopic expression of Twist enabled these cells to migrate significantly further distances in larval tunics, suggestive of Twist-mediated migratory behavior in vertebrate cranial neural crest cells [78]. Finally, the bipolar tail neurons (BTNs) in the larval trunk of Ciona tadpoles also share similarities with neural crest cells [93]. BTN precursors at the neural plate border co-express Snail, Msx, Pax3/7 and Zic and then migrate within the trunk before differentiating into sensory neurons. The shared expression of Islet and Neurogenin, as well as functional and ultrastructural similarities, suggests that BTNs are homologous to dorsal root ganglia, a neural crest-derived component of the PNS in vertebrates [93].
All of this provides strong evidence that tunicates possess cells that have some molecular and cellular hallmarks of the vertebrate neural crest. However, similar to amphioxus, much remains to be learned about the GRNs controlling the early development of these cells, particularly in the blastula. Based on gene expression patterns alone, it seems that tunicates, like vertebrates, express homologs of Tfap2a, FoxD3, SoxB1 and Myc in animal pole cells of early cleavage stages [94]. On the other hand, tunicates lack recognizable homologs of Ventx/Nanog and Oct4, two vertebrate factors that are instrumental to vertebrate pluripotency [89, 91].
1.3.4. Vertebrates—the origin of neural crest cells with multi-germ layer potential
The mechanisms underlying neural crest development are broadly conserved across the two major lineages of vertebrates, the jawed (gnathostome) and jawless (cyclostome) clades. Work over the past 20 years has revealed that, despite marked differences in morphology, physiology, and behavior, representatives of the jawed and jawless lineages nonetheless share a great deal in common at multiple tiers of neural crest development [95–99]. For example, both groups produce migratory neural crest cells that originate in the dorsal neural tube along the anterior-posterior axis and which give rise to both ectomesenchyme (cartilage, smooth muscle) and non-ectomesenchyme (pigment cells, neurons, glia) [100–107]. These similarities also extend the level of gene expression patterns and functions, with a common set of signaling molecules (WNT, BMP, FGF, Delta/Notch) and transcription factors (SoxE, Zic, Snail, Tfap2a, Id, Endothelins, Myc, Pax3) orchestrating a shared neural crest GRN that likely dates back to the last common ancestor of all vertebrates [95–99, 108–110]. More recent work has revealed that these shared features are also hardwired into the genome in the form of an ancestral cis-regulatory code that directs shared gene expression patterns throughout neural crest development, including cross-species conservation of Hox, Pbx, Meis and SoxE enhancers [109, 111].
With the origin of the vertebrates, we see for the first time the formation of bona fide neural crest cells with the broad potential to produce a diverse range of derivatives—features notably absent from the proto-neural crest-like cells of invertebrate chordates. What drove this increase in developmental potential? As described above, recent studies of the neural crest in jawed vertebrates have found that blastula-associated pluripotency factors (Ventx/Nanog, Klf4, Myc, Pou5F, SoxB1) are expressed in neural crest progenitors. This suggests that neural crest cells either retain or re-activate a pluripotency GRN from the blastula [6, 8]. Interestingly, in jawless vertebrates such as lampreys, we see some evidence for this in the form of pluripotency factors being co-expressed early in the blastula and later in the NPB and neural crest. For example, lamprey SoxB1a, and SoxB1b are expressed in animal pole cells of the blastula, similar to that of other vertebrates and invertebrate chordates [88]. At later stages, these same factors along with Myc are all expressed in the neural plate and extend laterally into the NPB, overlapping with expression of Msx1/2, Snail, Pax3/7, and Zic1 [88, 98, 108, 112]. Thus, in jawless vertebrates we see the first evidence of at least three core pluripotency genes (SoxB1a, SoxB1b, Myc) being co-expressed with canonical members of the neural crest GRN, similar to jawed vertebrates (Fig. 3). Although this suggests retention of a pluripotent state in the NPB and neural crest of jawless vertebrates, we still lack key information on the expression patterns of several genes that maintain a pluripotent state in blastula stem cells and neural crest cells of jawed vertebrates—Ventx/Nanog, Oct4, and Klf4.
Figure 3. Current relationships between expression of pluripotency factors and neural crest potential mapped onto a chordate phylogeny.
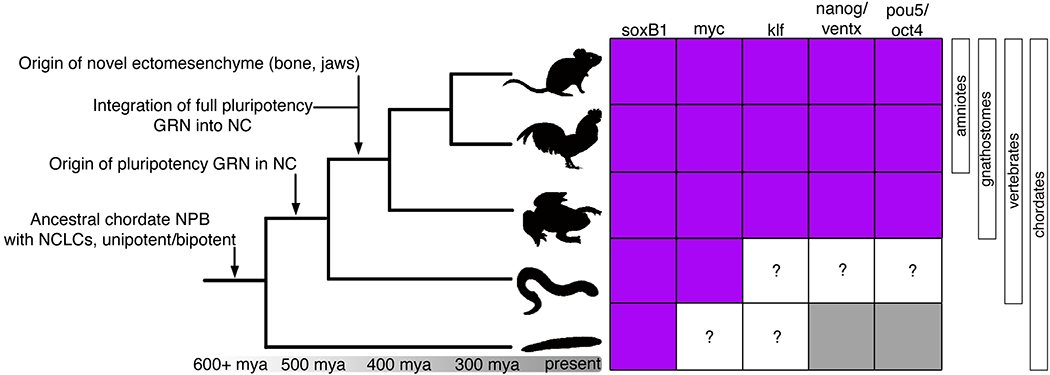
Pluripotency factors that are expressed in the blastula and in the neural plate border and/or neural crest are colored purple. White indicates absence of gene expression in these populations, whereas boxes with a question mark indicate that expression is unknown. Grey boxes indicate genes that are not expressed because they are not present in the indicated organism’s genome. Organisms from top: Mouse, Chick, Xenopus, lamprey, chordate ancestor.
