Abstract
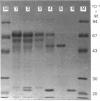
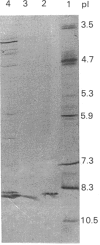
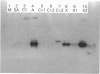
Trichoderma koningii G-39 produced xylanases in submerged culture using oat spelt xylan or crystalline cellulose, Avicel, as the sole carbon source. A low-Mr xylanase was purified from the culture filtrate by ion-exchange chromatography on SP-Trisacryl-M and gel filtration on Fractogel TSK HW-50F. It was homogeneous on SDS/PAGE and isoelectric focusing. A typical procedure provided about 11-fold purification with 4.5% protein yield and 50% activity recovery. The purified enzyme has an Mr value of about 21,500 and a pI of 8.9. Its specific activity was 6100 units/mg of protein, with optimal activity towards 0.5% xylan at about pH 5.5 and 60 degrees C. The purified enzyme had no activity against CM-cellulose with a degree of substitution of 0.63. It also showed no beta-xylosidase activity. The Km and Vmax. values, as determined with the soluble fraction of oat spelt xylan as substrate, were 0.70 mg/ml and 1.85 x 10(6) mumol/min per mg of enzyme respectively. Hg2+ (1 mM) and SDS (10 mM) completely inhibited xylanase activity, whereas Ca2+ showed no significant effect on the enzyme activity at 1 mM, but gave 80% inhibition at 10 mM. The enzyme contained about 4.4% carbohydrate and showed an immunological relationship to a cellobiohydrolase from the same fungal strain.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Grépinet O., Chebrou M. C., Béguin P. Nucleotide sequence and deletion analysis of the xylanase gene (xynZ) of Clostridium thermocellum. J Bacteriol. 1988 Oct;170(10):4582–4588. doi: 10.1128/jb.170.10.4582-4588.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellett L. E., Poole D. M., Ferreira L. M., Durrant A. J., Hazlewood G. P., Gilbert H. J. Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem J. 1990 Dec 1;272(2):369–376. doi: 10.1042/bj2720369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee S. F., Forsberg C. W., Rattray J. B. Purification and Characterization of Two Endoxylanases from Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 1987 Apr;53(4):644–650. doi: 10.1128/aem.53.4.644-650.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenecourt B. S., Eveleigh D. E. Preparation of mutants of Trichoderma reesei with enhanced cellulase production. Appl Environ Microbiol. 1977 Dec;34(6):777–782. doi: 10.1128/aem.34.6.777-782.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read S. M., Northcote D. H. Minimization of variation in the response to different proteins of the Coomassie blue G dye-binding assay for protein. Anal Biochem. 1981 Sep 1;116(1):53–64. doi: 10.1016/0003-2697(81)90321-3. [DOI] [PubMed] [Google Scholar]
- Wong K. K., Tan L. U., Saddler J. N. Multiplicity of beta-1,4-xylanase in microorganisms: functions and applications. Microbiol Rev. 1988 Sep;52(3):305–317. doi: 10.1128/mr.52.3.305-317.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]