1.3.5. Origin and evolution of cellular potential in the chordate neural plate border and the emergence of pluripotency in vertebrates
From the evidence presented above, both comparative genomics, and gene expression analysis in amphioxus and tunicates suggests that neural crest cells were not present in the last common ancestor of chordates (Fig. 3). Although some of the regulatory genes involved in vertebrate pluripotency were present, these likely served in capacities unrelated to pluripotency. For example, SoxB1 and Myc factors are present in the blastula of amphioxus and tunicates and gradually resolve to the neural plate and neural plate border where they are co-expressed with homologs of Msx, Zic, Snail, Pax3/7, Dlx and Tfap2a [88]. However, these gene expression patterns are likely part of an ancient neuroectodermal patterning module carried over from early bilaterians [77, 79, 112, 113]. Thus, in invertebrates, SoxB1 and Myc should be viewed in the context of their ancient role in neural stem cell proliferation and neuroectodermal patterning rather than maintenance of pluripotency.
After the split from the cephalochordate lineage, this ancient neuroectoderm module likely underwent further modifications in the last common ancestor of tunicates and vertebrates to create a “proto-neural crest” GRN. This event would have been characterized by introduction of novel genes such as FoxD3, as well downstream targets involved in the short range migration of a subset of sensory neurons and pigments cells [78, 93]. Conspicuously absent from these ancestral proto-neural crest cells, however, would have been the broad developmental potential needed to produce a range of specialized cell types. Indeed, rather than having multi-germ layer potential, invertebrate precursors of vertebrate neural crest cells were unipotent or bipotent and did not produce mesenchymal derivatives such as cartilage, bone, and smooth muscle [7, 8]. This is most likely explained by the absence of a functional pluripotency GRN, without which the potential of these cells was restricted to only cell types associated with the ancient neuroectoderm module
One possible explanation for the restricted potential of cells at the NPB of invertebrate chordates is that these embryos lack key pluripotency factors. Other than SoxB1, orthologs of transcription factors essential for maintenance of pluripotency in vertebrate blastulae—particularly Ventx/Nanog and Pou5/Oct4—appear to be absent from invertebrate chordate genomes [88, 89, 91]. Moreover, work in tunicates, the sister group to vertebrates, shows that most lineages of the early (112-cell) embryo have already been restricted to a single fate [114]. This suggests that somatic cell pluripotency may itself be an evolutionary innovation of the vertebrate clade.
The origins of somatic cell pluripotency likely involved evolution of novel gene families in vertebrates, in particular Ventx/Nanog and Pou5f/Oct4, and their introduction into an ancestral neuroectoderm GRN that included SoxB1, Myc, and Klf factors. Retention of this pluripotency GRN would have endowed cells at the NPB in early vertebrates with the capacity to produce both ectomesenchymal and non-ectomesenchymal derivatives (Fig. 3) [7]. Coupled with the evolution of novel genes and signaling pathways (e.g., Endothelins) [115, 116] that direct lineage diversification from this initial pluripotent state, these features together distinguish blastula and neural crest stem cells in vertebrates from the precursor cells with restricted developmental potential in tunicates. Evidence that Ventx/Nanog and Oct4 are co-expressed with Klf and SoxB1 factors in the blastula and neural crest of jawless vertebrates such as lamprey would support this hypothesis, and is an important area of future investigation.
1.3.6. Expansion of pluripotency in jawed vertebrates
Although some pluripotency factors are expressed in the neural crest of both jawed and jawless vertebrates, there are also key differences in cellular potential between these two lineages. Unlike jawed vertebrates, lamprey neural crest cells do not give rise to myelin sheaths, the jaw skeleton, bone, or sympathetic chain ganglia [117]. Moreover, lampreys have only cranial and trunk neural crest and do not display the more complex subdivisions of the neural crest domain along the anterior-posterior axis characteristic of jawed vertebrates (e.g., cranial, vagal, cardiac, sacral) [100, 110]. A similar lack of complexity in neural crest formation is observed at the molecular and cellular levels, with cranial neural crest in lampreys expressing some but not all of the cranial neural crest-specific GRN in jawed vertebrates [110]. These differences in regulatory complexity may reflect fundamental differences in neural crest cell potential between jawed and jawless vertebrates. Evidence for this idea comes from comparative genomics. Similar to invertebrate chordates, the genomes of jawless vertebrates encode only a fraction of the Klfs present in jawed vertebrates, and have also been reported to lack Pou5f/Oct4 and Ventx/Nanog, although some recent work suggests that some of these factors may indeed be present in lampreys and hagfishes [7, 89, 91]. Thus, evolution of novel pluripotency factors and their expression in presumptive neural crest—either by retention from the blastula, or through reactivation—would have endowed cranial neural crest cells with the capacity to produce a broad range of mesenchymal and non-mesenchymal derivatives [7, 8] (Fig. 3). Going forward it will be crucial to compare the regulatory components driving the developmental potential of blastula and neural crest stem cells in jawless vertebrates, such as lamprey, with those of jawed vertebrates.
1.4. Future directions
The advent of genomic technologies has provided exciting new insights into the genesis and development of the neural crest. However, much remains to be learned, including the mechanisms utilized to maintain broad multi-germ layer potential. Cross species comparisons of the transcriptomes and epigenomes of blastula stem cells, neural plate border/neural crest precursors, and the emerging neural crest will provide critical insights into the origins of cellular potential in these important cells. Furthermore, single cell sequencing studies have revealed significant gene expression heterogeneity in these cells [8, 29, 72] raising the possibility that individual neural crest cells may, at a given time, have different intrinsic propensities towards either ectomesenchymal or non-ectomesenchymal fates, and that this might shift dynamically even within individual cells. For example, during iPSC reprogramming, the relative levels and stichometry of Oct3/4 to Sox2 impact expression of mesendodermal genes vs ectodermal genes with high Oct3/4 biasing towards mesendoderm and high Sox2 biasing towards ectoderm [118]. Going forward it will be essential to rigorously quantify the expression levels of pluripotency GRN components in individual cells and relate this directly to the epigenome. Finally, as we seek to further understand the relationship between the core pluripotency and neural crest GRNs, functional studies in vivo or ex vivo are essential to determine if individual GRN components are essential for neural crest cells to give rise to their full range of derivatives.
Studies in traditional vertebrate research organisms will continue to be important for understanding the origins of the neural crest’s broad developmental potential but are also likely to continue to reveal species-specific differences in gene expression patterns, transcription factor functions, and more, owing to either adaptation or developmental drift. This highlights the importance of taking a comparative (evolutionary) approach involving analysis of neural crest potential in jawed vertebrates, jawless vertebrates, and invertebrate chordate taxa. With the advent of modern genomic, transcriptomic, and epigenomic tools, and their recent use in evolutionarily informative chordate models (lamprey, amphioxus, tunicates), it is now possible to make genome-wide comparisons of cellular potential in neural crest and neural crest-like cells across the chordate tree of life. By placing the results of such studies within a phylogenetic framework it will be possible to identify the universal features underlying the origins of neural crest potential and how re-wiring of pluripotency GRNs in early vertebrate history might have driven the evolution of novel phenotypic traits.
1.5. Funding statements
The authors acknowledge funding support from the NIH (1F32DE029113-ENS and R01GM116538-CL), the National Science Foundation (1764421, CL), and Simons Foundation (SFARI 597491-RWC, CL). JRY is a Walder Foundation Fellow of the Life Sciences Research Foundation.
Abbreviations:
- GRN
gene regulatory network
- NPB
neural plate border
- PNS
peripheral nervous system
- BTN
bipolar tail neuron
References
- [1].Waddington CH, The Strategy of Genes, George Unwin & Unwin; 1957. [Google Scholar]
- [2].Bronner ME, LeDouarin NM, Development and evolution of the neural crest: an overview, Dev Biol 366(1) (2012) 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bronner-Fraser M, Fraser SE, Cell lineage analysis reveals multipotency of some avian neural crest cells, Nature 335(6186) (1988) 161–4. [DOI] [PubMed] [Google Scholar]
- [4].Le Douarin NM, Dupin E, Multipotentiality of the neural crest, Curr Opin Genet Dev 13(5) (2003) 529–36. [DOI] [PubMed] [Google Scholar]
- [5].Briggs JA, Weinreb C, Wagner DE, Megason S, Peshkin L, Kirschner MW, Klein AM, The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution, Science 360(6392) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Buitrago-Delgado E, Nordin K, Rao A, Geary L, LaBonne C, Shared regulatory programs suggest retention of blastula-stage potential in neural crest cells, Science 348(6241) (2015) 1332–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Scerbo P, Monsoro-Burq AH, The vertebrate-specific VENTX/NANOG gene empowers neural crest with ectomesenchyme potential, Science Advances 6(18) (2020) eaaz1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zalc A, Sinha R, Gulati GS, Wesche DJ, Daszczuk P, Swigut T, Weissman IL, Wysocka J, Reactivation of the pluripotency program precedes formation of the cranial neural crest, Science 371(6529) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Takahashi K, Yamanaka S, A decade of transcription factor-mediated reprogramming to pluripotency, Nat Rev Mol Cell Biol 17(3) (2016) 183–93. [DOI] [PubMed] [Google Scholar]
- [10].Takahashi K, Yamanaka S, Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors, Cell 126(4) (2006) 663–76. [DOI] [PubMed] [Google Scholar]
- [11].Scesa G, Adami R, Bottai D, iPSC Preparation and Epigenetic Memory: Does the Tissue Origin Matter?, Cells 10(6) (2021) 1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chambers I, Smith A, Self-renewal of teratocarcinoma and embryonic stem cells, Oncogene 23(43) (2004) 7150–60. [DOI] [PubMed] [Google Scholar]
- [13].Niwa H, How is pluripotency determined and maintained?, Development 134(4) (2007) 635–46. [DOI] [PubMed] [Google Scholar]
- [14].Silva J, Smith A, Capturing pluripotency, Cell 132(4) (2008) 532–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liang G, Zhang Y, Embryonic stem cell and induced pluripotent stem cell: an epigenetic perspective, Cell Res 23(1) (2013) 49–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Smith ZD, Sindhu C, Meissner A, Molecular features of cellular reprogramming and development, Nat Rev Mol Cell Biol 17(3) (2016) 139–54. [DOI] [PubMed] [Google Scholar]
- [17].Kim J, Chu J, Shen X, Wang J, Orkin SH, An extended transcriptional network for pluripotency of embryonic stem cells, Cell 132(6) (2008) 1049–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liu X, Huang J, Chen T, Wang Y, Xin S, Li J, Pei G, Kang J, Yamanaka factors critically regulate the developmental signaling network in mouse embryonic stem cells, Cell Res 18(12) (2008) 1177–89. [DOI] [PubMed] [Google Scholar]
- [19].Heng J-CD, Feng B, Han J, Jiang J, Kraus P, Ng J-H, Orlov YL, Huss M, Yang L, Lufkin T, The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells, Cell stem cell 6(2) (2010) 167–174. [DOI] [PubMed] [Google Scholar]
- [20].Hanna LA, Foreman RK, Tarasenko IA, Kessler DS, Labosky PA, Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo, Genes & development 16(20) (2002) 2650–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Teng L, Mundell NA, Frist AY, Wang Q, Labosky PA, Requirement for Foxd3 in the maintenance of neural crest progenitors, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bellmeyer A, Krase J, Lindgren J, LaBonne C, The protooncogene c-myc is an essential regulator of neural crest formation in xenopus, Dev Cell 4(6) (2003) 827–39. [DOI] [PubMed] [Google Scholar]
- [23].Krishnakumar R, Chen AF, Pantovich MG, Danial M, Parchem RJ, Labosky PA, Blelloch R, FOXD3 Regulates Pluripotent Stem Cell Potential by Simultaneously Initiating and Repressing Enhancer Activity, Cell Stem Cell 18(1) (2016) 104–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang WD, Melville DB, Montero-Balaguer M, Hatzopoulos AK, Knapik EW, Tfap2a and Foxd3 regulate early steps in the development of the neural crest progenitor population, Dev Biol 360(1) (2011) 173–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pastor WA, Liu W, Chen D, Ho J, Kim R, Hunt TJ, Lukianchikov A, Liu X, Polo JM, Jacobsen SE, Clark AT, TFAP2C regulates transcription in human naive pluripotency by opening enhancers, Nat Cell Biol 20(5) (2018) 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lim LS, Loh Y-H, Zhang W, Li Y, Chen X, Wang Y, Bakre M, Ng H-H, Stanton LW, Zic3 is required for maintenance of pluripotency in embryonic stem cells, Molecular biology of the cell 18(4) (2007) 1348–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yang S-H, Andrabi M, Biss R, Baker SM, Iqbal M, Sharrocks AD, ZIC3 controls the transition from naive to primed pluripotency, Cell reports 27(11) (2019) 3215–3227.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lavial F, Acloque H, Bertocchini F, Macleod DJ, Boast S, Bachelard E, Montillet G, Thenot S, Sang HM, Stern CD, Samarut J, Pain B, The Oct4 homologue PouV and Nanog regulate pluripotency in chicken embryonic stem cells, Development 134(19) (2007) 3549–63. [DOI] [PubMed] [Google Scholar]
- [29].Lignell A, Kerosuo L, Streichan SJ, Cai L, Bronner ME, Identification of a neural crest stem cell niche by Spatial Genomic Analysis, Nat Commun 8(1) (2017) 1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lukoseviciute M, Gavriouchkina D, Williams RM, Hochgreb-Hagele T, Senanayake U, Chong-Morrison V, Thongjuea S, Repapi E, Mead A, Sauka-Spengler T, From pioneer to repressor: bimodal foxd3 activity dynamically remodels neural crest regulatory landscape in vivo, Dev. Cell 47(5) (2018) 608–628.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lignell A, Kerosuo L, Streichan SJ, Cai L, Bronner ME, Identification of a neural crest stem cell niche by Spatial Genomic Analysis, Nature communications 8(1) (2017) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rao A, LaBonne C, Histone deacetylase activity has an essential role in establishing and maintaining the vertebrate neural crest, Development 145(15) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Geary L, LaBonne C, FGF mediated MAPK and PI3K/Akt Signals make distinct contributions to pluripotency and the establishment of Neural Crest, Elife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Minoux M, Holwerda S, Vitobello A, Kitazawa T, Kohler H, Stadler MB, Rijli FM, Gene bivalency at Polycomb domains regulates cranial neural crest positional identity, Science 355(6332) (2017). [DOI] [PubMed] [Google Scholar]
- [35].Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG, Chromatin signatures of pluripotent cell lines, Nat Cell Biol 8(5) (2006) 532–8. [DOI] [PubMed] [Google Scholar]
- [36].Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES, A bivalent chromatin structure marks key developmental genes in embryonic stem cells, Cell 125(2) (2006) 315–26. [DOI] [PubMed] [Google Scholar]
- [37].Saunders A, Huang X, Fidalgo M, Reimer MH Jr, Faiola F, Ding J, Sánchez-Priego C, Guallar D, Sáenz C, Li D, The SIN3A/HDAC corepressor complex functionally cooperates with NANOG to promote pluripotency, Cell reports 18(7) (2017) 1713–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yamamoto T, Takenaka C, Yoda Y, Oshima Y, Kagawa K, Miyajima H, Sasaki T, Kawamata S, Differentiation potential of Pluripotent Stem Cells correlates to the level of CHD7, Sci Rep 8(1) (2018) 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Schnetz MP, Handoko L, Akhtar-Zaidi B, Bartels CF, Pereira CF, Fisher AG, Adams DJ, Flicek P, Crawford GE, Laframboise T, Tesar P, Wei CL, Scacheri PC, CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet 6(7) (2010) e1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T, Wysocka J, CHD7 cooperates with PBAF to control multipotent neural crest formation, Nature 463(7283) (2010) 958–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gandhi S, Hutchins EJ, Maruszko K, Park JH, Thomson M, Bronner ME, Bimodal function of chromatin remodeler Hmga1 in neural crest induction and Wnt-dependent emigration, Elife 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Shah SN, Kerr C, Cope L, Zambidis E, Liu C, Hillion J, Belton A, Huso DL, Resar LM, HMGA1 reprograms somatic cells into pluripotent stem cells by inducing stem cell transcriptional networks, PLoS One 7(11) (2012) e48533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Fujita K, Ogawa R, Kawawaki S, Ito K, Roles of chromatin remodelers in maintenance mechanisms of multipotency of mouse trunk neural crest cells in the formation of neural crest-derived stem cells, Mech Dev 133 (2014) 126–45. [DOI] [PubMed] [Google Scholar]
- [44].Exner CRT, Kim AY, Mardjuki SM, Harland RM, sall1 and sall4 repress pou5f3 family expression to allow neural patterning, differentiation, and morphogenesis in Xenopus laevis, Dev Biol 425(1) (2017) 33–43. [DOI] [PubMed] [Google Scholar]
- [45].Diener J, Baggiolini A, Pernebrink M, Dalcher D, Lerra L, Cheng PF, Varum S, Hausel J, Stierli S, Treier M, Studer L, Basler K, Levesque MP, Dummer R, Santoro R, Cantu C, Sommer L, Epigenetic control of melanoma cell invasiveness by the stem cell factor SALL4, Nat Commun 12(1) (2021) 5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bhattacharya D, Rothstein M, Azambuja AP, Simoes-Costa M, Control of neural crest multipotency by Wnt signaling and the Lin28/let-7 axis, Elife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Aoki Y, Saint-Germain N, Gyda M, Magner-Fink E, Lee YH, Credidio C, Saint-Jeannet JP, Sox10 regulates the development of neural crest-derived melanocytes in Xenopus, Dev Biol 259(1) (2003) 19–33. [DOI] [PubMed] [Google Scholar]
- [48].O’Donnell M, Hong CS, Huang X, Delnicki RJ, Saint-Jeannet JP, Functional analysis of Sox8 during neural crest development in Xenopus, Development 133(19) (2006) 3817–26. [DOI] [PubMed] [Google Scholar]
- [49].Spokony RF, Aoki Y, Saint-Germain N, Magner-Fink E, Saint-Jeannet JP, The transcription factor Sox9 is required for cranial neural crest development in Xenopus, Development 129(2) (2002) 421–32. [DOI] [PubMed] [Google Scholar]
- [50].Hong CS, Saint-Jeannet JP, Sox proteins and neural crest development, Seminars in cell & developmental biology 16(6) (2005) 694–703. [DOI] [PubMed] [Google Scholar]
- [51].Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MS, Niwa H, Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells, Nature cell biology 9(6) (2007) 625–35. [DOI] [PubMed] [Google Scholar]
- [52].Corsinotti A, Wong FC, Tatar T, Szczerbinska I, Halbritter F, Colby D, Gogolok S, Pantier R, Liggat K, Mirfazeli ES, Hall-Ponsele E, Mullin NP, Wilson V, Chambers I, Distinct SoxB1 networks are required for naive and primed pluripotency, Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Tai A, Cheung M, Huang YH, Jauch R, Bronner ME, Cheah KS, SOXE neofunctionalization and elaboration of the neural crest during chordate evolution, Sci Rep 6 (2016) 34964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Bell DM, Leung KK, Wheatley SC, Ng LJ, Zhou S, Ling KW, Sham MH, Koopman P, Tam PP, Cheah KS, SOX9 directly regulates the type-II collagen gene, Nat Genet 16(2) (1997) 174–8. [DOI] [PubMed] [Google Scholar]
- [55].Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B, Sox9 is required for cartilage formation, Nat Genet 22(1) (1999) 85–9. [DOI] [PubMed] [Google Scholar]
- [56].Healy C, Uwanogho D, Sharpe PT, Expression of the chicken Sox9 gene marks the onset of cartilage differentiation, Ann N Y Acad Sci 785 (1996) 261–2. [DOI] [PubMed] [Google Scholar]
- [57].Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B, The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6, Genes Dev 16(21) (2002) 2813–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M, The transcription factor Sox10 is a key regulator of peripheral glial development, Genes Dev 15(1) (2001) 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Southard-Smith EM, Kos L, Pavan WJ, Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model, Nat Genet 18(1) (1998) 60–4. [DOI] [PubMed] [Google Scholar]
- [60].Schock EN, LaBonne C, Sorting Sox: Diverse Roles for Sox Transcription Factors During Neural Crest and Craniofacial Development, Front Physiol 11 (2020) 606889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S, Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts, Nat Biotechnol 26(1) (2008) 101–6. [DOI] [PubMed] [Google Scholar]
- [62].Buitrago-Delgado E, Schock EN, Nordin K, LaBonne C, A transition from SoxB1 to SoxE transcription factors is essential for progression from pluripotent blastula cells to neural crest cells, Dev Biol 444(2) (2018) 50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Roellig D, Tan-Cabugao J, Esaian S, Bronner ME, Dynamic transcriptional signature and cell fate analysis reveals plasticity of individual neural plate border cells, Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lin Y, Li XY, Willis AL, Liu C, Chen G, Weiss SJ, Snail1-dependent control of embryonic stem cell pluripotency and lineage commitment, Nat Commun 5 (2014) 3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Tarashansky AJ, Musser JM, Khariton M, Li P, Arendt D, Quake SR, Wang B, Mapping single-cell atlases throughout Metazoa unravels cell type evolution, Elife 10 (2021) e66747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Prasad MS, Uribe-Querol E, Marquez J, Vadasz S, Yardley N, Shelar PB, Charney RM, Garcia-Castro MI, Blastula stage specification of avian neural crest, Dev Biol 458(1) (2020) 64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Barriga EH, Trainor PA, Bronner M, Mayor R, Animal models for studying neural crest development: is the mouse different?, Development 142(9) (2015) 1555–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].McGonnell IM, Graham A, Trunk neural crest has skeletogenic potential, Curr Biol 12(9) (2002) 767–71. [DOI] [PubMed] [Google Scholar]
- [69].Chibon P, [Nuclear labelling by tritiated thymidine of neural crest derivatives in the amphibian Urodele Pleurodeles waltlii Michah], J Embryol Exp Morphol 18(3) (1967) 343–58. [PubMed] [Google Scholar]
- [70].Nakamura H, Ayer-le Lievre CS, Mesectodermal capabilities of the trunk neural crest of birds, J Embryol Exp Morphol 70 (1982) 1–18. [PubMed] [Google Scholar]
- [71].Simoes-Costa M, Bronner ME, Reprogramming of avian neural crest axial identity and cell fate, Science 352(6293) (2016) 1570–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Soldatov R, Kaucka M, Kastriti ME, Petersen J, Chontorotzea T, Englmaier L, Akkuratova N, Yang Y, Haring M, Dyachuk V, Bock C, Farlik M, Piacentino ML, Boismoreau F, Hilscher MM, Yokota C, Qian X, Nilsson M, Bronner ME, Croci L, Hsiao WY, Guertin DA, Brunet JF, Consalez GG, Ernfors P, Fried K, Kharchenko PV, Adameyko I, Spatiotemporal structure of cell fate decisions in murine neural crest, Science 364(6444) (2019). [DOI] [PubMed] [Google Scholar]
- [73].LeDouarin NM, Kalcheim C, The Neural Crest, 2nd ed., Cambridge: University Press; 1999. [Google Scholar]
- [74].Gillis JA, Alsema EC, Criswell KE, Trunk neural crest origin of dermal denticles in a cartilaginous fish, Proc Natl Acad Sci U S A 114(50) (2017) 13200–13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Green SA, Bronner ME, The lamprey: A jawless vertebrate model system for examining origin of the neural crest and other vertebrate traits, Differentiation 87(1–2) (2014) 44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Shimeld SM, Donoghue PCJ, Evolutionary crossroads in developmental biology: cyclostomes (lamprey and hagfish), Development 139(12) (2012) 2091–2099. [DOI] [PubMed] [Google Scholar]
- [77].York JR, McCauley DW, The origin and evolution of neural crest cells, Open Biology 10(1) (2020) 190285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Abitua PB, Wagner E, Navarrete IA, Levine M, Identification of a rudimentary neural crest in a non-vertebrate chordate, Nature 492(7427) (2012) 104–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Zhao D, Chen S, Liu X, Lateral neural borders as precursors of peripheral nervous systems: A comparative view across bilaterians, Development, Growth & Differentiation 61(1) (2019) 58–72. [DOI] [PubMed] [Google Scholar]
- [80].Gans C, Northcutt RG, Neural crest and the origin of vertebrates: a new head, Science 220(4594) (1983) 268–274. [DOI] [PubMed] [Google Scholar]
- [81].Putnam NH, Butts T, Ferrier DEK, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu J-K, Benito-Gutiérrez EL, Dubchak I, Garcia-Fernàndez J, Gibson-Brown JJ, Grigoriev IV, Horton AC, de Jong PJ, Jurka J, Kapitonov VV, Kohara Y, Kuroki Y, Lindquist E, Lucas S, Osoegawa K, Pennacchio LA, Salamov AA, Satou Y, Sauka-Spengler T, Schmutz J, Shin-I T, Toyoda A, Bronner-Fraser M, Fujiyama A, Holland LZ, Holland PWH, Satoh N, Rokhsar DS, The amphioxus genome and the evolution of the chordate karyotype, Nature 453(7198) (2008) 1064. [DOI] [PubMed] [Google Scholar]
- [82].Gee H, Before the backbone: views on the origin of the vertebrates, Springer, New York, 1996. [Google Scholar]
- [83].Gee H, Across the bridge: understanding the origin of the vertebrates, University of Chicago Press, Chicago, 2018. [Google Scholar]
- [84].Delsuc F, Brinkmann H, Chourrout D, Philippe H, Tunicates and not cephalochordates are the closest living relatives of vertebrates, Nature 439(7079) (2006) 965–968. [DOI] [PubMed] [Google Scholar]
- [85].Bourlat SJ, Juliusdottir T, Lowe CJ, Freeman R, Aronowicz J, Kirschner M, Lander ES, Thorndyke M, Nakano H, Kohn AB, Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida, Nature 444(7115) (2006) 85. [DOI] [PubMed] [Google Scholar]
- [86].Yu J-K, Meulemans D, McKeown SJ, Bronner-Fraser M, Insights from the amphioxus genome on the origin of vertebrate neural crest, Genome research 18(7) (2008) 1127–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Yu J-K, Holland ND, Holland LZ, Tissue-specific expression of FoxD reporter constructs in amphioxus embryos, Developmental biology 274(2) (2004) 452–461. [DOI] [PubMed] [Google Scholar]
- [88].Cattell MV, Garnett AT, Klymkowsky MW, Medeiros DM, A maternally established SoxB1/SoxF axis is a conserved feature of chordate germ layer patterning, Evolution & Development 14(1) (2012) 104–115. [DOI] [PubMed] [Google Scholar]
- [89].Presnell JS, Schnitzler CE, Browne WE, KLF/SP transcription factor family evolution: expansion, diversification, and innovation in eukaryotes, Genome biology and evolution 7(8) (2015) 2289–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Ma Y, Zhang X, Ma H, Ren Y, Sun Y, Wang Q, Shi J, Bioinformatic analysis of the four transcription factors used to induce pluripotent stem cells, Cytotechnology 66(6) (2014) 967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Gold DA, Gates RD, Jacobs DK, The early expansion and evolutionary dynamics of POU class genes, Molecular biology and evolution 31(12) (2014) 3136–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Jeffery WR, Strickler AG, Yamamoto Y, Migratory neural crest- like cells form body pigmentation in a urochordate embryo, Nature 431(7009) (2004) 696–699. [DOI] [PubMed] [Google Scholar]
- [93].Stolfi A, Ryan K, Meinertzhagen IA, Christiaen L, Migratory neuronal progenitors arise from the neural plate borders in tunicates, Nature 527(7578) (2015) 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Imai KS, Hino K, Yagi K, Satoh N, Satou Y, Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks, Development 131 (2004) 4047–4058. [DOI] [PubMed] [Google Scholar]
- [95].Sauka-Spengler T, Bronner-Fraser M, Insights from a sea lamprey into the evolution of neural crest gene regulatory network, Biol. Bull 214(3) (2008) 303–314. [DOI] [PubMed] [Google Scholar]
- [96].Sauka-Spengler T, Bronner-Fraser M, A gene regulatory network orchestrates neural crest formation, Nat. Rev. Mol. Cell Biol 9(7) (2008) 557–568. [DOI] [PubMed] [Google Scholar]
- [97].Sauka-Spengler T, Bronner-Fraser M, Evolution of the neural crest viewed from a gene regulatory perspective, Genesis 46(11) (2008) 673–682. [DOI] [PubMed] [Google Scholar]
- [98].Sauka-Spengler T, Meulemans DM, Jones M, Bronner-Fraser M, Ancient evolutionary origin of the neural crest gene regulatory network, Dev. Cell 13(3) (2007) 405–420. [DOI] [PubMed] [Google Scholar]
- [99].Smith JJ, Timoshevskaya N, Ye C, Holt C, Keinath MC, Parker HJ, Cook ME, Hess JE, Narum SR, Lamanna F, Kaessmann H, Timoshevskiy VA, Waterbury CK, Saraceno C, Widemann LM, Robb SM, Baker C, EvanE Eichler, D. Hockman T. Sauka-Spengler M. Yandell R. Krumlauf G. Elgar CT. Amemiya, The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution, Nature Genetics 50(2) (2018) 270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Green SA, Uy BR, Bronner ME, Ancient evolutionary origin of vertebrate enteric neurons from trunk-derived neural crest, Nature 544(7648) (2017) 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Martin WM, Bumm LA, McCauley DW, Development of the viscerocranial skeleton during embryogenesis of the sea lamprey, Petromyzon marinus, Dev. Dyn 238(12) (2009) 3126–3138. [DOI] [PubMed] [Google Scholar]
- [102].McCauley DW, Bronner-Fraser M, Neural crest contributions to the lamprey head, Development 130(11) (2003) 2317–2327. [DOI] [PubMed] [Google Scholar]
- [103].McCauley DW, Bronner-Fraser M, Importance of SoxE in neural crest development and the evolution of the pharynx, Nature 441(7094) (2006) 750–752. [DOI] [PubMed] [Google Scholar]
- [104].York JR, Yuan T, McCauley DW, Evolutionary and Developmental Associations of Neural Crest and Placodes in the Vertebrate Head: Insights From Jawless Vertebrates, Frontiers in Physiology 11(986) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].York JR, Yuan T, Zehnder K, McCauley DW, Lamprey neural crest migration is Snail-dependent and occurs without a differential shift in cadherin expression, Developmental Biology 428(1) (2017) 176–187. [DOI] [PubMed] [Google Scholar]
- [106].Yuan T, York JR, McCauley DW, Gliogenesis in lampreys shares gene regulatory interactions with oligodendrocyte development in jawed vertebrates, Developmental Biology 441(1) (2018) 176–190. [DOI] [PubMed] [Google Scholar]
- [107].Yuan T, York JR, McCauley DW, Neural crest and placode roles in formation and patterning of cranial sensory ganglia in lamprey, Genesis 58 (2020) e23356. [DOI] [PubMed] [Google Scholar]
- [108].Nikitina N, Sauka-Spengler T, Bronner-Fraser M, Dissecting early regulatory relationships in the lamprey neural crest gene network, Proc. Natl. Acad. Sci. U. S. A 105(51) (2008) 20083–20088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Hockman D, Chong-Morrison V, Green SA, Gavriouchkina D, Candido-Ferreira I, Ling IT, Williams RM, Amemiya CT, Smith JJ, Bronner ME, A genome-wide assessment of the ancestral neural crest gene regulatory network, Nature Communications 10(1) (2019) 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Martik ML, Gandhi S, Uy BR, Gillis JA, Green SA, Simoes-Costa M, Bronner ME, Evolution of the new head by gradual acquisition of neural crest regulatory circuits, Nature 574(7780) (2019) 675–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Parker HJ, De Kumar B, Green SA, Prummel KD, Hess C, Kaufman CK, Mosimann C, Wiedemann LM, Bronner ME, Krumlauf R, A Hox-TALE regulatory circuit for neural crest patterning is conserved across vertebrates, Nature Communications 10(1) (2019) 1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].York JR, Zehnder K, Yuan T, Lakiza O, McCauley DW, Evolution of Snail-mediated regulation of neural crest and placodes from an ancient role in bilaterian neurogenesis, Developmental Biology 453(2) (2019) 180–190. [DOI] [PubMed] [Google Scholar]
- [113].Li Y, Zhao D, Horie T, Chen G, Bao H, Chen S, Liu W, Horie R, Liang T, Dong B, Conserved gene regulatory module specifies lateral neural borders across bilaterians, Proc. Natl. Acad. Sci. U. S. A 114(31) (2017) E6352–E6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Satou Y, A gene regulatory network for cell fate specification in Ciona embryos, Current Topics in Developmental Biology 139 (2020) 1–33. [DOI] [PubMed] [Google Scholar]
- [115].Square T, Jandzik D, Cattell M, Hansen A, Medeiros DM, Embryonic expression of endothelins and their receptors in lamprey and frog reveals stem vertebrate origins of complex Endothelin signaling, Scientific Reports 6 (2016) 34282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Square TA, Jandzik D, Massey JL, Romášek M, Stein HP, Hansen AW, Purkayastha A, Cattell MV, Medeiros DM, Evolution of the endothelin pathway drove neural crest cell diversification, Nature 585(7826) (2020) 563–568. [DOI] [PubMed] [Google Scholar]
- [117].York JR, McCauley DW, Functional genetic analysis in a jawless vertebrate, the sea lamprey: insights into the developmental evolution of early vertebrates, Journal of Experimental Biology 223(Supplement) (2020) jeb206433. [DOI] [PubMed] [Google Scholar]
- [118].Thomson M, Liu SJ, Zou LN, Smith Z, Meissner A, Ramanathan S, Pluripotency factors in embryonic stem cells regulate differentiation into germ layers, Cell 145(6) (2011) 875–89. [DOI] [PMC free article] [PubMed] [Google Scholar]


